Abstract
Mus81-Mms4 (Mus81-Eme1 in some species) is a heterodimeric DNA structure-specific endonuclease that has been implicated in meiotic recombination and processing of damaged replication forks in fungi. We generated and characterized mutations in Drosophila melanogaster mus81 and mms4. Unlike the case in fungi, we did not find any role for MUS81-MMS4 in meiotic crossing over. A possible role for this endonuclease in repairing double-strand breaks that arise during DNA replication is suggested by the finding that mus81 and mms4 mutants are hypersensitive to camptothecin; however, these mutants are not hypersensitive to other agents that generate lesions that slow or block DNA replication. In fungi, mus81, mms4, and eme1 mutations are synthetically lethal with mutations in genes encoding RecQ helicase homologs. Similarly, we found that mutations in Drosophila mus81 and mms4 are synthetically lethal with null mutations in mus309, which encodes the ortholog of the Bloom Syndrome helicase. Synthetic lethality is associated with high levels of apoptosis in proliferating tissues. Lethality and elevated apoptosis were partially suppressed by a mutation in spn-A, which encodes the ortholog of the strand invasion protein Rad51. These findings provide insights into the causes of synthetic lethality.
DNA repair and recombination processes involve formation of special DNA structures such as flaps, D-loops, and four-stranded Holliday junctions, which are processed by enzymes that recognize these specific structures. Mus81-Mms4 (or Mus81-Eme1) is one such enzyme. Boddy et al. (2001) reported that Schizosaccharomyces pombe Mus81-Eme1 displays endonuclease activity on Holliday junctions, but other studies have found that this enzyme and orthologs from yeast and mammalian cells have a higher affinity for other branched structures, such as 3′ flaps or substrates that mimic replication forks, and have the highest affinity for nicked Holliday junctions (Kaliraman et al. 2001; Constantinou et al. 2002; Doe et al. 2002; Ciccia et al. 2003; Gaillard et al. 2003; Ogrunc and Sancar 2003; Osman et al. 2003).
The in vitro substrate specificity of Mus81-Mms4 (or Mus81-Eme1) reflects the in vivo functions of this enzyme. In S. pombe, mus81 and eme1 mutations severely reduce the yield of viable meiotic spores (Boddy et al. 2001) and surviving spores display a near-complete absence of crossing over (Osman et al. 2003; Smith et al. 2003; Cromie et al. 2006). In Saccharomyces cerevisiae, mus81 and mms4 mutations eliminate the subset of meiotic crossovers that do not exhibit interference (de los Santos et al. 2003). These phenotypes presumably result from failure to cleave some recombination intermediate. Thus, Mus81 is required for a meiotic crossover pathway in S. pombe and S. cerevisiae, although both organisms also have Mus81-independent crossover pathways.
Yeast mus81, mms4, and eme1 mutants are hypersensitive to agents that produce lesions that can block progression of replication forks, such as ultraviolet (UV) light and the alkylating agent methyl methanesulfonate (MMS) (Boddy et al. 2000; Interthal and Heyer 2000). In S. pombe, mus81 mutants are moderately sensitive to prolonged exposure to hydroxyurea (HU) (Boddy et al. 2000), an agent that causes replication fork stalling, although they are not highly sensitive to acute HU treatment (Kai et al. 2005). Yeast mutants are also hypersensitive to camptothecin (CPT), a topoisomerase inhibitor that generates double-strand breaks (DSBs) during replication (Doe et al. 2002; Bastin-Shanower et al. 2003; Kai et al. 2005). These sensitivities suggest that Mus81 has a role in processing a DNA intermediate that arises during repair of replication forks or during replication restart.
An additional finding suggestive of a role for Mus81-Mms4 (or Mus81-Eme1) in replication fork repair comes from genetic interactions with RecQ helicases. In fungi, mus81, mms4, and eme1 mutations are synthetically lethal with mutations in the RecQ helicase genes SGS1 (S. cerevisiae) (Mullen et al. 2001) and rqh1 (S. pombe) (Boddy et al. 2000). RecQ helicases are thought to have important roles in repairing damaged, stalled, or blocked replication forks (Harmon and Kowalczykowski 1998; Courcelle and Hanawalt 1999; Doe et al. 2002; Liberi et al. 2005). One well-studied example is human BLM. Mutations in BLM cause Bloom Syndrome (BS), a rare hereditary disease characterized by short stature, severe sun sensitivity, and predisposition to a wide spectrum of cancers (reviewed in German 1993). A predominant feature of cells from BS patients is highly elevated levels of sister-chromatid exchange (Chaganti et al. 1974). These exchanges are believed to be the result of a change in processing of a DNA intermediate involved in replication fork metabolism. The synthetic lethality suggests that cleavage by Mus81-Mms4 is an alternative pathway for processing this intermediate. This interpretation is supported by the fact that the lethality of budding yeast sgs1Δ mus81Δ mutants is rescued in strains, such as rad51 mutants, that are unable to exchange DNA strands for homologous recombination (Fabre et al. 2002; Bastin-Shanower et al. 2003).
Genetic studies of murine Mus81 and Eme1 have revealed some functional differences from fungal studies. Mus81−/− mice are fully fertile and show no defects in gametogenesis (McPherson et al. 2004; Dendouga et al. 2005), suggesting that this nuclease has little or no role in vertebrate meiotic recombination. In addition, the constellation of genotoxic agents to which Mus81−/− mouse cells are sensitive differs from that of S. pombe and S. cerevisiae mutants. While the fungal mutants are highly sensitive to MMS, UV, and CPT, the mouse mutants are only mildly sensitive to these agents, but are hypersensitive to agents that generate interstrand crosslinks (ICLs), such as cisplatin and mitomycin C (Abraham et al. 2003; McPherson et al. 2004; Dendouga et al. 2005; Hanada et al. 2006).
To better understand the roles of Mus81-Mms4 in metazoans, we generated mutations in the mus81 and mms4 genes of Drosophila melanogaster. We did not detect any defects in meiotic crossing over in these mutants. The mutants showed mild sensitivity to some DNA-damaging agents, but no differences from wild type for most agents tested. We also found that mus81 and mms4 mutations cause lethality when combined with null mutations in mus309, which encodes the Drosophila ortholog of the BLM helicase. This lethality was suppressed in triple mutants with spn-A, which encodes the Drosophila ortholog of the strand invasion protein Rad51. The mus81 and mms4 mutations were viable in combination with mus309N2, a partial separation-of-function allele, providing novel insights into the cause of the lethality.
