Abstract
Purpose
In glaucoma, the optic nerve head (ONH) is the likely site of initial injury and elevated intraocular pressure (IOP) is the best-known risk factor. This study determines global gene expression changes in the pressure-injured ONH.
Methods
Unilateral sustained IOP elevation (glaucoma, n = 46) or optic nerve transection (n = 10) was produced in rats. ONHs were removed, and the retrobulbar optic nerves were graded for degeneration. Gene expression in the glaucomatous ONH with extensive injury was compared with that in the fellow ONH (n = 6/group), by using cDNA microarrays. Data from 12 arrays were normalized, significant differences in gene expression determined, and significantly affected gene classes identified. For the remaining ONH, grouped by experimental condition and degree of injury, quantitative reverse transcriptase-PCR (qPCR) and ANOVA were used to compare selected message levels.
Results
Microarray analysis identified more than 2000 significantly regulated genes. For 225 of these genes, the changes were greater than twofold. The most significantly affected gene classes were cell proliferation, immune response, lysosome, cytoskeleton, extracellular matrix, and ribosome. A 2.7-fold increase in ONH cellularity confirmed glaucoma model cell proliferation. By qPCR, increases in levels of periostin, collagen VI, and transforming growth factor β1 were linearly correlated to the degree of IOP-induced injury. For cyclinD1, fibulin 2, tenascin C, TIMP1, and aquaporin-4, correlations were significantly nonlinear, displaying maximum change with focal injury.
Conclusions
In the ONH, pressure-induced injury results in cell proliferation and dramatically altered gene expression. For specific genes, expression levels were most altered by focal injury, suggesting that further array studies may identify initial, and potentially injurious, altered processes.
Glaucoma is the second leading cause of blindness worldwide, affecting approximately 60 million people.1 Although many risk factors are associated with glaucoma, IOP is the most widely recognized, and lowering IOP is the goal of current glaucoma therapies. When IOP is experimentally elevated in nonhuman primates, the pattern of optic nerve head (ONH) cupping, optic nerve axon degeneration, and selective loss of retinal ganglion cells replicates the pathologic features of human glaucoma. Clinically, human glaucomatous optic neuropathy is characterized by optic disc cupping and a pattern of visual field loss.2 Cupping results from the loss of optic nerve axons and posterior bowing and remodeling of the support structures of the ONH.3,4 Often, these changes are most pronounced in the superior and inferior parts of the nerve head. The most characteristic visual field defect is the arcuate scotoma, which arches above or below central fixation and follows the pathways of the nerve fiber bundles as they converge on the superior and inferior poles of the ONH.2,5,6
Regional variation in laminar structure suggests less support and protection for axons, possibly explaining the apparent increased susceptibility of axons that pass through the superior and inferior ONH. In the face of increased or fluctuating IOP, movement of the lamina may result in preferential mechanical injury to neural tissues in these regions.7 In addition, because the vascular supply to the ONH tissue lies within the laminar beams, this pattern could result in regionally compromised blood flow.8 Current experimental evidence supporting either of these mechanisms is limited, and the cellular events that connect the regional pattern of glaucomatous optic nerve damage with the known structural anatomy of the ONH are still largely unknown. Regardless of mechanism, the variation in the structure of the ONH provides the only anatomic correlation with the characteristic pattern of glaucomatous optic nerve axon loss.
The apparent vulnerability of the ONH to pressure-induced axonal injury has led many investigators to examine changes in the composition of glaucomatous human and experimental monkey ONH tissues. These ONHs are characterized by axon loss, gliotic scaring, increased expression of matrix metalloproteinases, and abnormal deposition of extracellular matrix (ECM) materials, including collagens, tropoelastin, tenascin, and proteoglycans.9–20
In the rat, glaucoma can be modeled by sclerosing aqueous outflow pathways to produce a sustained elevation of IOP.21 In this model, pressure causes a selective loss of retinal ganglion cells and a characteristic pattern of axon degeneration that begins in the superior quadrant of the optic nerve.21,22 In addition, immunohistochemical studies of ONH reveal that pressure-induced injury is accompanied by deposition of collagens and other ECM components, similar to that in human glaucoma.23 This deposition of ECM is preceded by a loss of gap junctional connexin 43 immunolabeling and evidence of astrocytic proliferation.24 By immunohistochemical analysis, the initial evidence of ECM deposition coincided with a decrease in ONH labeling for neurotrophins and astrocytic GFAP.24
Although the ONH is recognized as the likely site of initial injury in human glaucoma and in experimental IOP elevation glaucoma models, very little is known about the changes in gene expression that accompany this injury. In this study, we used microarray analysis to identify the genes and functional gene classes most altered in expression in the rat ONH after exposure to experimentally elevated pressure that results in extensive and ongoing optic nerve degeneration. Then, we used real-time quantitative (q)PCR to verify selected changes in gene expression initially identified by microarray analysis and to examine several genes not included on the arrays. To expand this study, we included qPCR analyses of ONHs from eyes with focal regions of degeneration in optic nerve cross-sections. Focal injury occurs in eyes with more mild pressure elevations or those of shorter duration and probably reflects earlier responses to pressure-induced nerve injury. We compared injury in these ONHs to those in ONHs with more extensive injury due to elevated IOP and to those after optic nerve transection, to evaluate ONH responses to simple loss of axons. The expansion of the qPCR study to nerves with focal injuries allows us to explore the potential of discovering, by future microarray analyses, unique or more dramatic alterations in gene expression that occur early in the injury process. To our knowledge, this is the first genome-wide analysis of expression changes in the ONH in response to elevated IOP.
Methods
Glaucoma Model
All animal experiments were performed in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by the Oregon Health Sciences University (OHSU) Animal Care and Use Committee. Sustained IOP elevation was produced in Brown Norway rats by unilateral episcleral vein injection of hypertonic saline. IOP was measured 4 days per week with a handheld tonometer (Tono-PenXL; Medtronic, Minneapolis, MN) in awake animals housed in low-level, constant light to stabilize the circadian IOP oscillations that maximize during the dark phase.25,26 At 5 weeks after injection, the eyes were removed and rapidly chilled in cold phosphate-buffered saline, and the retinas were removed. Using a trephine, the ONH was removed from the posterior pole of the globe. The dissected ONH was then cut to 1 mm in length, a segment that included the optic disc, all the unmyelinated, anterior ONH, and the initial (approximately 0.5 mm) of the myelinated optic nerve, as illustrated in Figure 1A. The ONH was frozen on dry ice and stored at −80°C before extraction. RNA was isolated (Pico-Pure RNA Isolation Kit; Arcturus Bioscience, Inc., Mountain View, CA) by sonicating the frozen nerve heads in the kit extraction buffer using an MS 0.5 probe (UP50H Ultrasonic Processor; Hielscher USA, Inc., Ringwood, NJ) and then, after the instructions provided by the Arcturus kit, including DNase treatment to remove any DNA. Purified RNA was quantified (RiboGreen RNA Quantitation Kit; Invitrogen, Carlsbad, CA).
Figure 1.
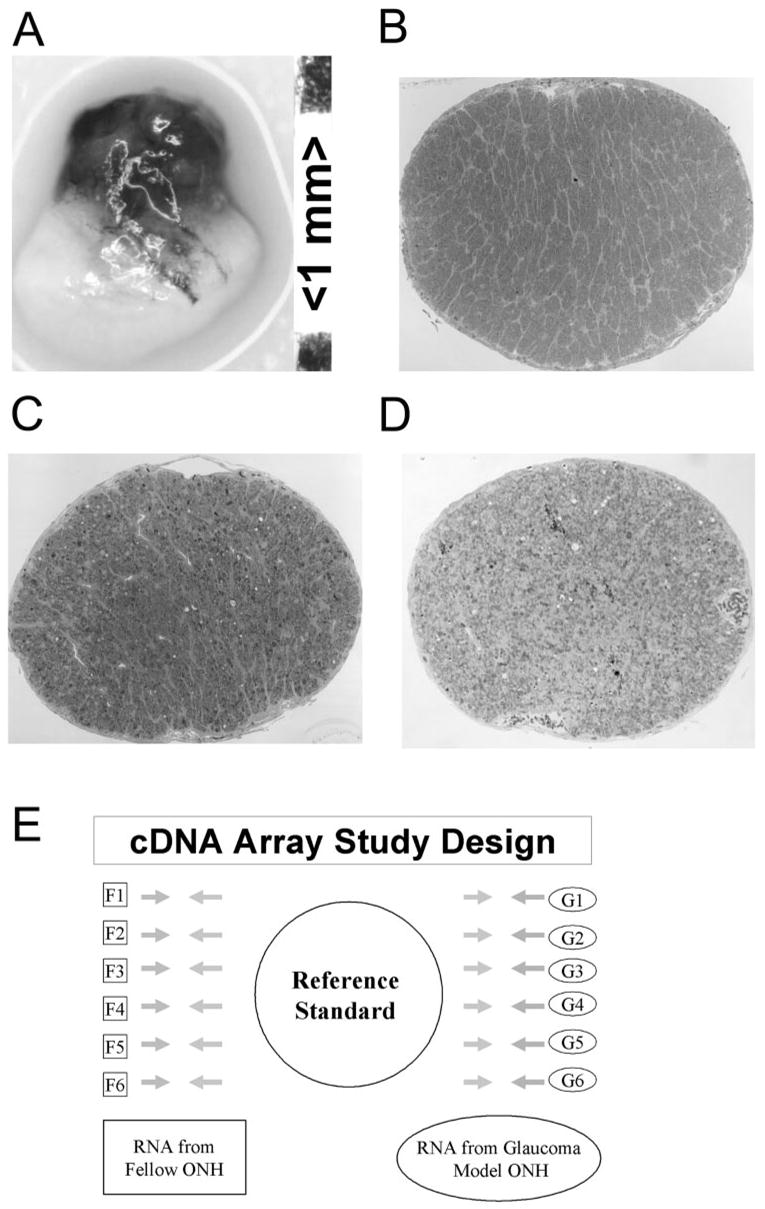
(A) Example of the dissected ONH used in this study. Each 1-mm-long ONH consisted of the optic disc and the unmyelinated and the initial myelinated portions of the nerve. A representative optic nerve cross-section from a normal, fellow eye (control) is shown in (B). All fellow nerves were graded 1.0. For the microarray study, glaucoma model nerve cross-sections exhibited extensive axonal degeneration (C). This nerve was graded 4.7. In the microarray study, glaucoma model ONH had nerves with grades between 4.6 and 4.95, indicating ongoing and extensive axonal degeneration affecting approximately 50% of the axons. ONH with grade-5 optic nerves that demonstrated more extensive gliosis (D) were excluded from the microarray study. The experimental design of the microarray study is illustrated in (E). For each gene, RNA from each of six fellow and six glaucomatous model eyes were independently compared by using 12 separate arrays with an ONH RNA reference standard.
All corresponding optic nerves were postfixed in glutaraldehyde, embedded in plastic, cross-sectioned, and evaluated by light microscopy. Optic nerve cross-sections were graded from 1 (no injury) to 5 (degeneration involving the total nerve area) by five masked observers.25 Our grading scale was designed to be sensitive to early pressure-induced axonal degeneration. By correlating counts of intact axons on transmission electron micrographs to the scale, we have determined that each grade unit corresponds to an approximate 12% increase in the number of degenerating axons.22 At grade 5, at least 50% of the axons are degenerating, whereas many other axons with intact, apparently normal, axoplasm remain.
Microarray Experimental Design and Analysis
Microarray analysis was used to compare gene expression in untreated, fellow ONHs to that in ONHs with extensive optic nerve injury due to elevated IOP. For these studies, the fellow eyes (n = 6) had normal optic nerves confirmed by optic nerve grading (Fig. 1B),25 whereas the glaucoma model ONHs (n = 6) were isolated from eyes with optic nerve injury grades just below 5 (4.64–4.95; Fig. 1C). Data from this group reflect extensive and ongoing elevated IOP-induced nerve injury affecting approximately 50% of the axons. We anticipated that the greatest number of genes and cellular processes within the nerve head would be affected by this injury paradigm. ONH from eyes with more advanced optic nerve gliotic scar formation, indicated by a relative reduction of axonal and myelin debris compared with the proportion of glial cell processes and extracellular matrix (Fig. 1D) were not included in the microarray study.
For each of the 12 ONHs, an aliquot of 50 ng total RNA underwent two rounds of linear amplification, was examined for integrity (Bio-Analyzer; Agilent, Palo Alto, CA), reverse transcribed and dye labeled at the OHSU Spotted Microarray Core Facility http://www.ohsu.edu/gmsr/smc/index.html). The facility also prepared and processed the cDNA arrays and compiled the data. For each sample, two cDNA arrays, SMCmou8400A and SMCmou6600A, were used with a combined total of 15,400 mouse cDNAs probes, representing the entire initial release of the NIA (National Institute on Aging) mouse library (http://lgsun.grc.nia.nih.gov/cDNA/15k.html) and approximately half the genome. Each probe was represented by duplicate spots on two duplicate slides, yielding four technical replicates per probe. As illustrated in the reference standard experimental design (Fig. 1E), mRNA expression patterns from each of six fellow and six grade-5 ONHs were compared separately to an ONH RNA reference standard. The standard was derived by pooling aliquots from all samples. Therefore, data represent two groups of six independent biological replicates and a total of 24 array comparisons, 12 each of the SMCmou8400A and SMCmou6600A arrays. Gene expression data are reported as the ratios of the average signal intensity (spot intensity minus local background) for each sample relative to the reference standard and were normalized by using a modified Lowess procedure.27 For each of the two array sets, Significance Analysis of Microarrays, (SAM, version 2.5, www-stat.stanford. edu/~tibs/SAM/provided in the public domain by Stanford University, Stanford, CA) was used to determine the significance of changes in gene expression due to elevated pressure injury. The parameters for SAM were set at 1000 permutations in the two-class, unpaired format, using the nearest number imputer of 10, a false-discovery rate of 2%, and a minimum change of 1.3-fold, to limit the called genes to those with potential biological significance.
