Abstract
Most phenotypic differences between human and chimpanzee are likely to result from differences in gene regulation, rather than changes to protein-coding regions. To date, however, only a handful of human–chimpanzee nucleotide differences leading to changes in gene regulation have been identified. To hone in on differences in regulatory elements between human and chimpanzee, we focused on 10 genes that were previously found to be differentially expressed between the two species. We then designed reporter gene assays for the putative human and chimpanzee promoters of the 10 genes. Of seven promoters that we found to be active in human liver cell lines, human and chimpanzee promoters had significantly different activity in four cases, three of which recapitulated the gene expression difference seen in the microarray experiment. For these three genes, we were therefore able to demonstrate that a change in cis influences expression differences between humans and chimpanzees. Moreover, using site-directed mutagenesis on one construct, the promoter for the DDA3 gene, we were able to identify three nucleotides that together lead to a cis regulatory difference between the species. High-throughput application of this approach can provide a map of regulatory element differences between humans and our close evolutionary relatives.
IN addition to substitutions at the protein level, changes in gene regulation are likely to underlie many phenotypes of interest, including adaptations and human diseases (Britten and Davidson 1971; King and Wilson 1975; Jin et al. 2001; Carroll 2003; Abzhanov et al. 2004; Iftikhar et al. 2004; Shapiro et al. 2004; Taron et al. 2004). Regulation of gene expression may be achieved by a large number of transcriptional and translational mechanisms (reviewed in Wray et al. 2003). At the transcription level, regulatory mechanisms include transcriptional initiation, chromatin condensation, DNA methylation, alternative splicing of RNA, and mRNA stability (Wray et al. 2003). For most genes, however, transcriptional initiation appears to be the principal determinant of the overall mRNA gene expression profile (Lemon and Tjian 2000; White 2001; Wray et al. 2003).
Transcriptional initiation is regulated by a combination of trans elements binding to cis regulatory sequences. The relative contribution of changes in cis and trans regulatory elements to the evolution of gene regulation remains largely unknown. However, accumulating evidence suggests that changes in cis may underlie many of the mRNA expression differences observed between individuals, strains, or species (Dickinson 1988). For example, Cowles et al. (2002) observed that of 69 genes that are differentially expressed in four different mice strains, at least 4 (6%) show large allelic difference in expression level (>1.5-fold) in F1 hybrids, indicative of differences in the cis regulatory regions. In yeast, Yvert et al. (2003) found that a minimum of 25% of expression differences between strains are due to changes in cis regulatory regions. Wittkopp et al. (2004) demonstrated that 28/29 studied differences in gene expression between Drosophila melanogaster and D. simulans can be at least partially explained by differences in cis regulatory regions. In humans, when Morley et al. (2004) mapped gene expression phenotypes, they found that 19% of significant associations mapped in cis. Thus, although the fraction of variation in gene expression levels explained by variation in cis remains unknown, the proportion is likely to be substantial (more examples can be found in a review by Gibson and Weir 2005).
From a theoretical perspective, changes in cis regulatory elements are thought to underlie a large number of adaptive phenotypes because mutations in these elements may be more likely to produce circumscribed expression pattern changes without deleterious pleiotropic effects (Stern 2000; Carroll et al. 2004; Gompel et al. 2005). Consistent with this view, cis-regulatory mutations, through their effect on gene expression levels, were found to underlie important phenotypes in a range of organisms, including beak morphology in Darwin's finches (Abzhanov et al. 2004), bristle patterns and wing pigmentation in fruit flies (Stern 1998; Gompel et al. 2005), branching structure in maize (Clark et al. 2006), skeletal patterning and pelvic reduction in sticklebacks (Cresko et al. 2004; Shapiro et al. 2004), and parental care in rodents (Hammock and Young 2005). In humans, mutations in putative cis regulatory regions have been associated with well over 100 phenotypes including diverse aspects of behavior, physiology, and disease (reviewed in Kleinjan and van Heyningen 2005 and Wray 2007).
In primates, interspecies gene expression studies suggest that extensive regulatory changes have occurred, with 10–20% of genes (depending on the tissue) found to be significantly differentially expressed between humans and chimpanzees (Khaitovich et al. 2005; Gilad et al. 2006). A subset of these genes exhibits patterns of interspecies expression consistent with the action of positive (directional) selection on gene regulation in humans (Gilad et al. 2006), suggesting that changes in expression in these genes are functionally important. However, while many human-specific adaptations in gene copy number and protein sequence have been documented, there are only a few known examples of differences in cis regulation between humans and other apes (Huby et al. 2001; Rockman et al. 2003, 2005). The lack of examples of nucleotide substitutions between human and chimpanzee in functional cis regulatory elements is unlikely to reflect their lack of importance to human adaptations or disease. Instead, it probably stems from the difficulty of identifying specific regulatory elements that may underlie the interspecies expression differences (Wray et al. 2003; Wray 2007). In particular, cis regulatory elements can be located up to hundreds of kilobases away from genes (i.e., long-range cis regulatory elements (Pastinen et al. 2006; Prabhakar et al. 2006), complicating their identification.
