Abstract
Meiotic checkpoints monitor chromosome status to ensure correct homologous recombination, genomic integrity, and chromosome segregation. In Drosophila, the persistent presence of double-strand DNA breaks (DSB) activates the ATR/Mei-41 checkpoint, delays progression through meiosis, and causes defects in DNA condensation of the oocyte nucleus, the karyosome. Checkpoint activation has also been linked to decreased levels of the TGFα-like molecule Gurken, which controls normal eggshell patterning. We used this easy-to-score eggshell phenotype in a germ-line mosaic screen in Drosophila to identify new genes affecting meiotic progression, DNA condensation, and Gurken signaling. One hundred eighteen new ventralizing mutants on the second chromosome fell into 17 complementation groups. Here we describe the analysis of 8 complementation groups, including Kinesin heavy chain, the SR protein kinase cuaba, the cohesin-related gene dPds5/cohiba, and the Tudor-domain gene montecristo. Our findings challenge the hypothesis that checkpoint activation upon persistent DSBs is exclusively mediated by ATR/Mei-41 kinase and instead reveal a more complex network of interactions that link DSB formation, checkpoint activation, meiotic delay, DNA condensation, and Gurken protein synthesis.
AS cells divide, checkpoints delay the transition to the next phase of each cycle until the previous phase is completed to ensure the genomic stability of the daughter cells (Kuzminov 2001). During meiosis in yeast, this surveillance allows the correct reduction of the DNA content into fully functional gametes (Roeder and Bailis 2000). Similarly, in Drosophila, the activation of a meiotic checkpoint is thought to delay meiotic progression in the oocyte (Huynh and St Johnston 2000).
Drosophila oogenesis begins at the anterior tip of the germarium, as cystoblasts divide synchronously to form cysts of 16 interconnected germ cells (Figure 1) (Spradling 1993; de Cuevas et al. 1997). Several cells in each cyst enter meiotic prophase, condense their chromosomes, form synapses between the homologs, and repair double-strand breaks (DSB) in the DNA, but only one cystocyte reaches the full pachytene state (Carpenter 1979; Page and Hawley 2001, 2004; Jang et al. 2003). Mutations in meiotic genes, such as the DSB repair genes okra/dRad54 (okr) and spindle-A/dRad51 (spn-A), delay this restriction. In later stages, DSB repair mutants show fragmented or thread-like chromatin organization within the oocyte nucleus instead of condensing into a hollow spherical “karyosome” as in the wild type (Gonzalez-Reyes et al. 1997; Huynh and St Johnston 2000; Staeva-Vieira et al. 2003).
Figure 1.—
Schematic of the onset of Drosophila female meiosis. The germarium is the most anterior structure of the Drosophila ovariole, where germ-line stem cells produce cystoblasts by asymmetric division. Germaria are divided into three regions. In region 1, cystoblast formation is followed by four rounds of mitosis, which give rise to cysts of 16 interconnected cells. In region 2, several cells per cyst initiate the assembly of synaptic chromosomes and form DSBs through the activity of the Drosophila SPO11 homologs mei-W68 and mei-P22. In region 3A, cysts become surrounded by follicle cells (not shown) and reorient so that each oocyte is placed at the posterior pole of each cluster where it remains for the rest of oogenesis. At stage 2, meiotic restriction to the oocyte (oo) is completed. The other “synaptic” cystocytes regress from meiosis and become nurse cells (nc). As the oocyte leaves pachytene, the meiotic chromatin releases the synaptonemal complex components and condenses into a karyosome (blue doughnut). In mid-oogenesis, the oocyte nucleus moves anteriorly. Tightly associated with it, Grk will be secreted to the adjacent follicle cells (gray grid), which will acquire a dorsal fate and later synthesize the dorsal appendages of the chorion (not shown). Persistent DSB in spn-A and okr mutants activate Mei-41, which causes a decrease in Grk production and consequent ventralization of the eggshell. The link between this activation and delays in meiotic progression is unclear (blue question mark).
In addition to delays in meiotic restriction, females with mutant DSB repair enzymes lay eggs with dorsal–ventral (DV) defects known as the spindle phenotype (Gonzalez-Reyes et al. 1997; Ghabrial et al. 1998; Morris and Lehmann 1999; Staeva-Vieira et al. 2003). The persistence of DSBs in these mutants activates the ATR-like kinase Mei-41 and the Drosophila checkpoint protein 2 (dChk2) (Ghabrial and Schupbach 1999; Abdu et al. 2002; Staeva-Vieira et al. 2003). This activation causes modification of Vasa (Vas), a germ-line-specific ATP-dependent helicase required for translation of a number of mRNAs, including that of the TGFα-like molecule gurken (grk) (Neuman-Silberberg and Schupbach 1994; Styhler et al. 1998; Ghabrial and Schupbach 1999). Grk is required during mid-oogenesis to establish the DV axis of the future eggshell (Ghiglione et al. 2002). Therefore, the activation of the Mei-41 checkpoint can prevent oocyte development.
Little is known about the effectors of the meiotic checkpoint that lead to delays in meiotic progression or oocyte polarity defects. Previous genetic screens based on female sterility and the spindle phenotype also identified DV polarity genes with functions other than DSB repair (Schupbach and Wieschaus 1989, 1991; Morris et al. 2003; Staeva-Vieira et al. 2003). These included genes required for grk mRNA transport (Swan and Suter 1996; Swan et al. 1999; Brendza et al. 2000; Huynh and St Johnston 2000; Navarro et al. 2004; Abdu et al. 2006), genes required for Grk processing (Styhler et al. 1998; Miura et al. 2006), and genes regulating the stability and trafficking of Grk protein (Saunders and Cohen 1999; Kennerdell et al. 2002; Findley et al. 2003; Wilhelm et al. 2005; Bokel et al. 2006).
In addition to DNA repair, different checkpoint pathways may monitor other meiotic steps (Bhalla and Dernburg 2005). Here we describe a large-scale mutagenesis screen to isolate mutations in genes linking the control of meiotic progression with oocyte development. Using Grk expression in the oocyte, oocyte nuclear markers, and genetic analysis, we characterize eight loci and group them into phenotypic classes according to their effect on meiotic chromatin condensation, DSB formation, meiotic checkpoint pathway activation, and DV polarity. Among DSB repair candidate genes, we identify the Drosophila cohesin-related gene dPds5 and the novel Tudor-domain gene montecristo (mtc). The characterization of their phenotypes provides evidence that additional mechanisms independent of the meiotic checkpoint kinase, Mei-41, delay meiosis and affect oocyte polarity. Two other mutants, indios (nds) and trinidad (trin), suggest the possibility that the process of meiotic chromatin condensation can be uncoupled from the Grk-mediated signaling pathway.
MATERIALS AND METHODS
Fly stocks:
The following alleles identified in the screen were used for phenotypic analysis: khcpgs1, khcpgs2, eight dPds5cohiba alleles (1–8), blv1, blv 2, nds2, nds 4, trin1, trin 3, three mtc alleles (1–3), three srpkcuaba alleles (1–3), and bha1 and bha 2. mei-41D3, mei-W681, mei-P221, spn-A1, spn-A093A, okrRU, and okrAA were present in the laboratory. khc27, P{lacW}l(2)k1223, PBac{WH}CG15707f06583, the 2R deficiencies, and all the starting lines used in the screen (see below) were obtained from Bloomington Stock Center. All flies were raised at 25° unless otherwise indicated.