Functional Classes of Significantly Altered Genes
DAVID gene ontology analysis tools (http://apps1.niaid.nih.gov/david/provided in the public domain by the National Institute of Allergies and Infectious Disease, Bethesda, MD) were used to identify and determine the number of altered genes in each significantly affected class for the categories of biological process, cellular component, and molecular function, as defined by the controlled vocabulary of the Gene Ontology consortium (http://www.geneontology.org/). AMIGO (http://www.godatabase.org/cgi-bin/amigo/go.cgi) was used to construct gene ontology hierarchies for the classes of affected ONH genes. In addition, GenMAPP was used to visualize gene expression changes in affected KEGG pathways (http://www.genmapp.org/).
ONH mRNA Quantitation by qPCR
Sustained IOP elevation was produced in an additional 40 Brown Norway rats,21,22 and ONHs were collected 5 weeks later, as has been described. Optic nerve transection was produced in 10 more rats to provide a comparison group of ONHs with injury due solely to retrograde axonal degeneration.28 Nerves were transected approximately 2 mm behind the globe and the ONHs collected 14 days later. ONHs from eyes exposed to elevated IOP with optic nerve injury grades between 1.5 and 4.5 demonstrated focal lesions of increasing size, from less than 15% to ~50% of axons degenerating.22 In nerves with injury grades of 5, the lesion filled the entire optic nerve cross-section and 50% or more of the axons were degenerating. In some grade-5 lesions, glial scar formation was evident. For the transection group, the average grade was 4.9.
For reverse transcription of total RNA, 30 ng of ONH mRNA from each sample, along with a pooled ONH RNA standard curve (2–480 ng) was reverse transcribed.29 The cDNA was amplified using a thermocycler (LightCycler, LightCycler Software 3.5, and DNA Master SYBR Green 1 kit; Roche, Indianapolis, IN) according to the manufacturer’s protocol, with a final concentration of 4 mM MgCl2 and 0.25 μM each of the relevant primers. The primers used for each cDNA were designed (Primer Designer 3 software for Windows; Sci-ed Software, State Line, PA) and sequences for rat mRNAs were obtained from the NIH National Center for Biotechnology Information (Bethesda, MD) databases (http://www.ncbi.nlm.nih.gov/entrez). Glyceraldehyde phosphate dehydrogenase (GAPDH) was used as the housekeeping gene for normalization, because its level was not significantly changed in the microarray study (P = 0.8) and it showed no significant correlation to either IOP or optic nerve injury grade by qPCR. GAPDH was measured by triplicate assay in each experimental sample and the mean value used for normalization. Table 1 lists the primers used and PCR product sizes. PCR was followed by the generation of melting curves for the amplified products to verify amplification specificity. All products were also verified by sequencing. Quantitation of the PCR product was made using the fit point method (LightCycler Manual, version 3.5; Roche).
Table 1.
Primers Sequences for qPCR
| Target | Forward Primer | Reverse Primer | bp |
|---|---|---|---|
| Aquaporin-4 | CATGAATCCAGCTCGATCCT | TTCTCGTCTCCACGGTCAAT | 297 |
| Collagen IV (α2) | AGGGATACCCGGCATAAC | GGCCTCTGCTTCCTTTCT | 119 |
| Collagen VI (α1) | CCAGGTCGAGCGTCTCTTCA | ACGTCGTACTCGGCGGTCTT | 332 |
| Connexin 43 | CGTGCCGCAATTACAACAAG | TTGGTCCACGATGGCTAATG | 194 |
| Cyclin D1 | CTGCGATGCAAGGCCTGAAC | GCGCAGGCTTGACTCCAGAA | 152 |
| Fibulin-2 | CGCTGTCTGCGCTTCGATTG | GGCCGATGCGGAAGATATGG | 175 |
| GAPDH | TGCCACTCAGAAGACTGTGGATG | GCCTGCTTCACCACCTTCTTGAT | 249 |
| GFAP | GCGGAGAACAACCTTGGCTGTGTA | GCAGTTGGCGGCGATAGTCATTA | 376 |
| Microglia (IBA1) | TCCGAGGAGACGTTCAGTTA | CTGGTCTCACAACTGCTTCTT | 237 |
| Periostin | GGAAGAACGCATCATCACAG | CATCGCCACCTTCAATGAAC | 137 |
| Tenascin C | GAGTCTGTGGATGGTACAGT | ATGTATAGAGGCCAGAGGT | 236 |
| TGFβ1 | TTGGAGCCTGGACACACA | TTGCGACCCACGTAGTAGAC | 132 |
| TGFβ2 | CATGCCCTTATCTGTGGAGTTC | TGGGCGTATTGCCAATGT | 150 |
| TIMP1 | GTTCCCTGGCATAATCTGAG | GGGATGAGGATCTGATCTGT | 155 |
qPCR Data Analysis
All data are expressed as a percentage of the mean (±SEM) value obtained for uninjected, fellow-eye ONHs. Statistical analyses were performed using commercial software (Excel; Microsoft, Redmond, WA; Prism; GraphPad, San Diego, CA) statistical software packages. Elevated IOP and transection groups were compared with the fellow-eye group by using ANOVA with the Dunnett multiple comparison post test. In addition, linear and nonlinear regression analyses were used to correlate mRNA expression levels to nerve injury grade and to compare regression fits by F test. Analysis of data based on IOP history, rather than optic nerve injury grade, produced equivalent results.
ONH DNA Measurement
Total ONH cell count was determined by extracting and measuring total DNA from seven fellow and five grade-5 ONH (Picopure DNA Extraction kit; Molecular Devices, Sunnyvale, CA; and Quant-iT PicoGreen dsDNA Assay kit). Cell count was determined by dividing total ONH DNA by the DNA content per rat cell (Gregory TR (2005). Animal Genome Size Database. http://www.genomesize.com/). Significance of difference was determined by Student’s t-test.
Immunohistochemistry
For immunohistochemistry, eight rats with unilateral IOP elevation were transcardially perfused with 4% phosphate-buffered paraformaldehyde and embedded in paraffin. Longitudinal sections through the ONH were prepared, and optic nerve cross-sections graded for degeneration.24,25 Immunohistochemistry was performed using the avidin-biotin technique with diaminobenzidine (DAB) chromogen with rabbit IBA-1 antibodies (Wako Chemicals USA, Richmond, VA) at 0.1 μg/mL with purified rabbit IgG serving as a negative control.24
Results
Significant Changes in Gene Expression by SAM Analysis
Using SAM analysis with a false-discovery rate of 2% and a difference of 1.3-fold, 1539 upregulated and 867 downregulated significantly altered genes were identified. All these genes have a q value (probability adjusted for multiple genes) of <0.02. Of these, 177 genes were upregulated more than twofold, 32 of which were increased greater than threefold (Table 2). Predominant among the most upregulated genes were those associated with mitotic cell division, cell adhesion, ECM, and immune responses. Forty-eight genes were downregulated by 50% or more. The most affected of these are listed in Table 3. These genes were primarily associated with synaptic and axonal mRNA,30–32 lipid and steroid metabolism, and cell signaling.
Table 2.
Genes Upregulated More Than Threefold in ONHs with Elevated-IOP-Induced Injury
| Change (x-Fold) | GenBank Accession | Unigene | Name | Gene Ontology Summary |
|---|---|---|---|---|
| 12.8 | AW553287 | Mm.236067 | Periostin, (fasciclin I related) | Integrin-mediated cell adhesion (beta-3), ECM, enhances motility |
| 6.1 | AW550270 | Mm.980 | Tenascin C | Extracellular space, localized in tendons, expressed by astrocytes, disrupts adhesion, supports migration |
| 5.9 | BG083139 | Mm.6856 | Securin, pituitary tumor-transforming 1 | Cell cycle, chromosome segregation, cysteine protease inhibitor activity |
| 5.3 | BG083522 | Mm.222228 | CDC28 protein kinase regulatory subunit 2 | Cell cycle, cyclin-dependent protein kinase activity; cytokinesis |
| 4.7 | AU018403 | Mm.6856 | Nuclear receptor subfamily 5, group A, member 2 | Regulates cholesterol degradation, regulation of transcription, DNA dependent |
| 4.6 | BG085568 | Mm.29798 | CD34 antigen | Cell adhesion, external side of plasma membrane |
| 4.4 | AW547306 | Mm.3453 | Complement component 1, q subcomponent, gamma polypeptide | Complement activation |
| 4.4 | BG079311 | mm.113278 | Potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 | Potassium ion transport, voltage-gated ion channel activity |
| 4.3 | BG080700 | Mm.28518 | Tumor necrosis factor receptor superfamily, member 12a, TWEAK receptor | Migration of endothelial cells, positive regulation of axon extension, adhesion |
| 4.3 | BG069098 | Mm.274086 | Kif18a: kinesin family member 18A | ATP binding, microtubule-based movement, motor activity |
| 4.3 | BG079289 | Mm.373656 | Riken cDNA 2700099C18 gene | Cell division and chromosome segregation |
| 4.2 | BG063624 | Mm.24337 | PDZ binding kinase | Phosphorylates MAP kinase p38. Seems to be active only in mitosis. |
| 4.1 | BG076632 | Mm.288206 | Adenylate cyclase 7 | Adenylate cyclase activation, guanylate cyclase activity, integral to membrane |
| 4.0 | BG080803 | Mm.9749 | B-cell linker | B-cell differentiation humoral immune response |
| 4.0 | BG079781 | Mm.181237 | Ribonuclease T2 | Mitochondrial RNAse activity |
| 4.0 | BG079350 | Mm.225956 | Kinetochore associated 2 | Actin binding, chromosome, pericentric region, cytoskeleton, mitotic metaphase |
| 3.8 | BG087868 | Mm.2570 | Complement component 1, q subcomponent, beta polypeptide | Complement activation |
| 3.7 | BG087390 | Mm.4237 | Topoisomerase (DNA) II alpha | Chromosome segregation |
| 3.7 | BG074248 | Mm.286488 | Kinesin family member 22 | Microtubule-based process, anaphase, nucleus |
| 3.5 | BG067419 | Mm.281298 | Growth arrest and DNA-damage-inducible 45 gamma | Activation of MAPKK, negative regulation of protein kinase activity, regulation of cell cycle |
| 3.5 | BG083088 | Mm.273049 | Cyclin D1 | Cyclin-dependent protein kinase holoenzyme complex, protein kinase activity |
| 3.5 | BG063496 | Mm.9916 | Centrosomal protein 55 | Cell division and chromosome partitioning |
| 3.4 | BG074814 | Mm.370 | Complement component 1, q subcomponent, alpha polypeptide | Complement activation |
| 3.5 | BG085131 | Mm.22673 | Fc receptor, IgE, high affinity 1, gamma polypeptide | Antigen presentation, neutrophil chemotaxis, positive regulation of immune response, positive regulation of interleukin-10 biosynthesis, transmembrane receptor activity |
| 3.4 | BG070808 | NA | Immunoglobulin | |
| 3.3 | BG073145 | Mm.22448 | Cytoskeleton-associated protein 2 | Unknown |
| 3.3 | BG064593 | Mm.12508 | Karyopherin (importin) alpha 2 | Nuclear protein import as an adapter protein for nuclear receptor KPNB1 |
| 3.2 | BG085966 | Mm.249146 | Fibulin 2 | Calcium-dependent binding to ECM components, elastic fiber assembly, elastosis |
| 3.2 | BG069532 | Mm.282556 | Niemann Pick type C2 | Egress of lipids from the lysosome |
| 3.2 | BG076991 | Mm.3468 | Suppressor of cytokine signaling 3 | Negative regulation of cytokine signaling via the JAK/STAT pathway, activated by insulin-like growth factor-1 receptor, blocks IL-6 signaling. |
| 3.1 | BG078426 | Mm.380027 | Homologous to G2/mitotic-specific cyclin B1 | Control of the cell cycle at the G2/M (mitosis) transition |
| 3.1 | BG064098 | Mm.171335 | Golgi phosphoprotein 2 | Transport of protein cargo through the Golgi |
Table 3.