Promoter regions, which are located just upstream from transcription start sites (TSS) of genes, may be the simplest cis regulatory elements to identify (Trinklein et al. 2003; Cooper et al. 2006; Pastinen et al. 2006). That said, predicting the effect of sequence variation in promoter regions on gene regulation is not straightforward (Wray et al. 2003; Wray 2007). While few nucleotide changes in promoter sequences can have a substantial effect on gene regulation (e.g., Storgaard et al. 1993; Haudek 1998), many sites in promoter regions can change without a discernable effect on the gene expression profile (e.g., Takahashi et al. 1999; Wolff et al. 1999). In humans, only 10–20% of polymorphic sites within promoters are estimated to have an effect on gene regulation (Buckland et al. 2004a,b). One approach to confirm putative cis regulatory variation is to test the ability of different variants to enhance transcription using reporter gene assays (Trinklein et al. 2003; Wray 2007). By this approach, 70–90% of putative human promoters, predicted on the basis of the TSS, can be empirically confirmed in human cell lines (Trinklein et al. 2003; Cooper et al. 2006).
Here, we used reporter gene assays to test for differences in transcriptional activity between human and chimpanzee promoters for 10 genes. We chose this set of genes because, in a previous study (Gilad et al. 2006), their expression levels in livers were similar among individuals from three nonhuman primate species, but were consistently elevated or reduced in humans—a pattern consistent with stabilizing selection on expression in nonhuman apes and with directional selection in the human lineage. We hypothesized that interspecies differences in promoter activity of these genes may underlie the observed gene expression patterns and could point to cis regulatory changes that were under selection in humans.
MATERIALS AND METHODS
Quantitative RT–PCR:
Of the 19 genes whose regulation has been previously inferred to evolve under directional selection in humans (Gilad et al. 2006), we chose to study the promoter activity of 13 genes, selected randomly among them. (We originally chose 10 genes, but we failed to amplify a PCR product for the predicted promoters of three of those genes, which were then replaced; see supplemental Table 1 at http://www.genetics.org/supplemental/.) Our general approach to study differences in promoter activity is similar to that of Heissig et al. (2005), who studied differences in activity between 12 human and chimpanzee promoters. However, Heissig et al. (2005) chose their genes on the basis of interspecies gene expression data from a single-species microarray, which can lead to a high error rate (Gilad et al. 2005), and did not confirm their original observations using an alternative approach, making it difficult to interpret their results. Instead, we started our study by using TaqMAN (Applied Biosystems, Foster City, CA) quantitative RT–PCR to validate the microarray results for the 10 genes in which we successfully obtained amplifications of their putative promoters (see below). More specifically, we designed PCR primers and TaqMAN probes for gene regions that are identical between human and chimpanzee (a list of PCR primers and TaqMAN probes is available in supplemental Table 1). As templates, we used total RNA from livers of three humans and three chimpanzees, which are different from the individuals that were originally used in the microarray study (Gilad et al. 2006). We synthesized first-strand cDNA using 5 μg of each RNA sample and pooled together the three cDNA samples from each species. Quantitative RT–PCR was performed in a 25-μl reaction containing 2× JumpStart Taq ReadyMix (Sigma, St. Louis), 0.2 pmol each primer, 100 pmol dual-labeled probe (BHQ-1 and FAM) (Sigma–Genosys), and 1 μl cDNA template. PCR was performed in a 7900HT Fast Real-Time PCR system (Applied Biosystems), in three technical replicates for each sample of pooled cDNA. The detection threshold cycle for every reaction was determined using a standard curve, after normalization of the results using quantitative RT–PCR with primers for the POLR2C gene, which was shown to have constant expression levels in livers of humans and chimpanzees (Gilad et al. 2006). The significance of differences in transcript levels between species was assessed by a (one-tailed) t-test.
Reporter gene assays:
For each of the 13 genes, we used the database of transcription start sites (DBTSS; http://dbtss.hgc.jp/index.html) to identify the TSS on the basis of their longest known transcript. Using the database information, we designed PCR primers to amplify a product from ∼100 bp downstream of the putative TSS to ∼900 bp upstream of it, from both human and chimpanzee genomic DNA (the list of all primers and PCR conditions is available in supplemental Table 1 at http://www.genetics.org/supplemental/). We ligated the PCR products into the Luciferase reporter gene vector pGL4.14 (Promega, Madison, WI) and cloned them in JM109 competent cells. We used touchdown PCR to amplify and then sequence (using an ABI3730 automated sequencer) the insert from individual colonies to confirm that no Taq-generated errors were incorporated. We did so by comparing the sequence of the individual inserts to the available human and chimpanzee genomic sequence (found at http://genome.ucsc.edu/).
Once the sequence of the insert from individual colonies was confirmed, we proceeded by extracting the plasmid and using it in transfections of human liver HEP cells by using Lipofectamine 2000 (Invitrogen, San Diego) with 200 ng of each plasmid. The HEP cells were also transfected with 20 ng of the Renilla vector pGL4.73 (Promega). The cotransfection allows us to normalize across experiments for transfection efficiency. Luciferase and Renilla activity were measured 24 hr after transfection, using the Dual-glo Luciferase kit (Promega) in a Veritas 96-well plate luminometer (Turner Biosystems).