Mutagenesis:
The screen was carried out as previously described for the 3R chromosome (Yohn et al. 2003). Males of the genotype w P{w+faf-LacZ}; P{w+ FRT 42B} were starved and treated with 25 or 35 mm ethylmethane sulfonate (EMS) (Sigma, St. Louis) in 1% sucrose for 16–24 hr as described (Ashburner 1989). Lac-Z expression from the P{w+faf-LacZ} transgene localizes to the pole plasm and persists in the primordial germ cells throughout embryogenesis. P{w+ FRT 42B} carries the FRT sequence in region 42B of a recently isogenized second chromosome. Mutagenized males were crossed to females of the genotype w P{w+faf-LacZ}; If/CyO hs-hid, in which CyO hs-hid is a balancer CyO bearing the heat-shock-inducible proapoptotic transgene head involution defective (hid). This cross was set up in bottles and flipped daily to fresh food four times, after which males were discarded. Twenty thousand single virgin females carrying a mutagenized P{w+ FRT 42B} chromosome in trans to CyO hs-hid were crossed to males of the genotype y w P{ry+ hs-FLP22}/Y; P{w+ FRT 42B} P{w+ ovoD}/CyO hs-hid (see Figure 2A for a schematic of the screen). The P{ry+ hs-FLP22} transgene allows the production of the yeast FRT-specific recombinase FLP, whereas P{w+ ovoD} inserts the dominant female sterile ovoD allele in the FRT42B chromosome (Chou and Perrimon 1992). On the fifth or sixth day after mating, the parents were discarded and the F1 larvae were heat-shocked in a 37° water bath for 2 hr to induce mitotic recombination and death of individuals carrying the balancer. F1 adults were transferred to fresh yeasted food for 3 days before egg collection. Eggs derived from germ-line clones were collected twice. The first collection was stained for β-galactosidase activity to visualize germplasm and germ cells by virtue of the P{w+faf-LacZ} transgene (Moore et al. 1998). This was used to detect defects in germ cell formation or migration (data not shown). The second collection was screened directly on the egg deposition plate for eggshell phenotypes (Staeva-Vieira et al. 2003). The lines of interest were established by crossing F1 w P{w+faf-LacZ}/Y; P{w+ FRT 42B}*/P{w+ FRT 42B} P{w+ ovoD} males to females of the genotype w P{w+faf-LacZ}; Sp P{w+ hs-hid}/CyO, P{ry+faf-LacZ} and heat-shocking the F2 larvae. To generate stable lines, single F2 Cy males were crossed back to w P{w+faf-LacZ}; Sp P{w+ hs-hid}/CyO, P{ry+faf-LacZ} females and the progeny heat-shocked as larvae. Only the F2 males carrying the P{w+ FRT 42B}* chromosome would produce viable offspring.
Figure 2.—
Karyosome and Grk phenotypes in DV polarity mutant egg chambers. (A) Schematic of the clonal screen procedure. Each parental (P) female carried one mutagenized second chromosome with the FRT sequence proximally inserted in its right arm (FRT 42B*). Virgins were crossed to males carrying a second chromosome with the same FRT insertion and the dominant female sterile ovo allele (FRT42B ovoD). Heat-shock-induced mitotic recombination in the germ line (hs-FLP22) and death of balanced (CyO hs-hid) F1 larvae. Eggshell polarity defects in the resulting eggs were identified and stocks established (materials and methods). (B–F) Ventralization of the eggshell. (B) Wild-type egg from a dPds5cohiba/CyO control fly. (C) Mildly ventralized eggshell from a nds2/nds4 fly with DAs fused at the base (arrowhead). (D) Ventralized egg showing a single DA from a bha1/bha2 fly. (E) Extreme ventralization with absent DAs and elongated eggshell from dPds51 homozygous germ-line clone. (F) Example of a small/collapsed mutation derived from a srpk2/srpk4 heterozygous fly. (G–P) Localization of Grk (red) and DNA (green) in oogenesis. (G–H) Wild-type stages 6 (G) and 10 (H) egg chambers and higher magnifications of each oocyte nucleus (insets) from mtc/CyO flies. (I–K) Class I mutants showing normal karyosome morphology and Grk levels. (I) Stage 10 bha1/bha2 egg chamber. (J) Stage 6 chamber homozygous for srpk2 showing a misplaced oocyte (arrow). (K) khcpgs1 mutant stage 10 with arrow pointing to the oocyte nucleus, which is associated with Grk but is not anchored to the anterior cortex. (L) Class II nds2/nds4 stage 6 chamber showing decreased levels of Grk and normal karyosome. (M) Class III, trin1/trin2 stage 10 chamber with normal Grk but collapsed karyosome (arrow and inset). (N–P) Class IV mutations showing decreased Grk levels. (N) A blv1/blv3 stage 6/7 chamber showing a typical thread-like karyosome defect (arrow). (O) A stage 6 mtc2/mtc3 egg chamber with a similar karyosome defect (arrow). (P) Stage 6 dPds2 homozygous germ-line clone. The oocyte nucleus (arrow) shows a region of expanded chromatin apparently emanating from a more condensed core.
Germ-line clones homozygous for 310 mutations causing DV defects were rescreened. Lines producing wild-type eggshells (18 lines) were discarded as false positives. Mutations causing defects in the eggshell without affecting its DV patterning (174 lines) were sent to L. Cooley (Yale University). The penetrance of the DV phenotype of the remaining 118 lines was quantified in this secondary screen as frequency of ventralized eggshells (n > 300). Mutant lines were divided into “strong” (from 80 to 100% ventralized eggshells), “medium” (from 50 to 70% of ventralized eggshells), “weak” (∼30% ventralization), and “small/collapsed” (in addition to ventralized, eggshells were small and/or collapsed, Figure 2F) categories.
Complementation testing and deficiency mapping:
Mutants of the strong, medium, and small/collapsed categories were used in complementation crosses (Table 2). The criteria for lack of complementation were lethality or >30% ventralized eggshells. With these criteria, three lines failed to complement more than one complementation group, indicating the presence of double mutants srpkcuaba4 and veguero3, corona2 and fonseca2, and troya2 and vegueiro1.
TABLE 2.
2R complementation groups isolated in the screen
| Allele no. and categoriesa
|
|||||||
|---|---|---|---|---|---|---|---|
| Locus name | s | m | w | c | Lethalityb | Mapc | Gene |
| partagas (pgs) | 3 | 1 | — | — | L | 53A2 | CG7765, khc |
| bahia (bha) | — | 2 | — | — | V | 48E-50F | |
| cuaba | — | 2 | — | 2 | SL | 51F11-12 | CG8174, srpk |
| indios (nds) | 3 | 1 | — | — | V | 57D2-58D1 | |
| trinidad (trin) | 2 | 4 | — | 1 | V | 52B1-54D2 | |
| cohiba | 4 | 4 | — | — | L | 48D7-8 | CG17509, dPds5 |
| montecristo (mtc) | — | 2 | — | 1 | V | 53A1 | CG15707, Tudor domain |
| bolivar (blv) | 1 | 3 | — | — | SL | 59D11-60A7 | |
| troya | 1 | 6 | — | — | L | 51D3-52F9 | |
| diplomatico | — | 1 | — | 1 | L | ||
| sancho panza | — | 1 | — | 1 | V | ||
| romeo y julieta | — | 1 | — | 1 | V | ||
| guantanamera | — | — | — | 2 | L | ||
| corona | — | — | — | 2 | V | ||
| veguero | — | 2 | — | 1 | L | ||
| rey del mundo | — | 2 | — | — | L | ||
| fonseca | 1 | — | — | 1 | L | ||
| Single alleles | 3 | 17 | 31 | 10 | — | ||
Categories are based on frequency of ventralized eggshells (see materials and methods). n > 300.
