Abstract
The yeast transcription factors Stp1 and Stp2 are synthesized as latent cytoplasmic precursors. In response to extracellular amino acids, the plasma membrane SPS sensor endoproteolytically excises the N-terminal domains that mediate cytoplasmic retention, enabling the processed forms to efficiently enter the nucleus and induce gene expression. Cytoplasmic retention is not absolute, low levels of full-length Stp1 and Stp2 “leak” into the nucleus, and the concerted action of inner nuclear membrane proteins Asi1, Asi2, and Asi3 restricts their promoter access. In cells lacking Asi function, the precursor forms bind promoters and constitutively induce gene expression. To understand the requirement of Asi-dependent repression, spontaneous mutations in Required for Latent Stp1/2-mediated transcription (RLS) genes that abolish the constitutive expression of SPS sensor-regulated genes in an asi1Δ strain were selected. A single gene, allelic with DAL81, was identified. We show that Dal81 indiscriminately amplifies the transactivation potential of both full-length and processed Stp1 and Stp2 by facilitating promoter binding. In dal81Δ mutants, the repressing activity of the Asi proteins is dispensable, demonstrating that without amplification, the levels of full-length Stp1 and Stp2 that escape cytoplasmic retention are insufficient to activate transcription. Conversely, the high levels of processed Stp1 and Stp2 that accumulate in the nucleus of induced cells activate transcription in the absence of Dal81.
ALL cells sense discrete environmental signals and respond by making appropriate adjustments in patterns of gene expression. A basic problem in biology is how cells generate clear differences between “off” and “on” transcriptional states. A strategy to achieve this in eukaryotic cells is to control the movement of transcription factors across the nuclear envelope. In such instances, nuclear targeting physically transmits signals from non-nuclear compartments to specific promoter sequences and simultaneously provides the means to initiate transcription. There are a growing number of transcription factors that have been shown to be maintained as latent cytoplasmic factors requiring proteolytic processing prior to nuclear targeting (Brivanlou and Darnell 2002). Understanding transcription factor latency requires the elucidation of the mechanisms that not only direct these proteins to the nucleus but also establish and maintain the dormant or repressed state of gene expression in the absence of inducing signals.
Saccharomyces cerevisiae cells respond to the presence of extracellular amino acids by inducing the expression of several genes encoding amino acid permeases, a family of proteins that transport amino acids across the plasma membrane into cells (Forsberg and Ljungdahl 2001b). Extracellular amino acids are recognized by the integral plasma membrane protein Ssy1 (Jørgensen et al. 1998; Iraqui et al. 1999; Klasson et al. 1999; Wu et al. 2006), which functions together with two peripheral membrane proteins, Ptr3 and Ssy5, as core components of the SPS sensor of extracellular amino acids (Forsberg and Ljungdahl 2001a). All three core components are required for proper sensing; inactivating mutations in SSY1, PTR3, or SSY5 completely abolishes SPS signaling.
Two homologous zinc-finger transcription factors, Stp1 and Stp2, are redundant downstream effector components of the SPS-signaling pathway. Single deletions of STP1 or STP2 partially impair, whereas deletions of both fully eliminate SPS sensor-regulated gene expression (De Boer et al. 2000; Andréasson and Ljungdahl 2002). Stp1 and Stp2 bind to specific upstream activating sequences (UASaa) present within SPS sensor-regulated promoters (De Boer et al. 2000; Nielsen et al. 2001; Abdel-Sater et al. 2004b). Stp1 and Stp2 are synthesized as latent factors with N-terminal regulatory domains that function as cytoplasmic retention motifs (Andréasson and Ljungdahl 2002, 2004). In response to amino acids, the SPS sensor endoproteolytically cleaves the N-terminal regulatory domains in a processing event termed receptor-activated proteolysis (Andréasson and Ljungdahl 2002; Andréasson et al. 2006). The shorter processed forms of Stp1 and Stp2 efficiently target to and accumulate in the nucleus where they function to transactivate SPS sensor-regulated genes (Andréasson and Ljungdahl 2002). Thus, the mobilization of Stp1 and Stp2 transfers amino acid-induced regulatory information from the plasma membrane to the nucleus.
We have recently found that the mechanisms responsible for retaining latent forms of Stp1 and Stp2 in the cytoplasm are not fully efficient (Andréasson and Ljungdahl 2004; Boban et al. 2006; Zargari et al. 2007). The concerted action of three inner nuclear membrane proteins Asi1, Asi2, and Asi3 is required to restrict promoter access of unprocessed forms of Stp1 and Stp2 that escape cytoplasmic retention and inappropriately enter the nucleus. In contrast to wild-type cells, where only processed forms of Stp1 and Stp2 bind SPS-regulated promoters, in asi mutant cells unprocessed latent forms also bind promoters, resulting in constitutive activation of SPS sensor-regulated genes even in the absence of amino acids or a functional SPS sensor (Forsberg et al. 2001).
Critical to understanding the role of Asi proteins is the observation that Stp1 and Stp2 do not accumulate in the nucleus of asi mutant strains, thus eliminating the possibility that Asi proteins affect cytoplasmic retention mechanisms (Boban et al. 2006). Remarkably, the low levels of full-length Stp1 and Stp2 that enter the nucleus of uninduced asi mutants, or asi mutants lacking a functional SPS sensor, suffice to induce SPS sensor gene expression at levels indistinguishable to those observed in induced wild-type cells (Forsberg et al. 2001). Clearly, if given the opportunity, latent forms of Stp1 and Stp2 can efficiently bind promoters and induce transcription (Boban et al. 2006). These findings demonstrate that negative regulation of Stp1 and Stp2 activity is not limited to controlling cytoplasmic retention and that cells require the Asi proteins to maintain the repressed state of signaling in the absence of inducing amino acids. Thus, two independent mechanisms control the latent properties of Stp1 and Stp2, i.e., cytoplasmic retention that restricts nuclear targeting and Asi-dependent repression that restricts promoter access. Notably, both mechanisms exert their regulatory effects via the first 70 amino acids within the N-terminal regulatory domains of these factors (Andréasson and Ljungdahl 2004; Boban et al. 2006).