Reporter gene study design and analysis:
The Luciferase activity of each construct was measured using 5 replicates (independent transfections) or 15 replicates for the DDA3 constructs (see below). In addition, we measured Luciferase activity for an empty (i.e., with no promoter) pGL4.14 vector, in 5 replicates, to estimate background Luciferase transcription levels. For each replicate, we normalized Luciferase by Renilla luminescence values to control for transfection efficiency. We then standardized the normalized luminescence values by the background activity (of the empty vector). Constructs were identified as enhancing transcriptional activity when the average luminescence across the 5 replicates was at least twice as high as that of the empty vector. When both the human and chimpanzee putative promoters successfully enhanced transcription, we used a one-tailed t-test to test for differences in promoter activity between the species (the test is one-tailed because we have an a priori expectation from the microarray and quantitative RT–PCR results). With respect to the use of a t-test, we note that the data do not depart significantly from a normal distribution (using the Shapiro–Wilk test for normality; see supplemental Figure 1 at http://www.genetics.org/supplemental/ for examples of quantile–quantile plots). Unfortunately, since chimpanzee liver cell lines are not available, we could not perform the reciprocal experiment.
DDA3 constructs and analysis:
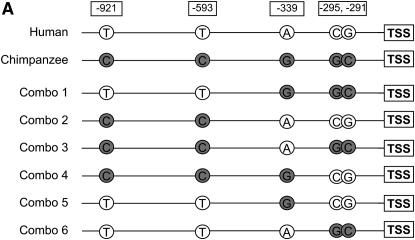
The human and chimpanzee DDA3 promoter constructs that we used differ by five nucleotides at positions −291, −295, −339, −593, and −921 (the “−” sign indicates that these sites are upstream of the TSS, which is designated position 0). To identify the nucleotides that underlie the difference in activity between the human and chimpanzee promoters, we built six constructs with different nucleotide compositions (see results). To do so, we initially used digestion with the ApoI restriction enzyme (New England Biolabs, Beverly, MA), followed by ligations of reciprocal ends of the human and chimpanzee promoters. This step resulted in two “combo” constructs, each containing approximately half the human and half the chimpanzee promoters. Next, we used the Quikchange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) to introduce individual nucleotide changes to each of the existing constructs. Reporter gene assays with all DDA3 plasmids were performed in 15 replicates to increase the power to detect subtle but consistent differences between constructs that differ by only one nucleotide substitution.
To fit a linear model to the measurements of the expression level in each experiment we used the R software environment for statistical computing (http://www.r-project.org). In contrast to the analysis of results from different pairs of constructs at a time (using a t-test; see above), when we considered the combined data from all the DDA3 combinations, we found that the residuals were not normally distributed (Shapiro–Wilk test for normality; P = 0.003). Thus, we transformed the data using the Box–Cox transformation 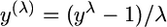 , where
, where 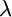 was estimated using the R function box.cox.powers (in the package “car”). The lm function was then used to fit the linear model
was estimated using the R function box.cox.powers (in the package “car”). The lm function was then used to fit the linear model
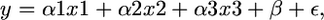 |
where y consists of the expression values from 60 measurements (15 replicates of the original human promoter, 15 replicates of constructs with nucleotide substitutions at positions −219 and −295, 15 replicates of constructs with nucleotide substitution at position −339, and 15 replicates of constructs with all three nucleotide substitutions), and x1, x2, and x3 are categorical variables that represent the effect of substitutions at positions −219 and −295 only, the effect of substitution at position −339 only, or an interaction effect of all three substitutions, respectively.
RESULTS
Using gene expression estimates from a multispecies cDNA microarray (Gilad et al. 2005), we previously identified 19 genes whose regulation in liver has likely evolved under positive selection in the human lineage (Gilad et al. 2006). Specifically, the expression levels of these genes are similar in the livers of chimpanzees, orangutans, and rhesus macaques, but are significantly elevated or reduced in human livers. Our aim here was to ask whether nucleotide differences in the putative promoters of these genes might contribute to the observed difference in expression levels between humans and other primates.
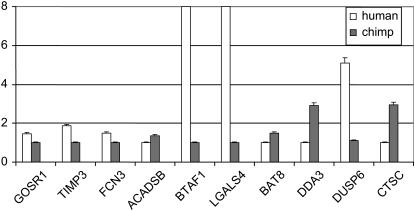
To address this question, we focused on 10 genes that show either elevated (DUSP6, BAT8, LGALS4, BTAF1, TIMP3, FCN3, and GOSR1) or reduced (CTSC, DDA3, and ACADSB) expression levels in human livers relative to the nonhuman primates on the basis of the microarray results. As a first step, we validated the microarray observations of interspecies differences in gene expression by performing quantitative RT–PCR using RNA from human and chimpanzee livers as a template (see materials and methods). Importantly, we did not use RNA from the same individuals that were used in the microarray study, so that we are corroborating the inference of interspecies expression patterns, as well as the reliability of the microarray observations. For 9 of the 10 genes (all but the gene BAT8), we confirmed a significant difference in expression levels between the species, in the same direction as seen on the array (Figure 1). Confirming the microarray results for 9 of 10 genes is consistent with the original false discovery rate (FDR) (Benjamini and Hochberg 1995) that was used to identify differentially expressed genes from the microarray data [i.e., an FDR of 0.05 (Gilad et al. 2006)]. Another possibility is that the expression levels of BAT8 in the liver are polymorphic in humans or chimpanzees.
Figure 1.—
Quantitative RT–PCR results. Mean fold differences (y-axis) and standard errors for three replicates are given for either the human (open bars) or the chimpanzee (shaded bars) RNA templates. For each gene (x-axis), results were standardized on the basis of the species with the lower expression level (set to one). All gene expression differences between human and chimpanzee are significant at P < 0.05.
