Abstract
SYP-3 is a new structural component of the synaptonemal complex (SC) required for the regulation of chromosome synapsis. Both chromosome morphogenesis and nuclear organization are altered throughout the germlines of syp-3 mutants. Here, our analysis of syp-3 mutants provides insights into the relationship between chromosome conformation and the repair of meiotic double-strand breaks (DSBs). Although crossover recombination is severely reduced in syp-3 mutants, the production of viable offspring accompanied by the disappearance of RAD-51 foci suggests that DSBs are being repaired in these synapsis-defective mutants. Our studies indicate that once interhomolog recombination is impaired, both intersister recombination and nonhomologous end-joining pathways may contribute to repair during germline meiosis. Moreover, our studies suggest that the conformation of chromosomes may influence the mode of DSB repair employed during meiosis.
DURING meiosis, chromosomes undergo programmed DNA double-strand breaks (DSBs) generated by the conserved topoisomerase-like protein Spo11 (Bergerat et al. 1997; Keeney et al. 1997). The existence of a conserved enzymatic function set in place to generate DSBs in fungi, plants, and animals underscores the significance of undergoing recombination during meiosis. Specifically, the reciprocal exchange of DNA that occurs during crossover recombination, after formation of programmed meiotic DSBs, is important for genetic diversity given that meiosis results in the formation of haploid gametes required for sexual reproduction. However, crossover recombination is also essential to establish physical attachments between homologous chromosomes during prophase, allowing for their proper subsequent alignment and accurate segregation to opposite sides of the spindle at meiosis I.
The importance of meiotic DSB formation and crossover recombination have led to intense studying of the various components and mechanisms operating in this process (for reviews, see Paques and Haber 1999; Zickler and Kleckner 1999; Keeney 2001). However, far less is known about the effects of chromosome configuration in DSB repair. Therefore, we exploited the existence of two different syp-3 (synapsis defective) alleles with distinct differences both in the nuclear organization and conformation of chromosomes throughout prophase to investigate how changes in chromosome configuration interface with the progression of meiotic recombination.
In an accompanying article (Smolikov et al. 2007, this issue), we report the identification of syp-3, which encodes for a structural component of the central region of the synaptonemal complex (SC) in Caenorhabditis elegans. The SC is a proteinaceous structure that forms between paired and aligned homologous chromosomes during meiosis from yeast to mammals. It is composed of a pair of lateral elements assembled along the axes of a pair of homologous chromosomes, interconnected by transverse filaments that form the central region of this structure (Page and Hawley 2004). Our analysis of syp-3 mutants revealed that SYP-3 restricts the loading of central region components of the SC promoting their assembly only in an appropriate context, bridging the axes of paired homologs. As a result, SYP-3 is required to stabilize homologous pairing interactions and for chiasma formation. However, we also uncovered differences in chromosome organization between a syp-3 null (ok758) and C-terminal truncation (me42) mutants. Specifically, in the syp-3 null mutant, as in syp-1 and syp-2 null mutants (MacQueen et al. 2002; Colaiácovo et al. 2003), SC formation is not observed by transmission electron microscopy (TEM), central region components SYP-1 and SYP-2 do not associate onto chromosomes, and chromosomes fail to redisperse upon entrance into pachytene, remaining instead in a clustered organization until late pachytene. In the C-terminal truncation mutant, SC formation is also not observed by TEM analysis. However, SYP-1 and SYP-2 are observed associating with unsynapsed chromosomes once they redisperse in pachytene. Interestingly, chromosomes redisperse prematurely in this mutant compared to wild type, suggesting that association of central region components, albeit an incorrect association along unsynapsed chromosomes, may serve as a trigger for redispersal. Moreover, our analysis suggested that this association may effectively truncate or shorten the extent of time during which chromosomes search for homology, driving them out from the clustered organization and resulting in reduced pairing levels.