Viability of transheterozygous adults. L, lethal; SL, semilethal or lower than expected number of adults; V, viable.
Minimal cytological interval containing each gene.
Lethal complementation groups were directly mapped using the Bloomington 2R deficiency kit. Each deficiency failing to complement lethality was then confirmed by crossing it with all the other mutants in the same complementation group. Deficiencies of the kit were also used to confirm the mapping of viable complementation groups after rough SNP mapping (see below). Complementation tests with known mutations mapped in each region were also performed for khcpartagas, srpkcuaba, dPds5cohiba, mtc, and blv.
Genetic mapping with single nucleotide polymorphism markers:
Nonessential loci were mapped to an approximate resolution of 2 Mb, similar to assays previously described (Berger et al. 2001). Single nucleotide polymorphisms (SNPs) causing restriction fragment-length polymorphism (RFLP) defining 10 evenly spaced intervals along 2R were selected (Table 1). The SNPs were defined between the P{w+ FRT 42B} chromosome (used for mutagenesis) and a divergent isogenized chromosome containing the distal marker If (used to generate recombinants for mapping). The SNP map was constructed by a light shotgun sequencing approach (Berger et al. 2001). Primers were designed to PCR amplify nine genomic fragments of ∼1 kb along both chromosomes (Table 1). The sequence of each FRT 42B PCR product was then compared to the If chromosome using SeqMan and MapDraw software (DNASTAR, Madison, WI) to find SNPs affecting the restriction site of common endonucleases (Table 1). These sites were tested by digesting PCR products generated from 10 single fly genomic DNA preparations of the genotypes P{w+ FRT 42B}/P{w+ FRT 42B} and P{w+ FRT 42B}/If as well as from genomic DNA isolated from If/If embryos. All enzymes were commercially available (NEB, Ipswich, MA).
TABLE 1.
SNPs defining a rough molecular map of 2R
| Primer namea | Primer sequenceb | Cytologic regionc | Enzyme used |
|---|---|---|---|
| pnut 4 L | GCGGCATGAGGGATGCTGAA | 44C1 | ClaI |
| pnut 4 R | TGCGCGAAAATTCCAACCGA | ||
| egr 8 L | CGGCTTTGCCTCGCTTCGTT | 46F4 | AvaI |
| egr 8 R | CGGTGGCAGATTCGTCCTGCT | ||
| jeb 2 L | CGGGAAAGGGGAGGACGCAG | 48E2 | PvuII |
| jeb 2 R | TATTGGGGGCGGCGAAAGGT | ||
| mam 4L | ACGCTGCCGCCTCTGTTGCT | 50D1 | AluII |
| mam 4R | CGCCCTCCCGCTCTGCATTT | ||
| Flo 4 L | GCACGGGTTGATTGACCGGAA | 52B1 | HhaI |
| Flo 4 R | CTTGTCCGCCGCTCCCCTCT | ||
| rhi 3 L | CGTGTGTGAAGGGGAAGGGCA | 54D3 | MwoI |
| rhi 3 R | GCTTCGGTGCTCATTGCGG | ||
| hts 4 L | CGGTCGGGTCGAAAGCGAGA | 56D8 | TaqI |
| hts 4 R | CGCTGGTGGCTGTGTGTATGCC | ||
| clt 3 L | GCACGTCCATCCGCCAAGTGA | 57F2 | RsaI |
| clt 3 R | TGCCACTCAGCTCCCCAGCA | ||
| bw 1 L | GCCTCCATCGGCGTTTCGCT | 59D11 | BsaJI |
| bw 1 R | TGTGAGGGGGTGTGGTGGGG |
Primers and SNPs were named after known 2R genes molecularly mapped to each interval. pnut, peanut; egr, eiger; jeb, jelly belly; mam, mastermind; Flo, Flotillin; rhi, rhino; hts, hu li tai shao; clt, cricklet; and bw, brown.
5′–3′ sequence.
Region containing the SNP–RFLP.
The mapping cross was as follows: w; If/P{w+ FRT 42B} m1 females crossed to w/Y; P{Δw FRT 42B} m2/CyO males, where m1 and m2 represent any two alleles of the complementation group m. P{Δw FRT 42B} was derived from a P{w+ FRT 42B} chromosome from which the w+ marker was removed by intramolecular recombination leaving the intact FRT sequence on the chromosome. Recombination events on 2R were recovered over P{Δw FRT 42B} m2 in the following generation and identified by their nonparental phenotypes: w If+ Cy+ and w+ If Cy+. Combined with the eggshell phenotype of each single recombinant female, the SNP analysis allowed the linkage of each locus to 1 of 10 intervals defined by the centromere proximal w+ transgene of the FRT42B insertion, the nine SNPs, and the centromere distal If (Tables 1 and 2).
Allele sequencing and molecular cloning:
Preparation of genomic DNA and sequencing reactions were carried out as described (Staeva-Vieira et al. 2003). Each mutant sequence from P{w+ FRT 42B} m/Df(2R) was aligned with that of the homozygous P{w+ FRT 42B} chromosome using DNASTAR.
To molecularly map the P{lacW}l(2)k1223 insertion, inverse PCR was carried out according to the Berkeley Drosophila Genome Project Resources website but using only the Sau3AI restriction enzyme. Sequences in the 3′ and 5′ ends of the P element obtained from two independent trials were compared with the Drosophila genome release 3, and the adjacent coding region CG17509 was sequenced in P{w+ FRT 42B} dPds5cohiba/Df(2R) mutants as above.
Immunostaining Drosophila ovaries:
Ovaries were processed for immunofluorescence as described (Navarro et al. 2004). The monoclonal anti-Grk antibody 1D12 (Developmental Studies Hybridoma Bank) and the rabbit polyclonal anti-C(3)G antibody were diluted 1:50 and 1:1000, respectively (Queenan et al. 1999; Navarro et al. 2004). Cy3-conjugated (Jackson Immunoresearch, West Grove, PA) and Alexa 488-conjugated (Molecular Probes, Eugene, OR) secondary antibodies were used at a dilution of 1:500. DNA was stained with either Oligreen or DAPI (Molecular Probes) diluted 1:5000 and 0.3 μm, respectively, according to the company's instructions. Ovaries were mounted in Vectashield (Vector Labs, Burlingame, CA) and visualized with a Leica TCS NT confocal microscope (Leica, Bannockburn, IL).
For visualization of mutant germ-line clones, 2- or 3-day-old adult females of the genotype y w P{ry+ hs-FLP22}; P{w+ FRT 42B} P{w+ FRT nls-GFP}/CyO hs-hid were heat-shocked on two consecutive days for 1 hr each. Heat-shocked adults were transferred to fresh food for five additional days, fattened on fresh yeast on the sixth day, and dissected on the seventh day as described. The autofluorescence of nuclear GFP was always preferred to its indirect immunolabeling using commercially available antibodies.