Reporter gene assays:
Next, we tested whether the interspecies difference in gene expression could be explained, at least in part, by sequence differences between human and chimpanzee in the region immediately upstream of the TSS, i.e., in the putative promoter. To do so, we used the DBTSS (http://dbtss.hgc.jp/index.html) to identify the TSS of each of the 10 genes, on the basis of their longest known transcript. On the basis of that information, we designed PCR primers to amplify a product from ∼100 bp downstream of the putative TSS to ∼900 bp upstream of it, from both human and chimpanzee genomic DNA. These ∼1-kb segments likely contain the proximal promoter as well as some of the cis regulatory elements for the 10 genes (Trinklein et al. 2003; Cooper et al. 2006; Wray 2007). We ligated these products into a Luciferase reporter gene vector and confirmed the sequence of the insert by direct sequencing (see materials and methods). The sequence divergence between human and chimpanzee in these 10 putative promoters was found to be 0.55–1.3%, consistent with genomewide estimates (The Chimpanzee Sequencing and Analysis Consortium 2005).
We used the Luciferase constructs to transfect human liver cell lines (Hep) with the empty (control) vector, the vector containing the human promoter, or the one with the chimpanzee promoter. We performed five independent replicates of each transfection. As a measure of transfection efficiency, the Luciferase plasmids were cotransfected along with a vector containing a Renilla gene downstream of an SV40 constitutive promoter. We then measured Luciferase activity in each sample, normalized by Renilla activity, and tested the ability of the different constructs to increase Luciferase activity (by at least twofold) beyond the background activity of the control empty vector (Table 1). If both the chimpanzee and the human promoters were found to be active, we examined whether the relative activity of the promoters from both species is in the same direction as seen in the microarray experiments (Gilad et al. 2006).
TABLE 1.
Reporter gene assays of 10 human and chimpanzee promoters
| Gene | Human averagea | Human SD | Chimpanzee averagea | Chimpanzee SD |
|---|---|---|---|---|
| BTAF1 | 265.8 | 42.1 | 331.1 | 50.7 |
| CTSC* | 151.9 | 10.1 | 177.4 | 18.7 |
| GOSR1* | 122.5 | 8 | 164.4 | 12.5 |
| DUSP6* | 83.2 | 2.3 | 75.4 | 6.4 |
| DDA3* | 38.5 | 2.87 | 45.9 | 5.4 |
| BAT8 | 3.5 | 0.18 | 3.5 | 0.2 |
| LGALS4 | 3.3 | 0.5 | 3.1 | 0.6 |
| ACADSB | 2.1 | 0.25 | 1.3 | 0.38 |
| FCN3 | 1.9 | 0.3 | 1.4 | 0.38 |
| TIMP3 | 0.9 | 0.1 | 0.9 | 0.1 |
The difference in activity between the human and the chimpanzee promoters is significant at P < 0.05 (see Figure 2).
Average luminescence for each construct over 5 replicates (15 in the case of DDA3) was standardized by the average luminescence of the control (empty Luciferase vector).
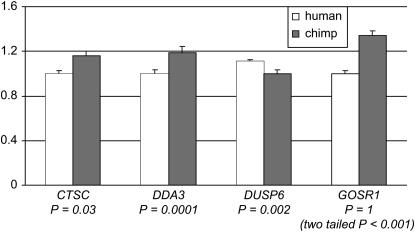
As can be seen in Table 1, promoters for three of the genes (TIMP3, FCN3, and ACADSB) failed to increase Luciferase activity twofold above that of the control (i.e., the empty vector). We therefore excluded these genes from subsequent analysis. Of the seven promoters that increased Luciferase activity beyond twofold of the control levels, three (the promoters for BAT8, LGALS4, and BTAF1) did not show a significant difference in activity between the human and the chimpanzee promoters (Table 1). Finally, for four genes, we observed a significant difference (P < 0.05) in activity between the human and the chimpanzee promoters (Table 1 and Figure 2). In one case (GOSR1), the difference in activity between the two promoters was opposite to our previous observation based on the microarray results, which we had confirmed by quantitative RT–PCR (see above). Although the GOSR1 gene was found to be highly expressed in humans compared to chimpanzees, the human promoter has lower activity in the HEP human liver cell line compared to the chimpanzee promoter.
Figure 2.—
Differences in promoter activity between human and chimpanzee. Mean fold differences (y-axis) and standard errors for 5 replicates (or 15 in the case of DDA3) are given for either the human (open bars) or the chimpanzee (shaded bars) promoters. For each gene (x-axis), results were standardized on the basis of the species with the lower promoter activity level (set to one). P-values are given below the gene names for a one-tailed t-test of the difference in activity between the human and the chimpanzee promoters (see materials and methods).
In the three remaining cases (the promoters for CTSC, DDA3, and DUSP6), we found a significant difference in activity between the human and chimpanzee promoters that recapitulated the previously observed interspecies difference in gene expression. Thus, it is likely that changes in cis regulatory elements contribute to the observed difference in gene expression levels between human and chimpanzee in 3 of the 10 genes that we examined.