The 30 Genes Most Downregulated in Expression in ONHs with Elevated-IOP-Induced Injury
| Change (x-Fold) | GenBank Accession | Unigene | Name | Gene Ontology Summary |
|---|---|---|---|---|
| 0.04 | BG069400 | Mm.41035 | Unc-13 homolog C (C. elegans) | Synaptic and impulse transmission, neurotransmitter release |
| 0.12 | BG069739 | Mm.61526 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | Cholesterol biosynthesis |
| 0.14 | BG076743 | Mm.200203 | RUN and TBC1 domain containing 2 | GTPase activation, vesicle-mediated transport |
| 0.17 | BG076766 | Mm.2108 | Transthyretin | Retinol and thyroxine transport from the bloodstream to the brain |
| 0.26 | BG086656 | Mm.250866 | Aldehyde dehydrogenase (NAD) | Aldehyde dehydrogenase (NAD) retinoic acid metabolism |
| 0.27 | BG074458 | Mm.133370 | 24-dehydrocholesterol reductase | Cholesterol biosynthesis, inactivation of MAPK |
| 0.27 | BG065170 | Mm.173337 | Mitogen-activated protein kinase 8 interacting protein 2 | JNK cascade, kinase activity, protein binding |
| 0.30 | BG083949 | Mm.30119 | Sterol-C4-methyl oxidase-like | Sterol biosynthesis |
| 0.30 | BG076999 | Mm.223744 | Kinesin family member 5B, Kinesin 1 | Microtubule-based movement, heavy chain, axonal transport |
| 0.32 | BG068358 | Mm.181166 | Rabphilin 3A | Rab GTPase binding, recruitment of synaptic vesicles for exocytosis |
| 0.33 | BG079889 | Mm.29847 | Isopentenyl-diphosphate delta isomerase | Cholesterol biosynthesis |
| 0.35 | BG073378 | Mm.360538 | Acetyl-coenzyme A acetyltransferase 2 | Acyltransferase activity, transferase activity, cholesterol biosynthesis |
| 0.35 | BG084178 | Mm.45372 | Protein phosphatase 1, regulatory (inhibitor) subunit 1B | Protein phosphatase inhibitor activity, dopaminergic signal transduction |
| 0.36 | BG069211 | Mm.371560 | Farnesyl diphosphate farnesyl transferase 1 | Cholesterol biosynthesis |
| 0.38 | BG063992 | Mm.3213 | Low-density lipoprotein receptor | Cholesterol metabolism |
| 0.38 | BG080229 | Mm.7729 | Aldolase 3, C isoform | Glycolysis |
| 0.39 | BG068310 | Mm.229532 | Endothelin receptor type B | G-protein-coupled receptor activity, nonspecific endothelin receptor, regulation of blood pressure |
| 0.42 | BG070284 | Mm.292102 | RAB30 | Small GTP-binding protein of the RAB family |
| 0.42 | BG071079 | Mm.228798 | Patched homolog 1 | Hedgehog receptor activity, negative regulation of mitosis, Wnt and TGFβsignaling |
| 0.42 | BG076141 | Mm.257073 | Chimerin (chimaerin) 1 | Rho GTPase activating protein, is transcriptionally upregulated after exposure to neurotrophins, enhance neurite outgrowth |
| 0.43 | BG079550 | Mm.272361 | Protease (rosome, macropain) 26S subunit, ATPase 5 | Protein catabolism, thyroid hormone receptor interactor 1 |
| 0.43 | BG082257 | Mm.273108 | Abhydrolase domain containing 3 | Biological process unknown, catalytic activity, hydrolase activity, integral to membrane |
| 0.43 | BG088107 | Mm.55143 | Dickkopf homolog 3 (Xenopus laevis) | Negative regulation of Wnt receptor signaling pathway |
| 0.43 | BG087831 | Mm.320691 | Potassium voltage-gated channel, Shal-related family, member 2 | Repolarization phase of the action potential, outward K+ currents |
| 0.44 | BG071519 | Mm.70950 | SRY-box containing gene 21 | Repression of NGF-induced neurite outgrowth in PC12 cells |
| 0.44 | BG071963 | Mm.54126 | IPLA2-2 | Membrane-associated, calcium-independent phospholipase A2 gamma, fatty acid metabolism |
| 0.44 | BG078816 | Mm.316652 | 3-Hydroxy-3-methylglutaryl-coenzyme A reductase | Cholesterol biosynthesis |
| 0.45 | BG073755 | Mm.130063 | Riken cDNA 9330182L06 gene | Receptor activity |
| 0.45 | BG066372 | Mm.1287 | Microtubule-associated protein tau | Negative regulation of microtubule depolymerization, axonal microtubules |
| 0.45 | BG068973 | Mm.13148 | Riken cDNA 1110064P04 gene, mRNA | Homology to AHA1, activator of heat shock 90-kDa protein ATPase homolog 2 |
Functional Classes of Significantly Altered Genes
The DAVID gene ontology analysis tool (http://apps1.niaid.nih.gov/david) was used to identify significantly affected gene classes using the controlled vocabulary of the Gene Ontology Consortium (http://www.geneontology.org/) from the SAM analysis lists of significantly regulated genes. This tool gives a probability (EASE score) for the significance of gene expression changes for each gene class. The significantly affected gene classes are listed for all regulated genes (Table 4), all upregulated genes (Table 5), and all downregulated genes (Table 6).
Table 4.
Significantly Regulated Gene Classes by DAVID Analysis of All Genes Identified by SAM as Significantly Altered in Expression
| Biological Process GO Term: Number of Genes (P) | Cell Component GO Term: Number of Genes (P) | Molecular Function GO Term: Number of Genes (P) |
|---|---|---|
| Cell proliferation: 113 (<0.0002) | Lysosome: 26 (<0.000009) | Structural constituent of ribosome: 56 (<0.00001) |
| Immune response: 38 (<0.0004) | Cytoskeleton: 72 (<0.0006) | ECM structural constituent: 13 (<0.002) |
| Ribosome biogenesis: 23 (<0.02) | ECM: 34 (<0.002) | Cytoskeletal protein binding: 39 (<0.002) |
| Sterol synthesis: 7 (<0.02) | Ribonucleoprotein complex: 66 (<0.002) | Transmembrane receptor activity: 36 (<0.01) |
| Spindle: 9 (<0.02) | Peroxidase activity: 7 (<0.02) | |
| Cell surface: 7 (<0.04) | Glycosaminoglycan binding: 9 (<0.03) | |
| Rho small monomeric GTPase activity: 6 (<0.03) | ||
| Metal ion binding: 97 (<0.04) | ||
| Cytokine binding: 6 (<0.05) | ||
| Transferase activity, transferring hexosyl groups: 12 (<0.05) | ||
| Long-chain-fatty-acid-CoA ligase activity: 4 (<0.05) |
Listed in order of ascending probabilities, P < 0.05 for all listed classes. For nested gene ontology (GO) terms, the most descriptive parent term is listed.
Table 5.
Significantly Upregulated Gene Classes by DAVID Analysis of Significantly Upregulated Genes Identified by SAM
| Biological Process GO Term: Number of Genes (P) | Cell Component GO Term: Number of Genes (P) | Molecular Function GO Term: Number of Genes (P) |
|---|---|---|
| Immune response: 34 (<0.00001) | Lysosome: 25 (<0.0000001) | Structural constituent of ribosome: 52 (<0.00000001) |
| Cell proliferation: 87 (<0.0001) | Ribonucleoprotein: complex: 57 (<0.00002) | ECM structural constituent: 12 (<0.0003) |
| Ribosome biogenesis and assembly: 21 (<0.0003) | ECM: 28 (<0.0006) | Glycosaminoglycan binding: 9 (<0.003) |
| Protein metabolism: 182 (<0.001) | Cytoskeleton: 48 (<0.02) | Transferase activity, transferring glycosyl groups: 19 (<0.003) |
| Cell adhesion: 35 (<0.03) | Cell surface: 6 (<0.04) | Kinase regulator activity: 10 (<0.02) |
| Spindle: 7 (<0.04) | Cytoskeletal protein binding: 27 (<0.02) | |
| Golgi apparatus: 33 (<0.05) | Transmembrane receptor activity: 26 (<0.04) | |
| Hydrolase activity, acting on glycosyl bonds: 11 (<0.04) | ||
| Rho small monomeric GTPase activity: 5 (<0.04) | ||
| Peptidase activity: 39 (<0.05) |
Data are as described in Table 4.
Table 6.
Significantly Downregulated Gene Classes by DAVID Analysis of Downregulated Genes Identified by SAM
| Biological Process GO Term: Number of Genes (P) | Cell Component GO Term: Number of Genes (P) | Molecular Function GO Term: Number of Genes (P) |
|---|---|---|
| Lipid biosynthesis: 11 (<0.01) | Cytoskeleton: 23 (<0.04) | Transferase activity, transferring alkyl or aryl (other than methyl) groups: 8 (<0.002) |
| Main pathways of carbohydrate metabolism: 9 (<0.04) | Ligase activity, forming carbon-sulfur bonds: 4 (<0.01) | |
| Amino acid derivative metabolism: 6 (<0.05) | Metal ion binding: 38 (<0.02) |
Data are as described in Table 4.
Elevated IOP Results in Significant ONH Cellular Proliferation
DAVID analysis of significantly regulated genes was dominated by altered expression of cell cycle and cell proliferation–associated messages (Table 4), implying that cellular proliferation occurs in the pressure-injured ONH. DAVID identified 113 proliferation-associated genes with altered expression. Eighty-seven of these genes were significantly upregulated (Table 5) including the neural progenitor cell marker, nestin (1.8-fold).33 Mapping the genes to a KEGG cell cycle map revealed that every stage of cell division was affected (not shown). Among upregulated cell cycle genes was cyclin D1. Cyclin D regulates the initiation of the cell cycle. We used qPCR, to confirm the upregulation of cyclin D1 and extend it by examining its expression levels in ONH from eyes with increasing grades of optic nerve injury and in ONH after optic nerve transection (Fig. 2A). We found that the correlation between cyclin D1 and optic nerve injury due to elevated IOP was significantly nonlinear (F test for comparison of linear and third-order polynomial fit, P < 0.0001) with the highest levels of cyclin D1 mRNA occurring in focally injured ONH. These ONHs were from eyes with approximately 5-mm Hg mean pressure elevation and nerve injury grades between 1.5 and 3.5 (Fig. 2B). This, along with other evidence,24,34 suggested that ONH cellular proliferation accompanies early pressure-induced nerve injury. Because cell cycle genes also can be activated during apoptosis,35,36 we used DNA content to determine the number of cells in additional fellow and grade-5 ONHs. In fellow eye rat ONHs, we found an average content of 4,894 ± 196 cells, whereas the number of cells in grade-5 ONHs had increased to 13,266 ± 2,200 cells (271%, P < 0.000002, Fig. 2C).
Figure 2.
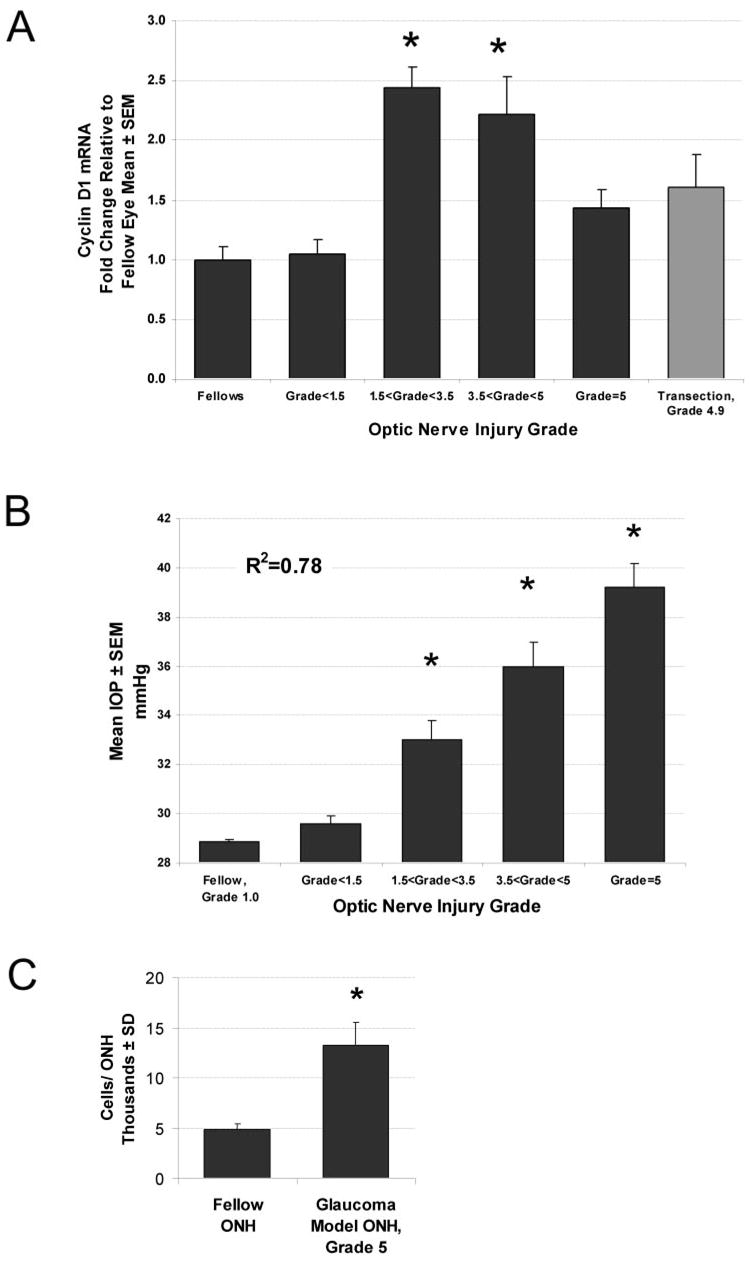
Elevated IOP results in ONH cellular proliferation. mRNA levels of cyclin D, the key regulator for cell-cycle progression, are shown in (A) for ONH with graded optic nerve injury due to elevated IOP. ONHs were grouped according to injury grade (■; n = 7–11/group). (
 ) For comparison, ONH from eyes with transected optic nerves (n = 7) are shown. Each bar represents the mean ± SEM for that group relative to the mean for untreated fellow eyes (n = 14). *Significant increases compared with fellow eye levels (ANOVA with the Dunnett posttest, P < 0.01). (B) In a pressure-injured ONH, the relationship of mean IOP to optic nerve injury grade is shown over the 5-week experimental period, by group. Mean IOP differences between groups are significant for all injury groups (*P < 0.05), except that between fellow and injury grade <1.5. For two separate groups of fellow and grade-5 ONH, total DNA measurements are shown in (C), illustrating the approximately threefold increase in cellularity in ONH with grade-5 optic nerve damage due to elevated IOP (*P < 0.000002).