Genetic interactions:
The DV phenotype in viable or semilethal complementation groups was tested in a mei-41 mutant background by comparing the frequency of ventralized eggshells in mei-41D3/mei-41D3; P{w+ FRT 42B} m1/P{w+ FRT 42B} m2 and mei-41D3/FM7; P{w+ FRT 42B} m1/P{w+ FRT 42B} m2 flies. To assess genetic interactions with mutants defective in DSB (mei-P22), the frequency between P{w+ FRT 42B} m1/P{w+ FRT 42B} m2; mei-P221/mei-P221 and P{w+ FRT 42B} m1/P{w+ FRT 42B} m2; mei-P221/TM3 flies was compared. For the alleles of each locus used in this study, see supplemental Table 1 (http://www.genetics.org/supplemental/). Interactions with lethal groups and the checkpoint were tested by heat-shock induction of germ-line clones in mei-41D3/mei-41D3; P{w+ FRT 42B} l(2R)/P{w+ FRT 42B} P{w+ ovoD}; MKRS P{ry+ hs-FLP22} and in mei-41D3/FM7; P{w+ FRT 42B} l(2R)/P{w+ FRT 42B} P{w+ ovoD}; MKRS P{ry+ hs-FLP22} flies. l(2R) is any allele of the lethal groups tested, khcpgs and dPds5cohiba. dPds5 mutants were tested in a DSB-free background by inducing germ-line clones in y w P{ry+ hs-FLP22}; mei-41D3/mei-41D3; P{w+ FRT 42B} dPds5cohiba/P{w+ FRT 42B} P{w+ ovoD} and in y w P{ry+ hs-FLP22}; mei-41D3/FM7; P{w+ FRT 42B} dPds5cohiba/P{w+ FRT 42B} P{w+ ovoD}. As a positive control for each type of suppression, the allelic pair okrRU/okrAA was used except for the dPds5, mei-W68 interaction experiments, for which spn-A1/spn-A093A was used (Ghabrial et al. 1998; Staeva-Vieira et al. 2003).
RESULTS
A screen for genes controlling meiosis and oocyte patterning:
The Grk-mediated EGF receptor pathway is a sensitive readout for two fundamental processes of early oogenesis: meiosis and oocyte polarity. Abrogation of this pathway causes a characteristic ventralized eggshell phenotype that can be easily recognized by fusion or lack of the two dorsal appendages in the eggs of mutant females (Figure 2, B–F). We used this phenotype to identify new germ-line-specific genes on the right arm of chromosome 2 (2R) involved in meiotic progression and Grk ligand production. To isolate both lethal and viable EMS-derived mutations, we employed the FRT/ovoD technique to produce germ-line clones homozygous for 2R in an otherwise heterozygous adult (Figure 2A) (Chou and Perrimon 1992). Among 8179 independent lines, we isolated 310 potential mutations, of which 118 were kept for further analysis after a secondary screen (materials and methods).
The final 118 lines were divided into four categories—strong (18 lines), medium (47 lines), weak (31 lines), and small/collapsed (22 lines)—based on penetrance of the mutant phenotype (materials and methods and Table 2). Only lines with the most penetrant phenotypes were used for complementation tests. This allowed the identification of 17 complementation groups with two or more alleles among 57 lines tested (Table 2). Ten complementation groups were lethal or semilethal, suggesting that the corresponding genes have essential somatic functions and would not have been identified in previous maternal screens (Schupbach and Wieschaus 1989, 1991). Due to their shared eggshell phenotype, we named the complementation groups after brands of Cuban cigars. We have determined the genomic location of nine loci by combining SNP mapping with complementation analysis using the 2R deficiency kit (materials and methods). In this study, we describe the phenotypic characterization of eight of these genes (Table 3).
TABLE 3.
Characterization of eight loci on 2R required for DV patterning of the eggshell
| Gurkena
|
Suppression of DV defectsc
|
||||||
|---|---|---|---|---|---|---|---|
| Classes | Gene | Protein | Localized | Karyosome | C(3)G restrictionb | mei-41 | mei-P22 or mei-W68d |
| I | khcpgs | + | No | Normal | — | No | — |
| bha | + | Yes | Normal | Normal | No | No | |
| srpkcuaba | + | Yes | Normal | Normal | No | No | |
| II | nds | Low | Yes | Normal | Delayed | Yes | No |
| III | trin | + | Yes | Abnormal | Normal | No | No |
| IV | dPds5cohiba | Low | Yes | Abnormal | Delayed | No | Yes |
| mtc | Low | Yes | Abnormal | Delayed | No | No | |
| blv | Low | Yes | Abnormal | Normal | No | No | |
| spnAe | Low | Yes | Abnormal | Delayed | Yes | Yes | |
Qualitative analysis of Grk production. Protein: +, normal; low, decreased amount detected by fluorescence; localized, correct localization of Grk in stages 6–10.
Timing of restriction of C(3)G to one germ cell in stage 2 egg chambers. Delayed, frequency of C(3)G in more than one cell is >30%; normal, C(3)G always in one cell, n = 25.
Frequency of DV polarity defects in eggshells from doubles with mei-41D3 and with DSB formation mutants (n > 300). No, frequency comparable to controls; yes, frequency significantly decreased.
All tests were carried out with DSB formation mutant mei-P221, except for dPds5cohiba, which was tested with mei-W681.
Phenotypic characteristics of dRad51/spnA for comparison (see Staeva-Vieira et al. 2003).
Classification of new DV polarity mutations based on Grk protein distribution and oocyte chromatin condensation:
As an initial phenotypic assay, we used oocyte nuclear morphology and Grk protein distribution to characterize the effect of each complementation group on meiosis and oocyte polarity (Gonzalez-Reyes et al. 1997) (Figure 2, G–P, and Table 3). By stage 6 of wild-type oogenesis, the DNA within the oocyte nucleus is fully condensed to form a dense karyosome (Figure 2G). At this stage, Grk protein is tightly associated with the oocyte nucleus. Subsequently, at stage 9 of oogenesis, grk RNA and protein, together with the oocyte nucleus, move to an anterior corner of the oocyte, where high levels of Grk induce dorsal cell fates in the overlaying follicle cells; at this point, the karyosome takes on a more “relaxed” morphology (Figure 2H) (Neuman-Silberberg and Schupbach 1996). According to our analysis, the new mutants fall into four phenotypic classes. (For a summary of mutant phenotypes and genetic interactions, see Table 3.)
Class I—normal karyosome and Grk protein levels (Figure 2, I–K):
Three complementation groups fall into this class (Table 3)—bahia (bha), cuaba, and partagas (pgs). The two viable bha alleles had apparently normal Grk distribution and karyosome morphologies (Figure 2I). This phenotype resembles that of mutations affecting Grk processing or secretion (Valcarcel et al. 1999; Bokel et al. 2006; Miura et al. 2006). In contrast to bha mutations, cuaba and pgs mutants showed defects in egg chamber morphology and oocyte nuclear positioning, respectively (Figure 2, J and K). cuaba mutations produced egg chambers with the normal number of germ cells, but the oocyte was abnormally positioned within the oocyte-nurse cell cluster in cuaba mutants, not posterior to the nurse cells as in wild type (Figure 2J). This phenotype resembles that caused by DV mutations in genes required for oocyte adhesion to the follicle cells, such as cadherin, dicephalic, and brainiac (Goode et al. 1996; Godt and Tepass 1998; Gonzalez-Reyes and St Johnston 1998; McCaffrey et al. 2006). We mapped cuaba to the coding region CG8174, and all four cuaba alleles carry mutations in this gene. CG8174 is predicted to encode the Drosophila homolog of human SR protein kinase 2 (SRPK2). SRPK2 affects alternative splicing of specific RNAs by regulating the function or subcellular localization of SR proteins (Tenenbaum and Aguirre-Ghiso 2005). In pgs mutant egg chambers, the oocyte was positioned correctly with respect to the nurse cells. However, at stage 10, when the oocyte nucleus has normally moved to the anterior dorsal side of the oocyte, the oocyte nucleus in pgs mutants was found misplaced (Figure 2K). We mapped pgs to the genomic region of kinesin heavy chain (khc), and all pgs mutations failed to complement the lethality of a khc allele (khc27) (Table 2). Since germ-line clones of khc27 also show a nuclear migration phenotype similar to that of our pgs alleles, we conclude that pgs mutations affect khc (Brendza et al. 2002).