Identifying a specific cis regulatory change:
Next, we wanted to identify specific nucleotide substitution that underlie differences in activity between the human and chimpanzee promoters. To do so, we focused on the promoter for the DDA3 gene, for which the human and chimpanzee sequence differs by only five nucleotide changes (Figure 3A). As a first step, we constructed (see materials and methods) two combinations of approximately half of the human and half of the chimpanzee promoters (combo 1 and combo 2; see Figure 3A). We then tested the ability of the combo constructs to enhance Luciferase activity using the same approach that was described above.
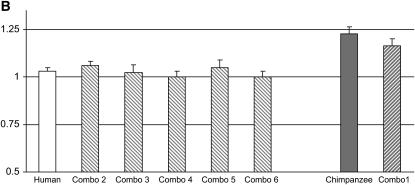
Figure 3.—
Reporter gene assays with DDA3 constructs. (A) The promoter constructs of the human, the chimpanzee, and the different combinations are shown. (B) Mean fold differences (y-axis) and standard errors for 15 replicates are given for the human (open bar), the chimpanzee (shaded bar), or the different combo promoters. The results were standardized on the basis of the combination with the lower promoter activity level (i.e., combo 4 was set to one). All pairwise comparisons between the chimpanzee or combination 1 mean activity levels on the one hand and the human or combinations 2–6 mean activity levels on the other are significant at P < 0.01. The raw data for all 15 replicates of all the DDA3 constructs are available in supplemental Table 2 at http://www.genetics.org/supplemental/.
The results of this experiment led us to exclude two of the five nucleotide differences (at positions −593 and −921 upstream of the TSS), as they did not contribute significantly to the difference in activity between the human and chimpanzee promoters (P = 0.33; Figure 3B and supplemental Table 2 at http://www.genetics.org/supplemental/). Two of the remaining three nucleotide substitutions between human and chimpanzee are only 4 bp apart (at positions −291 and −295 upstream of the TSS). To test whether these two substitutions underlie the difference in expression between the human and chimpanzee DDA3 gene in the liver, we proceeded to substitute these two nucleotides, by site-directed mutagenesis, on the human, the chimpanzee, or the combination backgrounds, resulting in four additional constructs (Figure 3A). We then tested the ability of each of the four constructs to enhance Luciferase activity. We did not find significant differences in activity associated with any of the individual constructs (minimum P = 0.31; Figure 3B and supplemental Table 2), suggesting a nonadditive effect of the different substitutions. This result was confirmed when we applied a linear model to estimate the individual effects of the substitution at position −339 and the effect of the two substitutions at positions −291 and −295, as none of these effects were found to be significant (minimum P < 0.12). In contrast, we found a significant interaction effect, suggesting that the effects of substitutions at different sites are not simply additive (P = 0.037; see materials and methods for details). Thus, on the basis of these results, it appears that nucleotide differences in three sites (at positions −291, −295, and −339 upstream of the TSS) are needed to attain the higher level of chimpanzee promoter activity in DDA3.
DISCUSSION
To identify specific nucleotide differences between human and chimpanzee that led to changes in cis regulatory elements, we focused on 10 genes that were previously identified as differentially expressed between humans and chimpanzees (Gilad et al. 2006). Using reporter gene assays in a human liver cell line, we confirmed the ability of 7 of the 10 promoters to enhance transcription beyond background levels.
The proportion of predicted promoters that failed to enhance Luciferase activity in this experiment (30%) is consistent with previous observations using reporter gene assays in human cell lines (Trinklein et al. 2003; Cooper et al. 2006) and might be explained by poor prediction of the TSS or the existence of alternative promoters in different tissues. An additional explanation is that some of the transcription factors that are expressed in human livers are not expressed in the particular human liver cell line with which we worked (HEP). If so, the three promoters may not be active in this cell line because of missing trans regulatory elements. One would then predict that the genes downstream of the endogenous promoters may not be expressed in the cell line either. We tested this hypothesis by attempting to amplify a cDNA product for each of the 10 genes using RNA extracted from the cell line as template (we used the quantitative RT–PCR primers for the 10 genes; see supplemental Table 1 at http://www.genetics.org/supplemental/). We failed to amplify a product only for 2 of the 10 genes: FCN3 and ACADSB. These genes are two of the three cases for which no promoter activity was observed. Hence, it is likely that in these two cases, lack of promoter activity can be explained by cell-line-specific artifacts.
Although none of the human–chimpanzee promoter pairs are identical at the sequence level (the range of sequence divergence is 0.55–1.3%), we did not find significant differences in transcriptional activity between the human and the chimpanzee promoter constructs for three of the seven active promoter pairs. An obvious explanation for this observation is that the three genes are differentially expressed between the species due to changes outside of the promoter regions examined in this study (e.g., in cis elements that are further upstream or downstream from the TSS) or to changes in trans regulatory elements that bind to the promoters of these three genes.
Differences in promoter activity:
In one case (the promoter for the gene GOSR1), the reporter gene assays were not consistent with the observation from the microarray. While the GOSR1 gene shows elevated expression in humans compared to chimpanzees, there is significantly higher transcriptional activity for the chimpanzee promoter compared to the human promoter. This discrepancy is unlikely to reflect a spurious result of the microarray analysis, as we confirmed the array observations by using quantitative RT–PCR (see results). Instead, it might be explained by compensatory changes in regulatory elements in chimpanzee (either in cis or in trans), which are missing from the human cell line or are not located within the 1-kb chimpanzee segment that was used in the construct. Compensatory changes in transcription-factor binding sites have been observed previously in fruit flies (Ludwig et al. 1998, 2005) and have been inferred from a comparison of human and mouse regulatory sequences (Dermitzakis and Clark 2002). In theory, this hypothesis could be tested by a reciprocal experiment in which the activity of both human and chimpanzee promoters is tested in chimpanzee liver cell lines. Unfortunately, there are no chimpanzee liver cell lines available, so this approach is not feasible at present.