) For comparison, ONH from eyes with transected optic nerves (n = 7) are shown. Each bar represents the mean ± SEM for that group relative to the mean for untreated fellow eyes (n = 14). *Significant increases compared with fellow eye levels (ANOVA with the Dunnett posttest, P < 0.01). (B) In a pressure-injured ONH, the relationship of mean IOP to optic nerve injury grade is shown over the 5-week experimental period, by group. Mean IOP differences between groups are significant for all injury groups (*P < 0.05), except that between fellow and injury grade <1.5. For two separate groups of fellow and grade-5 ONH, total DNA measurements are shown in (C), illustrating the approximately threefold increase in cellularity in ONH with grade-5 optic nerve damage due to elevated IOP (*P < 0.000002).
Effect of Elevated IOP on ONH Expression of Immune Response Genes and Evidence of Microglial Activation
Genes associated with the biological process of immune response were also significantly regulated (Table 4). Of 38 significantly regulated genes in this category, 34 of these were upregulated in expression. Predominant in this class were all five components of the complement 1 complex (range, 2- to 4.4-fold), FC receptor (3.4-fold), beta 2 microglobulin (2.5-fold), and several histocompatibility class II antigens (1.8–2.3 fold), all genes expressed by activated microglia in the nervous system.37–42 Complement receptor related protein, Crry, was also significantly upregulated (1.55-fold). Crry inhibits the activation of the third component of complement (CR3), protects from complement-mediated autoimmune damage, and is abundant in microglia.43,44 Many of these messages are associated with the activation of microglia, and markers for activated microglia have been identified in the pressure-injured ONH.45,46 In activated microglia, the upregulation of ionized calcium-binding adaptor molecule 1 (IBA1), also known as allograft inflammatory factor 1, is associated with the formation of lamellipodia and membrane ruffles.47 Using qPCR to measure IBA1 mRNA levels in the ONH described in Figures 2A and 2B, we found that IBA1 mRNA levels in the ONH demonstrated a positive linear correlation with the degree of optic nerve injury after 5 weeks of elevated IOP (R2 = 0.44, P < 0.0001) By group, IBA1 mRNA levels were significantly elevated in ONH with injury grades greater than 3.5 (Fig. 3A). Immunostaining for IBA1 in elevated-IOP–injured nerve heads suggested that increases in IBA1 mRNA levels reflected endogenous microglia activation, without ruling out a contribution due to proliferation (Fig. 3B).
Figure 3.
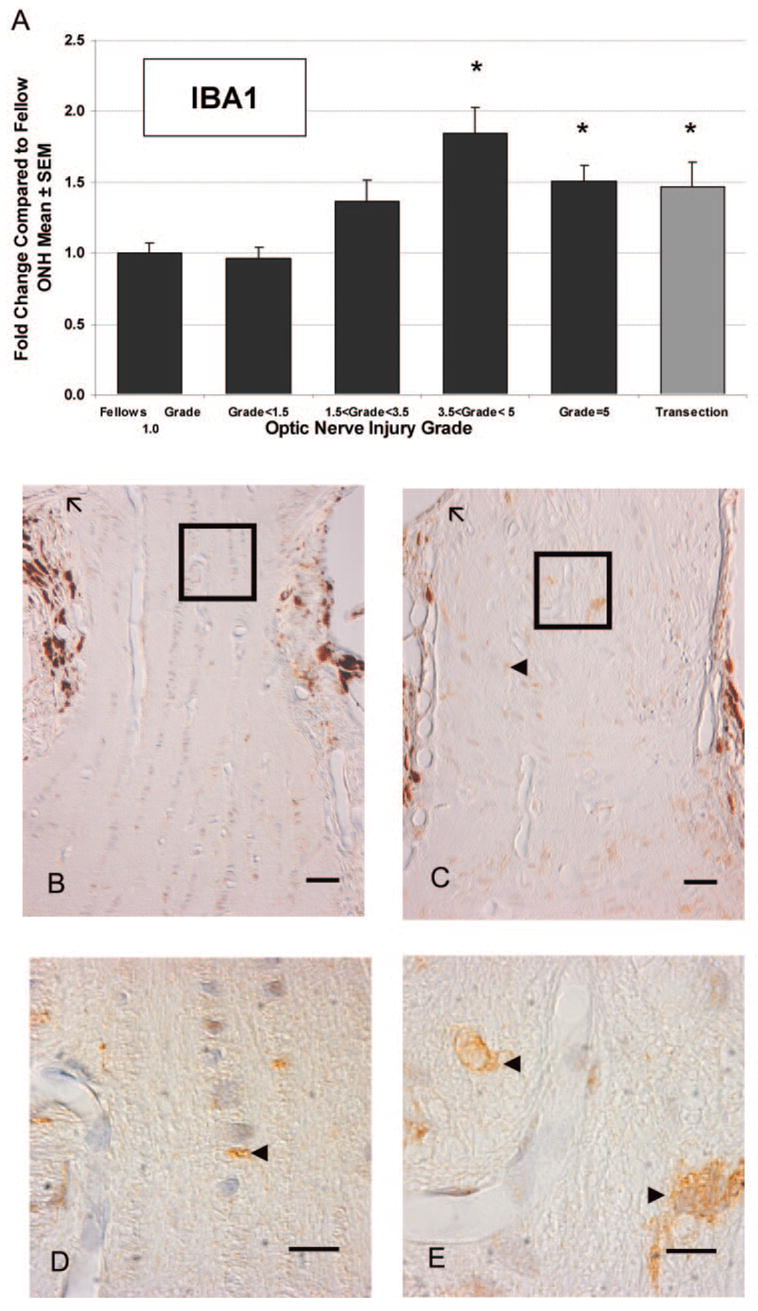
Microglial activation in response to elevated IOP. In the same nerve injury groups as described in Figure 2, mRNA levels for the microglial marker, IBA1, were measured (A). *Significantly different from fellow eye (P < 0.05). IBA1 mRNA levels were significantly elevated in ONHs damaged by elevated IOP (■) with optic nerve injury grades greater than 3.5, as well as in ONHs at 2 weeks after optic nerve transection (
 ). Immunohistochemical labeling indicated activation and potential proliferation of ONH microglia. Representative fellow ONH (B) and glaucoma model ONH with an optic nerve injury grade of 3.6 (C). (D, E) Higher magnifications of the boxed regions in (B) and (C), respectively. Arrowheads: labeled cells; arrows: Bruch’s membrane. Brown: chromogen labeling; pale blue: counterstained nuclei. Scale bars: (B, C) 10 μm; (D, E) 3 μm.
). Immunohistochemical labeling indicated activation and potential proliferation of ONH microglia. Representative fellow ONH (B) and glaucoma model ONH with an optic nerve injury grade of 3.6 (C). (D, E) Higher magnifications of the boxed regions in (B) and (C), respectively. Arrowheads: labeled cells; arrows: Bruch’s membrane. Brown: chromogen labeling; pale blue: counterstained nuclei. Scale bars: (B, C) 10 μm; (D, E) 3 μm.
Upregulation of ONH Ribosomal Gene Classes
All three gene ontology components of DAVID analysis (Table 4) identified significant effects on ribosomal gene expression in ONH injured by pressure. For genes associated with the biological process of ribosome biogenesis and assembly, 21 of 23 significantly affected genes were upregulated (1.3- to 1.6-fold). For ribosomes and associated components (ribosonucleoprotein complex), 57 of 66 significantly regulated genes were upregulated (1.3- to 3.5-fold). Finally, for genes that function as structural constituents of ribosomes, 52 of 56 genes were upregulated (1.3- to 3.5-fold).
Dramatic Upregulation of Lysosomal Genes
The lysosome was both the most significantly affected cell component (Table 4) and the most significantly upregulated cell component (Table 5). Among upregulated lysosomal genes were cathepsin-D, -B, -Z, -L, -8, -C, and -H (2.2- to 1.4-fold); lysosomal-associated protein transmembrane 5, a protein activated in retinal microglia in response to optic nerve transection48 (2.4-fold); and ganglioside degrading enzymes: beta galactosidase and beta hexosaminidase-A and -B (2.2-, 2.2- and 2.0-fold, respectively).
ECM Gene Expression
Elevated IOP resulted in the upregulation of 28 ECM components including periostin (12.8-fold); fibulin 2 (3.2-fold); tenascin C (2.7-fold); isoforms of collagens I, III, IV, V, and VI (1.5-to 2.5-fold); fibronectin 1 (2.3-fold); cartilage-associated protein (2.0-fold); and decorin (1.8-fold) as well as the matrix proteases ADAMTS-1 (2.9-fold) and matrix metalloproteinase-7 (MMP7, 2.8-fold) and -2 (MMP2, 2.1-fold). In addition, increased processing of ECM and other exported proteins was suggested by the upregulation of 33 genes associated with the Golgi complex.
To confirm some of these changes, we used qPCR to measure the expression levels of ECM messages in the ONH samples from Figures 2A and 2B. Because our previous study of altered gene expression in retinas exposed to elevated IOP had shown that the gene most altered in expression was an MMP inhibitor, tissue inhibitor of metalloproteinase (TIMP)-1 (46-fold increase49), we examined ONH TIMP-1 expression as well. This probe was not present on the cDNA arrays used in this study, whereas two other TIMPs were downregulated (described later).
For periostin, collagen IV and collagen VI mRNA, we found a significant positive linear correlation between mRNA level and optic nerve injury grade in ONHs injured by elevated IOP (R2 = 0.51, 0.11, and 0.37, respectively). When grouped by injury grade, levels of periostin and collagen VI were significantly elevated in ONH with optic nerve injury grades greater than 3.5, as well as in ONH from eyes after optic nerve transection (Fig. 4A). Type 2α-collagen IV demonstrated a similar pattern, although only the increase after ONH transection was significant. By our microarray analysis, another isoform of collagen IV (type1α) was increased 2.3-fold.
Figure 4.
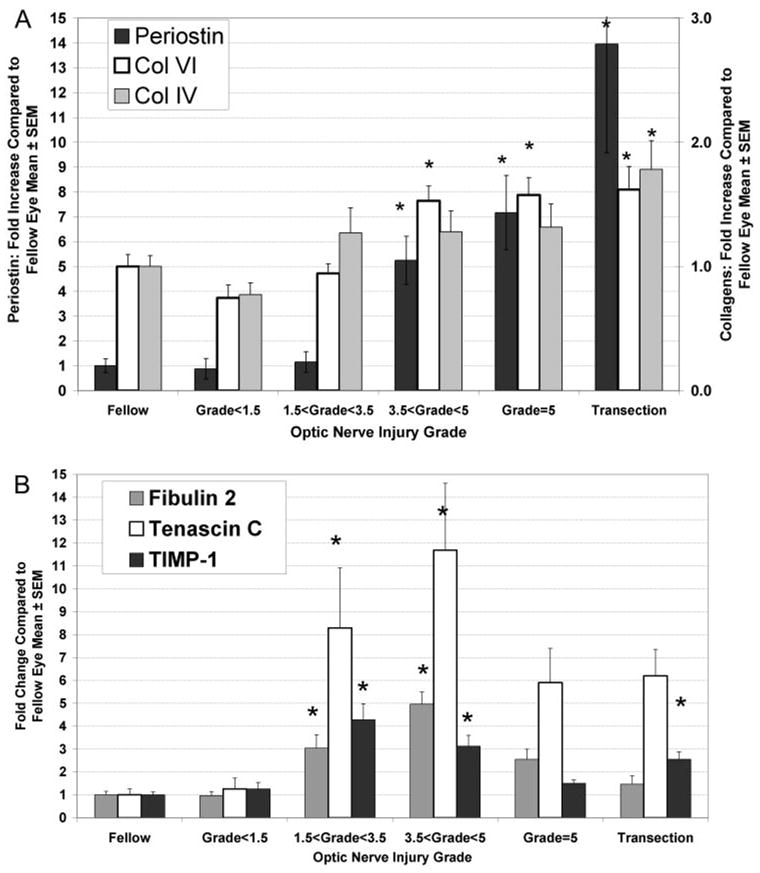
Two response patterns for altered ONH ECM message expression accompanying pressure-induced injury. For the ECM components, periostin, collagen IV, and collagen VI, increasing optic nerve injury was accompanied by linear increases in ONH message level (A). For periostin and collagen VI, ONH groups with injury grades greater than 3.5 demonstrated significant increases compared with the fellow eye group (*P < 0.05). Note that the magnitude of increase for periostin is much greater, as indicated by the fivefold difference in y-axis scale labels. Collagen IV demonstrated a similar pattern, although the increase was only statistically significant in the transection group. In contrast, message levels for fibulin2, tenascin c, and TIMP-1 were highest in ONHs from eyes with focal optic nerve injury (B). *Significant elevation relative to untreated, fellow ONH (P < 0.05). In addition, for fibulin 2, the grade-3.5 to less than grade-5 group, had significantly greater responses than all other groups. For TIMP-1, the grade-1.5 to less than grade-3.5 group, had significantly greater values than all groups, except the grade-3.5 to less than grade-5 group. ANOVA with the Tukey multiple comparison posttest.
However, the pattern expression of other ECM-related messages demonstrated a different relationship to optic nerve injury. Using the same samples, as described in Figures 2A and 2B, we found that fibulin 2, tenascin C, and TIMP-1 mRNA levels showed a significantly nonlinear response to elevated IOP (F test for comparison of linear versus third order polynomial fit, P < 0.03), similar to that demonstrated by cyclin D1. For all three ECM messages, we found that the upregulation of expression in the experimental ONH groups with focal optic nerve injury was greater than in groups from eyes with more extensive injury due either to elevated IOP or transection (Fig. 4B).