Here, we examined DSB repair in our syp-3 mutants and observed that SYP-3 is required for crossover recombination given its requirement for appropriate synapsis between homologs. We also observed both sister chromatid-mediated recombination and nonhomologous end joining (NHEJ) operating in meiotic DSB repair once accessibility to a homologous partner or intersister recombination is respectively abrogated. Comparisons between different syp-3 alleles support a model in which two temporal windows (one early, the other late) for DSB repair during prophase are tightly related to chromosome configuration and structural constraints. Altogether, our analysis revealed how DSB repair is modulated by changes in both chromosome organization and structural constraints resulting from alterations in the assembly of the central region of the SC.
MATERIALS AND METHODS
Genetics:
C. elegans strains were cultured at 20° under standard conditions as described in brenner (1974). Bristol N2 worms were utilized as the wild-type background. The following mutations and chromosome rearrangements were used (Riddle et al. 1997; Dernburg et al. 1998; MacQueen et al. 2002; Colaiácovo et al. 2003; Martin et al. 2005; this article):
LGI: syp-3(me42, ok758), hT2[bli-4(e937) qIs48] (I;III)
LGIII: lig-4(ok716)
LGIV: spo-11(ok79), nT1 [unc-?(n754) let-?(m435)] (IV, V)
LGV: syp-2(ok307), syp-1(me17)
LGX: dpy-3(e27) unc-3(e151)
DAPI analysis and immunostaining:
DAPI staining, immunostaining, and analysis of stained meiotic nuclei were carried out as in Colaiácovo et al. (2003) and as in Rogers et al. (2002). Antibodies were used at the following dilutions: α-RAD-51 (1:100) and α-REC-8 (1:100). Secondary antibodies used were: Cy3 anti-rabbit (Jackson Immunochemicals) at 1:200 and Alexa 488 anti-mouse (Molecular Probes, Eugene, OR) at 1:400.
RNA interference:
Production of double-stranded RNA for rec-8(RNAi) and the RNAi procedure were carried out as described in Colaiácovo et al. (2003). REC-8 depletion was assayed by REC-8 antibody staining of whole mounted gonads and monitoring for oocytes carrying 24 univalents at diakinesis.
Time-course analysis for RAD-51 foci:
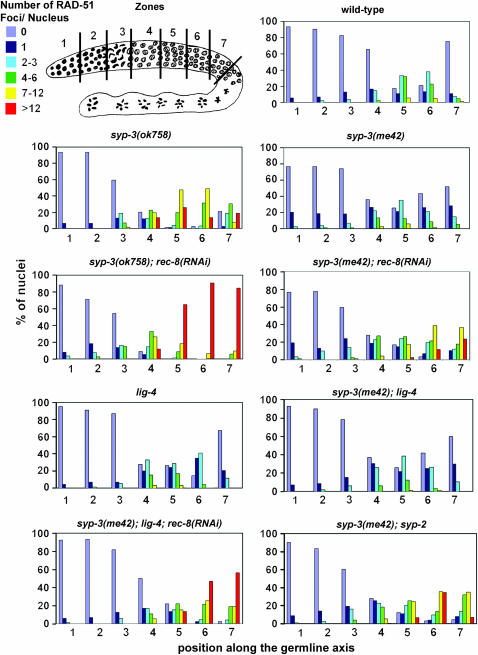
Quantitation of RAD-51 foci was performed as described in Colaiácovo et al. (2003), except that all seven zones composing the divided germline were quantitated. The average number of nuclei scored per zone (n) for wild type, syp-3(ok758), syp-3(me42), syp-3(me42); syp-2, syp-3(ok758); rec-8(RNAi), syp-3(me42); rec-8(RNAi), lig-4(ok716), syp-3(me42); lig-4(ok716), syp-3(me42); rec-8(RNAi); lig-4(ok716) were: zone 1 (n = 123), zone 2 (n = 128), zone 3 (n = 115), zone 4 (n = 135), zone 5 (n = 109), zone 6 (n = 90), and zone 7 (n = 79).