Class II—defects in Grk protein synthesis:
A single complementation group, indios (nds), falls into this class. In nds mutant egg chambers, Grk protein levels were clearly reduced, although no defects in karyosome morphology were observed (Figure 2L and Table 3). This phenotype resembles that of grk, suggesting that nds specifically affects the synthesis or stability of grk protein or RNA (Gonzalez-Reyes et al. 1995; Volpe et al. 2001).
Class III—defects in karyosome morphology:
Seven viable alleles of trinidad (trin) were identified on the basis of their ventralized and flaccid eggshell phenotype. In these mutant oocytes, chromatin condensation appeared irregular (Figure 2M, inset), while Grk protein levels seemed normal (Figure 2M). It remains unclear how this apparently germ-line-specific gene affects both nuclear morphology and Grk function. A similar phenotype has been observed in mutants defective in actin dynamics such as Src64, Tec29, and Kelch (Dodson et al. 1998; Djagaeva et al. 2005).
Class IV—defects in both Grk production and karyosome formation (Figure 2, N–P):
Three complementation groups fall into this class (Table 3)—bolivar (blv), montecristo (mtc), and cohiba. All three genes seemed to specifically affect karyosome morphology and Grk distribution. These mutants did not alter other aspects of egg chamber development, such as oocyte determination, the number of nurse cells per egg chamber, and the positioning of the oocyte posterior to the nurse cells (data not shown). blv and mtc oocytes showed a thread-like chromatin morphology typical of other meiotic mutants (Figure 2, N and O, respectively) (Gonzalez-Reyes et al. 1997). We used SNP recombination and lack of complementation for female sterility with the P-element PBac{WH}CG15707f06583 to map mtc to CG15707. All mtc alleles carry mutations in this gene, which encodes a 746-aa protein predicted to contain a Tudor domain near its carboxyl terminus (Ponting 1997). BLAST searches using the predicted Mtc protein sequence found significant alignments with a Tudor-domain protein in Anopheles (XM_312463) and several mammalian Tudor-domain proteins. These homologies are, however, restricted to the Tudor domain. In contrast to the karyosome defects observed in mtc and blv, >50% of cohiba karyosomes showed regions of “open” chromatin apparently emerging from a condensed core (Figure 2P, arrow). We mapped cohiba by deficiency mapping and complementation analysis with candidate mutants and found that the previously uncharacterized P-element l(2)k13312 failed to complement the lethality of all cohiba alleles. Using inverse PCR, we identified the insertion site of l(2)k13312 upstream of the start codon of CG17509 (materials and methods). CG17509 encodes a Drosophila homolog of the yeast protein Pds5p (Celniker et al. 2002; Dorsett et al. 2005). All cohiba alleles carried mutations in the CG17509 open reading frame, confirming the identity of cohiba as Drosophila Pds5 (dPds5). A more detailed description of both mtc and dPds5cohiba phenotypes will be presented elsewhere.
Classification of new DV polarity mutations based on restriction of meiosis to the oocyte:
To determine whether any of the newly identified DV genes may control meiotic progression, we analyzed the mutants for a block or delay in meiosis. One readout for meiotic progression is the restriction of the synaptonemal complex (SC) component C(3)G to the oocyte in region 3 of the germarium (Huynh and St Johnston 2000). In wild type, meiosis initiates in more than one cell per cyst in the germarial region 2 as described in electron micrographs of the SC and by fluorescently labeling C(3)G (Carpenter 1979; Page and Hawley 2001). As the cyst matures, the nuclear C(3)G signal restricts from the two pro-oocytes to one cell and synapses are resolved in all nurse cells (Figures 1 and 3, A and E). Mutations in genes controlling RNA and protein transport into the oocyte, such as egl and BicD, as well as mutations in DSB repair genes such as spn-A delay this restriction (Huynh and St Johnston 2000; Staeva-Vieira et al. 2003; Navarro et al. 2004).
Figure 3.—
Effect of mutations on the timing of meiotic restriction to the oocyte. (A–D) Localization of the SC protein C(3)G (red) with respect to DNA (green) in meiotic nuclei in germaria (arrows) and stage 2 egg chambers (arrowheads). (A) Control germarium of the genotype nds/CyO showing more than one germ cell marked with C(3)G in region 2b and only one marked in the stage 2 chamber. (B) nds and (C) mtc ovaries showing delays in restriction of C(3)G to the oocyte in stage 2 chambers. (D) bha germarium and stage 2 showing normal meiotic restriction of C(3)G (n > 70). (E and F) Germ-line clones induced in flies of the genotype FRT 42B dPds5cohiba/FRT 42B nls-GFP (green), C(3)G (red). (E) Control germarium and stage 2 with all cells expressing nuclear GFP. (F) dPds5cohiba mutant germ-line clones lack GFP and show delays in C(3)G restriction to stage 2 oocytes (arrowheads).
We assayed the progression of meiosis in the new DV mutants using C(3)G staining (Table 3 and Figure 3). Mutations in three genes, nds, mtc, and dPds5, delayed the restriction of meiosis to the oocyte as evidenced by stage 2 egg chambers that have two C(3)G-positive cells. nds mutants showed delays in ∼30% of stage 2 egg chambers (n = 25, Figure 3B), mtc caused delays in 49% of mutant egg chambers (n = 25, Figure 3C), and two dPds5cohiba mutant alleles, dPds52 and dPds56, showed delays in 29% (n =15) and 40% (n = 27) of egg chambers, respectively (Figure 3, E and F). In wild type and egg chambers mutant for bahia, khcpgs, srpkcuaba, trin, and blv, delays in meiotic restriction were rarely observed (3–5%, Figure 3D).
In summary, mutants in mtc and dPds5 behave like “classical spindle mutants” that decrease Grk production, affect karyosome morphology, and delay meiotic restriction. Similar to vas mutants, blv mutants also affect Grk levels and karyosome morphology but do not show evident delays in meiotic restriction (Tomancak et al. 1998; Huynh and St Johnston 2000). On the other hand, the nds phenotype, with decreased Grk protein levels and delayed meiotic restriction but an apparently normal karyosome morphology, and the trin phenotype, with normal Grk protein levels, abnormal karyosome condensation, and no evident delays in meiotic restriction, suggest that condensation of meiotic chromatin, timing of meiotic restriction, and control of Grk levels can be uncoupled.
Classification of new DV polarity mutations based on meiotic checkpoint activation and defects in DSB repair:
We next determined the genetic relationship of each complementation group with genes affecting DSB repair and meiotic checkpoint activation. As previously shown, mutations in genes controlling checkpoint activation and DSB formation can suppress eggshell ventralization in DSB-repair mutants by restoring Grk protein levels (Ghabrial and Schupbach 1999; Abdu et al. 2002; Staeva-Vieira et al. 2003).