In addition, microarray analysis indicated that elevated IOP led to the significant downregulation of some ECM components. Included were osteonectin/SPARC and two other SPARC-related genes (0.53–0.68-fold): collagen XVIII, which contains anti-angiogenic endostatin (0.7-fold) and two other TIMPs, TIMP-3, and -4 (both 0.7-fold). SPARC influences collagen fibril formation50 and has recently been shown to promote retinal ganglion cell survival.51
Differential Regulation of TGFβ Isoforms
Various studies have associated increased expression TGFβ1 and -β2 with human glaucoma or have found responses to experimentally elevated IOP and the deposition of ECM proteins in the ONH.45,52–56 Only the TGFβ2 isoform was present on our microarrays. By microarray analysis, we found a 29% decrease in the TGFβ2 mRNA level, compared with a nonsignificant 19% decrease in grade-5 ONHs by qPCR, using the ONH samples from Figure 2. When samples were grouped by injury mechanism and grade, there were no significant differences between groups (Fig. 5). However, a comparison of linear and nonlinear (third-order polynomial) fits to the expression data indicated a significant nonlinear fit (P < 0.02) in the pressure-injured ONHs. In contrast, the expression of TGFβ1 demonstrated a significant positive linear correlation with optic nerve injury in the ONHs damaged by elevated IOP (P < 0.001), and ANOVA analysis found that the increase was significant for the group with grade-5 injury (Fig. 5; P < 0.01). Two-way ANOVA revealed significant differences in the level of TGFβ1 and -β2 isoform expression in all elevated IOP injury groups with grades greater than 1.5. Together these data indicate that elevated IOP injury to the ONH results in differential regulation of TGFβ isoforms, with an upregulation of TGFβ1 and a relative downregulation of TGFβ2.
Figure 5.
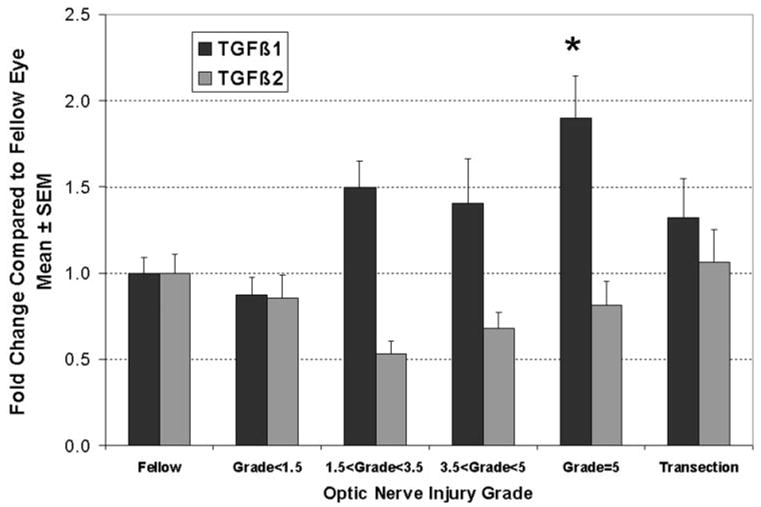
TGFβ isoform mRNAs were differentially regulated in pressure-injured ONH. For TGFβ1, there was a significant increase in mRNA level that correlated with the degree of optic nerve injury, so that levels were significantly elevated in the group of pressure-injured ONHs with grade 5 (*P < 0.01). In contrast, expression levels for TGFβ2 demonstrated a significantly nonlinear correlation to optic nerve injury grade, with lowest levels in ONH with focal optic nerve injury. This resulted in a significant (approximately twofold) difference in the ratio of TGFβ1 to TGFβ2 in all ONHs with pressure-induced optic nerve injury grades greater than 1.5.
Microarray analysis also implicated significantly altered expression of TGFβ receptors and signaling pathway intermediates. Upregulated were TGFβ receptor 1 (2.3-fold), TGFβ receptor 2 (1.5-fold), SMAD3 (1.4-fold), and SMAD-specific E3 ubiquitin protein ligase 1 (SMURF1; 1.5-fold), whereas Pura 1 and Spectrin β2, both regulators of SMAD signaling, were downregulated 0.73- and 0.64-fold, respectively.
Differential Regulation of Cytoskeletal Messages
The cytoskeleton was also a significantly affected cell component with upregulation of 48 and downregulation of 24 genes. Upregulated cytoskeletal genes were primarily associated with mitotic cell division (kinesin family member 22, protein regulator of cytokinesis 1, lamin B1, transforming acidic coiled-coil–containing protein 3, myosin heavy chain IX, centromere protein E, nucleolar and spindle-associated protein 1, and chromosome-transforming acidic coiled-coil–containing protein 3 (3.6- to 2-fold). Other classes were actin cytoskeletal assembly including gene actin-related 2/3 complex proteins, capping proteins, and dynein-1 (1.3- to 1.8 fold).
Among the downregulated genes, the only statistically significant cell component was the cytoskeleton (Table 6). Down-regulated cytoskeletal genes included many retinal ganglion cell–associated genes (kinesin family members 5B and 1B, microtubule-associated protein tau, myosin 1B, adducin 3 (gamma), tropomodulin 2, neurofilament L, and spectrin β2 (0.17- to 0.66-fold). This indicates that the loss of axonal mRNA,30–32 due to axon degeneration, was readily detectible in this study.
Downregulation of Lipid Biosynthesis
In addition to effects on the cytoskeletal cell component, elevated IOP resulted in the downregulation of genes associated with myelin and other lipid biosynthetic processes, including 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1, 24-dehydrocholesterol reductase, sterol-C4-methyl oxidase-like, isopentenyl-diphosphate delta isomerase, farnesyl diphosphate farnesyl transferase 1, low-density lipoprotein receptor, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, and ELOVL family member 6 (0.12- to 0.5-fold). Elevated IOP also decreased transferases, including glutathione transferases (0.5- to 0.65-fold).
Downregulation of Aquaporin-4 mRNA
Preliminary data from our examination of ONH injured by elevated IOP, using transmission electron microscopy, found vacuoles or swellings associated with ONH astrocytic endfeet (Morrison JC et al. IOVS. 2002;43:ARVO E-abstract 2885). Aquaporin-4, the principle water channel in astrocytes, is associated with astrocytic endfeet and is implicated in neural edema. After traumatic brain injury, aquaporin-4 mRNA levels are altered.57–59 Because aquaporin-4 was not on our arrays, we used qPCR to determine the aquaporin-4 mRNA levels in the ONH samples described in Figure 2. We found that mRNA for the water channel protein was significantly decreased (to approximately 50% of that in the fellow eye values in all groups with optic nerve injury; Fig. 6). In addition, there was a significant nonlinear correlation between aquaporin level and the amount of nerve injury in the pressure injured ONHs (P < 0.01), with lowest levels in the ONHs with focal injuries.
Figure 6.
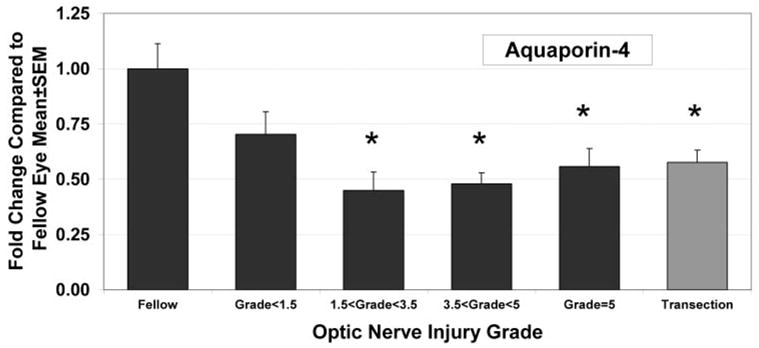
ONH Aquaporin-4 mRNA levels decreased in a pattern similar to that of TGFβ2. For all injury groups, except that with injury grades <1.5, there was a significant decrease in aquaporin-4 mRNA expression (*P < 0.05) compared with levels in the fellow eye. Correlation of expression levels with optic nerve injury grade demonstrated a significant, nonlinear fit, with the lowest expression levels in ONH from eyes with focal optic nerve injury (F test; P < 0.01).
Expression Levels of Connexin 43 and GFAP in ONH are Unaffected by Elevated IOP or Transection
Probes for Connexin 43, the astrocyte gap junctional protein, and GFAP, the principal astrocyte intermediate filament protein, were not present on our microarrays. Our previous immunohistochemical study of ONHs injured by elevated IOP demonstrated early loss of gap junctional labeling and an apparent initial dilution or decrease in GFAP label intensity with injury followed by intensification of GFAP labeling as glial scars were formed.24 We used qPCR and the samples from Figure 2 to measure the levels of these messages in ONH injured by either elevated IOP or transection. There was no significant correlation between optic nerve injury grade and the mRNA levels for these proteins. In addition, when grouped by injury grade, there were no significant differences between groups (Fig. 7). Whereas there appears to be a decrease in these messages in nerves with focal injuries that is similar to that seen for aquaporin-4 and TGFβ2, fit by nonlinear correlation was not significantly better than a linear fit by regression analysis (F test; P > 0.05).
Figure 7.
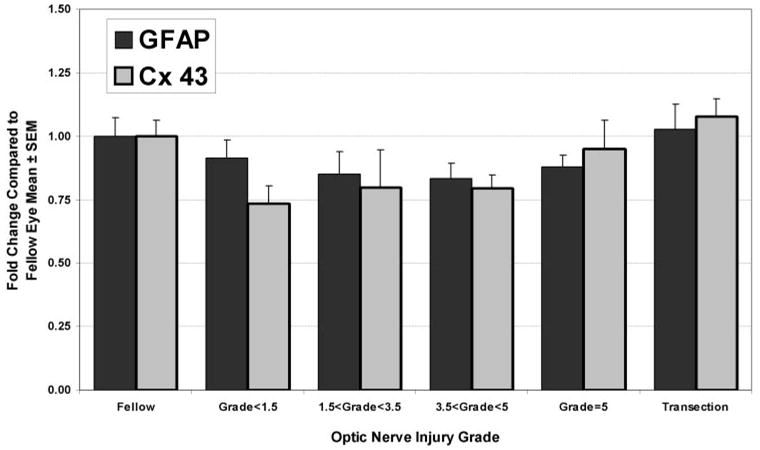
Expression levels of two other proteins associated with differentiated astrocytes, GFAP and connexin43, were not altered in the ONH by either elevated IOP or transection. Although the pattern of expression in the various optic nerve injury groups may appear similar to that seen for TGFβ2 and aquaporin-4, levels in the optic nerve injury groups were not significantly different from fellow eye levels. Regression analysis also indicated no significant correlation.
Discussion
This microarray analysis revealed that the dramatic reorganization of the ONH resulting from pressure-induced injury and associated axonal degeneration is reflected in extensive alterations in gene expression. These alterations significantly affect the processes of cell proliferation, immune response, and mRNA and protein synthesis and the lysosomal, cytoskeletal, ECM, and ribonucleoprotein cellular components. The measurement of selected message levels by qPCR both confirmed and extended these microarray analysis findings. In interpreting these alterations, it is important to recognize that the dissected ONH included portions of both the unmyelinated and the initial myelinated segment of the optic nerve. The myelinated optic nerve is well known to contain progenitor cells that proliferate in response to nerve injury.60–62 In addition, the microarray studies were limited to ONH with substantial damage due to elevated IOP.
Global Patterns of Altered ONH Gene Expression in Glaucoma Model
The most dramatic alteration is the increased cellularity of the nerve head that occurs in response to elevated IOP and ongoing axonal degeneration. Our data indicate that in extensively damaged ONH, the number of cells increased to almost three times the initial value. The large number of regulated cell cycle genes and their association with all cycle phases suggests that the increase in cellularity occurs by proliferation of endogenous nerve head cells, most likely glia or glial precursors. Our previous analysis of time course of nerve head alterations using immunohistochemistry suggested extensive cell division within the ONH glial columns based on heavy labeling with antibodies to proliferating cell nuclear antigen.24 In this study, we used qPCR to examine the message levels for the key regulator of cell cycle progression, cyclin D1, in nerve heads with a wide range of pressure-induced injury. We found that the initiation of cell division appears to be at its maximum in nerve heads that have been exposed to mild IOP elevation and have focal axonal degeneration. In these ONH segments, astrocytes, oligodendroglia, as well as adult optic nerve progenitor cells, potentially contribute to the proliferative response resulting from elevated IOP exposure.60–69 In addition, moderate upregulation of the mRNA for microglial calcium-binding protein, IBA1, the increased immunostaining with IBA1 antibodies and the increased mRNA levels for complement components all suggest microglial activation and a potential for microglial proliferation as well, as has been suggested by other observations.37–42,45,46,70 Studies designed to identify these proliferating ONH cells are currently under way in our laboratory.