RESULTS
SYP-3 is required for meiotic crossover recombination:
Cytological analysis of germline chromosome morphology in both syp-3 mutants revealed that in addition to altered chromosome morphogenesis and nuclear organization earlier in prophase, both carried 12 univalents instead of 6 bivalents in diakinesis oocytes (Smolikov et al. 2007 (accompanying article). Therefore, we examined whether this lack of chiasmata results from an inability to form crossovers. We measured crossover frequencies for a genetic interval spanning 80% of the X chromosome and observed a reduction to <0.9 and 2.5% of the wild-type level in ok758 and me42, respectively (Table 1).
TABLE 1.
Reduced crossover recombination in syp-3 mutants
| Genotype | Recombinant progeny | Total progeny | Map distance (cM) |
|---|---|---|---|
| +/(ok758 or +); dpy-3 unc-3/++ | 405 hermaphrodites | 1407 hermaphrodites | 35 |
| +/(me42 or +); dpy-3 unc-3/++ | 238 hermaphrodites | 901 hermaphrodites | 32 |
| ok758/ok758; dpy-3 unc-3/++ | 1 hermaphrodite | 290 hermaphrodites | <0.3 |
| 0 males | 137 males | ||
| me42/me42; dpy-3 unc-3/++ | 2 hermaphrodites | 300 hermaphrodites | <0.8 |
| 2 males | 165 males |
Recombination analysis was performed as in Kelly et al. (2000).
Formation and repair of DSBs in syp-3 mutants:
The lack of chiasmata and the severely reduced levels of crossover recombination observed in the syp-3 mutants could reflect either an inability to form meiotic DSBs or an inability to repair DSBs as crossovers. To distinguish between these possibilities, we utilized an antibody against RAD-51 (a protein involved in DNA strand exchange during DSB repair; Sung 1994) and examined the levels of RAD-51 foci in nuclei throughout meiotic prophase (Figures 1 and 2). We observed that levels of RAD-51 foci rose upon entrance into transition zone (zones 2 and 3) in both syp-3 mutants with wild-type kinetics, indicating that SYP-3 is not required for the initiation of meiotic recombination.
Figure 1.—
Quantitative time-course analysis of RAD-51 foci during meiotic prophase. Germline diagram indicates the zones along the germline throughout which RAD-51 foci were scored for all nuclei. Zone 1 and most of zone 2 consist of nuclei undergoing mitotic divisions (premeiotic tip). Zone 3 consists of nuclei entering meiosis (transition zone). Zones 4–7 consist of nuclei in early through late pachytene. Levels of observed RAD-51 foci are indicated by the color code. Histograms depict the quantitation of RAD-51 foci in germlines of the indicated genotypes. Positions along the x-axis correspond to the zones along the germline as indicated in the diagram. The y-axis indicates the percentage of nuclei falling into each corresponding category indicated by the color code.
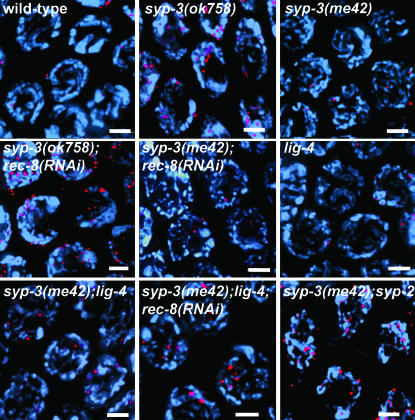
Figure 2.—
Sister chromatid-mediated recombination and NHEJ are alternative modes of meiotic DSB repair in syp-3 mutants. Elevated levels of RAD-51 foci are apparent in syp-3(ok758) and further accentuated if sister chromatid cohesion is disrupted in syp-3(ok758); rec-8(RNAi). In syp-3(me42), levels of RAD-51 foci are comparable to wild-type, lig-4, or syp-3(me42); lig-4. However, they are elevated in syp-3(me42); lig-4; rec-8(RNAi) and in syp-3(me42); syp-2. Images are projections halfway through 3D data stacks of mid-pachytene (zone 5) nuclei. DAPI-stained chromosomes (blue); α-RAD-51 (red). Bars, 2 μm.