Here we have directly tested the necessity of Asi-dependent control in repressing SPS sensor-regulated gene expression under noninducing conditions. Using an unbiased genetic approach, we selected spontaneous mutations in Required for Latent Stp1/2-mediated transcription (RLS) genes that abolish the constitutive expression of SPS sensor-regulated genes in an asi1Δ strain. The RLS selection identified a single gene that is allelic to DAL81. Dal81 is a pleiotropic nuclear factor that is required for full induction of SPS sensor-regulated AAP gene expression (Iraqui et al. 1999; Bernard and André 2001; Abdel-Sater et al. 2004b), induction of genes involved in utilization of urea and allantoin (Jacobs et al. 1981; Turoscy and Cooper 1982), and γ-aminobutyric acid (GABA) (Vissers et al. 1989). We show that Dal81 amplifies Stp1- and Stp2-dependent transactivation by indiscriminately facilitating the binding of both latent and processed forms to SPS sensor-regulated promoters. Consistent with its function merely as an amplifier, Dal81 does not by itself activate SPS sensor-regulated gene expression. Strikingly, in dal81Δ mutants, the repressing activity of Asi proteins is not required to maintain the off state of SPS sensor-regulated gene expression. Our findings illuminate important aspects of the SPS-sensing pathway that puts the requirement of the Asi “backup” system in biological context.
MATERIALS AND METHODS
Media and strains:
Standard media, including YPD and ammonia-based synthetic minimal dextrose (SD) supplemented as required to enable growth of auxotrophic strains, were prepared as described (Burke and Stewart 2002). Ammonia-based synthetic complex dextrose (SC) was prepared as described (Andréasson and Ljungdahl 2002). Where indicated, L-leucine was added at a concentration of 1.3 mm to induce the SPS sensor. When required, 5-fluoroortic acid (FOA) was added to SC (1 g/liter). Media were made solid with 2% (w/v) Bacto Agar (Difco, Detroit, MI). Antibiotic selections were made on solid YPD supplemented with 200 mg/liter G418 (Invitrogen, Carlsbad, CA), 100 mg/liter clonNAT (Werner Bioagents, Jena, Germany), or 300 mg/liter Hygromycin B (Duchefa, Haarlem, The Netherlands). Sensitivity to 1 mm L-azetidine-2-carboxylic acid (AzC) was tested on SD supplemented with L-leucine (1.3 mm) and L-glutamic acid (1 mm). YPD containing 0.5 mg/ml 2-{[({[(4-methoxy-6-methyl)-1,3,5-triazin-2-yl]-amino}carbonyl)amino-]-sulfonyl}-benzoic acid (MM) was prepared as described (Jørgensen et al. 1998).
The yeast strains used in data collection are listed in Table 1. The sequences of primers used in yeast strain construction are available on request. All strains are isogenic descendants of the S288c derived from strain AA255 (Antebi and Fink 1992). Strain CAY118 is a meiotic segregant of a cross between CAY62 and CAY117. Strain YMH117 was obtained from a cross between AA255 and HKY20. Strain YMH173 was obtained from a cross between YMH117 and PLY1016 (MATα ura3-52 lys2Δ201 ptr3Δ14∷hisG-URA3-kanR-hisG). Strain MBY3 was generated from a cross between CAY224 and PLY1314. Strains MBY4 and MBY5 were generated from a cross between MBY3 and YMH173. Strains MBY13 and MBY14 are ura− derivatives of strains MBY4 and MBY5, respectively, which were passaged on FOA. As described in the Isolation of mutations in RLS1 section, strains MBY15 and MBY16 were generated from MBY13 and MBY14, respectively. MBY17 and MBY18 were constructed by introducing dal81Δ76∷kanMX4 null allele into strains MBY13 and MBY14; dal81Δ76∷kanMX4 was generated by PCR using prMB12F and prMB12R primers and genomic DNA isolated from the strain Y07303 (European S. cerevisiae archive for functional analysis, http://web.uni-frankfurt.de/fb15/mikro/euroscarf/) as a template. Strain MBY40 was constructed by introducing the ssy1Δ77∷natMX4 null allele into strain CAY28; the ssy1Δ77∷natMX4 was generated by PCR using primers prMB75/76 and pAG25 (Goldstein and McCusker 1999) as a template. The dal81Δ77∷natMX4 cassette was constructed by PCR using primers prMB130/131 and pAG25 (Goldstein and McCusker 1999) as a template. Strains MBY61, MBY62, MBY64, MBY66, and MBY67 were generated by introducing the dal81Δ77∷natMX4 null allele into strains CAY119, CAY123, CAY150, CAY206, and PLY1314, respectively. Strain MBY79 was generated from a cross between strain CAY28 and MBY62. CAY151 is a ura− derivative of a meiotic segregant obtained from a cross between CAY47 and CAY126, which was passaged on FOA. Strains MBY80 and MBY82 are meiotic segregants from a cross between strain CAY151 and MBY62. Strain MBY101 is a meiotic segregant obtained from a cross between HKY93 (MATα ura3-52 ssy5Δ1∷hisG-URA3-kanR-hisG) (Forsberg and Ljungdahl 2001a), and CAY152 and strains MBY93 and MBY102 are ura− derivatives of meiotic segregants obtained from the same cross. Strain MBY124 is an ura− derivative of a meiotic segregant obtained from a cross between MBY82 and MBY101.