MATERIALS AND METHODS
Drosophila stocks:
Flies were maintained on standard medium at 25°. Experiments with mei-9 used the allele mei-9A2, and experiments with spn-A used the spn-A093A allele. Both are nonsense mutations that eliminate detectable protein expression (Staeva-Vieira et al. 2003; Yildiz et al. 2004). The mei-4129D mutation deletes 975 bp of coding sequence and has a 1-bp insertion resulting in a frameshift; it is also genetically null (Laurencon et al. 2003). Several alleles of mus309 were used; these are described in the text.
Mutations in mus81 and mms4:
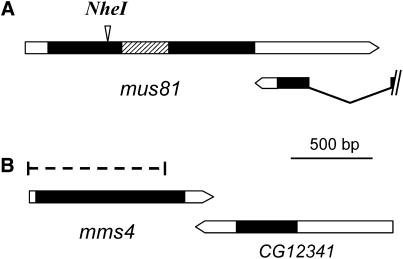
A 3.9-kb fragment containing mus81 and portions of the adjacent genes was cloned into pTV2. The 16-bp sequence between two PstI sites in mus81 was replaced with an I–SceI recognition sequence. A 16-bp sequence containing an in-frame stop codon and an NheI site was inserted into a BglII site at codon 130. Ends-in targeting was conducted to generate integrations into the mus81 locus, resulting in a tandem duplication in which one copy of mus81 carried the insertion. Collapse of the tandem duplication, as in Rong and Golic (2000), produced the mus81Nhe allele (Figure 1A).
Figure 1.—
Mutations in mus81 and mms4. Schematics of (A) mus81 and (B) mms4 are shown. Protein-coding sequences are in solid. The region encoding the nuclease domain of MUS81 is stippled. The 16-bp insertion in mus81Nhe is indicated, and the 3′-end of the adjacent, overlapping gene is shown. In B, the region deleted in mms4ex1 is denoted with a dashed bar. Both mus81 and mms4 lack introns. The 3′-UTR of each overlaps with the adjacent gene, which is transcribed in the opposite direction.
To generate a mutation in mms4, we obtained the P-element line P{SUPor-P, y+w+}KG06402 from the Bloomington Drosophila Stock Center. We generated males that carried P{SUPor-P, y+w+}KG06402/CyO, H{w+, Δ2-3} to induce germline excision and crossed them to y w1118 females. Cy+ male progeny were screened for those that had lost the y+ and/or w+ markers on the P element. A total of 228 males, each from a different vial and therefore independent of one another, were crossed to generate progeny and then subjected to PCR to identify lines carrying deletions extending into mms4. We obtained three deletions: mms4ex1, mms4ex2, and mms4ex3, which delete 839, 757, and 602 bp, respectively, of 958 coding base pairs in the gene (Figure 1B). We used mms4ex1 in the experiments described below.
Meiotic crossing over:
Virgin females heterozygous for a series of markers on chromosome 3 (cu sr e Pr ca) were crossed to tester males (cu sr e ca) and progeny were scored for the five phenotypes. The female parents were w1118 (control), y mus81Nhe w1118, w1118 mei-9A2, or y mus81Nhe w1118 mei-9A2. The white-eye phenotype of w1118 precluded scoring the claret-eye phenotype in males, so only female progeny were scored. Statistical comparisons were done for each pair of genotypes. For each interval, a χ2 test was conducted to compare the number of progeny with a crossover in that interval vs. the number of progeny without a crossover.
Sensitivity to DNA-damaging agents:
Crosses were designed to give a 1:1 ratio of mutant:control progeny. In most of the experiments, mus81Nhe mutant males were crossed to C(1)DX, y f/Y virgin females to generate mus81Nhe (mutant) males and C(1)DX, y f/Y (control) females. In some cases, mus81Nhe males were crossed to Df(1)AD11/FM7 or y cv v f virgin females, and female progeny, which were either mus81Nhe/Df(1)AD11 (hemizygous mutant) or mus81Nhe/+ (control), were counted. For mms4, homozygous mms4ex1 virgin females were crossed to Df(2R)E3363/CyO males. Parents were allowed to mate and lay embryos for 3 days and then transferred to new vials and allowed to lay for 2 days before being removed. The first brood was left untreated. One day after removal of the parents from the second brood, 250 μl of a solution of the agent being tested was added to the food and ingested by feeding larvae. Adults were counted 9–17 days after removal of parents. To determine relative survival, the ratio of mutant:control was determined and then normalized to the mutant:control ratio found in untreated vials. All agents were dissolved in water except CPT. A stock solution of 5 mg/ml CPT in DMSO was made and this was diluted into 10% ethanol/2% Tween 20 in water. Mock treatment solution containing everything except CPT was added to the control brood. UV sensitivity assays were performed by irradiating third instar larvae in a Stratalinker (Stratagene, La Jolla, CA) as in Radford et al. (2005).
Synthetic lethality:
Crosses were set up with parents homozygous for mus81Nhe or mms4ex1 and heterozygous for mus309 (mus309 alleles were carried over a TM3 balancer chromosome in the mus81Nhe background and MKRS in the mms4ex1 background). The progeny were scored for survival to adulthood. Genotypes for experimental crosses are shown in Table 2. Control crosses were set up using females with the same genotype as those used in the experimental crosses and males that had wild-type mus81 and were heterozygous for mus309 (carried over the same balancer as used in the experimental crosses). Since mus81 is on the X chromosome, the male progeny of the control cross were mus81Nhe mutants; therefore, only female progeny were used in calculations. Crosses to test for suppression of synthetic lethality by spn-A093A were identical except for the addition of spn-A093A to the mus309N1 and mus309D2 chromosomes.
TABLE 2.