In contrast to the GOSR1 gene, a comparison of the transcriptional activity of promoters for three other genes (CTSC, DDA3, and DUSP6) yielded results that are in agreement with the observations from the microarray. It is difficult to compare fold changes across the different techniques (microarrays, quantitative RT–PCR, and reporter gene assays). This caveat notwithstanding, our results strongly suggest that the expression of the genes CTSC, DDA3, and DUSP6 differs between human and chimpanzee due, at least in part, to changes in cis regulatory elements that reside within a segment 100 bp downstream of the TSS to 900 bp upstream of the TSS.
Identifying cis regulatory changes:
Ultimately, our goal was to identify particular nucleotide substitutions between human and chimpanzee that contribute to differences in gene regulation between the species. We chose to focus on the DDA3 gene as a test case because it has the fewest nucleotide substitutions between human and chimpanzee among the three genes for which our experiments suggested an interspecies difference in cis regulation. Using site-directed mutagenesis, we were able to identify three nucleotides that underlie the difference in transcriptional activity between the human and the chimpanzee promoters of the DDA3 gene. Interestingly, we observed a significant difference in promoter activity only when all three nucleotides were substituted. It remains possible that single-nucleotide substitutions have subtle effects on DDA3 promoter activity, which our reporter gene assays (even using 15 replicates) were underpowered to detect.
The expression level of the DDA3 gene was originally inferred to be under directional selection in the human lineage (Gilad et al. 2006). This conclusion was based on the observation that DDA3 expression levels were found to be relatively constant in nonhuman apes, yet consistently reduced in humans. On the basis of this result, we expected the three nucleotides that underlie the cis regulatory difference between human and chimpanzee DDA3 promoters to be derived in the human lineage. However, when we aligned the human and chimpanzee DDA3 promoters with the corresponding rhesus macaque sequence, the three nucleotides substitutions were inferred to have occurred on the chimpanzee lineage. Hence, although we were able to identify individual nucleotides that underlie a cis regulatory difference between human and chimpanzee, our observations do not point to the genetic basis for adaptive changes in DDA3 expression level in humans. Instead, our observations suggest that a regulatory change (outside of the 1-kb segment used in the reporter construct or in trans) occurred in the ancestor of humans and chimpanzees, which reduced levels of expression of DDA3. This regulatory change was then compensated in the chimpanzee lineage by the three nucleotide substitutions, but not in the human lineage, where the reduced levels of DDA3 expression may have been advantageous (Gilad et al. 2006).
Possible DDA3 regulatory mechanism:
DDA3 has been shown to be a downstream target of P53 and to be involved in activation of the β-catenin pathway (Hsieh et al. 2007), which has a role in regulating a large number of cellular processes including cell growth and circadian rhythm (Meijer et al. 2004). We hypothesized that the three nucleotide substitutions in the DDA3 promoter region are part of at least two transcription-factor binding sites, with one binding site that includes the locus at position −339 and another that includes the loci at positions −291 and −295. If so, and assuming the relevant transcription factor(s) did not change between human and chimpanzee (i.e., the protein is identical) and therefore have similar binding properties, we would expect the nucleotide differences between the species to change the affinity with which the transcription factor(s) binds to the DDA3 promoter.
To find candidate transcription factors that are consistent with this hypothesis, we used the TRANSFAC database (BioBase biological databases) to identify all known transcription-factor binding sites that overlap these two locations (using a matrix P-value cutoff for the match of the predicted binding site of 0.8 for the core and 0.7 for the extended consensus element). We note that it is unclear how to assign significance to the identification of transcription-factor binding sites on the basis of a single sequence (Vavouri and Elgar 2005). In particular, since transcription-factor binding sites are short (6–12 mers), multiple false positives are expected at nearly every locus. That said, we were able to identify only one transcription factor (albumin D-box binding protein, DBP) that met our criteria. The DBP transcription factor can bind to both locations in the human DDA3 promoter. In chimpanzee, DBP is expected to bind much less efficiently to both locations: The two nucleotide substitutions at positions −291 and −295 change the binding site for DBP such that it is no longer recognized using our statistical cutoff. Similarly, the substitution at position −339 is expected to disrupt the chimpanzee binding site for DBP (see supplemental Table 3 at http://www.genetics.org/supplemental/). In addition, there are no differences at the protein level between the human and the chimpanzee DBP orthologous genes. Thus, it seems reasonable to assume that there are no interspecies differences in DBP DNA-binding properties. Finally, DBP has been demonstrated to regulate circadian gene expression in the liver and kidney (Wuarin et al. 1992; Ripperger et al. 2000), consistent with our limited knowledge regarding the role of DDA3 (Hsieh et al. 2007). In summary, these observations, although clearly speculative, suggest that DBP is a repressor of the DDA3 gene and that both its binding sites were weakened or disrupted in chimpanzee.