TABLE 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| AA255 | MATα ura3-52 ade2 his3Δ200 lys2Δ201 leu2-3,112 | Antebi and Fink (1992) |
| CAY28 | MATα ura3-52 | Andréasson and Ljungdahl (2002) |
| CAY47 | MATα ura3-52 stp1Δ50∷CaURA3MX3 | Boban et al. (2006) |
| CAY60 | MATα ura3-52 stp1Δ51∷Agleu2 | Boban et al. (2006) |
| CAY62 | MATα ura3-52 lys2Δ201 stp1Δ51∷Agleu2 | Boban et al. (2006) |
| CAY118 | MATα ura3-52 stp2Δ50∷hphMX4 | This work |
| CAY117 | MATaura3-52 stp2Δ50∷hphMX4 | Andréasson and Ljungdahl (2002) |
| CAY119 | MATaura3-52 stp2Δ50∷hphMX4 | Andréasson and Ljungdahl (2002) |
| CAY123 | MATaura3-52 stp1Δ51∷Agleu2 stp2Δ50∷hphMX4 | Andréasson and Ljungdahl (2002) |
| CAY126 | MATaura3-52 asi1Δ8∷kanMX stp2Δ50∷hphMX4 | Boban et al. (2006) |
| CAY150 | MATaura3-52 asi1Δ8∷kanMX stp1Δ51∷Agleu2 | Boban et al. (2006) |
| CAY151 | MATα ura3-52 stp1Δ51∷Agleu2 stp2Δ50∷hphMX4 asi1Δ8∷kanMX | This work |
| CAY152 | MATa ura3-52 stp1Δ51∷Agleu2 stp2Δ50∷hphMX4 asi1Δ8∷kanMX | Andréasson and Ljungdahl (2004) |
| CAY206 | MATaura3-52 ssy1Δ13∷hisG asi1Δ8∷kanMX | Andréasson and Ljungdahl (2004) |
| CAY224 | MATaura3-52 gap1Δ∷PAGP1-LacZ | Andréasson and Ljungdahl (2004) |
| HKY20 | MATaura3-52 lys2Δ201 ssy1Δ13∷hisG | Klasson et al. (1999) |
| MBY3 | MATα ura3-52 asi1Δ80∷hphMX4 gap1Δ∷PAGP1-LacZ | This work |
| MBY4 | MATα ura3-52 his3Δ200 ptr3Δ14∷hisG-URA3-kanR-hisG asi1Δ80∷ hphMX4 gap1Δ∷PAGP1-LacZ | This work |
| MBY5 | MATaura3-52 ade2 ptr3Δ14∷hisG-URA3-kanR-hisG asi1Δ80∷ hphMX4 gap1Δ∷PAGP1-LacZ | This work |
| MBY13 | MATα ura3-52 his3Δ200 ptr3Δ15∷hisG asi1Δ80∷hphMX4 gap1Δ∷ PAGP1-LacZ | This work |
| MBY14 | MATaura3-52 ade2 ptr3Δ15∷hisG asi1Δ80∷hphMX4 gap1Δ∷PAGP1-LacZ | This work |
| MBY15 | MATα ura3-52 his3Δ200 ptr3Δ15∷hisG asi1Δ80∷hphMX4 dal81-101 gap1Δ∷ PAGP1-LacZ | This work |
| MBY16 | MATaura3-52 ade2 ptr3Δ15∷hisG asi1Δ80∷hphMX4 dal81-102 gap1Δ∷ PAGP1-LacZ | This work |
| MBY17 | MATα ura3-52 his3Δ200 ptr3Δ15∷hisG asi1Δ80∷hphMX4 gap1Δ∷ PAGP1-LacZ dal81Δ76∷kanMX4 | This work |
| MBY18 | MATaura3-52 ade2 ptr3Δ15∷hisG asi1Δ80∷hphMX4 gap1Δ∷ PAGP1-LacZ dal81Δ76∷kanMX4 | This work |
| MBY40 | MATα ura3-52 ssy1Δ77∷natMX4 | This work |
| MBY61 | MATaura3-52 stp2Δ50∷hphMX4 dal81Δ77∷natMX4 | This work |
| MBY62 | MATaura3-52 stp1Δ51∷Agleu2 stp2Δ50∷hphMX4 dal81Δ77∷natMX4 | This work |
| MBY64 | MATaura3-52 asi1Δ8∷kanMX stp1Δ51∷Agleu2 dal81Δ77∷natMX4 | This work |
| MBY66 | MATaura3-52 ssy1Δ13∷hisG asi1Δ8∷kanMX dal81Δ77∷natMX4 | This work |
| MBY67 | MATα ura3-52 asi1Δ80∷hphMX4 dal81Δ77∷natMX4 | This work |
| MBY79 | MATα ura3-52 stp1Δ51∷Agleu2 dal81Δ77∷natMX4 | This work |
| MBY80 | MATaura3-52 asi1Δ8∷kanMX stp1Δ51∷Agleu2 stp2Δ50∷hphMX4 dal81Δ77∷natMX4 | This work |
| MBY82 | MATaura3-52 stp1Δ51∷Agleu2 stp2Δ50∷hphMX4 dal81Δ77∷natMX4 | This work |
| MBY83 | MATα ura3-52 stp1Δ51∷Agleu2 stp2Δ50∷hphMX4 dal81Δ77∷natMX4 | This work |
| MBY91 | MATaura3-52 ssy5Δ1∷hisG-URA3-kanR-hisG stp1Δ51∷Agleu2 stp2Δ50∷ hphMX4 | This work |
| MBY93 | MATaura3-52 ssy5Δ1∷hisG stp1Δ51∷Agleu2 stp2Δ50∷hphMX4 | This work |
| MBY101 | MATα ura3-52 ssy5Δ1∷hisG-URA3-kanR-hisG asi1Δ8∷kanMX stp1Δ51∷ Agleu2 stp2Δ50∷hphMX4 | This work |
| MBY102 | MATα ura3-52 ssy5Δ1∷hisG asi1Δ8∷kanMX stp1Δ51∷Agleu2 stp2Δ50∷ hphMX4 | This work |
| MBY124 | MATaura3-52 stp1Δ51∷Agleu2 stp2Δ50∷hphMX4 ssy5Δ1∷hisG asi1Δ8∷ anMX dal81Δ77∷natMX4 | This work |
| PLY1313 | MATaura3-52 asi1Δ80∷hphMX4 | Boban et al. (2006) |
| PLY1314 | MATα ura3-52 asi1Δ80∷hphMX4 | Boban et al. (2006) |
| YMH117 | MATaura3-52 ade2 his3Δ200 lys2Δ201 leu2-3,112 | This work |
| YMH173 | MATaura3-52 ade2 his3Δ200 lys2Δ201 ptr3Δ14∷hisG-URA3-kanR-hisG | This work |
Plasmids:
Plasmids used are listed in Table 2. Plasmid pMB31 was generated by homologous recombination in yeast by cotransforming StuI/XmnI-restricted pMB10 and KpnI/SacI-restricted pRS202 (Connelly and Hieter 1996). The Stp1-HA epitope-tagged allele of Stp1 in plasmid pMB10 encodes a fully functional protein on the basis of the following criteria. First, a stp1Δ stp2Δ double mutant transformed with pMB10 is unable to grow in the presence of AzC on SD medium containing 1.3 mm leucine and grows well on YPD medium containing MM. Second, pMB10-encoded Stp1 is endoproteolytically processed and activates a PAGP1-lacZ reporter gene expression in an amino acid-dependent manner. Finally, when pMB10 is introduced into an ssy1 leu2 strain, which is not able to grow on amino acid rich SC medium owing to a defect in amino acid uptake, the pMB10-encoded Stp1 does not confer growth, indicating that the unprocessed pMB10-encoded Stp1 is not constitutively active. Plasmids pMB44 and pMB45 were isolated from yeast cotransformed with PCR product amplified by primers prMB163-F and 167-R using plasmid pCA120 (Andréasson and Ljungdahl 2004) as a template and MluI/BglII-restricted pMB10 or pMB31, respectively. Generation of plasmid pMB71 is described in the following section.
TABLE 2.