Synthetic lethality between mus81Nhe and mus309
| Double mutantc
|
|||||
|---|---|---|---|---|---|
| Maternal genotypea | Paternal genotypea | Balancedb | Expected no.d | Observed no. | Survival (%) |
| mus81; mus309D2/TM3 | mus81; mus309D3/TM3 | 1024 | 974 | 0 | 0 |
| mus81; mus309N1/TM3 | mus81; mus309D3/TM3 | 2489 | 2200 | 0 | 0 |
| mus81; mus309N1/TM3 | mus81; mus309D2/TM3 | 590 | 666 | 0 | 0 |
| mus81; mus309N1/TM3 | mus81; mus309N2/TM3 | 1924 | 1128 | 938 | 83 |
| mms4; mus309N1/TM3 | mms4; mus309D2/TM3 | 3984 | 1353 | 0 | 0 |
| mus81; mus309N1 spn-A/TM3 | mus81; mus309D2 spn-A/TM3 | 1710 | 1094 | 735 | 67 |
Alleles used were mus81Nhe, mms4ex1, and spn-A093. mus309 alleles are indicated.
Number of progeny carrying the TM3 balancer and therefore heterozygous for the mus309 chromosome.
Number of double-mutant progeny (triple mutant, in the case of mus81; mus309 spn-A).
The expected number is based on the number of balanced progeny, normalized to the ratio measured in the control crosses (identical except that the fathers did not carry a mus81 or mms4 mutation).
Apoptosis measurements:
Imaginal discs were dissected from wandering third instar larvae in Ringer's solution and fixed for 45 min in 4% formaldehyde and PBS with 0.1% Tween 20 (PBT). Discs were washed and blocked in PBT with 5% bovine serum albumin. Discs were incubated with 1:500 dilution of rabbit anti-human cleaved caspase-3 (Asp175) antibody (Cell Signaling Technology) in PBT overnight at 4°. Different lot numbers of the antibody stained different numbers of cells in wild-type discs; the experiments reported here were all done with the same lot number. Discs were incubated for 2 hr at room temperature with 1:1000 secondary goat anti-rabbit rhodamine-conjugated antibody (Molecular Probes, Eugene, OR) or secondary goat anti-rabbit fluorescein-conjugated antibody (Molecular Probes), stained with 10 μg/ml DAPI in PBT, and mounted with Flouromount-G (Southern Biotechnology Associates). Discs were visualized using TRIT-C and FIT-C filter of a Nikon Eclipse E800 fluorescent microscope. Quantification was performed by counting the number of antibody-stained cells in each of 5–15 wing discs of each genotype. Counts from each pair of genotypes were compared through an unpaired t-test with Welch's correction, using InStat (Graphpad) statistical software; two-tailed P-values are reported.
RESULTS
Mutations in mus81 and mms4:
The Drosophila ortholog of mus81 maps to 1D2, near the tip of the X chromosome. We used the targeted mutagenesis method of Rong and Golic (2001) to introduce a mutation into mus81 (see materials and methods for details). A 16-bp fragment harboring a unique NheI site and an in-frame stop codon was inserted at a position predicted to terminate translation upstream of sequences encoding the conserved nuclease domain (Figure 1A); we refer to this allele as mus81Nhe.
Since there is little sequence similarity between S. pombe Eme1 and S. cerevisiae Mms4, we used iterative BLAST searches to identify a Drosophila ortholog. This strategy pointed to CG12936, located in 47C1. After confirming that the product of this gene interacts with MUS81 in a yeast two-hybrid assay (J. LaRocque and J. Sekelsky, unpublished data), we renamed the gene mms4. During the course of these studies, human EME1 was identified (Ciccia et al. 2003; Ogrunc and Sancar 2003). Eme1 proteins from sequenced mammalian genomes are predicted to be 570–590 residues and are highly similar throughout. The predicted Drosophila MMS4 protein is only 309 residues; this protein shares sequence similarity (25% identity and 43% similarity) with the carboxy-terminal 300 residues of human Eme1. A full-length Drosophila mms4 cDNA sequence has been reported (RE20777, GenBank accession no. AY071164), indicating that the size difference between human EME1 and Drosophila MMS4 is not due to an annotation error (there are no introns in the mms4 gene, and there is another gene, CG7637, transcribed in the opposite direction, only 200 bp upstream of mms4). The N-terminal 200 residues of human Eme1 is conserved in other mammalian species but not in other vertebrates. S. cerevisiae Mms4 and S. pombe Eme1 are also longer at their amino termini, but conservation at the level of primary sequence is not evident. It is not known what function, if any, this N-terminal region has.
To generate mutations in mms4, we used the P-element insertion P[SUPor-P]KG06402, which is inserted 23 bp upstream of the ATG corresponding to the predicted start codon of mms4. No mutant phenotypes were observed in flies homozygous for P[SUPor-P]KG06402, so we conducted an excision screen to generate deletions (see materials and methods for details). In the experiments described below, we used the deletion allele mms4ex1, which removes 839 of 927 protein-coding base pairs (Figure 1B).
MUS81 is not required to generate meiotic crossovers:
Yeast mus81, mms4, and eme1 mutants have meiotic defects that include reductions in crossovers and spore viability (Boddy et al. 2001; de los Santos et al. 2001; Kaliraman et al. 2001). In S. pombe, all meiotic crossovers require Mus81-Eme1 (Osman et al. 2003; Smith et al. 2003), whereas in S. cerevisiae Mus81-Mms4 contributes to formation of the subset of crossovers that do not exhibit interference (de los Santos et al. 2003). We found that mus81Nhe flies have normal fertility and wild-type levels of meiotic nondisjunction of the X chromosome (data not shown). We measured meiotic crossing over along chromosome 3 (Table 1). Differences between mus81 and wild type were significant in one of the four intervals, e–Pr (P = 0.005). Because this was the largest interval surveyed, spanning more than a third of the total genetic distance, this led to a slight decrease in the total map length of the region assayed; however, the map distances for both genotypes are within the range seen in different wild-type strains (Kidwell 1977).
TABLE 1.