Analysis of nuclei in early and mid-pachytene (zones 4 and 5) revealed that levels of RAD-51 foci were higher at these stages in syp-3(ok758) than in wild type and remained elevated throughout late pachytene. While only 13% of nuclei still bear more than one RAD-51 focus by late pachytene (zone 7) in wild type, 61% carry more than one focus in syp-3(ok758). Furthermore, RAD-51 foci persisted in nuclei upon entrance into diplotene (one to two foci per nucleus), a stage at which RAD-51 foci are no longer observed in wild type. This result suggests that SYP-3 is required for the proper progression of recombination. Meanwhile, in syp-3(me42), both the levels of RAD-51 foci throughout prophase and the kinetics of foci formation and disappearance closely paralleled those observed in wild type. Moreover, RAD-51 foci were not present in diplotene nuclei in syp-3(me42). This suggests that DSBs are being repaired with near-wild-type efficiency in syp-3(me42), although repair is not resulting in crossovers.
We also examined the levels of germ cell apoptosis observed in syp-3 mutants (Table 2). Previous studies have shown that late pachytene nuclei undergo apoptosis triggered by unrepaired meiotic DSBs that are sensed as DNA damage (Gartner et al. 2000). Utilizing the levels of germ cell corpses observed in late pachytene as a readout, we observed a 4.5-fold increase in syp-3(ok758) compared to either age-matched wild-type worms or spo-11 worms where the initiation of meiotic DSB formation is abrogated. This increase is similar to that observed in the syp-1 and syp-2 mutants, where levels of RAD-51 foci also remain elevated throughout late pachytene (MacQueen et al. 2002; Colaiácovo et al. 2003). Meanwhile, levels of germ cell apoptosis were not as significantly increased in syp-3(me42) compared with either wild type or spo-11. This suggests that in syp-3(ok758), DSBs are either failing to be repaired or are engaging in aberrant intermediates leading to germ cell apoptosis via the activation of a pachytene DNA damage checkpoint. In contrast, DSB repair is more effective in syp-3(me42), resulting in lower levels of germ cell corpses.
TABLE 2.
Germ cell apoptosis in syp-3 mutants
| Genotype | Mean no. of germ cell corpses ± SE |
|---|---|
| Wild-type | 1.81 ± 0.12 |
| spo-11(ok79) | 1.6 ± 0.22 |
| syp-1(me17) | 8.94 ± 0.67 |
| syp-3(ok758) | 8.25 ± 0.43 |
| syp-3(me42) | 2.54 ± 0.21 |
Germ cell corpses were scored in adult hermaphrodites 20 hr post-L4 as in Kelly et al. (2000). Between 32 and 134 gonad arms were scored for each genotype. Statistical comparisons between genotypes were conducted using the two-tailed Mann-Whitney test. syp-3(ok758) differed significantly from wild type (P = <0.0001) and from spo-11 (P = <0.0001) but did not differ significantly from syp-1(me17) (P = 0.6254). Meanwhile, syp-3(me42) differed significantly from both syp-1(me17) and syp-3(ok758) (P = <0.0001, P = 0.0001); however, it also differed, albeit to a much lesser extent, from either spo-11 (P = 0.0061) or wild-type (P = 0.0062).