We placed mutations of each group in the background of the Drosophila checkpoint protein ATR kinase, mei-41 (mei-41D3), and scored the percentage of ventralized eggshells (see materials and methods) (Laurencon et al. 2003). Only one group, nds, showed significant suppression by mei-41D3 (Figure 4, A and B, and supplemental Table 1 for P-values; http://www.genetics.org/supplemental/). This suppression is dominant since the frequency of DV defects decreased from 45.3% (n = 546) in the progeny of nds females to 3.1% (n = 964) and 2.1% (n = 828) in the progeny of mei-41D3/+; indios and mei-41D3; indios females, respectively. Interestingly, reducing and eliminating ATR/Mei-41 function in group IV mutants either in germ-line clones homozygous for three lethal dPds5cohiba alleles or in combination with viable mutations in mtc and blv had no significant effect on the frequency of ventralized eggs generated by germ-line clones (Figure 4B).
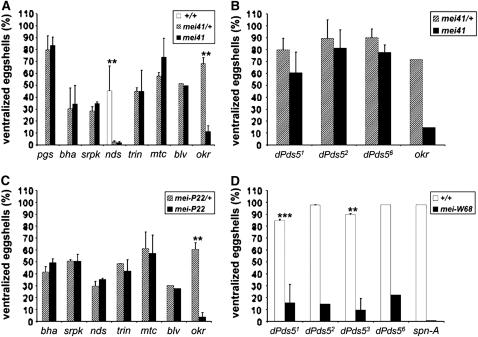
Figure 4.—
Interactions of mutants with the DSB formation and DNA repair meiotic checkpoint pathways. Frequency of ventralized eggshells produced by mutant flies in an otherwise wild-type (open bars), heterozygous (striped bars) or homozygous mutant (solid bars) background for mei-41D1 (A and B), mei-P221 (C), or mei-W681 (D). The error bars, when indicated, give one standard deviation after two independent trials. (A and D) Eggs were derived from trans-heterozygous mutant flies for viable allelic combinations (see materials and methods for the allelic combinations used in each experiment) or homozygous mutant germ-line clones for khcpgs. A value for ventralized eggs from individuals of the genotype nds2/nds4; mei-41+/mei-41+ was determined in parallel (open bar in A) to illustrate the dominant genetic interaction (see text). (B and D) Germ-line clones homozygous for dPds5cohiba alleles (dPds51, dPds52, dPds53, and dPds56). **P ≤ 0.05; ***P ≤ 0.001.
We subsequently tested the phenotype of our complementation groups in the absence of DSB. This was achieved by combining our mutants with a mutation in either the Spo11p ortholog mei-W68 or the meiotic chromatin component mei-P22 (see materials and methods) (Liu et al. 2002; Jang et al. 2003). Because the molecular mechanism of khcpgs in microtubule-based transport is already established, we excluded this group from the epistatic analysis (Table 3). Mutations from each of the viable complementation groups (blv, bha, srpkcuaba, nds, trin, and mtc) were tested with mei-P221. However, we did not see suppression of the eggshell phenotype in any of these double mutants (Figure 4C). Since the dPds5cohiba alleles are homozygous lethal, we tested dPds5 in combination with mei-W68, which is located on the same chromosome. We induced mei-W681 dPds5cohiba double mutant homozygous clones using the FRT/ovoD method. In the progeny of these clones, the frequency of ventralized eggshells was significantly reduced (Figure 4D and supplemental Table 1; http://www.genetics.org/supplemental/).
In summary, none of our new mutants behaves identically to mutants in the previously described spindle genes spn-A and okr (Figure 4), which encode enzymes required for DSB repair. Instead, our results suggest that: (1) mutations in nds cause Mei-41/ATR checkpoint activation independently of DSB formation; (2) dPds5 mutants are sensitive to DSBs but seem not to activate the Mei-41/ATR checkpoint; and (3) Blv, Mtc, and Trin may function downstream of, or in parallel to, the Mei-41/ATR checkpoint.
DISCUSSION
In this study, we used a clonal screen to identify genes regulating meiotic progression in Drosophila. Instead of testing directly for defects in meiosis, we used an easy-to-score eggshell phenotype that is produced when the levels or activity of the morphogen Grk are affected. This allowed us to efficiently screen a large number of mutant lines and to identify germ-line-specific genes as well as genes with essential functions. The number of new genes identified is likely less than the total number of 2R genes required for Grk synthesis and function since we discarded mutations that blocked oogenesis (Morris et al. 2003). Of the eight genes described in this study, five show meiotic phenotypes. dPds5, nds, and mtc delay meiotic restriction to the oocyte, although only dPds5 and nds genetically interact with mei-W68 and mei-41, respectively. trin and blv affect the morphology of the karyosome in spite of normal timing in meiotic restriction. This confirms the effectiveness of our screening method for meiotic genes. Genetic and developmental analysis of the newly identified genes provides evidence for new regulatory steps in a network that coordinates Drosophila meiosis and oocyte development.
Chromatin cohesion and DSB formation:
One of our complementation groups, cohiba, identifies the Drosophila homolog of Pds5p in Schizosaccharomyces pombe, Spo76 in Sordaria macrospore, and BimD in Aspergillus nidulans, which have been found associated with the cohesion complex of mitotic and meiotic chromosomes (van Heemst et al. 1999; Panizza et al. 2000; Storlazzi et al. 2003; Losada et al. 2005; Ding et al. 2006). More recently, it was shown that depletion of Pds5 affects not only cohesion but also condensation in meiotic prophase (van Heemst et al. 1999; Panizza et al. 2000; Storlazzi et al. 2003; Losada et al. 2005; Ding et al. 2006). The unique “open chromatin” karyosome defect we observe in dPds5cohiba mutants is consistent with a role of Pds5 in chromosome cohesion during Drosophila meiosis. Like Spo76, the dPds5cohiba phenotype is suppressed by Spo11 (mei-W68) mutations defective in DSB formation. This suggests that dPds5 is necessary to maintain the structure of the meiotic chromosomes after DSBs are induced (van Heemst et al. 1999; Panizza et al. 2000; Storlazzi et al. 2003; Losada et al. 2005; Ding et al. 2006). However, in contrast to known DSB repair genes, the meiotic delay and oocyte patterning defects of dPdscohiba mutants are not due to activation of ATR/Mei-41-dependent checkpoint. One possibility is that the ATR downstream effector kinase dChk2 is activated via an alternative pathway, such as the Drosophila ataxia-telangiectasia mutated (ATM) homolog, which indeed activates dChk2 in the early embryo independently of ATR (Brodsky et al. 2004). Alternatively, dPdscohiba mutants may activate a checkpoint that measures cohesion rather than DSB breaks. The only other cohesion protein characterized in Drosophila is the product of the orientation disruptor (ord). ORD plays a role in early prophase I by maintaining synaptic chromosomes and allowing interhomolog recombination (Webber et al. 2004). More importantly and perhaps similar to dPds5, ORD seems not to be required for DSB repair. However, in contrast to dPds5 mutants, karyosome morphology is normal in ord mutants, and an eggshell polarity phenotype has not been reported. Although required for chromatid cohesion, dPds5 and ORD might play complementary roles in SC dynamics: ORD may stabilize the SC in the oocyte, whereas dPds5 may be required for the disassembly of synapses as one of the pro-oocytes regresses from meiosis.