Meiotic crossing over in mus81 and mei-9 mutants
| Genetic distance (MU)
|
% of wild type | ||||||
|---|---|---|---|---|---|---|---|
| Genotype | cu–sr | sr–e | e–Pr | Pr–ca | Total | n | |
| w1118 | 9.8 | 9.0 | 20.4 | 14.2 | 53.4 | 100 | 1463 |
| mus81Nhe | 11.5 | 9.4 | 16.5 | 11.3 | 48.7 | 91 | 1967 |
| mei-9A2 | 1.5 | 0.7 | 2.4 | 1.9 | 6.5 | 12 | 538 |
| mus81Nhe mei-9A2 | 1.9 | 1.5 | 4.5 | 3.4 | 11.3 | 21 | 470 |
Previous work has shown that most crossovers in Drosophila require the nucleotide excision repair endonuclease MEI-9-ERCC1, in a complex with the novel protein MUS312 (Baker and Carpenter 1972; Yildiz et al. 2002; Radford et al. 2005). Null mutations in mei-9, which encodes the catalytic subunit, eliminate 90% of crossovers (Sekelsky et al. 1995). Interestingly, the nuclease domain in MEI-9 is related to that of MUS81 (Aravind et al. 1999). We hypothesized that MUS81-MMS4 might be involved in generating the residual crossovers observed in mei-9 mutants. To test this hypothesis, we assayed meiotic crossing over in mus81Nhe mei-9A2 double mutants (Table 1). Relative to wild-type or mus81 single-mutant females, crossovers were significantly reduced in both mei-9 single mutants and mus81 mei-9 double mutants (P < 0.0001 for each interval). The increased frequency of crossovers in the double mutant vs. the mei-9 single mutant is statistically significant over the entire interval (P = 0.01), but is not statistically significant in any single interval. We do not know whether the difference between mei-9 single mutants and mus81 mei-9 double mutants is biologically meaningful.
Sensitivity to DNA-damaging agents:
Insight into functions of proteins involved in DNA metabolism can be gained by examining sensitivities of mutants to agents that damage DNA or interfere with DNA metabolism. Yeast mus81, mms4, and eme1 mutants are hypersensitive to UV irradiation, MMS, HU, and CPT (Boddy et al. 2000; Interthal and Heyer 2000; Doe et al. 2002; Bastin-Shanower et al. 2003), whereas mouse Mus81−/− and Eme1−/− cells are hypersensitive to agents that cause ICLs (Abraham et al. 2003; Dendouga et al. 2005; Hanada et al. 2006). Although all these agents have multiple effects on cell metabolism, a commonality between them is that they have detrimental effects on replication forks: ICLs and damage induced by UV or MMS block DNA synthesis, HU treatment leads to stalled replication forks, and CPT causes replication-dependent DSBs (reviewed in Friedberg et al. 1995). Hypersensitivities to these agents have been taken as evidence for the involvement of the Mus81-Mms4/Eme1 endonuclease in responding to replication fork problems.
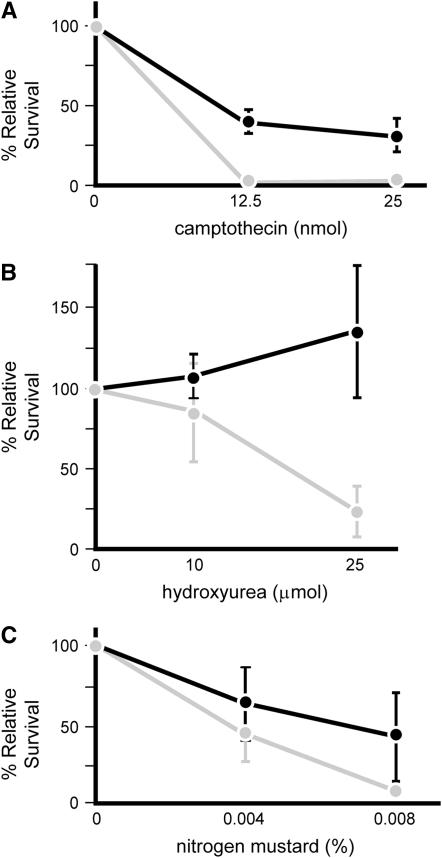
We assayed mus81Nhe mutant larvae for sensitivity to UV, MMS, HU, CPT, ionizing radiation (IR), and nitrogen mustard. As a positive control, we conducted simultaneous assays of sensitivity on mei-4129D mutants. This gene encodes the Drosophila ortholog of ATR, which is required for the DNA-damage-dependent cell cycle checkpoint (Hari et al. 1995); mei-41 mutants are hypersensitive to a wide range of DNA-damaging agents (Boyd et al. 1976; Nguyen et al. 1979; Mason et al. 1981; Banga et al. 1986). We did not detect hypersensitivity of mus81Nhe mutants to UV, MMS, or IR (data not shown). We did observe moderate hypersensitivity to CPT, which generates replication-dependent DSBs, and to the crosslinking agent nitrogen mustard (Figure 2). Surprisingly, mus81Nhe mutant larvae were less sensitive to HU than wild-type larvae (Figure 2). This was true for both hemizygous males (mus81Nhe/Y) and hemizygous females (mus81Nhe/Df), suggesting that the hyposensitivity to HU is due to mutation of mus81, rather than to some other mutation on the X chromosome; however, mms4ex1 mutants were not hyposensitive (data not shown).
Figure 2.—
(A–C) Sensitivity to DNA damage in mus81Nhe. Rates of survival to adulthood were calculated relative to control siblings in the same vial and normalized to ratios in untreated vials. Numbers on the x-axis represent the total quantity of agent added to the food (see materials and methods), except for nitrogen mustard, which indicates the percentage (v/v) of agent in the 250 μl added to ∼8 ml of food. Solid lines, mus81Nhe; shaded lines, mei-4129D (positive control to verify activity of the agents). Relative survival for each point is the weighted mean from four to seven independent experiments, each including 7–10 independent vials. Error bars show standard deviation. For each point shown, there is a significant difference between survival of mutant and control siblings (P < 0.05), with the exception of mus81Nhe at the lower hydroxyurea dose (P = 0.1221).
Mutations in mus81 and mms4 are synthetically lethal with null mutations in mus309:
Mutations in S. pombe mus81 are synthetically lethal with mutations in rqh1, which encodes a RecQ helicase (Boddy et al. 2000). Similarly, mutations in S. cerevisiae MUS81 or MMS4 are synthetically lethal with mutations in SGS1 (Mullen et al. 2001). There are three RecQ helicase genes in Drosophila: RecQ4, RecQ5, and mus309 (Sekelsky et al. 2000). Mutations in mus309, which encodes the Drosophila ortholog of the BLM helicase, DmBlm (Kusano et al. 1999), result in hypersensitivity to MMS and IR, defects in DSB repair, maternal-effect embryonic lethality, and decreased meiotic recombination (Boyd et al. 1981; Beall and Rio 1996; Kusano et al. 2001; Adams et al. 2003; Min et al. 2004; Johnson-Schlitz and Engels 2006; McVey et al. 2007, accompanying article in this issue).