To further examine the correlation between DSB repair, chromosome configuration, and structural constraints, we assessed progression of meiotic recombination via immunostaining of RAD-51 in syp-3(me42); syp-2 double mutants (Figures 1 and 2). In contrast to syp-3(me42) single mutants, in syp-3(me42); syp-2 double mutants, chromosomes remain in a clustered organization instead of redispersing prematurely and SYP proteins no longer associate with chromosomes, similar to syp-1, syp-2, and syp-3 null mutants (Figure 2; MacQueen et al. 2002; Colaiácovo et al. 2003; Smolikov et al. 2007, accompanying article). Whereas in syp-3(me42), disappearance of RAD-51 foci occurs with near wild-type kinetics, in syp-3(me42); syp-2, DSB repair is no longer completed by late pachytene (zone 7), with RAD-51 foci persisting through early diplotene (one to three foci per nucleus). Moreover, levels of RAD-51 foci per nucleus are elevated and resemble those observed for either syp-2 or syp-3(ok758). These results suggest that either progression into a dispersed configuration and/or SYP-1 and SYP-2 loading, even if along unsynapsed chromosomes, can promote DSB repair throughout early and mid-prophase.
Evidence that both intersister recombination and NHEJ contribute to meiotic DSB repair when interhomolog recombination is impaired:
The eventual disappearance of RAD-51 foci in syp-3 mutants coupled with the production of viable progeny (∼4%; Smolikov et al. 2007, accompanying article) suggests that DSB repair occurs in syp-3 mutants despite the absence of stable pairing between homologous chromosomes. Thus, we investigated the contribution of sister chromatid-directed repair by examining the consequences of depleting the meiotic cohesin REC-8 in syp-3 mutants.
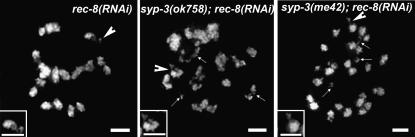
Depletion of REC-8 via RNAi in an otherwise wild-type background leads to the formation of 24 chromatid-sized DAPI-stained bodies and occasional fragments in diakinesis oocytes due to the inability of maintaining the attachments between the 12 pairs of sister chromatids (Figure 3; Pasierbek et al. 2001). REC-8 depletion in either syp-3 mutant did not alter the temporal progression of the nuclear reorganization characteristic of either single syp-3 mutant. However, it resulted in the formation of 24 DAPI-stained bodies in addition to severe chromosome fragmentation. Some chromosome clumping was also observed in syp-3(ok758); rec-8(RNAi). These results suggest that repair is occurring between sister chromatids rather than between homologous chromosomes in the syp-3 single mutants. Moreover, quantitative analysis of RAD-51 foci in syp-3(me42); rec-8(RNAi) reveals significantly lower levels of RAD-51 foci during mid- and late pachytene in this background compared to syp-3(ok758); rec-8(RNAi) (P < 0.0001 for zone 6 by the two-tailed Mann-Whitney test, 95% C.I.; Figure 1 and supplemental Figure 1 at http://www.genetics.org/supplemental/). Specifically, 83–94% of all nuclei in mid- through late pachytene (zones 5–7) carry seven or more RAD-51 foci in syp-3(ok758); rec-8(RNAi). This is similar to the 90–97% observed in rec-8(RNAi) (data not shown) but clearly different from the only 19–60% observed in syp-3(me42); rec-8(RNAi) throughout the same region. This last observation suggests that in addition to sister chromatid-mediated repair, chromosomes might be able to engage in additional modes of repair when they are dispersed as opposed to restrained in a clustered organization during early or mid-prophase.
Figure 3.—
Examining how changes in chromosome structure affect DSB repair. DAPI-stained oocytes at late diakinesis (projections encompass entire nuclei) are depicted. rec-8(RNAi) results in 24 separated sister chromatids with occasional fragments (indicated by arrowhead and enlarged in inset). In syp-3(ok758); rec-8(RNAi) and syp-3(me42); rec-8(RNAi), more frequent chromosome fragmentation is observed, as indicated by arrows (arrowhead indicates example depicted at a higher magnification in the inset). In addition, chromosome aggregation is observed in syp-3(ok758); rec-8(RNAi). Bars, 2 μm.