Regardless of the cell type(s) involved, the magnitude of the cell proliferation, at least in these extensively injured ONH, implies a similarly extensive disruption of cellular organization within the nerve head. The normal interposition of astrocytes between the vasculature and axons is very likely affected, disrupting the activity-dependent supply of glucose and oxygen to the axons and adversely affecting their ionic environment. Our previous immunohistochemical investigation of ONH after exposure to elevated IOP identified the loss of connexin 43 gap junctional labeling as an early event in pressure-induced nerve injury.24 Disruption of gap junctions implies a reduction in the ability of astrocytes to buffer the axonal environment and supply the metabolic demands of the nerve fibers. These compromised astrocytic functions may become critical factors in axon survival in the unmyelinated anterior portion of the ONH, where the energy demands are significantly higher than those for myelinated nerve.71,72
The proliferation of glial cells in response to injury is well documented in neural tissues, including the myelinated optic nerve.60–68 However, the extent of cellular proliferation in the ONH injured by glaucoma or experimentally elevated IOP, at least in the initial stages of injury, has not been generally recognized or extensively investigated. In rhesus optic nerves after 1 to 4 years of experimental glaucoma, the total number of glia appeared constant, although an increase in the optic nerve area occupied by glial tissue was reported.73 After optic nerve transection, glial cells in the optic nerve and retinal ganglion cell layer have been reported to increase 41% and 210%, respectively,74,75 whereas normal aging in rhesus monkeys results in a 50% increase in optic nerve glia.76 In addition, optic nerve astrocytes and lamina cribrosa cells proliferate in vitro in various experimental conditions.77–80
The formation of an astrocytic scar, labeled by GFAP, is characteristic of glaucomatous injury.24,73,81,82 However, in this study, we found that ONH GFAP mRNA levels were not significantly altered. This observation is consistent with our previous observation that, coincidentally with glial cell migration and disruption of ONH glial columns, GFAP label intensity appeared decreased or diluted.24 Intensified ONH immunolabeling for GFAP occurred later and accompanied glial scar formation. This labeling pattern could suggest the proliferation of adult glial progenitor cells in the pressure injured ONH. Although glial progenitor cells may not express glial cell type markers, they do express nestin, which was significantly upregulated in this study.83 Our observations suggest that glial proliferation and gliotic scar formation are sequential and distinct processes in the ONH response to pressure-induced injury. This sequence may offer windows of opportunity for therapeutic agents to affect the proliferative or scarring processes.
Previously, human glaucoma and experimental glaucoma in both monkeys and rats has been characterized by the deposition of ECM components, including collagens, elastin, tenascin, laminin, fibronectin, and proteoglycans, within the ONH.10 –13,15,17,20,23,24,84,85 It has been hypothesized that these materials increase the susceptibility of the remaining axons to elevated pressure by changing the material properties of the nerve head and may contribute to the inhibition of axon regeneration.22,23,82 Our microarray analysis confirms these previous observations by identifying significantly upregulated expression for the functional classes of ECM, ECM structural constituent, Golgi apparatus, and glycosaminoglycan binding. In addition to confirming the upregulation of collagens I, III, IV, and VI; fibronectin; and tenascin C, previously unidentified ECM components were also found to be significantly affected. Predominant among these was periostin, a fasciclin homologue that induces cell attachment and spreading and binds to heparin and alpha V integrin. Periostin upregulation is also associated with expression of TGFβ, vascular hypertrophy and stress overload.86–88 Another significantly upregulated ECM component identified by our study was fibulin 2, a protein associated with elastic fiber formation.89
Our previous studies in the rat suggested that there is a sequence of ECM deposition and that collagen VI deposition precedes the formation of the glial scar.24 In this study, using qPCR to examine changes in gene expression in relationship to the degree of optic nerve injury, we observed two patterns of gene expression response. For several ECM components, including periostin, collagen VI, and collagen IV, expression increased linearly with increasing optic nerve injury. However, for other ECM components, expression patterns were not linear, but demonstrated the highest expression in ONH with focal injuries. These components included fibulin 2, tenascin C, and the matrix metalloproteinase inhibitor TIMP-1. This nonlinear pattern was similar to that seen for cyclin D1, suggesting that the expression of these ECM components is associated with early injury and ONH cell proliferation.
TGFβ isoforms play important roles in the regulation of the expression of ECM components and in glial progenitor, astrocyte, and microglial proliferation.90–92 Therefore, we examined the changes in TGFβ isoform expression in the ONH in response to elevated IOP. Again, we found two distinct patterns of expression. TGFβ1 expression increased linearly with increasing nerve injury, whereas the nonlinear expression pattern of TGFβ2 resulted in the lowest expression values in focally injured nerves, such that the difference in expression ratios between the two TGFβ isoforms was significant in focally injured nerves. Astrocytes can produce both isoforms, whereas microglia are known to produce TGFβ1, and both isoforms inhibit microglial proliferation.93–95 Previously, Pena et al.52 have reported that whereas TGFβ2 mRNA levels appeared unaffected in glaucomatous ONH, protein levels were increased by both immunohistochemistry and bioassay, leading them to suggest a posttranslational regulation of this isoform. Together, these observations suggest that the differential regulation of TGFβ isoforms may play important roles in the regulation of ONH response to elevated IOP.
In a pattern similar to that of TGFβ2, the principle water channel protein in astrocytes, aquaporin-4, showed the greatest downregulation in ONH from focally injured nerves. Aquaporin-4 immunolocalizes to the astrocytic endfeet that abut brain microvessels and subarachnoidal spaces and is also expressed by activated microglia.96,97 Aquaporins function to maintain neural volume and water homeostasis.98 The astrocytic endfeet probably play a key role in the transfer of metabolites and water between the axons and the vasculature and subarachnoid space. Preliminary data from our electron microscopic studies of nerve heads exposed to elevated IOP identified these endfeet as sites of early injury (Morrison JC et al. IOVS. 2002;43:ARVO E-Abstract 2885). The downregulation of aquaporin-4 may be associated with the retraction of these endfeet accompanying the glial migration or proliferation that is associated with the disruption of the normal columnar arrangement of glial nuclei in the unmyelinated ONH. This morphologic alteration is a characteristic of exposure to elevated IOP in experimental monkey and rat glaucoma models.24,82 Our data confirm these observations and further point to astrocytic responses as early events in axonal injury.
For our qPCR studies, we included a group of ONH at 2 weeks after transection. Axon degeneration in the distal optic nerves from these eyes was similar to that in the highest pressure-induced injury group and gene expression levels were similar, so that no pressure-specific changes in gene expression were identified. However, this study was limited both by the number of genes measured and by the limitation of the transection ONH analysis to a single time point.
Our ONH microarray study also demonstrated downregulation of genes primarily associated with retinal ganglion cells. This most likely reflects a loss of optic nerve axons and the mRNA they contain.30–32 Because axonal mRNA levels are low, we feel this gives a strong indication of the sensitivity of the cDNA microarrays used in the study.
Comparison of This Study with Other Microarray Studies of Cultured ONH Cells
In other microarray studies gene expression has been examined in cells cultured from the normal and glaucomatous ONH or normal ONH cells exposed to conditions hypothesized to be important in glaucoma pathogenesis. Although differences in microarray platform, array genes, species, experimental design, and determination of significance of expression changes complicate comparisons of these studies to ours, some commonalities can be found in all cases. In microarray studies of astrocyte cultures from normal and glaucomatous human ONH, 150 genes were identified as changed in expression.99 These genes represented functional classes of signal transduction, cell adhesion, and proliferation, ECM synthesis, and degradation. In further studies of normal human ONH astrocytes exposed to hydrostatic pressure, approximately 600 genes were found to be altered in expression.100 In common with this study, we also found increased expression of Fos, Ki-67 antigen, AXL receptor tyrosine kinase, Smad3, and Kcnn4 potassium channel. GFAP-negative cells isolated from human ONH have been exposed to mechanical strain or TGFβ1, to model proposed glaucomatous processes. With mechanical strain, the functional classes of ECM, cell proliferation, growth factor activity, and signal transduction were affected in these cells.101 Exposure to TGFβ1 resulted in expression of genes for ECM components, cell proliferation, angiogenesis, and growth factor binding.102 To explore a potential role for epidermal growth factor in glaucoma, rat ONH astrocyte cultures were stimulated with EGF, and the upregulation of EGF receptor and other messages were evaluated.103 In common, we found an upregulation of leukemia inhibitory factor, versican, Cd44, TGFβ1, and endothelin converting enzyme 1 in the pressure-damaged ONH.
Comparison of This Study with Other Glial Cell Microarray Studies
Most microarray studies of neurodegenerative diseases focus on the gene expression changes in affected neuronal populations, rather than those in the glial cells associated with degenerating nerves. However, there are several microarray analyses of glial cell responses that may be compared with our study.
Demyelinating disease, such as multiple sclerosis, is the focus of many of these investigations.104–107 Genes upregulated in demyelinating disease that were also upregulated in our study included complement component 1q, β2 microglobulin, FC receptor, histocompatibility (HLA) antigens, and polyA-binding protein.
Other studies have been conducted to investigate the difference between glial cell environments permissive and restrictive to axon outgrowth and regeneration. Kubo et al.108 examined the changes in gene expression in the distal segment of peripheral nerves after transection, to identify genes associated with an environment supportive of nerve regeneration. In common with this peripheral nerve study, 22 of the same genes were similarly upregulated in the pressure-damaged ONH. These included tenascin C, CDC28 protein kinase regulatory subunit 2, complement component 1qb, prostaglandin E receptor 4, Ms4a6d, SKI-like, cathepsin D, tubulin β2b, and β4 thymosin.
For the study of characteristics of a central nervous system environment conducive to axonal regeneration, cultures of olfactory ensheathing cells (OECs) that promote axon outgrowth were compared to similar OEC cultures that do not.109 Genes associated with a permissive environment that were also upregulated in our study included matrix metalloproteinase 2, versican, brain acidic membrane protein, and calretinin, whereas those upregulated in our study that were associated with an inhibitory environment were insulin-like growth factor 2 receptor, benzodiazepine receptor (translocator protein), and annexin.
Although in all these other studies, the expression of specific genes or gene classes seemed to respond similarly to the changes we observed in the ONH damaged by elevated pressure, it is difficult to draw any overall conclusions. However, several studies of cultured ONH cells did identify cell proliferation, the most significantly affected gene category in our study, as among the affected functional classes of genes.99,101,102
Altered Expression of Specific ONH Genes in Focal Optic Nerve Injury
One unique feature of our study is the use of qPCR to compare selected gene expression changes with the degree of IOP damage. Using this powerful tool, we were able not only to confirm selected microarray findings but to extend them to discover two patterns of gene expression response in ONH injured by elevated IOP. One of these showed a positive correlation with the extent of nerve damage. The other pattern was distinctly nonlinear, demonstrating the greatest change in the eyes with focal injury and thus providing an opportunity to identify early gene changes that may play an important role in the pathogenesis of axon damage.
In ongoing work in our laboratory, we are extending the present study to isolate genes significantly regulated by early, focal injury in the unmyelinated portion of the optic nerve. By identifying these genes, we expect to identify specific processes by which the stress resulting from elevated IOP is translated to axonal injury within the ONH. The identification of these processes and determining their molecular pathways is likely to suggest therapeutic interventions that have the potential of limiting axonal injury in glaucoma.
Acknowledgments
Supported by National Eye Institute Grants R01EY-016866 and R01EY-010145; Alcon Research, Ltd.; and Research to Prevent Blindness.
Footnotes
Presented in part at the annual meeting of the Association for Research in Vision and Ophthalmology, Fort Lauderdale, Florida, May 2005 and May 2006.
Disclosure: E.C. Johnson, None; L. Jia, None; W.O. Cepruna, None; T. Doser, None; J.C. Morrison, Alcon (F).