We generated double mutants with mus81Nhe and various alleles of mus309. The mus309D2 and mus309D3 mutations were isolated in a screen for EMS-induced mutations causing hypersensitivity to MMS (Boyd et al. 1981); mus309D2 is a nonsense mutation and is null, whereas mus309D3 has a missense mutation that changes the glutamic acid residue in the DEAH motif to lysine, which should abolish helicase activity (Kusano et al. 2001). The mus309N1 mutation is a deletion beginning 110 bp upstream of the coding region and extending for 2480 bp into the helicase domain; this allele is genetically null (McVey et al. 2007, accompanying article in this issue). Double mutants between mus81Nhe or mms4ex1 and any of these null alleles of mus309 were synthetically lethal (Table 2). We also tested for synthetic lethality between mus81Nhe and mus309N2, a deletion that is predicted to truncate at least 566 residues from the amino terminus (McVey et al. 2007, accompanying article in this issue). The deletion does not extend into the helicase domain, which begins at residue 736. This allele behaves like a null allele in a DSB repair assay, but is hypomorphic with respect to early embryonic function. Double mutants of the genotype mus81Nhe; mus309N2/mus309N1 were viable (Table 2). In the cross reported in Table 2, survival was 83% relative to the control cross. In addition, these double-mutant adults are fertile. The mus309N2 mutation therefore uncouples the role of DmBlm in DSB repair from the function required for survival in the absence of MUS81.
We examined the synthetic lethality between mus81Nhe and null alleles of mus309 more carefully. Although we did not quantify lethality through all stages of development, we did observe many double-mutant pupae, often as pharate adults, indicating that double mutants can survive through embryonic and larval development. This observation is in agreement with a recent report by Johnson-Schlitz and Engels (2006), who found that lethality of mus81; mus309 double mutants occurred primarily at the pupal stage. Pupal lethality often results from defects in cell proliferation (Gatti and Baker 1989). This occurs when maternally deposited mRNA or protein is sufficient for embryonic development, but not for postembryonic proliferation of diploid tissues. Most larval growth is attributed to increases in cell size and ploidy rather than cell proliferation, so the mutant larvae grow and undergo pupariation; however, proliferation of diploid imaginal tissues and cells in the central nervous system is impaired, leading to the absence or reduction in size of the imaginal discs and the ventral ganglion. We dissected double-mutant larvae and did not detect any reductions in size of imaginal discs or the ventral ganglion.
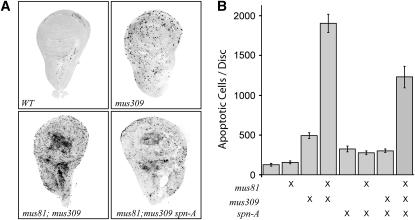
Pupal lethality may also result from elevated cell death. We quantified the number of cells in wing imaginal discs that were undergoing apoptosis (Figure 3). There was no significant difference between wild-type and mus81 single mutants (P = 0.26), but apoptosis was increased 3- to 4-fold in mus309N1 single mutants (P < 0.0001 compared to wild type or mus81). Apoptosis was highly elevated in mus81; mus309N1 double mutants, ∼15-fold over wild-type and 4-fold over mus309N1 single mutants (P < 0.0001 for both comparisons). We did not detect any developmental delay in the double mutants, suggesting that the increased apoptosis is due to genome damage rather than to sampling of different developmental stages.
Figure 3.—
Apoptosis in proliferating tissues. (A) Wing imaginal discs from mature third instar larvae, stained with an antibody that recognized cells undergoing apoptosis. Representative discs from four different genotypes are shown. The mus309N1 null allele was used in all cases. (B) Quantification of apoptosis in wing discs from larvae of various genotypes. Genotypes are given below the graph, with an “X” indicating a null mutation in the corresponding gene. Each bar represented the mean number of apoptotic cells (n = 10, 5, 15, 12, 12, 11, 13, and 8, left to right). Error bars are standard error of the mean.
Synthetic lethality is suppressed by preventing homologous recombination:
In S. cerevisiae, synthetic lethality between sgs1 and mus81 is suppressed by mutations in RAD51 (Fabre et al. 2002; Bastin-Shanower et al. 2003), which encodes a strand exchange protein essential for DSB repair through homologous recombination. One interpretation of this result is that Rad51-mediated strand invasion allows formation of a repair intermediate that must be processed by either the Sgs1 helicase or the Mus81-Mms4 endonuclease. In this model, the absence of Rad51 results in use of a different repair pathway that does not require Sgs1 or Mus81-Mms4. The Drosophila ortholog of Rad51 is encoded by spn-A (Staeva-Vieira et al. 2003). We found that mutation of spn-A partially suppressed the synthetic lethality between mus81 and mus309, resulting in 67% of the mutants surviving to adulthood (Table 2).
We also quantified the effects of spn-A mutations on apoptosis (Figure 3). Apoptosis was elevated in spn-A single mutants relative to wild type (P = 0.0003), but not significantly different among spn-A single mutants, mus309N1 spn-A double mutants, and mus81; spn-A double mutants (P > 0.3 for each comparison). Notably, apoptosis was lower in spn-A single mutants and mus309N1 spn-A double mutants than in mus309N1 single mutants (P = 0.0049 and 0.0003, respectively), indicating that spn-A is epistatic to mus309. Consistent with this interpretation, we found that apoptosis was decreased in mus81; mus309N1 spn-A triple mutants, relative to mus81; mus309N1 double mutants (P = 0.0017). However, the level in the triple mutants was still significantly higher than that in spn-A single mutants (P < 0.0001), suggesting that spn-A is only partially epistatic to mus81 and mus309. Interpretations of this finding are discussed below.
DISCUSSION
We generated mutations in Drosophila mus81 and mms4 and examined the effects of these mutations on processes in which fungal and mammalian Mus81, Eme1, and Mms4 have been implicated. In fungi, Mus81 is required to generate meiotic crossovers that do not display interference (Boddy et al. 2001; de los Santos et al. 2003; Argueso et al. 2004; Cromie et al. 2006). We observed a small decrease in crossing over in mus81 mutants relative to the wild-type strain that we used as a control, but crossing over in the mutants was still within the range seen in different wild-type strains (Kidwell 1977). Our sample size was not large enough to rule out the possibility that Drosophila MUS81-MMS4 has a role in generating a subset of crossovers, such as those that do not exhibit interference. However, mathematical models of interference are consistent with a model in which all crossovers in Drosophila exhibit interference (Copenhaver et al. 2002).