To examine the contribution of NHEJ to repair in the syp-3(me42) mutants, we observed the levels of RAD-51 foci throughout meiotic prophase in a syp-3(me42); lig-4 double mutant (ligase IV is involved in religating DNA ends during NHEJ; Figures 1 and 2). The levels of RAD-51 foci in syp-3(me42); lig-4 followed a trend similar to that observed in either a lig-4 or wild-type background, indicating that NHEJ does not play a primary role in meiotic DSB repair in C. elegans, consistent with previous studies (Martin et al. 2005; Clejan et al. 2006). However, the contribution of NHEJ to DSB repair becomes more evident when access to sister chromatids as a template for repair is no longer available. Thus, in a syp-3(me42); lig-4; rec-8(RNAi) background, levels of RAD-51 foci increased significantly in late pachytene compared to syp-3(me42), syp-3(me42); lig-4, or syp-3(me42); rec-8(RNAi) mutants (P < 0.0001 for all pairwise combinations in zone 6, two-tailed Mann-Whitney test, 95% C.I.; Figure 1 and supplemental Figure 1 at http://www.genetics.org/supplemental/). Moreover, RAD-51 foci (one to three per nucleus) were still apparent in mid-diakinesis oocytes in syp-3(me42);lig-4; rec-8(RNAi) mutants, while they are no longer present at that stage in any of the other aforementioned backgrounds. Altogether, these results indicate that NHEJ can also contribute to meiotic DSB repair when homologous chromosomes are unavailable.
DISCUSSION
Relationship between chromosome organization and mode of DSB repair:
Although the stable association between homologs is impaired in both syp-3 mutants, these mutants differ in their kinetics of disappearance of RAD-51 foci, which may reflect differences in the timing and/or mode of repair. In the case of syp-3(ok758), and similar to syp-1 and syp-2 null mutants, high levels of both RAD-51 foci and apoptosis are observed in late prophase, correlating with a persistent clustered organization of chromosomes. In contrast, in syp-3(me42), a timely disappearance of RAD-51 foci and near wild-type levels of apoptosis correlates with chromosomes dispersing upon exit from transition zone. Moreover, in syp-3(me42); syp-2 double mutants where chromosomes remain in an extended polarized nuclear organization, elevated levels of RAD-51 are observed in late prophase [once again similar to syp-2 and syp-3(ok758) null mutants], supporting the idea that DSB repair correlates with chromosome dispersal and loading of SC components. Together, these data raise the possibility that chromosome configuration (i.e., clustered vs. dispersed) may affect the capacity for and/or timing of DSB repair. Under this scenario, the clustered organization would inhibit repair, whereas the dispersed organization would permit progression of repair.
Whereas the idea that progression of DSB repair is influenced by the clustered vs. dispersed state of the chromosomes accounts nicely for some aspects of our observations, it does not, however, readily explain all features of progression of DSB repair in the syp-3 mutants. During wild-type meiosis, a barrier to sister chromatid repair is proposed to operate so that repair occurs between nonsister chromatids from homologous chromosomes (Hollingsworth et al. 1995; Schwacha and Kleckner 1997; Wan et al. 2004; Webber et al. 2004; Niu et al. 2005). However, in the syp-3 mutants, given that DSBs are formed but homologs are not stably paired, interhomolog repair is also impaired. In syp-3(ok758) mutants, repair may occur in a delayed fashion when chromosomes finally redisperse in late pachytene and the relationship between sister chromatids is proposed to become less stringent (Colaiácovo et al. 2003). In syp-3(me42) mutants, the timing of disappearance of RAD-51 foci suggests that the barrier preventing sister chromatid recombination may be lifted prematurely. It is possible that in addition to promoting the dispersal of chromosomes, the inappropriate loading of SC central region proteins in the syp-3(me42) mutant also affects the relationship between sister chromatids in a manner that permits intersister recombination during early-mid prophase when interhomolog recombination is favored during wild-type meiosis. In the accompanying article (Smolikov et al. 2007), we demonstrated that some of the SYP-1 protein appears localized between sister chromatids in the syp-3(me42) mutant; this localization may allow it to function in promoting intersister recombination events.