Plasmids
| Plasmid | Description | Reference |
|---|---|---|
| pCA029 | STP1 in pRS316 | Andréasson and Ljungdahl (2002) |
| pHK027 | PTR3 in pRS316 | Klasson et al. (1999) |
| pMB10 | 13xMYC-STP1-6xHA in pRS316 | Boban et al. (2006) |
| pMB31 | 13xMYC-STP1-6xHA in pRS202 | This work |
| pMB44 | 13xMYC-STP1-133-6xHA in pRS316 | This work |
| pMB45 | 13xMYC-STP1-133-6xHA in pRS202 | This work |
| pMB71 | DAL81 in pSEY18 | This work |
| pRS202 | 2μ URA3 | Connelly and Hieter (1996) |
| pRS316 | CEN URA3 | Sikorski and Hieter (1989) |
| pSEY18 | 2μ URA3 | Emr et al. (1986) |
Isolation of mutations in RLS1:
Twenty independent colonies from ptr3Δ asi1Δ strains of both mating types (MBY13 and MBY14) were separately inoculated in 40 tubes containing SD medium supplemented with 1 mm glutamic acid, 1.3 mm leucine, and standard concentrations of uracil, adenine, and histidine, and the cultures were grown to OD600 of 8. Aliquots (150 μl) of each culture were individually spread on separate SD plates containing 1 mm AzC and supplemented as above. Plates were incubated at 30° for 6 days, and the β-galactosidase activity in AzC resistant colonies was assayed using an X-Gal overlay. Fifty out of 100 AzC-resistant and β-galactosidase negative colonies were picked for further analysis.
The mutants were backcrossed to the appropriate RLS+ starting strain of the opposite mating type (either MBY13 or MBY14). All of the resulting diploid strains were AzC sensitive and exhibited high levels of β-galactosidase activity, indicating that the mutations were recessive. Complementation analysis was carried out by crossing all possible combinations of MATa and MATα rls mutants; the ability of the diploids to grow on SD medium containing AzC and the levels of β-galactosidase activity were determined. No complementation was observed in any of the crosses, all diploids were AzC resistant, and they did not express detectable β-galactosidase activity. One strain of each mating type MBY15 and MBY16 was backcrossed to MBY14 and MBY13, respectively. The resulting diploids were subjected to tetrad analysis, and in both cases a 2:2 (AzCr β-gal−:AzCs β-gal+) segregation pattern was observed, indicating that the phenotypes were due to mutations in a single RLS1 gene.
Cloning of RLS1:
To clone RLS1 we used YPD medium containing MM, an inhibitor of branched chain amino acid synthesis. On this medium (YPD + MM), growth is dependent on the ability of cells to express two SPS sensor-controlled permeases, Bap2 and Bap3 (Jørgensen et al. 1998). Because of their inability to express these permeases, ptr3Δ asi1Δ rls1 mutant strains MBY15 and MBY16 are not able to grow on YPD + MM. Strains MBY15 and MBY16 were transformed with a genomic plasmid library (Thompson et al. 1993). Several transformants were found to confer partial complementation, i.e., slow growth on YPD + MM and low levels of β-galactosidase activity. Plasmids rescued from these transformants contained PTR3. Our failure to obtain a fully complementing plasmid prompted us to use an alternative genomic library kindly provided by Michael N. Hall (Biozentrum, University of Basel, Basel, Switzerland). This second library is comprised of plasmids carrying large 15–20-kb inserts. Using this pSEY18-based library, we obtained two populations of transformants, one exhibiting partial and the other complete complementation. Plasmid pMB71 conferred complete complementation, and sequencing revealed that this plasmid contained DAL81. We constructed ptr3Δasi1Δdal81Δ strains in both mating types (MBY17 and MBY18). Subsequent genetic analysis confirmed that dal81Δ and rls1 are allelic.
β-galactosidase activity overlay assay:
Semiquantitative measurements of β-galactosidase activity were determined with N-lauroyl-sarcosine permeabilized cells (Kippert 1995). Low melting-point agarose (0.5%) was melted in 0.4 m potassium phosphate buffer (pH 7.0) and, after slight cooling, 0.2% N-lauroyl sarcosine, 0.05% β-mercaptoethanol, and 0.2 mg/ml X-Gal (from 100 mg/ml stock in dimethyl formamide) were added. Approximately 10 ml of the final agarose mixture (37°) was poured over cells grown on the solid medium. Plates were incubated at 30° until a blue precipitate was visible and kept at 4° until photographed.
Chromatin immunoprecipitation:
Chromatin immunoprecipitation (ChIP) analysis was performed according to Strahl-Bolsinger et al. (1997) with minor modifications. Cells were grown to an OD600 of 0.8 and fixed for 30 min at room temperature in the presence of 1% formaldehyde. The formaldehyde was added directly to the cultures. Cells were harvested by centrifugation, resuspended in lysis buffer, and disrupted with glass beads by beating 6 × 40 sec in a beadbeater (Biospec Products, Bartlesville, OK). The resulting lysate was sonicated twice for 10 sec using a Branson Sonifier 250 (Branson Ultrasonics, Plainview, NY) with output control set to 5 (average size of DNA fragments was 0.5 kb). Sonicated lysates were clarified by centrifugation (twice for 10 min at 15,000 × g). The protein content was measured, and the samples were adjusted to 10 mg/ml in 1200 μl, and 10 μl of the total lysate was put aside to control input levels. The remaining lysates were split in two equal fractions. Magnetic beads with covalently attached sheep anti-rat IgG (Dynabeads M-450, Dynal Biotech, Carlsbad, CA) were incubated with rat monoclonal anti-HA antibody (clone 3F10, Roche, Basel, Switzerland). A total of 50 μl of coated beads were used in immunoprecipitation reactions. Immunoprecipitates were sequentially washed in lysis buffer, lysis buffer containing 500 mm NaCl, washing buffer (10 mm Tris Cl pH 8.0, 500 mm LiCl, 1% Nonidet-P-40, 1% Na-deoxycholate, and 1 mm EDTA), and TE. Bound protein was eluated by incubating beads twice for 10 min at 65° in 75 μl of eluation buffer (50 mm Tris Cl pH 8.0, 10 mm EDTA, and 1% SDS). Crosslinking of immunoprecipitates and input samples was reversed by an overnight incubation at 65°, after which DNA was extracted (PCR Purification kit, Qiagen GmbH, Hilden, Germany). PCR was carried out with primers that amplify promoter regions of AGP1 (PrMB23/24), GNP1 (PrMB31/32), and ACT1 (PrMB41/42) (sequence of primers is available on request). Taq polymerase (Invitrogen) and corresponding buffer system were used. Hot start was achieved by using TaqStart Antibody (Clontech, Mountain View, CA). The appropriate dilution of template DNA and the number of cycles (25–30) were empirically determined. Samples were first incubated for 3 min at 94°, and the amplification cycle was as follows: 45 sec at 94°, 45 sec at 50°, and 20 sec at 72°. The reactions were stopped in the logarithmic phase of amplification, and the PCR products were separated on 2.3% agarose gel and visualized by ethidium bromide. The quantity of PCR products was determined using the LAS1000 system and Image Gauge software V.4.22 (Fuji Photo Film, Tokyo, Japan). The intensities of bands from immunoprecipitations (minus background) were normalized to the bands obtained from input DNA.