References
- 1.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sommer A, Katz J, Quigley HA, et al. Clinically detectable nerve fiber atrophy precedes the onset of glaucomatous field loss. Arch Ophthalmol. 1991;109:77–83. doi: 10.1001/archopht.1991.01080010079037. [DOI] [PubMed] [Google Scholar]
- 3.Burgoyne CF, Downs JC, Bellezza AJ, Suh JK, Hart RT. The optic nerve head as a biomechanical structure: a new paradigm for understanding the role of IOP-related stress and strain in the pathophysiology of glaucomatous optic nerve head damage. Prog Retin Eye Res. 2005;24:39–73. doi: 10.1016/j.preteyeres.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Quigley HA, Addicks EM, Green WR, Maumenee AE. Optic nerve damage in human glaucoma. II. The site of injury and susceptibility to damage. Arch Ophthalmol. 1981;99:635–649. doi: 10.1001/archopht.1981.03930010635009. [DOI] [PubMed] [Google Scholar]
- 5.Tuulonen A, Airaksinen PJ. Initial glaucomatous optic disk and retinal nerve fiber layer abnormalities and their progression. Am J Ophthalmol. 1991;111:485–490. doi: 10.1016/s0002-9394(14)72385-2. [DOI] [PubMed] [Google Scholar]
- 6.Quigley HA, Green WR. The histology of human glaucoma cupping and optic nerve damage: clinicopathologic correlation in 21 eyes. Ophthalmology. 1979;86:1803–1830. doi: 10.1016/s0161-6420(79)35338-6. [DOI] [PubMed] [Google Scholar]
- 7.Quigley HA, Addicks EM. Regional differences in the structure of the lamina cribrosa and their relation to glaucomatous optic nerve damage. Arch Ophthalmol. 1981;99:137–143. doi: 10.1001/archopht.1981.03930010139020. [DOI] [PubMed] [Google Scholar]
- 8.Flammer J, Orgul S. Optic nerve blood-flow abnormalities in glaucoma. Prog Retin Eye Res. 1998;17:267–289. doi: 10.1016/s1350-9462(97)00006-2. [DOI] [PubMed] [Google Scholar]
- 9.Tengroth B, Ammitzboll T. Changes in the content and composition of collagen in the glaucomatous eye: basis for a new hypothesis for the genesis of chronic open angle glaucoma—a preliminary report. Acta Ophthalmol (Copenh) 1984;62:999–1008. doi: 10.1111/j.1755-3768.1984.tb08452.x. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez MR, Andrzejewska WM, Neufeld AH. Changes in the extracellular matrix of the human optic nerve head in primary open-angle glaucoma. Am J Ophthalmol. 1990;109:180–188. doi: 10.1016/s0002-9394(14)75984-7. [DOI] [PubMed] [Google Scholar]
- 11.Morrison JC, Dorman-Pease ME, Dunkelberger GR, Quigley HA. Optic nerve head extracellular matrix in primary optic atrophy and experimental glaucoma. Arch Ophthalmol. 1990;108:1020–1024. doi: 10.1001/archopht.1990.01070090122053. [DOI] [PubMed] [Google Scholar]
- 12.Quigley HA, Brown A, Dorman-Pease ME. Alterations in elastin of the optic nerve head in human and experimental glaucoma. Br J Ophthalmol. 1991;75:552–557. doi: 10.1136/bjo.75.9.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quigley HA, Dorman-Pease ME, Brown AE. Quantitative study of collagen and elastin of the optic nerve head and sclera in human and experimental monkey glaucoma. Curr Eye Res. 1991;10:877–888. doi: 10.3109/02713689109013884. [DOI] [PubMed] [Google Scholar]
- 14.Fukuchi T, Sawaguchi S, Hara H, Shirakashi M, Iwata K. Extracellular matrix changes of the optic nerve lamina cribrosa in monkey eyes with experimentally chronic glaucoma. Graefes Arch Clin Exp Ophthalmol. 1992;230:421–427. doi: 10.1007/BF00175926. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez MR. Ultrastructural immunocytochemical analysis of elastin in the human lamina cribrosa: changes in elastic fibers in primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 1992;33:2891–2903. [PubMed] [Google Scholar]
- 16.Fukuchi T, Sawaguchi S, Yue BY, Iwata K, Hara H, Kaiya T. Sulfated proteoglycans in the lamina cribrosa of normal monkey eyes and monkey eyes with laser-induced glaucoma. Exp Eye Res. 1994;58:231–243. doi: 10.1006/exer.1994.1012. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez MR, Ye H, Roy S. Collagen type IV gene expression in human optic nerve heads with primary open angle glaucoma. Exp Eye Res. 1994;59:41–51. doi: 10.1006/exer.1994.1079. [DOI] [PubMed] [Google Scholar]
- 18.Pena JD, Roy S, Hernandez MR. Tropoelastin gene expression in optic nerve heads of normal and glaucomatous subjects. Matrix Biol. 1996;15:323–330. doi: 10.1016/s0945-053x(96)90135-3. [DOI] [PubMed] [Google Scholar]
- 19.Agapova OA, Kaufman PL, Lucarelli MJ, Gabelt BT, Hernandez MR. Differential expression of matrix metalloproteinases in monkey eyes with experimental glaucoma or optic nerve transection. Brain Res. 2003;967:132–143. doi: 10.1016/s0006-8993(02)04234-8. [DOI] [PubMed] [Google Scholar]
- 20.Pena JD, Varela HJ, Ricard CS, Hernandez MR. Enhanced tenascin expression associated with reactive astrocytes in human optic nerve heads with primary open angle glaucoma. Exp Eye Res. 1999;68:29–40. doi: 10.1006/exer.1998.0577. [DOI] [PubMed] [Google Scholar]
- 21.Morrison JC, Moore CG, Deppmeier LM, Gold BG, Meshul CK, Johnson EC. A rat model of chronic pressure-induced optic nerve damage. Exp Eye Res. 1997;64:85–96. doi: 10.1006/exer.1996.0184. [DOI] [PubMed] [Google Scholar]
- 22.Morrison JC, Johnson EC, Cepurna W, Jia L. Understanding mechanisms of pressure-induced optic nerve damage. Prog Retin Eye Res. 2005;24:217–240. doi: 10.1016/j.preteyeres.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Johnson EC, Morrison JC, Farrell S, Deppmeier L, Moore CG, McGinty MR. The effect of chronically elevated intraocular pressure on the rat optic nerve head extracellular matrix. Exp Eye Res. 1996;62:663–674. doi: 10.1006/exer.1996.0077. [DOI] [PubMed] [Google Scholar]
- 24.Johnson EC, Deppmeier LM, Wentzien SK, Hsu I, Morrison JC. Chronology of optic nerve head and retinal responses to elevated intraocular pressure. Invest Ophthalmol Vis Sci. 2000;41:431–442. [PubMed] [Google Scholar]
- 25.Jia L, Cepurna WO, Johnson EC, Morrison JC. Patterns of intraocular pressure elevation after aqueous humor outflow obstruction in rats. Invest Ophthalmol Vis Sci. 2000;41:1380–1385. [PubMed] [Google Scholar]
- 26.Jia L, Cepurna WO, Johnson EC, Morrison JC. Effect of general anesthetics on IOP in rats with experimental aqueous outflow obstruction. Invest Ophthalmol Vis Sci. 2000;41:3415–3419. [PubMed] [Google Scholar]
- 27.Yang YH, Dudoit S, Luu P, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang IH, Johnson EC, Jia L, et al. Evaluation of inducible nitric oxide synthase in glaucomatous optic neuropathy and pressure-induced optic nerve damage. Invest Ophthalmol Vis Sci. 2005;46:1313–1321. doi: 10.1167/iovs.04-0829. [DOI] [PubMed] [Google Scholar]
- 29.Schlamp CL, Johnson EC, Li Y, Morrison JC, Nickells RW. Changes in Thy1 gene expression associated with damaged retinal ganglion cells. Mol Vis. 2001;7:192–201. [PubMed] [Google Scholar]
- 30.Piper M, Holt C. RNA translation in axons. Annu Rev Cell Dev Biol. 2004;20:505–523. doi: 10.1146/annurev.cellbio.20.010403.111746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willis D, Li KW, Zheng JQ, et al. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J Neurosci. 2005;25:778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng JQ, Kelly TK, Chang B, et al. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiese C, Rolletschek A, Kania G, et al. Nestin expression: a property of multi-lineage progenitor cells? Cell Mol Life Sci. 2004;61:2510–2522. doi: 10.1007/s00018-004-4144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grozdanic SD, Betts DM, Sakaguchi DS, Allbaugh RA, Kwon YH, Kardon RH. Laser-induced mouse model of chronic ocular hypertension. Invest Ophthalmol Vis Sci. 2003;44:4337–4346. doi: 10.1167/iovs.03-0015. [DOI] [PubMed] [Google Scholar]
- 35.Becker EB, Bonni A. Cell cycle regulation of neuronal apoptosis in development and disease. Prog Neurobiol. 2004;72:1–25. doi: 10.1016/j.pneurobio.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 36.Herrup K, Neve R, Ackerman SL, Copani A. Divide and die: cell cycle events as triggers of nerve cell death. J Neurosci. 2004;24:9232–9239. doi: 10.1523/JNEUROSCI.3347-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnum SR. Complement biosynthesis in the central nervous system. Crit Rev Oral Biol Med. 1995;6:132–146. doi: 10.1177/10454411950060020301. [DOI] [PubMed] [Google Scholar]
- 38.Lynch NJ, Willis CL, Nolan CC, et al. Microglial activation and increased synthesis of complement component C1q precedes blood-brain barrier dysfunction in rats. Mol Immunol. 2004;40:709–716. doi: 10.1016/j.molimm.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Song X, Shapiro S, Goldman DL, Casadevall A, Scharff M, Lee SC. Fcgamma receptor I- and III-mediated macrophage inflammatory protein 1alpha induction in primary human and murine microglia. Infect Immun. 2002;70:5177–5184. doi: 10.1128/IAI.70.9.5177-5184.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanguri P. Interferon-gamma-inducible genes in primary glial cells of the central nervous system: comparisons of astrocytes with microglia and Lewis with brown Norway rats. J Neuroimmunol. 1995;56:35–43. doi: 10.1016/0165-5728(94)00131-7. [DOI] [PubMed] [Google Scholar]
- 41.O’Keefe GM, Nguyen VT, Benveniste EN. Regulation and function of class II major histocompatibility complex, CD40, and B7 expression in macrophages and microglia: Implications in neurological diseases. J Neurovirol. 2002;8:496–512. doi: 10.1080/13550280290100941. [DOI] [PubMed] [Google Scholar]
- 42.Lehrmann E, Christensen T, Zimmer J, Diemer NH, Finsen B. Microglial and macrophage reactions mark progressive changes and define the penumbra in the rat neocortex and striatum after transient middle cerebral artery occlusion. J Comp Neurol. 1997;386:461–476. [PubMed] [Google Scholar]
- 43.Molina H. The murine complement regulator Crry: new insights into the immunobiology of complement regulation. Cell Mol Life Sci. 2002;59:220–229. doi: 10.1007/s00018-002-8418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davoust N, Nataf S, Holers VM, Barnum SR. Expression of the murine complement regulatory protein crry by glial cells and neurons. Glia. 1999;27:162–170. doi: 10.1002/(sici)1098-1136(199908)27:2<162::aid-glia6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 45.Yuan L, Neufeld AH. Activated microglia in the human glaucomatous optic nerve head. J Neurosci Res. 2001;64:523–532. doi: 10.1002/jnr.1104. [DOI] [PubMed] [Google Scholar]
- 46.Lam TT, Kwong JM, Tso MO. Early glial responses after acute elevated intraocular pressure in rats. Invest Ophthalmol Vis Sci. 2003;44:638–645. doi: 10.1167/iovs.02-0255. [DOI] [PubMed] [Google Scholar]
- 47.Ohsawa K, Imai Y, Kanazawa H, Sasaki Y, Kohsaka S. Involvement of Iba1 in membrane ruffling and phagocytosis of macrophages/microglia. J Cell Sci. 2000;113:3073–3084. doi: 10.1242/jcs.113.17.3073. [DOI] [PubMed] [Google Scholar]
- 48.Origasa M, Tanaka S, Suzuki K, Tone S, Lim B, Koike T. Activation of a novel microglial gene encoding a lysosomal membrane protein in response to neuronal apoptosis. Brain Res Mol Brain Res. 2001;88:1–13. doi: 10.1016/s0169-328x(01)00005-5. [DOI] [PubMed] [Google Scholar]
- 49.Ahmed F, Brown KM, Stephan DA, Morrison JC, Johnson EC, Tomarev SI. Microarray analysis of changes in mRNA levels in the rat retina after experimental elevation of intraocular pressure. Invest Ophthalmol Vis Sci. 2004;45:1247–1258. doi: 10.1167/iovs.03-1123. [DOI] [PubMed] [Google Scholar]
- 50.Bradshaw AD, Sage EH. SPARC, a matricellular protein that functions in cellular differentiation and tissue response to injury. J Clin Invest. 2001;107:1049–1054. doi: 10.1172/JCI12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bampton ET, Ma CH, Tolkovsky AM, Taylor JS. Osteonectin is a Schwann cell-secreted factor that promotes retinal ganglion cell survival and process outgrowth. Eur J Neurosci. 2005;21:2611–2623. doi: 10.1111/j.1460-9568.2005.04128.x. [DOI] [PubMed] [Google Scholar]
- 52.Pena JD, Taylor AW, Ricard CS, Vidal I, Hernandez MR. Transforming growth factor beta isoforms in human optic nerve heads. Br J Ophthalmol. 1999;83:209–218. doi: 10.1136/bjo.83.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuchshofer R, Birke M, Welge-Lussen U, Kook D, Lütjen-Drecoll E. Transforming growth factor-beta 2 modulated extracellular matrix component expression in cultured human optic nerve head astrocytes. Invest Ophthalmol Vis Sci. 2005;46:568–578. doi: 10.1167/iovs.04-0649. [DOI] [PubMed] [Google Scholar]
- 54.Guo L, Moss SE, Alexander RA, Ali RR, Fitzke FW, Cordeiro MF. Retinal ganglion cell apoptosis in glaucoma is related to intraocular pressure and IOP-induced effects on extracellular matrix. Invest Ophthalmol Vis Sci. 2005;46:175–182. doi: 10.1167/iovs.04-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirwan RP, Crean JK, Fenerty CH, Clark AF, O’Brien CJ. Effect of cyclical mechanical stretch and exogenous transforming growth factor-beta1 on matrix metalloproteinase-2 activity in lamina cribrosa cells from the human optic nerve head. J Glaucoma. 2004;13:327–334. doi: 10.1097/00061198-200408000-00011. [DOI] [PubMed] [Google Scholar]