Most crossovers in Drosophila require the MEI-9-ERCC1 endonuclease (Baker and Carpenter 1972; Sekelsky et al. 1995; Yildiz et al. 2004; Radford et al. 2005). Importantly, the residual crossing over that occurs in mei-9 mutants was not eliminated by mutation of mus81. Our results do demonstrate, however, that MUS81-MMS4 does not have a major role in generating meiotic crossovers. This is similar to the case in other metazoans, such as mice and Caenorhabditis elegans, but differs from S. cerevisiae and S. pombe, where Mus81 is important in generating some or all meiotic crossovers. In other aspects of meiotic recombination, vertebrates appear to be more similar to S. cerevisiae than to Drosophila (McKim et al. 2002; Blanton and Sekelsky 2004). These similarities and differences reveal unexpected variability in this crucial process.
We measured the effect of various DNA-damaging agents on survival of mus81 mutant larvae to adulthood. We detected hypersensitivity to some DNA-damaging agents that suggest a function in repairing damaged or stalled replication forks. Although these phenotypes were relatively mild, the spectrum of agents to which Drosophila mus81 mutants are hypersensitive is more similar to that of mouse MUS81 mutants than to that of S. cerevisiae and S. pombe mus81 mutants. Nonetheless, each of these model organisms exhibits hypersensitivities that suggest a role in responding to damaged or stalled replication forks, and thus it appears that some important function is conserved, but that different response pathways are used to different extents in different species.
We found that mus81 and mms4 mutations are synthetically lethal with null mutations in mus309. Although we did not find any severe phenotypes in mus81 single mutants, null mutants of mus309 have several severe phenotypes, including strong hypersensitivity to MMS and IR, defects in DSB repair and meiotic recombination, and maternal-effect embryonic lethality (Boyd et al. 1981; Beall and Rio 1996; Kusano et al. 2001; Adams et al. 2003; McVey et al. 2007, accompanying article in this issue). Synthetic lethality cannot be caused by defects in meiotic recombination or, given the late stage at which the lethal phenotype manifests, to the essential early embryonic function of DmBlm. Synthetic lethality is also not due to the function of DmBlm in DSB repair, since the mus309N2 allele is not synthetically lethal with mus81, despite the fact that mus309N2 behaves like a null allele in DSB repair assays (McVey et al. 2007, accompanying article in this issue).
Insights into the etiology of synthetic lethality come from studies of the effects of spn-A on lethality and apoptosis. Similar to the case in fungi, we found that a mutation in spn-A, which encodes the Drosophila ortholog of the strand invasion protein Rad51, suppresses synthetic lethality. This result differs from a recent report by Johnson-Schlitz and Engels (2006), who were not able to suppress synthetic lethality with a spn-A mutation. A likely explanation is that Johnson-Schlitz and Engels (2006) used spn-A1, which is a hypomorphic allele (Gonzalez-Reyes et al. 1997), whereas we used the protein-null allele spn-A093 (Staeva-Vieira et al. 2003). Another difference is that the mus81 deletion used by Johnson-Schlitz and Engels (2006) also removes the adjacent gene, CG3703. The function of this gene is unknown, but the protein it encodes is highly conserved (49 and 54% similarity to the human homolog, RUNDC1, over stretches of 379 and 204 residues, respectively) and contains a RUN domain characteristic of proteins involved in Ras-like GTPase signaling (Callebaut et al. 2001). In our experiments, suppression of synthetic lethality was incomplete (67% of the expected number of adults eclosed), suggesting that the mus81; mus309N1 spn-A triple-mutant genotype is near the threshold of viability. Additional mutations, such as deletion of CG3703, may decrease overall fitness so as to preclude survival of any individuals to adulthood.
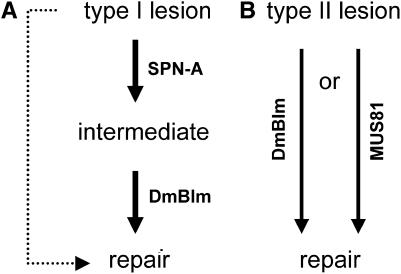
We found that mus309N1 single mutants had elevated apoptosis. Apoptosis is also elevated in spn-A single mutants, but to a lesser degree. SPN-A is epistatic to DmBlm, since mus309N1 spn-A double mutants had the same amount of apoptosis as spn-A single mutants. We propose that there is some type of spontaneous lesion that is processed into a repair intermediate by SPN-A and then by DmBlm (type I lesions, Figure 4A). In the absence of SPN-A, there is a secondary pathway for repair of this type of lesion. This secondary pathway is not as efficient as the SPN-A-dependent pathway, or is error-prone, leading to some increased apoptosis in spn-A single mutants. Inability to process the SPN-A-dependent intermediate by DmBlm, however, results in more severe defects than using the SPN-A-independent pathway, so apoptosis is correspondingly higher in mus309N1 single mutants than in spn-A single mutants or mus309N1 spn-A double mutants.
Figure 4.—
Conceptual model for roles of SPN-A, DmBlm, and MUS81 in repair of spontaneous lesions. (A) Type I lesions are normally processed by SPN-A into an intermediate that is further processed by DmBlm. Our data do not reveal any evidence for a role of MUS81 in repairing this type of lesion. (B) Type II lesions are processed by either of two pathways, one requiring MUS81 and the other requiring DmBlm. Our data do not reveal a role for SPN-A in repairing this type of lesion.
In mus81; mus309N1 double mutants, apoptosis is dramatically elevated. This suggests that there is a second type of spontaneous lesion (type II) that can be repaired by either of two pathways, one requiring DmBlm and the other requiring MUS81 (Figure 4B). Synthetic lethality may be due to the sum of the apoptosis resulting from failure to repair type I and type II lesions. In the mus81; mus309N1 spn-A triple mutant, apoptosis is significantly reduced relative to the mus81; mus309N1 double mutant, but is still quite high. During proliferation of imaginal tissues, cell death caused by DNA damage is compensated for by increased proliferation, at least up to some threshold beyond which the tissue cannot regenerate (Haynie and Bryant 1977; Jaklevic and Su 2004). The amount of apoptosis in the triple mutants may be near the threshold for viability, so that only a fraction of individuals can survive to adulthood. Alternatively, there may be tissue-specific effects on apoptosis, and the tissue that we examined (wing discs) may not be affected to the same extent as tissues that contribute to inviability (e.g., central nervous system).