Microscopy:
Cells were grown to an OD600 of 0.8 and processed for indirect immunofluorescence analysis essentially as described in (Burke et al. 2000). Cells were fixed by the addition of an aliquot of 37% formaldehyde directly to the cultures to a final concentration of 4.5% and incubated 45 min at 30°. To detect HA-tagged proteins, the primary antibody used was the 3F10 anti-HA monoclonal antibody diluted 1:300. To detect myc-tagged proteins, the primary antibody used was the 9E10 anti-myc monoclonal antibody diluted 1:300. The secondary antibody was Alexa Fluor 488 conjugated to goat anti-mouse or donkey anti-rat IgG (H + L; Molecular Probes, Eugene, OR), diluted 1:500. Cells were viewed using a Zeiss Axiophot microscope with a Plan-Apochromat 63×/1.40 objective. Digital images of cells examined using Nomarski optics, and antibody-dependent and DAPI fluorescence (standard filter sets) were captured using a C4742-95 CCD camera (Hamamatsu Photonics, Hamamatsu, Japan) and QED Imaging software (Media Cybernetics, Bethesda, MD). Image files were incorporated into figures using Adobe Photoshop CS.
RESULTS
Genetic analysis of constitutive Stp1- and Stp2-dependent gene expression in asi1Δ mutants:
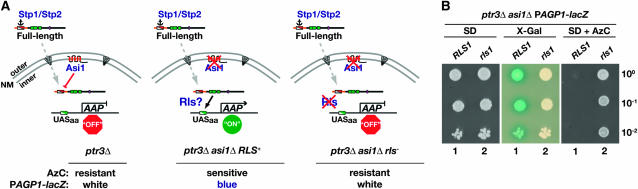
To investigate the necessity of the Asi proteins in the maintenance of the repressed state of SPS sensor-regulated gene expression under noninducing conditions, we selected spontaneous mutations in RLS genes (Figure 1A). Briefly, to prevent Stp1 and Stp2 processing, the asi1Δ starting strain lacked a functional SPS sensor (ptr3Δ). Because of loss of Asi1 function, SPS sensor-regulated promoters are constitutively active, and consequently the strain is sensitive to the toxic proline analog AzC. AzC is efficiently transported into cells by two SPS sensor-regulated permeases, Agp1 and Gnp1 (Andréasson et al. 2004). The effective uptake of AzC by multiple permeases provides the basis of an extremely tight selection for mutations conferring resistance. Such mutations must affect factors required for the functional expression of both Agp1 and Gnp1. The redundant function of Stp1 and Stp2 minimized the possibility of isolating mutations in these genes. Finally, to identify mutations affecting transcription and to differentiate those from mutations affecting post-transcriptional events, we monitored the activity of β-galactosidase expressed from a PAGP1-lacZ construct integrated in the genome at the GAP1 locus (gap1Δ∷PAGP-lacZ). Colonies carrying mutations in RLS genes were identified on the basis of their ability to grow in the presence of AzC and the lack of β-galactosidase activity (Figure 1B). All analyzed rls mutations were found to be recessive and to belong to the same rls1 complementation group. Details regarding the rls selection are provided in materials and methods and summarized in Table 3.
Figure 1.—
Mutations in RLS1 repress asi1Δ-induced constitutive SPS sensor-regulated gene expression. (A) Schematic of downstream events in the SPS-sensing pathway. In cells lacking a functional SPS sensor (left), the latent unprocessed forms of transcription factors Stp1 and Stp2 are primarily localized to the cytosol. The presence of cytoplasmic retention signals (anchor) restricts their entry to the nucleus (Andréasson and Ljungdahl 2004). The low levels of latent forms of Stp1 and Stp2 that enter the nucleus (dashed arrow) are prevented from binding SPS sensor-regulated promoters of amino acid permease genes (AAP) by the combined action of Asi proteins (Asi1, Asi2, and Asi3) localized to the inner nuclear membrane (NM) (Boban et al. 2006; Zargari et al. 2007). The ability of the Asi proteins to prevent transcription is dependent on the presence of sequences of Stp1 and Stp2 (Region I) in the N-terminal regulatory domain (red/white diagonal box). Consequently, there are low levels of AAP gene expression; cells are AzC resistant and remain white when incubated in the presence of X-Gal because of lack of PAGP1-lacZ expression. In cells lacking Asi1 (middle), the latent forms of Stp1 and Stp2 that enter the nucleus constitutively induce the expression of SPS-regulated AAP genes, and cells are AzC sensitive and turn blue in the presence of X-Gal (Andréasson and Ljungdahl 2004; Boban et al. 2006). Mutations in RLS genes (right) prevent the latent forms of Stp1 and Stp2 from gaining access to SPS sensor-regulated promoter; mutant cells are AzC resistant and remain white in the presence of X-Gal. (B) Strains MBY13 (ptr3Δasi1Δ) and MBY15 (ptr3Δasi1Δrls1−01) were grown on SD medium. Cells were resuspended in water to obtain identical cell densities. Aliquots of 10-fold serial dilutions were spotted on SD supplemented with glutamate, leucine, adenine, uracil, histidine (SD), and SD containing AzC (SD + AzC). The plates were grown at 30° for 2 days, after which the SD plate was overlaid with X-Gal substrate (materials and methods).
TABLE 3.
Summary of the RLS selection
| Step | Quantity |
|---|---|
| Cells analyzed | 2.5 × 108 |
| AzCr colonies | 2000 |
| β-Gal− colonies (white) | 100 |
| rls mutants analyzed for complementation | 50 |
| Complementation groups | 1 |
RLS1 is allelic with DAL81 and is required for constitutive SPS sensor gene expression in asi1Δ mutants:
Plasmids from two genomic libraries were identified on the basis of their ability to complement the recessive rls1 mutation (see materials and methods). Interestingly, two classes of complementing plasmids were isolated. The first class comprised plasmids that only partially complemented the rls1 mutation. These plasmids were found to carry PTR3. The second class of plasmids, which fully complemented the rls1 mutation, contained DAL81. The ability of these plasmids to fully complement suggested that RLS1 is in fact DAL81. To rigorously test this possibility, we constructed ptr3Δ asi1Δ dal81Δ strains MBY17 (MATα) and MBY18 (MATa). These strains were crossed with the ptr3Δ asi1Δ rls1 strains MBY16 (MATa) and MBY15 (MATα), respectively. Subsequent genetic analysis confirmed that dal81Δ and rls1 are allelic; the mutations did not complement, and after sporulation, all meiotic segregants were resistant to AzC.