The syp-3(me42) mutant has also enabled us to investigate the contribution of NHEJ to DSB repair in the germline. Although NHEJ is not a primary mode of repair in the germline (Martin et al. 2005; Clejan et al. 2006), NHEJ has recently been observed to participate in meiotic DSB repair in brc-2 mutants where homologous recombination is impaired due to the misregulation of RAD-51 (Martin et al. 2005). In this work, we demonstrate that NHEJ can participate in germline DSB repair when both homolog-mediated and sister chromatid-mediated recombination are impaired due to synapsis defects and defective sister chromatid cohesion. This suggests that NHEJ may be simply outcompeted by interhomolog or intersister-mediated homologous recombination. Alternatively, NHEJ may be under regulatory constraints that are released concomitant with changes in chromosome configuration and structural constraints.
DSB repair does not drive early changes in chromosome configuration during C.elegans meiosis:
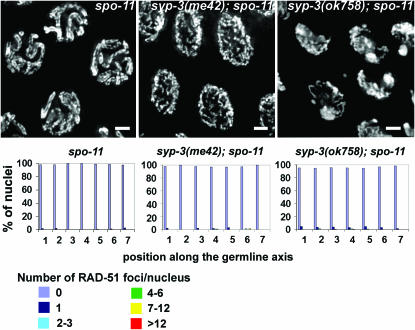
Our studies are consistent with a model by which changes in chromosome configuration affect DSB repair progression. However, we also addressed whether the reverse also applied, which is whether DSB repair drives early changes in chromosome configuration during meiosis. While this is the case for budding yeast and Sordaria (Trelles-Sticken et al. 1999; Storlazzi et al. 2003), it does not seem to be the case in C. elegans (MacQueen et al. 2002; Colaiácovo et al. 2003; this article). First, the formation of DSBs per se is not required for chromosomes to acquire a polarized nuclear organization upon entrance into meiosis, for SC assembly between homologs or for redispersal of chromosomes upon progression into pachytene, as determined through the analysis of spo-11 mutants (Dernburg et al. 1998). Second, in syp-1, syp-2, and syp-3(ok758) null mutants, chromosomes persist in a polarized nuclear organization that is not altered when meiotic DSB formation is abrogated in the absence of SPO-11 (Figure 4; MacQueen et al. 2002; Colaiácovo et al. 2003). Third, in the case of syp-3(me42), the premature chromosome redispersal characteristic of this mutant is also not altered in a syp-3(me42); spo-11 mutant (Figure 4). Fourth, while the levels of RAD-51 are increased in syp-3(me42); rec-8(RNAi) double mutants compared to syp-3(me42), the altered progression of DSB repair does not lead to a change in chromosome organization within the nuclei and chromosomes still redisperse prematurely as in syp-3(me42). Therefore, initiation and/or progression of DSB repair are not directly driving early changes in chromosome configuration during meiosis. This is, however, distinct from a later wave of chromosome organization that is observed to occur between the early and late pachytene substages and that has been proposed to be under recombination-dependent regulation in C. elegans (Carlton et al. 2006).
Figure 4.—
Uncoupling the exit from a clustered organization from meiotic DSB formation. DAPI-stained mid-pachytene nuclei (projections are halfway through 3D data stacks). syp-3(me42); spo-11 and syp-3(ok758); spo-11 are indistinguishable from the corresponding syp-3 single mutants. Bars, 2 μm. Histograms depict the quantitation of RAD-51 foci in germlines of the indicated genotypes (as described in Figure 1). The average number of nuclei scored per zone (n) for spo-11, syp-3(me42); spo-11, syp-3(ok758); spo-11 were: zone 1 (n = 138), zone 2 (n = 158), zone 3 (n = 145), zone 4 (n = 138), zone 5 (n = 127), zone 6 (n = 102), and zone 7 (n = 90).