- 56.Fukuchi T, Ueda J, Hanyu T, Abe H, Sawaguchi S. Distribution and expression of transforming growth factor-beta and platelet-derived growth factor in the normal and glaucomatous monkey optic nerve heads. Jpn J Ophthalmol. 2001;45:592–599. doi: 10.1016/s0021-5155(01)00414-2. [DOI] [PubMed] [Google Scholar]
- 57.Ke C, Poon WS, Ng HK, Pang JC, Chan Y. Heterogeneous responses of aquaporin-4 in oedema formation in a replicated severe traumatic brain injury model in rats. Neurosci Lett. 2001;301:21–24. doi: 10.1016/s0304-3940(01)01589-0. [DOI] [PubMed] [Google Scholar]
- 58.Sun MC, Honey CR, Berk C, Wong NL, Tsui JK. Regulation of aquaporin-4 in a traumatic brain injury model in rats. J Neurosurg. 2003;98:565–569. doi: 10.3171/jns.2003.98.3.0565. [DOI] [PubMed] [Google Scholar]
- 59.Vizuete ML, Venero JL, Vargas C, et al. Differential upregulation of aquaporin-4 mRNA expression in reactive astrocytes after brain injury: potential role in brain edema. Neurobiol Dis. 1999;6:245–258. doi: 10.1006/nbdi.1999.0246. [DOI] [PubMed] [Google Scholar]
- 60.Shi J, Marinovich A, Barres BA. Purification and characterization of adult oligodendrocyte precursor cells from the rat optic nerve. J Neurosci. 1998;18:4627–4636. doi: 10.1523/JNEUROSCI.18-12-04627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Privat A, Valat J, Fulcrand J. Proliferation of neuroglial cell lines in the degenerating optic nerve of young rats: a radioautographic study. J Neuropathol Exp Neurol. 1981;40:46–60. [PubMed] [Google Scholar]
- 62.Wren D, Wolswijk G, Noble M. In vitro analysis of the origin and maintenance of O-2A adult progenitor cells. J Cell Biol. 1992;116:167–176. doi: 10.1083/jcb.116.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang H, Lu P, McKay HM, et al. Endogenous neurogenesis replaces oligodendrocytes and astrocytes after primate spinal cord injury. J Neurosci. 2006;26:2157–2166. doi: 10.1523/JNEUROSCI.4070-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zai LJ, Wrathall JR. Cell proliferation and replacement following contusive spinal cord injury. Glia. 2005;50:247–257. doi: 10.1002/glia.20176. [DOI] [PubMed] [Google Scholar]
- 65.Tatsumi K, Haga S, Matsuyoshi H, et al. Characterization of cells with proliferative activity after a brain injury. Neurochem Int. 2005;46:381–389. doi: 10.1016/j.neuint.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 66.Vroemen M, Aigner L, Winkler J, Weidner N. Adult neural progenitor cell grafts survive after acute spinal cord injury and integrate along axonal pathways. Eur J Neurosci. 2003;18:743–751. doi: 10.1046/j.1460-9568.2003.02804.x. [DOI] [PubMed] [Google Scholar]
- 67.Kernie SG, Erwin TM, Parada LF. Brain remodeling due to neuronal and astrocytic proliferation after controlled cortical injury in mice. J Neurosci Res. 2001;66:317–326. doi: 10.1002/jnr.10013. [DOI] [PubMed] [Google Scholar]
- 68.Noble M, Wren D, Wolswijk G. The O-2A (adult) progenitor cell: a glial stem cell of the adult central nervous system. Semin Cell Biol. 1992;3:413–422. doi: 10.1016/1043-4682(92)90012-k. [DOI] [PubMed] [Google Scholar]
- 69.Wolswijk G, Noble M. Cooperation between PDGF and FGF converts slowly dividing O-2Aadult progenitor cells to rapidly dividing cells with characteristics of O-2Aperinatal progenitor cells. J Cell Biol. 1992;118:889–900. doi: 10.1083/jcb.118.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neufeld AH. Microglia in the optic nerve head and the region of parapapillary chorioretinal atrophy in glaucoma. Arch Ophthalmol. 1999;117:1050–1056. doi: 10.1001/archopht.117.8.1050. [DOI] [PubMed] [Google Scholar]
- 71.Hildebrand C, Remahl S, Persson H, Bjartmar C. Myelinated nerve fibres in the CNS. Prog Neurobiol. 1993;40:319–384. doi: 10.1016/0301-0082(93)90015-k. [DOI] [PubMed] [Google Scholar]
- 72.Barron MJ, Griffiths P, Turnbull DM, Bates D, Nichols P. The distributions of mitochondria and sodium channels reflect the specific energy requirements and conduction properties of the human optic nerve head. Br J Ophthalmol. 2004;88:286–290. doi: 10.1136/bjo.2003.027664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Furuyoshi N, Furuyoshi M, May CA, Hayreh SS, Alm A, Lütjen-Drecoll E. Vascular and glial changes in the retrolaminar optic nerve in glaucomatous monkey eyes. Ophthalmologica. 2000;214:24–32. doi: 10.1159/000027470. [DOI] [PubMed] [Google Scholar]
- 74.Vaughn JE, Hinds PL, Skoff RP. Electron microscopic studies of Wallerian degeneration in rat optic nerves, I: the multipotential glia. J Comp Neurol. 1970;140:175–206. doi: 10.1002/cne.901400204. [DOI] [PubMed] [Google Scholar]
- 75.Gellrich MM, Gellrich NC. Quantitative relations in the retinal ganglion cell layer of the rat: neurons, glia and capillaries before and after optic nerve section. Graefes Arch Clin Exp Ophthalmol. 1996;234:315–323. doi: 10.1007/BF00220707. [DOI] [PubMed] [Google Scholar]
- 76.Sandell JH, Peters A. Effects of age on the glial cells in the rhesus monkey optic nerve. J Comp Neurol. 2002;445:13–28. doi: 10.1002/cne.10162. [DOI] [PubMed] [Google Scholar]
- 77.Desai D, He S, Yorio T, Krishnamoorthy RR, Prasanna G. Hypoxia augments TNF-alpha-mediated endothelin-1 release and cell proliferation in human optic nerve head astrocytes. Biochem Biophys Res Commun. 2004;318:642–648. doi: 10.1016/j.bbrc.2004.04.073. [DOI] [PubMed] [Google Scholar]
- 78.Prasanna G, Krishnamoorthy R, Clark AF, Wordinger RJ, Yorio T. Human optic nerve head astrocytes as a target for endothelin-1. Invest Ophthalmol Vis Sci. 2002;43:2704–2713. [PubMed] [Google Scholar]
- 79.Lambert WS, Clark AF, Wordinger RJ. Effect of exogenous neurotrophins on Trk receptor phosphorylation, cell proliferation, and neurotrophin secretion by cells isolated from the human lamina cribrosa. Mol Vis. 2004;10:289–296. [PubMed] [Google Scholar]
- 80.Agapova OA, Malone PE, Hernandez MR. A neuroactive steroid 5alpha-androstane-3alpha,17beta-diol regulates androgen receptor level in astrocytes. J Neurochem. 2006;98:355–363. doi: 10.1111/j.1471-4159.2006.03879.x. [DOI] [PubMed] [Google Scholar]
- 81.Varela HJ, Hernandez MR. Astrocyte responses in human optic nerve head with primary open-angle glaucoma. J Glaucoma. 1997;6:303–313. [PubMed] [Google Scholar]
- 82.Hernandez MR. The optic nerve head in glaucoma: role of astrocytes in tissue remodeling. Prog Retin Eye Res. 2000;19:297–321. doi: 10.1016/s1350-9462(99)00017-8. [DOI] [PubMed] [Google Scholar]
- 83.Quinn SM, Walters WM, Vescovi AL, Whittemore SR. Lineage restriction of neuroepithelial precursor cells from fetal human spinal cord. J Neurosci Res. 1999;57:590–602. [PubMed] [Google Scholar]
- 84.Pena JD, Agapova O, Gabelt BT, et al. Increased elastin expression in astrocytes of the lamina cribrosa in response to elevated intraocular pressure. Invest Ophthalmol Vis Sci. 2001;42:2303–2314. [PubMed] [Google Scholar]
- 85.Gong H, Ye W, Freddo TF, Hernandez MR. Hyaluronic acid in the normal and glaucomatous optic nerve. Exp Eye Res. 1997;64:587–595. doi: 10.1006/exer.1996.0245. [DOI] [PubMed] [Google Scholar]
- 86.Litvin J, Blagg A, Mu A, et al. Periostin and periostin-like factor in the human heart: possible therapeutic targets. Cardiovasc Pathol. 2006;15:24–32. doi: 10.1016/j.carpath.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 87.Wang D, Oparil S, Feng JA, et al. Effects of pressure overload on extracellular matrix expression in the heart of the atrial natriuretic peptide-null mouse. Hypertension. 2003;42:88–95. doi: 10.1161/01.HYP.0000074905.22908.A6. [DOI] [PubMed] [Google Scholar]
- 88.Horiuchi K, Amizuka N, Takeshita S, et al. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res. 1999;14:1239–1249. doi: 10.1359/jbmr.1999.14.7.1239. [DOI] [PubMed] [Google Scholar]
- 89.Hunzelmann N, Nischt R, Brenneisen P, Eickert A, Krieg T. Increased deposition of fibulin-2 in solar elastosis and its colocalization with elastic fibres. Br J Dermatol. 2001;145:217–222. doi: 10.1046/j.1365-2133.2001.04337.x. [DOI] [PubMed] [Google Scholar]
- 90.Verrecchia F, Mauviel A. Transforming growth factor-beta signaling through the Smad pathway: role in extracellular matrix gene expression and regulation. J Invest Dermatol. 2002;118:211–215. doi: 10.1046/j.1523-1747.2002.01641.x. [DOI] [PubMed] [Google Scholar]
- 91.Wachs FP, Winner B, Couillard-Despres S, et al. Transforming growth factor-beta1 is a negative modulator of adult neurogenesis. J Neuropathol Exp Neurol. 2006;65:358–370. doi: 10.1097/01.jnen.0000218444.53405.f0. [DOI] [PubMed] [Google Scholar]
- 92.Gomes FC, Sousa Vde O, Romao L. Emerging roles for TGF-beta1 in nervous system development. Int J Dev Neurosci. 2005;23:413–424. doi: 10.1016/j.ijdevneu.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 93.Dhandapani KM, Hadman M, De Sevilla L, Wade MF, Mahesh VB, Brann DW. Astrocyte protection of neurons: role of transforming growth factor-beta signaling via a c-Jun-AP-1 protective pathway. J Biol Chem. 2003;278:43329–43339. doi: 10.1074/jbc.M305835200. [DOI] [PubMed] [Google Scholar]
- 94.Constam DB, Philipp J, Malipiero UV, ten Dijke P, Schachner M, Fontana A. Differential expression of transforming growth factor-beta 1, -beta 2, and -beta 3 by glioblastoma cells, astrocytes, and microglia. J Immunol. 1992;148:1404–1410. [PubMed] [Google Scholar]
- 95.Jones LL, Kreutzberg GW, Raivich G. Transforming growth factor beta’s 1, 2 and 3 inhibit proliferation of ramified microglia on an astrocyte monolayer. Brain Res. 1998;795:301–306. doi: 10.1016/s0006-8993(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 96.Nielsen S, Nagelhus EA, Amiry-Moghaddam M, Bourque C, Agre P, Ottersen OP. Specialized membrane domains for water transport in glial cells: high-resolution immunogold cytochemistry of aquaporin-4 in rat brain. J Neurosci. 1997;17:171–180. doi: 10.1523/JNEUROSCI.17-01-00171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Inoue M, Wakayama Y, Liu JW, Murahashi M, Shibuya S, Oniki H. Ultrastructural localization of aquaporin 4 and alpha1-syntrophin in the vascular feet of brain astrocytes. Tohoku J Exp Med. 2002;197:87–93. doi: 10.1620/tjem.197.87. [DOI] [PubMed] [Google Scholar]
- 98.Nagelhus EA, Mathiisen TM, Ottersen OP. Aquaporin-4 in the central nervous system: cellular and subcellular distribution and coexpression with KIR4.1. Neuroscience. 2004;129:905–913. doi: 10.1016/j.neuroscience.2004.08.053. [DOI] [PubMed] [Google Scholar]
- 99.Hernandez MR, Agapova OA, Yang P, Salvador-Silva M, Ricard CS, Aoi S. Differential gene expression in astrocytes from human normal and glaucomatous optic nerve head analyzed by cDNA microarray. Glia. 2002;38:45–64. doi: 10.1002/glia.10051. [DOI] [PubMed] [Google Scholar]
- 100.Yang P, Agapova O, Parker A, et al. DNA microarray analysis of gene expression in human optic nerve head astrocytes in response to hydrostatic pressure. Physiol Genomics. 2004;17:157–169. doi: 10.1152/physiolgenomics.00182.2003. [DOI] [PubMed] [Google Scholar]
- 101.Kirwan RP, Fenerty CH, Crean J, Wordinger RJ, Clark AF, O’Brien CJ. Influence of cyclical mechanical strain on extracellular matrix gene expression in human lamina cribrosa cells in vitro. Mol Vis. 2005;11:798–810. [PubMed] [Google Scholar]
- 102.Kirwan RP, Leonard MO, Murphy M, Clark AF, O’Brien CJ. Transforming growth factor-beta-regulated gene transcription and protein expression in human GFAP-negative lamina cribrosa cells. Glia. 2005;52:309–324. doi: 10.1002/glia.20247. [DOI] [PubMed] [Google Scholar]
- 103.Liu B, Chen H, Johns TG, Neufeld AH. Epidermal growth factor receptor activation: an upstream signal for transition of quiescent astrocytes into reactive astrocytes after neural injury. J Neurosci. 2006;26:7532–7540. doi: 10.1523/JNEUROSCI.1004-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ibrahim SM, Mix E, Bottcher T, et al. Gene expression profiling of the nervous system in murine experimental autoimmune enceph-alomyelitis. Brain. 2001;124:1927–1938. doi: 10.1093/brain/124.10.1927. [DOI] [PubMed] [Google Scholar]
- 105.Brand-Schieber E, Werner P, Iacobas DA, et al. Connexin43, the major gap junction protein of astrocytes, is down-regulated in inflamed white matter in an animal model of multiple sclerosis. J Neurosci Res. 2005;80:798–808. doi: 10.1002/jnr.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Renaud S, Hays AP, Brannagan TH, 3rd, et al. Gene expression profiling in chronic inflammatory demyelinating polyneuropathy. J Neuroimmunol. 2005;159:203–214. doi: 10.1016/j.jneuroim.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 107.Tajouri L, Mellick AS, Ashton KJ, et al. Quantitative and qualitative changes in gene expression patterns characterize the activity of plaques in multiple sclerosis. Brain Res Mol Brain Res. 2003;119:170–183. doi: 10.1016/j.molbrainres.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 108.Kubo T, Yamashita T, Yamaguchi A, Hosokawa K, Tohyama M. Analysis of genes induced in peripheral nerve after axotomy using DNA microarrays. J Neurochem. 2002;82:1129–1136. doi: 10.1046/j.1471-4159.2002.01060.x. [DOI] [PubMed] [Google Scholar]
- 109.Pastrana E, Moreno-Flores MT, Gurzov EN, Avila J, Wandosell F, Diaz-Nido J. Genes associated with adult axon regeneration promoted by olfactory ensheathing cells: a new role for matrix metalloproteinase 2. J Neurosci. 2006;26:5347–5359. doi: 10.1523/JNEUROSCI.1111-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