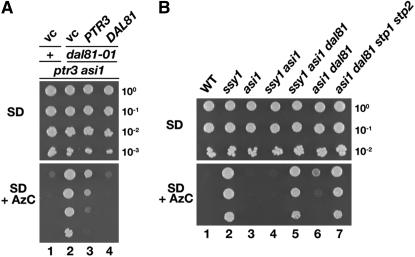
We examined the ability of plasmids carrying either PTR3 or DAL81 to complement the dal81Δ null allele on SD media supplemented with leucine and containing AzC (Figure 2A). The ptr3Δasi1Δ starting strain for the RLS selection, carrying an empty vector (vc), did not grow on this medium (vc; dilution 1), whereas the ptr3Δ asi1Δ dal81Δ carrying an empty vector exhibited robust growth (vc; dilution 2). Consistent with the partial complementation of rls1 mutations with plasmids containing PTR3, the introduction of PTR3 weakly suppressed the AzC sensitivity of dal81Δ strains; poor but detectable growth was observed (dilution 3). Thus, restoration of SPS sensor signaling partially complements the loss of DAL81. In contrast, introduction of DAL81 fully suppressed the AzC-resistant phenotype of this strain (dilution 4). These results strongly suggested that Dal81 affects the efficiency of Stp1- and Stp2-mediated gene expression.
Figure 2.—
Constitutive SPS sensor-regulated gene expression in asi1Δ mutants is strictly Dal81 dependent. (A) Strains ptr3Δ asi1Δ (MBY13; dilution series 1) and ptr3Δ asi1Δ dal81−101 (MBY15; dilution series 2–4) carrying plasmids pRS316 (vc), pHK027 (PTR3), or pMB71 (DAL81) were grown in SD supplemented with glutamate, leucine, and histidine (SD). Aliquots of 10-fold serial dilutions were spotted on SD and SD containing AzC (SD + AzC). Plates were incubated at 30°. (B) Wild-type (WT; CAY28) and mutant ssy1Δ (MBY40), asi1Δ (PLY1313), ssy1Δ asi1Δ (CAY206), ssy1Δ asi1Δ dal81Δ (MBY66), asi1Δ dal81Δ (MBY67), and asi1Δ dal81Δ stp1Δ stp2Δ (MBY80) strains were grown and growth characteristics analyzed as in A.
Amino acid-induced SPS sensor gene expression is not strictly Dal81 dependent:
We examined the role of Dal81 in the SPS sensor pathway (Figure 2B). As expected, on media containing AzC, a wild-type strain (WT; dilution 1) is unable to grow, whereas a mutant lacking a functional SPS sensor (ssy1Δ; dilution 2) grows well. Because of the constitutive expression of AGP1 and GNP1, asi1Δ (dilution 3) and ssy1Δ asi1Δ (dilution 4) mutants do not grow. Consistent with our previous findings (Figures 1B and 2A), the introduction of the dal81Δ null allele into the ssy1Δasi1Δ strain completely restored growth (dilution 5). However, the introduction of the dal81Δ into the asi1Δ strain possessing an intact and functional SPS sensor did not fully prevent AGP1 and GNP1 expression; weak growth was observed (dilution 6). This latter observation demonstrates that Dal81 is not absolutely essential for the expression of SPS sensor-regulated AAP genes under amino acid-inducing conditions. As expected, the introduction of null mutations in STP1 and STP2 conferred AzC resistance in an asi1Δdal81Δ mutant (dilution 7). Together, these results are consistent with the notion that Dal81 is strictly required for transcription of SPS sensor-regulated genes under conditions when there are low levels of Stp1 and Stp2 present in the nucleus, a situation that occurs in asi1Δ mutants lacking a functional SPS sensor or in asi1Δ mutants grown under noninducing conditions. In contrast, under amino acid-inducing conditions, the requirement for Dal81 is not strict, which indicates that when Stp1 and Stp2 are processed and efficiently targeted to the nucleus, the levels of these factors are sufficiently high to induce expression independently of Dal81. Thus, Dal81 appears to function analogously to an amplifier.
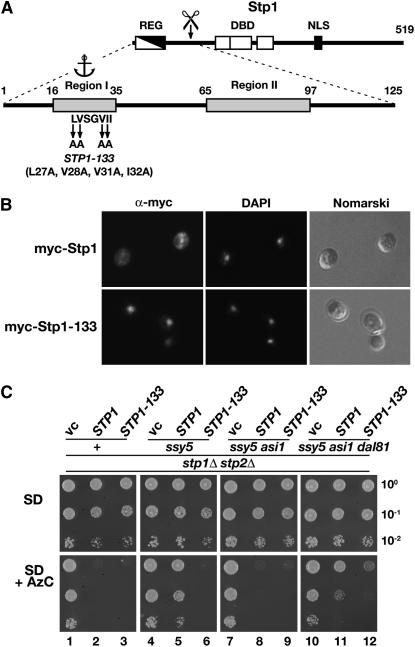
The constitutive STP1-133 mutation abolishes cytoplasmic retention and bypasses the strict requirement of Dal81 in SPS sensor-regulated gene expression:
The N-terminal regulatory domain of Stp1 possesses two conserved sequence motifs (Regions I and II) required to control the latent behavior of Stp1 (Figure 3A) (Andréasson and Ljungdahl 2004). When fused to the bacterial DNA binding protein lexA, the first 70 amino acids of the N terminus of Stp1 (encompassing the Region I motif) mediate both cytoplasmic retention and Asi1-dependent control (Boban et al. 2006). Distinct mutations within Region I (STP1-133) give rise to a constitutively active factor. The constitutive nature of this mutant allele is not due to enhanced processing (Andréasson and Ljungdahl 2004), thus, the STP1-133 mutations could either result in impaired cytoplasmic retention or loss of Asi1-dependent control. If the mutations impair cytoplasmic retention, we anticipated to find Stp1-133 constitutively accumulated in the nucleus. Alternatively, if the mutations specifically impair Asi1-dependent control and not cytoplasmic retention, then the bulk of Stp1-133 would remain in the cytoplasm and would not be found accumulated in the nuclei of uninduced cells. To distinguish between these two possibilities, we used epitope-tagged wild-type Stp1 and Stp1-133 constructs carrying the myc epitope at their extreme N termini and compared the intracellular localization of these constructs in cells grown in the absence of inducing amino acids. In cells expressing the wild-type myc-Stp1, we did not observe nuclear localized myc-dependent fluorescence (Figure 3B, top). In contrast, cells expressing myc-Stp1-133 displayed intense and highly focused fluorescence that colocalized with DAPI-stained DNA (Figure 3B, bottom). These results clearly demonstrate that the STP1-133 mutant allele gives rise to constitutive SPS sensor-regulated gene expression primarily because of impaired cytoplasmic retention of the mutant Stp1-133 protein.