In conclusion, SYP-3 is a novel SC component that plays a central role in the regulated formation of the SC central region between homologous chromosomes. Comparisons between different syp-3 mutants are consistent with a model in which changes in chromosome configuration may modulate the mode and timing of DSB repair employed during prophase progression. Therefore, our findings support the view that a complex system of checks and balances acts to coordinate these processes to achieve a successful outcome of meiosis.
Acknowledgments
Some strains were kindly provided by the Caenorhabditis Genetics Center, the Caenorhabditis Gene Knockout Consortium and Shawn Ahmed. We thank Fred Winston, JoAnne Engebrecht, and Kristina Schild-Prufert for critical reading of this manuscript and David Reich for helpful suggestions regarding the statistical treatment of the RAD-51 data. This work was supported by National Institutes of Health grants R01GM072551 to M.P.C. and R01GM53804 to A.M.V.
References
- Bergerat, A., B. de Massy, D. Gadelle, P. C. Varoutas, A. Nicolas et al., 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton, P. M., A. P. Farruggio and A. F. Dernburg, 2006. A link between meiotic prophase progression and crossover control. PLoS Genet. 2: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clejan, I., J. Boerckel and S. Ahmed, 2006. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics 173: 1301–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo, M. P., A. J. MacQueen, E. Martinez-Perez, K. McDonald, A. Adamo et al., 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474. [DOI] [PubMed] [Google Scholar]
- Dernburg, A. F., K. McDonald, G. Moulder, R. Barstead, M. Dresser et al., 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Gartner, A., S. Milstein, S. Ahmed, J. Hodgkin and M. O. Hengartner, 2000. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443. [DOI] [PubMed] [Google Scholar]
- Hollingsworth, N. M., L. Ponte and C. Halsey, 1995. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9: 1728–1739. [DOI] [PubMed] [Google Scholar]
- Keeney, S., 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53. [DOI] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kelly, K. O., A. F. Dernburg, G. M. Stanfield and A. M. Villeneuve, 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen, A. J., M. P. Colaiácovo, K. McDonald and A. M. Villeneuve, 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16: 2428–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, J. S., N. Winkelmann, M. I. Petalcorin, M. J. McIlwraith and S. J. Boulton, 2005. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol. Cell. Biol. 25: 3127–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, H., L. Wan, B. Baumgartner, D. Schaefer, J. Loidl et al., 2005. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell 16: 5804–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2004. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell. Dev. Biol. 20: 525–558. [DOI] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek, P., M. Jantsch, M. Melcher, A. Schleiffer, D. Schweizer et al., 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15: 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, D. L., T. Blumenthal, B. J. Meyer and J. R. Priess, 1997. C elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Rogers, E., J. D. Bishop, J. A. Waddle, J. M. Schumacher and R. Lin, 2002. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 157: 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha, A., and N. Kleckner, 1997. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135. [DOI] [PubMed] [Google Scholar]
- Smolikov, S., A. Eizinger, K. Schild-Prufert, A. Hurlburt, K. McDonald et al., 2007. SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics 176: 2015–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi, A., S. Tesse, S. Gargano, F. James, N. Kleckner et al., 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 17: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, P., 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265: 1241–1243. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken, E., J. Loidl and H. Scherthan, 1999. Bouquet formation in budding yeast: initiation of recombination is not required for meiotic telomere clustering. J. Cell Sci. 112(Pt 5): 651–658. [DOI] [PubMed] [Google Scholar]
- Wan, L., T. de los Santos, C. Zhang, K. Shokat and N. M. Hollingsworth, 2004. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol. Biol. Cell 15: 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber, H. A., L. Howard and S. E. Bickel, 2004. The cohesion protein ORD is required for homologue bias during meiotic recombination. J. Cell Biol. 164: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler, D., and N. Kleckner, 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33: 603–754. [DOI] [PubMed] [Google Scholar]