Outlook:
Our work demonstrates that, by using interspecies gene expression profiles followed by reporter gene assays, it is possible to hone in on specific cis regulatory differences between human and chimpanzee. High-throughout application of this approach will allow us to identify cis regulatory elements that are functionally important in humans, and, in particular, which have evolved under positive selection in the human lineage. In that respect, an important resource would be the development of chimpanzee cell lines from a large number of tissues. Such a resource will facilitate the elucidation of the relative contribution of cis and trans regulatory changes in humans and chimpanzees.
Acknowledgments
We thank Molly Przeworski, Paola De Candia, Alicia Oshlack, and Matthew Stephens for comments on the manuscript and helpful discussions. This research was supported by National Institutes of Health grant GM077959.
References
- Abzhanov, A., M. Protas, B. R. Grant, P. R. Grant and C. J. Tabin, 2004. Bmp4 and morphological variation of beaks in Darwin's finches. Science 305: 1462–1465. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Britten, R. J., and E. H. Davidson, 1971. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q. Rev. Biol. 46: 111–138. [DOI] [PubMed] [Google Scholar]
- Buckland, P. R., S. L. Coleman, B. Hoogendoorn, C. Guy, S. K. Smith et al., 2004. a A high proportion of chromosome 21 promoter polymorphisms influence transcriptional activity. Gene Expr. 11: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckland, P. R., B. Hoogendoorn, C. A. Guy, S. L. Coleman, S. K. Smith et al., 2004. b A high proportion of polymorphisms in the promoters of brain expressed genes influences transcriptional activity. Biochim. Biophys. Acta 1690: 238–249. [DOI] [PubMed] [Google Scholar]
- Carroll, S. B., 2003. Genetics and the making of Homo sapiens. Nature 422: 849–857. [DOI] [PubMed] [Google Scholar]
- Carroll, S. B., J. K. Grenier and S. D. Weatherbee, 2004. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Science, Malden, MA.
- The Chipmanzee Sequencing and Analysis Consortium, 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87. [DOI] [PubMed] [Google Scholar]
- Clark, R. M., T. N. Wagler, P. Quijada and J. Doebley, 2006. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 38: 594–597. [DOI] [PubMed] [Google Scholar]
- Cooper, S. J., N. D. Trinklein, E. D. Anton, L. Nguyen and R. M. Myers, 2006. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res 16: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles, C. R., J. N. Hirschhorn, D. Altshuler and E. S. Lander, 2002. Detection of regulatory variation in mouse genes. Nat. Genet. 32: 432–437. [DOI] [PubMed] [Google Scholar]
- Cresko, W. A., A. Amores, C. Wilson, J. Murphy, M. Currey et al., 2004. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc. Natl. Acad. Sci. USA 101: 6050–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis, E. T., and A. G. Clark, 2002. Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol. Biol. Evol. 19: 1114–1121. [DOI] [PubMed] [Google Scholar]
- Dickinson, W. J., 1988. On the architecture of regulatory systems: evolutionary insights and implications. BioEssays 8: 204–208. [DOI] [PubMed] [Google Scholar]
- Gibson, G., and B. Weir, 2005. The quantitative genetics of transcription. Trends Genet. 21: 616–623. [DOI] [PubMed] [Google Scholar]
- Gilad, Y., S. A. Rifkin, P. Bertone, M. Gerstein and K. P. White, 2005. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 15: 674–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad, Y., A. Oshlack, G. K. Smyth, T. P. Speed and K. P. White, 2006. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440: 242–245. [DOI] [PubMed] [Google Scholar]
- Gompel, N., B. Prud'homme, P. J. Wittkopp, V. A. Kassner and S. B. Carroll, 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487. [DOI] [PubMed] [Google Scholar]
- Hammock, E. A., and L. J. Young, 2005. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science 308: 1630–1634. [DOI] [PubMed] [Google Scholar]
- Haudek, S. B., B. E. Natmessnig, H. Redl, G. Schlag and B. P. Giroir, 1998. Genetic sequences and transcriptional regulation of the TNFA promoter: comparison of human and baboon. Immunogenetics 48: 202–207. [DOI] [PubMed] [Google Scholar]
- Heissig, F., J. Krause, J. Bryk, P. Khaitovich, W. Enard et al., 2005. Functional analysis of human and chimpanzee promoters. Genome Biol. 6: R57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, P. C., J. C. Chang, W. T. Sun, S. C. Hsieh, M. C. Wang et al., 2007. p53 downstream target DDA3 is a novel microtubule-associated protein that interacts with end-binding protein EB3 and activates beta-catenin pathway. Oncogene (in press). [DOI] [PubMed]
- Huby, T., C. Dachet, R. M. Lawn, J. Wickings, M. J. Chapman et al., 2001. Functional analysis of the chimpanzee and human apo(a) promoter sequences: identification of sequence variations responsible for elevated transcriptional activity in chimpanzee. J. Biol. Chem. 276: 22209–22214. [DOI] [PubMed] [Google Scholar]
- Iftikhar, R., R. D. Kladney, N. Havlioglu, A. Schmitt-Graff, I. Gusmirovic et al., 2004. Disease- and cell-specific expression of GP73 in human liver disease. Am. J. Gastroenterol. 99: 1087–1095. [DOI] [PubMed] [Google Scholar]