Meiotic restriction to the oocyte:
Our screen identified mutations in montecristo (mtc) that affect the restriction of meiosis to the oocyte. It has been proposed that this delay reflects the activation of the ATR/Mei-41 checkpoint pathway (Huynh and St Johnston 2000). Similar to dPds5, Mtc may control the regression from pachytene in those cyst cells that will not adopt the oocyte fate. The delayed meiotic restriction observed in mtc mutants occurs, however, independently of DSB formation or Mei-41 checkpoint activation (Table 3). Mtc contains a Tudor domain. In other Tudor-domain proteins, this domain has been shown to interact with methylated target proteins (Ponting 1997). Identification of specific Mtc targets may clarify its role in meiotic restriction and oocyte patterning.
Karyosome formation and Gurken activity:
A particularly intriguing and novel phenotype is uncovered by mutations in indios (nds). By delaying meiotic restriction and activating Mei-41 without affecting the karyosome morphology, nds mutants separate checkpoint activation leading to Grk decrease from checkpoint activation controlling karyosome compaction. The nds phenotype also occurs independently of DSBs, suggesting that the trigger that leads Nds to trigger checkpoint activation is not DNA breaks. The fact that nds mutants are extremely sensitive to Mei-41 dosage further suggests that Nds activity may specifically control a branch of the Mei-41 checkpoint regulating Grk activity. In contrast to nds, trin mutants do not delay meiotic restriction and show defects in the karyosome in spite of normal Grk levels. Like mutants in src64B and tec29, which show a similar phenotype, Trin may mediate chromatin remodeling in the oocyte by regulating the actin cytoskeleton (Simon et al. 1983; Guarnieri et al. 1998; Roulier et al. 1998; Djagaeva et al. 2005). In this context, the DV phenotype of eggs from trin mutants may be an indirect effect due to defects in actin cytoskeleton function (Djagaeva et al. 2005; Miralles and Visa 2006). The production of collapsed eggs by trin mutant germ-line clones is consistent with this idea (Table 2).
Finally, blv mutants show striking similarity to vas mutants with respect to lack of sensitivity to DSB formation, no evident delays of meiotic restriction, or karyosome and Grk phenotypes (Styhler et al. 1998; Tomancak et al. 1998; Ghabrial and Schupbach 1999; Huynh and St Johnston 2000). Blv may thus act downstream or independent of the Mei41/ATR checkpoint, and its further characterization may help to understand the effector side of the meiotic checkpoint pathway.
Previous knowledge pointed to Drosophila meiosis as a linear progression of events from homologous chromosome pairing and recombination to meiotic restriction, karyosome formation, and eggshell patterning, with DSB repair as the main checkpoint linking meiosis to Grk signaling. By uncoupling some of these events, our study suggests the existence of a more complex network that links the surveillance of meiotic progression to oocyte patterning.
Acknowledgments
We thank A. Arkov, Y. Arkova, P. Kunwar, T. Marty, A. Renault, H. Sano, and H. Zinszner for help in performing the mutagenesis screen; and the Lehmann lab for stimulating discussions. We also thank Trudi Schüpbach and Caryn Navarro for scientific advice, and Daria Siekhaus, Lilach Gilboa, and Jessica Seifert for critically reading the manuscript. This work was supported by the Howard Hughes Medical Institute and by Fundação para a Ciência e Tecnologia, Portugal.
References
- Abdu, U., M. Brodsky and T. Schupbach, 2002. Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr. Biol. 12: 1645–1651. [DOI] [PubMed] [Google Scholar]
- Abdu, U., D. Bar and T. Schupbach, 2006. spn-F encodes a novel protein that affects oocyte patterning and bristle morphology in Drosophila. Development 133: 1477–1484. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., 1989. Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Berger, J., T. Suzuki, K. A. Senti, J. Stubbs, G. Schaffner et al., 2001. Genetic mapping with SNP markers in Drosophila. Nat. Genet. 29: 475–481. [DOI] [PubMed] [Google Scholar]
- Bhalla, N., and A. F. Dernburg, 2005. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 310: 1683–1686. [DOI] [PubMed] [Google Scholar]
- Bokel, C., S. Dass, M. Wilsch-Brauninger and S. Roth, 2006. Drosophila Cornichon acts as cargo receptor for ER export of the TGF{alpha}-like growth factor Gurken. Development 133: 459–470. [DOI] [PubMed] [Google Scholar]
- Brendza, R. P., L. R. Serbus, J. B. Duffy and W. M. Saxton, 2000. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science 289: 2120–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza, R. P., L. R. Serbus, W. M. Saxton and J. B. Duffy, 2002. Posterior localization of dynein and dorsal-ventral axis formation depend on kinesin in Drosophila oocytes. Curr. Biol. 12: 1541–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, M. H., B. T. Weinert, G. Tsang, Y. S. Rong, N. M. McGinnis et al., 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell Biol. 24: 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A. T., 1979. Synaptonemal complex and recombination nodules in wild-type Drosophila melanogaster females. Genetics 92: 511–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker, S. E., D. A. Wheeler, B. Kronmiller, J. W. Carlson, A. Halpern et al., 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, T. B., and N. Perrimon, 1992. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics 131: 643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cuevas, M., M. A. Lilly and A. C. Spradling, 1997. Germline cyst formation in Drosophila. Annu. Rev. Genet. 31: 405–428. [DOI] [PubMed] [Google Scholar]
- Ding, D. Q., N. Sakurai, Y. Katou, T. Itoh, K. Shirahige et al., 2006. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J. Cell Biol. 174: 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djagaeva, I., S. Doronkin and S. K. Beckendorf, 2005. Src64 is involved in fusome development and karyosome formation during Drosophila oogenesis. Dev. Biol. 284: 143–156. [DOI] [PubMed] [Google Scholar]
- Dodson, G. S., D. J. Guarnieri and M. A. Simon, 1998. Src64 is required for ovarian ring canal morphogenesis during Drosophila oogenesis. Development 125: 2883–2892. [DOI] [PubMed] [Google Scholar]
- Dorsett, D., J. C. Eissenberg, Z. Misulovin, A. Martens, B. Redding et al., 2005. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 132: 4743–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley, S. D., M. Tamanaha, N. J. Clegg and H. Ruohola-Baker, 2003. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 130: 859–871. [DOI] [PubMed] [Google Scholar]
- Ghabrial, A., and T. Schupbach, 1999. Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat. Cell Biol. 1: 354–357. [DOI] [PubMed] [Google Scholar]
- Ghabrial, A., R. P. Ray and T. Schupbach, 1998. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 12: 2711–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione, C., E. A. Bach, Y. Paraiso, K. L. Carraway, III, S. Noselli et al., 2002. Mechanism of activation of the Drosophila EGF Receptor by the TGFalpha ligand Gurken during oogenesis. Development 129: 175–186. [DOI] [PubMed] [Google Scholar]
- Godt, D., and U. Tepass, 1998. Drosophila oocyte localization is mediated by differential cadherin-based adhesion. Nature 395: 387–391. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes, A., and D. St Johnston, 1998. The Drosophila AP axis is polarised by the cadherin-mediated positioning of the oocyte. Development 125: 3635–3644. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes, A., H. Elliott and D. St Johnston, 1995. Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature 375: 654–658. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes, A., H. Elliot and D. St Johnston, 1997. Oocyte determination and the origin of polarity in Drosophila: the role of the spindle genes. Development 124: 4927–4937. [DOI] [PubMed] [Google Scholar]