Figure 3.—
The STP1-133 mutation abolishes cytoplasmic retention and enables SPS sensor-regulated gene expression in the absence of Dal81. (A) Schematic of Stp1. The location of the inhibitory regulatory domain (REG), the DNA binding domains (DBD), and putative nuclear localization sequence are depicted in the full-length Stp1 (519 aa). Conserved Region I (aa 16–35) containing sequences important for cytoplasmic retention (anchor) and Region II (aa 65–97) are indicated in the enlargement of the N-terminal domain (1–125 aa). The alanine substitution mutations of the STP1-133 allele are shown (Andréasson and Ljungdahl 2004). (B) Indirect immunolocalization of myc-Stp1 (pMB31) and myc-Stp1-133 (pMB45) in strain CAY60 (stp1Δ). Cells were grown in SD medium under noninducing conditions to an OD600 of 0.8 and fixed. Left to right: α-myc monoclonal antibody (9E11)-dependent Alexa Fluor 488 fluorescence; DAPI staining; and cells viewed by Nomarski optics. (C) Phenotypic analysis of the constitutive active STP1-133 allele and the dal81Δ null mutation. Plasmids pRS316 (vc), pMB10 (STP1), and pMB44 (STP1-133) were introduced into a stp1Δ stp2Δ mutant (+, CAY123) and into stp1Δ stp2Δ strains carrying ssy5Δ (MBY93), ssy5Δ asi1Δ (MBY102), and ssy5Δ asi1Δ dal81Δ (MBY124). The strains were grown on SC (-ura) medium. Aliquots of 10-fold serial dilutions in water were spotted on SD medium containing leucine (SD) and SD-containing leucine and 0.33 mM AzC (SD + AzC). Plates were incubated at 30°.
The finding that unprocessed Stp1-133 constitutively accumulates in the nuclei of noninduced cells prompted us to use the STP1-133 allele to more rigorously examine the requirement of Dal81 in the SPS-sensing pathway. An empty plasmid vector or plasmids carrying STP1-133 and STP1 were independently introduced into a stp1Δ stp2Δ strain (CAY123) and into stp1Δ stp2Δ strains carrying ssy5Δ (MBY93), ssy5Δ asi1Δ (MBY102), and ssy5Δ asi1Δ dal81Δ (MBY124) mutations. The strains were grown under amino acid-inducing conditions in absence and presence of AzC (Figure 3C). The strain lacking Stp1 and Stp2, but with an intact functional SPS sensor (+), grew in the presence of AzC; the introduction of either STP1 or STP1-133 restored AzC sensitivity (dilutions 1–3). Inactivation of SSY5, encoding the Stp1- and Stp2-processing protease (Abdel-Sater et al. 2004a; Andréasson et al. 2006), conferred AzC resistance in the cells expressing STP1 but not STP1-133 (dilutions 5 and 6, respectively). These findings clearly confirm that unprocessed full-length Stp1-133 escapes cytoplasmic retention and targets to the nucleus (Figure 3B), where it constitutively activates SPS sensor-regulated genes. Inactivation of ASI1 in the ssy5Δ strain enabled full-length Stp1 to gain access to promoters, resulting in AzC sensitivity (Figure 3C, dilution 8). Consistent with our previous results (Figure 2), introduction of the dal81Δ mutation in the ssy5Δ asi1Δ strain impaired Stp1-mediated gene expression, and AzC resistant growth was observed (compare dilution 11 with 8). In contrast, the same strain expressing the constitutive STP1-133 allele exhibited clear AzC sensitivity (compare dilution 12 with 10). These results are fully consistent with Dal81 playing an important role in the expression of SPS sensor-regulated genes, however, it is not stringently required for SPS sensor signaling under inducing conditions when abundant amounts of Stp1 and Stp2 are present in the nucleus. Importantly, the slight, but noticeable AzC resistance of the dal81Δ strain expressing the STP1-133 allele (compare dilution 12 with 9) supports the notion that Dal81 functions to amplify SPS sensor-induced signals.
Dal81 facilitates binding of both processed and latent forms of Stp1 to SPS sensor-regulated promoters:
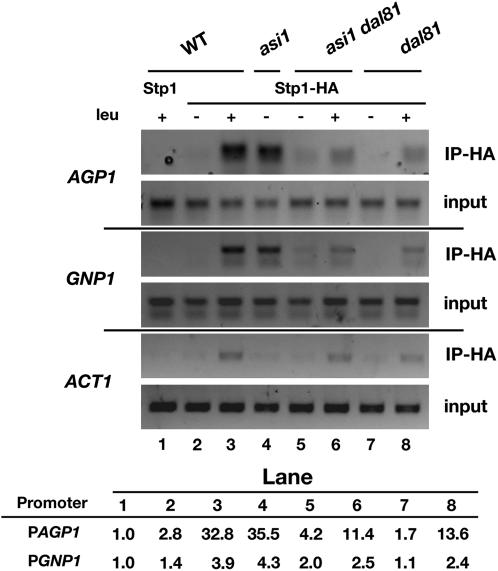
The finding that Dal81 is strictly required for SPS sensor-regulated gene expression under conditions when low levels of Stp1 and Stp2 enter the nucleus suggested that Dal81 may facilitate the binding of Stp1 and Stp2 to SPS sensor-regulated promoters. We used ChIP to examine this possibility by analyzing the association of Stp1 with two SPS sensor-regulated promoters AGP1 and GNP1. To facilitate the analysis, we used a plasmid encoding a Stp1 construct that carried an HA-epitope at the C terminus; this plasmid fully complements stp1Δ stp2Δ null mutant phenotypes, thus, the Stp1-HA protein is functional (see materials and methods). A plasmid encoding native Stp1 without an epitope tag was included in the experiment as a control for nonspecific immunoprecipitation, and the ability to amplify the ACT1 promoter was used to control the binding to nonspecific DNA sequences. Amino acid induction results in a slightly enhanced nonspecific immunoprecipitation of ACT1 promoter sequences; presumably this is a consequence of the greatly enhanced levels of nuclear localized Stp1 in induced cells (Andréasson and Ljungdahl 2002; Boban et al. 2006).