We speculate that type II lesions may be blocked replication forks, since both RecQ helicases and Mus81 have been implicated in processing blocked forks (reviewed in Branzei and Foiani 2007; Osman and Whitby 2007). Type I lesions may be DSBs associated with broken replication forks. Recombinational repair of these DSBs would require SPN-A and, subsequently, DmBlm. In spn-A mutants, repair could be accomplished through nonhomologous end joining. The spn-A mutation causes a similar reduction in apoptosis in mus309N1 single mutants and mus81; mus309N1 double mutants (34% vs. 35%, P = 0.81 by Fisher's exact test). The similar effect of loss of SPN-A on both genotypes can be explained if type II lesions that are not properly processed are converted into type I lesions; i.e., in the absence of MUS81 and DmBlm, blocked replication forks break, generating DSBs.
The mus309N2 mutation is not synthetically lethal with the mus81 mutation (Table 2). We hypothesize that the mus309N2 deletion destroys the ability of DmBlm to dissociate D-loops (“disruptase” activity), but does not eliminate the ability to promote Holliday junction branch migration. Thus, mus309N2 mutants are unable to repair type I lesions, consistent with the observed defects in DSB repair (McVey et al. 2007, accompanying article in this issue), but are competent to process type II lesions.
In conclusion, we found that Drosophila MUS81-MMS4 is not required to generate meiotic crossovers, but may have some functions in responding to lesions that arise during DNA replication. These functions are at least partially redundant with those of DmBlm, so that loss of both proteins is lethal. Lethality occurs in pupal stages, due to increased apoptosis. A subset of the damage leading to apoptosis is due to failure to process recombination intermediates generated by the SPN-A strand invasion protein.
Acknowledgments
The authors thank Jan Mullen and Suzanne Shanower for initiating the construction of mus81Nhe, Jeannine LaRocque for conducting a yeast two-hybrid assay with Drosophila MUS81 and MMS4 and for assisting with the apoptosis assay, and members of the Sekelsky lab for helpful comments on the manuscript. This work was supported by grants from the American Cancer Society (RSG DDC-104804) to K.M. and from the National Institutes of Health (GM067956 and GM61252) to S.J.B. and to J.S., respectively.
References
- Abraham, J., B. Lemmers, M. P. Hande, M. E. Moynahan, C. Chahwan et al., 2003. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 22: 6137–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, M. D., M. McVey and J. Sekelsky, 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267. [DOI] [PubMed] [Google Scholar]
- Aravind, L., D. R. Walker and E. V. Koonin, 1999. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 27: 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso, J. L., J. Wanat, Z. Gemici and E. Alani, 2004. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168: 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, B. S., and A. T. C. Carpenter, 1972. Genetic analysis of sex chromosomal meiotic mutants in Drosophila melanogaster. Genetics 71: 255–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga, S. S., R. Shenkar and J. B. Boyd, 1986. Hypersensitivity of Drosophila mei-41 mutants to hydroxyurea is associated with reduced mitotic chromosome stability. Mutat. Res. 163: 157–165. [DOI] [PubMed] [Google Scholar]
- Bastin-Shanower, S. A., W. M. Fricke, J. R. Mullen and S. J. Brill, 2003. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 23: 3487–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall, E. L., and D. C. Rio, 1996. Drosophila IRBP/Ku p70 corresponds to the mutagen-sensitive mus309 gene and is involved in P-element excision in vivo. Genes Dev. 10: 921–933. [DOI] [PubMed] [Google Scholar]
- Blanton, H., and J. Sekelsky, 2004. Unique invasions and resolutions: DNA repair proteins in meiotic recombination in Drosophila melanogaster. Cytogenet. Genome Res. 107: 172–179. [DOI] [PubMed] [Google Scholar]
- Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W. D. Heyer et al., 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20: 8758–8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates, III et al., 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548. [DOI] [PubMed] [Google Scholar]
- Boyd, J. B., M. D. Golino, T. D. Nguyen and M. M. Green, 1976. Isolation and characterization of X-linked mutants of Drosophila melanogaster which are sensitive to mutagens. Genetics 84: 485–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, J. B., M. D. Golino, K. E. S. Shaw, C. J. Osgood and M. M. Green, 1981. Third-chromosome mutagen-sensitive mutants of Drosophila melanogaster. Genetics 97: 607–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei, D., and M. Foiani, 2007. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Rep. 6: 994–1003. [DOI] [PubMed] [Google Scholar]
- Callebaut, I., J. de Gunzburg, B. Goud and J. P. Mornon, 2001. RUN domains: a new family of domains involved in Ras-like GTPase signaling. Trends Biochem. Sci. 26: 79–83. [DOI] [PubMed] [Google Scholar]
- Chaganti, R. S., S. Schonberg and J. German, 1974. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl. Acad. Sci. USA 71: 4508–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia, A., A. Constantinou and S. C. West, 2003. Identification and characterization of the human mus81-eme1 endonuclease. J. Biol. Chem. 278: 25172–25178. [DOI] [PubMed] [Google Scholar]
- Constantinou, A., X. B. Chen, C. H. McGowan and S. C. West, 2002. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 21: 5577–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver, G. P., E. A. Housworth and F. W. Stahl, 2002. Crossover interference in Arabidopsis. Genetics 160: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle, J., and P. C. Hanawalt, 1999. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet. 262: 543–551. [DOI] [PubMed] [Google Scholar]
- Cromie, G. A., R. W. Hyppa, A. F. Taylor, K. Zakharyevich, N. Hunter et al., 2006. Single Holliday junctions are intermediates of meiotic recombination. Cell 127: 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos, T., J. Loidl, B. Larkin and N. M. Hollingsworth, 2001. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159: 1511–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos, T., N. Hunter, C. Lee, B. Larkin, J. Loidl et al., 2003. The mus81/mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendouga, N., H. Gao, D. Moechars, M. Janicot, J. Vialard et al., 2005. Disruption of murine mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol. Cell. Biol. 25: 7569–7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe, C. L., J. S. Ahn, J. Dixon and M. C. Whitby, 2002. Mus81-eme1 and rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277: 32753–32759. [DOI] [PubMed] [Google Scholar]
- Fabre, F., A. Chan, W. D. Heyer and S. Gangloff, 2002. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99: 16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg, E. C., G. C. Walker and W. Siede, 1995. DNA Repair and Mutagenesis. American Society for Microbiology, Washington, DC.