- Goode, S., M. Morgan, Y. P. Liang and A. P. Mahowald, 1996. Brainiac encodes a novel, putative secreted protein that cooperates with Grk TGF alpha in the genesis of the follicular epithelium. Dev. Biol. 178: 35–50. [DOI] [PubMed] [Google Scholar]
- Guarnieri, D. J., G. S. Dodson and M. A. Simon, 1998. SRC64 regulates the localization of a Tec-family kinase required for Drosophila ring canal growth. Mol. Cell. 1: 831–840. [DOI] [PubMed] [Google Scholar]
- Huynh, J. R., and D. St Johnston, 2000. The role of BicD, Egl, Orb and the microtubules in the restriction of meiosis to the Drosophila oocyte. Development 127: 2785–2794. [DOI] [PubMed] [Google Scholar]
- Jang, J. K., D. E. Sherizen, R. Bhagat, E. A. Manheim and K. S. McKim, 2003. Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116: 3069–3077. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J. R., S. Yamaguchi and R. W. Carthew, 2002. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev. 16: 1884–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov, A., 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 98: 8461–8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurencon, A., A. Purdy, J. Sekelsky, R. S. Hawley and T. T. Su, 2003. Phenotypic analysis of separation-of-function alleles of MEI-41, Drosophila ATM/ATR. Genetics 164: 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., J. K. Jang, N. Kato and K. S. McKim, 2002. mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics 162: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada, A., T. Yokochi and T. Hirano, 2005. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J. Cell Sci. 118: 2133–2141. [DOI] [PubMed] [Google Scholar]
- McCaffrey, R., D. St Johnston and A. Gonzalez-Reyes, 2006. A novel mutant phenotype implicates dicephalic in cyst formation in the Drosophila ovary. Dev. Dyn. 235: 908–917. [DOI] [PubMed] [Google Scholar]
- Miralles, F., and N. Visa, 2006. Actin in transcription and transcription regulation. Curr. Opin. Cell Biol. 18: 261–266. [DOI] [PubMed] [Google Scholar]
- Miura, G. I., J. Buglino, D. Alvarado, M. A. Lemmon, M. D. Resh et al., 2006. Palmitoylation of the EGFR ligand Spitz by Rasp increases Spitz activity by restricting its diffusion. Dev. Cell 10: 167–176. [DOI] [PubMed] [Google Scholar]
- Moore, L. A., H. T. Broihier, M. Van Doren, L. B. Lunsford and R. Lehmann, 1998. Identification of genes controlling germ cell migration and embryonic gonad formation in Drosophila. Development 125: 667–678. [DOI] [PubMed] [Google Scholar]
- Morris, J., and R. Lehmann, 1999. Drosophila oogenesis: versatile spn doctors. Curr. Biol. 9: R55–58. [DOI] [PubMed] [Google Scholar]
- Morris, J. Z., C. Navarro and R. Lehmann, 2003. Identification and analysis of mutations in bob, Doa and eight new genes required for oocyte specification and development in Drosophila melanogaster. Genetics 164: 1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, C., H. Puthalakath, J. M. Adams, A. Strasser and R. Lehmann, 2004. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat. Cell Biol. 6: 427–435. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg, F. S., and T. Schupbach, 1994. Dorsoventral axis formation in Drosophila depends on the correct dosage of the gene gurken. Development 120: 2457–2463. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg, F. S., and T. Schupbach, 1996. The Drosophila TGF-alpha-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech. Dev. 59: 105–113. [DOI] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2001. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15: 1330–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2004. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20: 525–558. [DOI] [PubMed] [Google Scholar]
- Panizza, S., T. Tanaka, A. Hochwagen, F. Eisenhaber and K. Nasmyth, 2000. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 10: 1557–1564. [DOI] [PubMed] [Google Scholar]
- Ponting, C. P., 1997. Tudor domains in proteins that interact with RNA. Trends Biochem. Sci. 22: 51–52. [DOI] [PubMed] [Google Scholar]
- Queenan, A. M., G. Barcelo, C. Van Buskirk and T. Schupbach, 1999. The transmembrane region of Gurken is not required for biological activity, but is necessary for transport to the oocyte membrane in Drosophila. Mech. Dev. 89: 35–42. [DOI] [PubMed] [Google Scholar]
- Roeder, G. S., and J. M. Bailis, 2000. The pachytene checkpoint. Trends Genet. 16: 395–403. [DOI] [PubMed] [Google Scholar]
- Roulier, E. M., S. Panzer and S. K. Beckendorf, 1998. The Tec29 tyrosine kinase is required during Drosophila embryogenesis and interacts with Src64 in ring canal development. Mol. Cell 1: 819–829. [DOI] [PubMed] [Google Scholar]
- Saunders, C., and R. S. Cohen, 1999. The role of oocyte transcription, the 5′UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol. Cell. 3: 43–54. [DOI] [PubMed] [Google Scholar]
- Schupbach, T., and E. Wieschaus, 1989. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics 121: 101–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach, T., and E. Wieschaus, 1991. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129: 1119–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, M. A., T. B. Kornberg and J. M. Bishop, 1983. Three loci related to the src oncogene and tyrosine-specific protein kinase activity in Drosophila. Nature 302: 837–839. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., 1993. Developmental genetics of oogenesis, pp. 1–70 in Drosophila Development, edited by M. Bate and A. Martinez-Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Staeva-Vieira, E., S. Yoo and R. Lehmann, 2003. An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 22: 5863–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi, A., S. Tesse, S. Gargano, F. James, N. Kleckner et al., 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 17: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styhler, S., A. Nakamura, A. Swan, B. Suter and P. Lasko, 1998. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125: 1569–1578. [DOI] [PubMed] [Google Scholar]
- Swan, A., and B. Suter, 1996. Role of Bicaudal-D in patterning the Drosophila egg chamber in mid-oogenesis. Development 122: 3577–3586. [DOI] [PubMed] [Google Scholar]
- Swan, A., T. Nguyen and B. Suter, 1999. Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat. Cell Biol. 1: 444–449. [DOI] [PubMed] [Google Scholar]
- Tenenbaum, S. A., and J. Aguirre-Ghiso, 2005. Dephosphorylation shows SR proteins the way out. Mol. Cell 20: 499–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak, P., A. Guichet, P. Zavorszky and A. Ephrussi, 1998. Oocyte polarity depends on regulation of gurken by Vasa. Development 125: 1723–1732. [DOI] [PubMed] [Google Scholar]
- Valcarcel, R., U. Weber, D. B. Jackson, V. Benes, W. Ansorge et al., 1999. Sec61beta, a subunit of the protein translocation channel, is required during Drosophila development. J. Cell Sci. 112(Pt 23): 4389–4396. [DOI] [PubMed] [Google Scholar]
- van Heemst, D., F. James, S. Poggeler, V. Berteaux-Lecellier and D. Zickler, 1999. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell 98: 261–271. [DOI] [PubMed] [Google Scholar]
- Volpe, A. M., H. Horowitz, C. M. Grafer, S. M. Jackson and C. A. Berg, 2001. Drosophila rhino encodes a female-specific chromo-domain protein that affects chromosome structure and egg polarity. Genetics 159: 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber, H. A., L. Howard and S. E. Bickel, 2004. The cohesion protein ORD is required for homologue bias during meiotic recombination. J. Cell Biol. 164: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm, J. E., M. Buszczak and S. Sayles, 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev. Cell 9: 675–685. [DOI] [PubMed] [Google Scholar]
- Yohn, C. B., L. Pusateri, V. Barbosa and R. Lehmann, 2003. l(3)malignant brain tumor and three novel genes are required for Drosophila germ-cell formation. Genetics 165: 1889–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]