- Jin, W., R. M. Riley, R. D. Wolfinger, K. P. White, G. Passador-Gurgel et al., 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29: 389–395. [DOI] [PubMed] [Google Scholar]
- Khaitovich, P., I. Hellmann, W. Enard, K. Nowick, M. Leinweber et al., 2005. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309: 1850–1854. [DOI] [PubMed] [Google Scholar]
- King, M. C., and A. C. Wilson, 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107–116. [DOI] [PubMed] [Google Scholar]
- Kleinjan, D. A., and V. van Heyningen, 2005. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76: 8–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon, B., and R. Tjian, 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14: 2551–2569. [DOI] [PubMed] [Google Scholar]
- Ludwig, M. Z., N. H. Patel and M. Kreitman, 1998. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development 125: 949–958. [DOI] [PubMed] [Google Scholar]
- Ludwig, M. Z., A. Palsson, E. Alekseeva, C. M. Bergman, J. Nathan et al., 2005. Functional evolution of a cis-regulatory module. PLoS Biol. 3: e93.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, L., M. Flajolet and P. Greengard, 2004. Pharmacological inhibitors of glycogen synthase kinase 3. Trends Pharmacol. Sci. 25: 471–480. [DOI] [PubMed] [Google Scholar]
- Morley, M., C. M. Molony, T. M. Weber, J. L. Devlin, K. G. Ewens et al., 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430: 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastinen, T., B. Ge and T. J. Hudson, 2006. Influence of human genome polymorphism on gene expression. Hum. Mol. Genet. 15(Spec. No. 1): R9–16. [DOI] [PubMed] [Google Scholar]
- Prabhakar, S., F. Poulin, M. Shoukry, V. Afzal, E. M. Rubin et al., 2006. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 16: 855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger, J. A., L. P. Shearman, S. M. Reppert and U. Schibler, 2000. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 14: 679–689. [PMC free article] [PubMed] [Google Scholar]
- Rockman, M. V., M. W. Hahn, N. Soranzo, D. B. Goldstein and G. A. Wray, 2003. Positive selection on a human-specific transcription factor binding site regulating IL4 expression. Curr. Biol. 13: 2118–2123. [DOI] [PubMed] [Google Scholar]
- Rockman, M. V., M. W. Hahn, N. Soranzo, F. Zimprich, D. B. Goldstein et al., 2005. Ancient and recent positive selection transformed opioid cis-regulation in humans. PLoS Biol. 3: e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, M. D., M. E. Marks, C. L. Peichel, B. K. Blackman, K. S. Nereng et al., 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428: 717–723. [DOI] [PubMed] [Google Scholar]
- Stern, D. L., 1998. A role of Ultrabithorax in morphological differences between Drosophila species. Nature 396: 463–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, D. L., 2000. Evolutionary developmental biology and the problem of variation. Evol. Int. J. Org. Evol. 54: 1079–1091. [DOI] [PubMed] [Google Scholar]
- Storgaard, T., J. Christensen, B. Aasted and S. Alexandersen, 1993. cis-acting sequences in the Aleutian mink disease parvovirus late promoter important for transcription: comparison to the canine parvovirus and minute virus of mice. J. Virol. 67: 1887–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H., Y. Mitani, G. Satoh and N. Satoh, 1999. Evolutionary alterations of the minimal promoter for notochord-specific Brachyury expression in ascidian embryos. Development 126: 3725–3734. [DOI] [PubMed] [Google Scholar]
- Taron, M., R. Rosell, E. Felip, P. Mendez, J. Souglakos et al., 2004. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum. Mol. Genet. 13: 2443–2449. [DOI] [PubMed] [Google Scholar]
- Trinklein, N. D., S. J. Aldred, A. J. Saldanha and R. M. Myers, 2003. Identification and functional analysis of human transcriptional promoters. Genome Res. 13: 308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri, T., and G. Elgar, 2005. Prediction of cis-regulatory elements using binding site matrices–the successes, the failures and the reasons for both. Curr. Opin. Genet. Dev. 15: 395–402. [DOI] [PubMed] [Google Scholar]
- White, R. J., 2001. Gene Transcription: Mechanims and Control. Blackwell Science, Malden, MA.
- Wittkopp, P. J., B. K. Haerum and A. G. Clark, 2004. Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88. [DOI] [PubMed] [Google Scholar]
- Wolff, C., M. Pepling, P. Gergen and M. Klingler, 1999. Structure and evolution of a pair-rule interaction element: runt regulatory sequences in D. melanogaster and D. virilis. Mech. Dev. 80: 87–99. [DOI] [PubMed] [Google Scholar]
- Wray, G. A., 2007. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8: 206–216. [DOI] [PubMed] [Google Scholar]
- Wray, G. A., M. W. Hahn, E. Abouheif, J. P. Balhoff, M. Pizer et al., 2003. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20: 1377–1419. [DOI] [PubMed] [Google Scholar]
- Wuarin, J., E. Falvey, D. Lavery, D. Talbot, E. Schmidt et al., 1992. The role of the transcriptional activator protein DBP in circadian liver gene expression. J. Cell Sci. Suppl. 16: 123–127. [DOI] [PubMed] [Google Scholar]
- Yvert, G., R. B. Brem, J. Whittle, J. M. Akey, E. Foss et al., 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 35: 57–64. [DOI] [PubMed] [Google Scholar]