- Gaillard, P. H., E. Noguchi, P. Shanahan and P. Russell, 2003. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell 12: 747–759. [DOI] [PubMed] [Google Scholar]
- Gatti, M., and B. S. Baker, 1989. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 3: 438–453. [DOI] [PubMed] [Google Scholar]
- German, J., 1993. Bloom syndrome: a Mendelian prototype of somatic mutational disease. Medicine 72: 393–406. [PubMed] [Google Scholar]
- Gonzalez-Reyes, A., H. Elliot and D. St. Johnston, 1997. Oocyte determination and the origin of polarity in Drosophila: the role of the spindle genes. Development 124: 4927–4937. [DOI] [PubMed] [Google Scholar]
- Hanada, K., M. Budzowska, M. Modesti, A. Maas, C. Wyman et al., 2006. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 25: 4921–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari, K. L., A. Santerre, J. Sekelsky, K. S. McKim, J. B. Boyd et al., 1995. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell 82: 815–821. [DOI] [PubMed] [Google Scholar]
- Harmon, F. G., and S. C. Kowalczykowski, 1998. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 12: 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynie, J. L., and P. J. Bryant, 1977. The effects of X-rays on the proliferation dynamics of cells in the imaginal wing disc of Drosophila melanogaster. Rouxs Arch. Dev. Biol. 183: 85–100. [DOI] [PubMed] [Google Scholar]
- Interthal, H., and W. D. Heyer, 2000. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 263: 812–827. [DOI] [PubMed] [Google Scholar]
- Jaklevic, B. R., and T. T. Su, 2004. Relative contribution of DNA repair, cell cycle checkpoints, and cell death to survival after DNA damage in Drosophila larvae. Curr. Biol. 14: 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Schlitz, D., and W. R. Engels, 2006. Template disruptions and failure of double Holliday junction dissolution during double-strand break repair in Drosophila BLM mutants. Proc. Natl. Acad. Sci. USA 103: 16840–16845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai, M., M. N. Boddy, P. Russell and T. S. Wang, 2005. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev. 19: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman, V., J. R. Mullen, W. M. Fricke, S. A. Bastin-Shanower and S. J. Brill, 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15: 2730–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell, M. G., 1977. Reciprocal differences in female recombination associated with hybrid dysgenesis in Drosophila melanogaster. Genet. Res. 30: 77–88. [DOI] [PubMed] [Google Scholar]
- Kusano, K., M. E. Berres and W. R. Engels, 1999. Evolution of the RECQ family of helicases: a Drosophila homolog, Dmblm, is similar to the human Bloom syndrome gene. Genetics 151: 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano, K., D. M. Johnson-Schlitz and W. R. Engels, 2001. Sterility of Drosophila with mutations in the Bloom syndrome gene: complementation by Ku70. Science 291: 2600–2602. [DOI] [PubMed] [Google Scholar]
- Laurencon, A., A. Purdy, J. Sekelsky, R. S. Hawley and T. T. Su, 2003. Phenotypic analysis of separation-of-function alleles of MEI-41, Drosophila ATM/ATR. Genetics 164: 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi, G., G. Maffioletti, C. Lucca, I. Chiolo, A. Baryshnikova et al., 2005. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 19: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, J. M., M. M. Green, K. E. S. Shaw and J. B. Boyd, 1981. Genetic analysis of X-linked mutagen-sensitive mutants of Drosophila melanogaster. Mutat. Res. 81: 329–343. [DOI] [PubMed] [Google Scholar]
- McKim, K. S., J. K. Jang and E. A. Manheim, 2002. Meiotic recombination and chromosome segregation in Drosophila females. Annu. Rev. Genet. 36: 205–232. [DOI] [PubMed] [Google Scholar]
- McPherson, J. P., B. Lemmers, R. Chahwan, A. Pamidi, E. Migon et al., 2004. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science 304: 1822–1826. [DOI] [PubMed] [Google Scholar]
- McVey, M., S. L. Andersen, Y. Broze and J. Sekelsky, 2007. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176: 1979–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, B., B. T. Weinert and D. C. Rio, 2004. Interplay between Drosophila Bloom's syndrome helicase and Ku autoantigen during nonhomologous end joining repair of P element-induced DNA breaks. Proc. Natl. Acad. Sci. USA 101: 8906–8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen, J. R., V. Kaliraman, S. S. Ibrahim and S. J. Brill, 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T. D., J. B. Boyd and M. M. Green, 1979. Sensitivity of Drosophila mutants to chemical carcinogens. Mutat. Res. 63: 67–77. [DOI] [PubMed] [Google Scholar]
- Ogrunc, M., and A. Sancar, 2003. Identification and characterization of human MUS81–MMS4 structure-specific endonuclease. J. Biol. Chem. 278: 21715–21720. [DOI] [PubMed] [Google Scholar]
- Osman, F., and M. C. Whitby, 2007. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Rep. 6: 1004–1017. [DOI] [PubMed] [Google Scholar]
- Osman, F., J. Dixon, C. L. Doe and M. C. Whitby, 2003. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12: 761–774. [DOI] [PubMed] [Google Scholar]
- Radford, S. J., E. Goley, K. Baxter, S. McMahan and J. Sekelsky, 2005. Drosophila ERCC1 is required for a subset of MEI-9-dependent meiotic crossovers. Genetics 170: 1737–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2001. A targeted gene knockout in Drosophila. Genetics 157: 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky, J., K. S. McKim, G. M. Chin and R. S. Hawley, 1995. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141: 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky, J., M. H. Brodsky and K. C. Burtis, 2000. DNA Repair in Drosophila. Insights from the Drosophila genome sequence. J. Cell Biol. 150: F31–F36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. R., M. N. Boddy, P. Shanahan and P. Russell, 2003. Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics 165: 2289–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staeva-Vieira, E., S. Yoo and R. Lehmann, 2003. An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 22: 5863–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz, Ö., S. Majumder, B. C. Kramer and J. Sekelsky, 2002. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol. Cell 10: 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz, Ö., H. Kearney, B. C. Kramer and J. Sekelsky, 2004. Mutational analysis of the Drosophila repair and recombination gene mei-9. Genetics 167: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]