In wild-type cells, we readily detected the specific association of Stp1 with AGP1 and GNP1 promoters, but only after induction with amino acids and only in lysates prepared from cells expressing the HA-tagged construct (Figure 4, lanes 1–3). In lysates prepared from asi1Δ cells, anti-HA antibodies immunoprecipitated the AGP1 and GNP1 promoters even in cells grown in the absence of inducing amino acids (lane 4). This finding, consistent with our previously published results (Boban et al. 2006), indicates that in the absence of Asi1, full-length unprocessed Stp1 is able to gain access to SPS sensor-regulated promoters. This fully accounts for the constitutive expression of SPS sensor-regulated promoters observed in asi1Δ mutants. Strikingly, in lysates prepared from uninduced asi1Δ dal81Δ cells, the levels of immunoprecipitated AGP1 and GNP1 promoters were significantly decreased (compare lanes 4 and 5), clearly indicating that Dal81 is important for the association of Stp1 with SPS sensor-regulated promoters. Furthermore, the data show that Dal81 also facilitates the binding of processed Stp1; significantly lower levels of AGP1 and GNP1 promoters were immunoprecipitated from lysates prepared from amino acid-induced dal81Δ cells (compare lanes 6 and 8 with lane 3).
Figure 4.—
Dal81 is required for the efficient promoter binding of latent and processed forms of Stp1. ChIP analysis of Stp1 association with AGP1 and GNP1 promoters before and after amino acid-induced processing. Cultures of wild-type (WT; CAY60), asi1Δ (CAY150), asi1Δdal81Δ (MBY64), and dal81Δ (MBY79) strains carrying plasmid pMB10 (Stp1-HA) were grown in SD medium (− leu), and where indicated leucine was added 30 min prior to harvest (+ leu). Cell lysates were prepared and analyzed by ChIP using anti-HA antibody (3F10). Strain CAY60 (WT) carrying pCA029 (nontagged Stp1) was included to control nonspecific immunoprecipitation, and the ability to amplify the ACT1 promoter was assessed to control association with nonspecific DNA sequences. The size of amplified fragments are: AGP1, 246 bp; GNP1, 313 bp; and ACT1, 274 bp. The relative levels of immunoprecipitated promoter DNA are indicated (arbitrary units).
We note that the amounts of Stp1 found associated with AGP1 promoters (Figure 4) directly correlate with the observed AzC sensitivity of wild-type and mutant strains (Figures 1, 2, and 3C). Specifically, the lack of Stp1 binding in uninduced asi1Δ dal81Δ cells accounts for the AzC resistance of asi1Δ dal81Δ cells that lack a functional SPS sensor [ptr3Δ (Figures 1 and 2A); ssy1Δ (Figure 2B); and ssy5Δ (Figure 3C)]. The low levels of Stp1 associated with AGP1 promoters detected in amino acid-induced dal81Δ mutants (Figure 4, lanes 6 and 8) account for the reduced but still detectable AzC resistant growth (Figure 2A, dilution 3; Figure 2B dilution 6; Figure 3B, dilution 11). These results demonstrate that AzC sensitivity provides a highly sensitive measure of SPS sensor promoter activity, as barely detectable promoter binding (Figure 4, lanes 6 and 8) leads to clear AzC sensitivity (Figure 2B, dilution 6, Figure 3C, dilution 11).
Dal81 does not affect targeting of processed Stp1 to the nucleus:
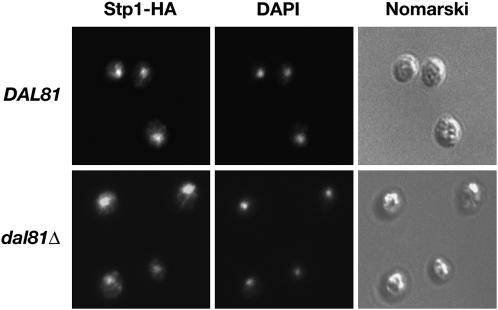
Since dal81Δ mutations greatly diminish the binding of both latent and processed forms of Stp1 to promoters of SPS sensor-regulated genes (Figure 4, lanes 5–8), we examined the possibility that the loss of Dal81 function interfered with mechanisms facilitating nuclear localization of Stp1. In this case, we anticipated that Stp1 would not target to the nucleus in dal81Δ mutants. Using immunofluorescence microscopy, we determined the intracellular location of processed Stp1 in wild-type (DAL81) and dal81Δ strains grown under inducing conditions in the presence of amino acids (Figure 5). In both strains, an intense and highly focused fluorescence, which colocalized with DAPI-stained DNA, was observed. These observations clearly demonstrate that Dal81 does not play a major role in nuclear targeting of Stp1.
Figure 5.—
Dal81 is not required for nuclear localization of Stp1. Indirect immunolocalization of Stp1-HA in DAL81 (CAY60) and dal81Δ (MBY79) cells was performed with anti-HA monoclonal antibodies. Strains carrying plasmid pMB31 (Stp1-HA) were grown in SD and induced 30 min with leucine. Left to right: α-HA monoclonal antibody-dependent Alexa Fluor 488 fluorescence; DAPI staining; and cells viewed by Nomarski optics.
DISCUSSION
Here, we used an unbiased genetic approach to identify components required for transcription mediated by unprocessed latent forms of Stp1 and Stp2 in asi1Δ mutants. We selected spontaneous mutations in RLS genes that suppressed the constitutive expression of SPS sensor-regulated genes in asi1Δ cells. We anticipated finding loss-of-function mutations in multiple genes, i.e., genes encoding transcriptional coactivators that function together with Stp1 or Stp2, but also mutations in genes encoding proteins facilitating nuclear import or stability of Stp1 and Stp2. However, we found that all rls mutations belonged to a single complementation group. The number of rls mutations analyzed strongly suggests that under the conditions used, the RLS screen is saturated. Our inability to identify a large set of RLS genes could be due to the extremely tight selection used that allowed isolation of only those mutations that completely abolish expression of AGP1 and GNP1. Alternatively, the processes governing the nuclear import or stability of Stp1 and Stp2 may be functionally redundant or may rely on proteins with essential functions required for cell viability.
RLS1 is identical to DAL81. DAL81 encodes a nuclear factor that pleiotropically contributes to the proper expression of multiple genes in at least three nitrogen assimilation and utilization pathways. Mutations in DAL81 prevent induction of genes involved in utilization of urea and allantoin (Jacobs et al. 1981; Turoscy and Cooper 1982; Coornaert et al. 1991) and GABA (Vissers et al. 1990). More recently, and consistent with our results presented here, it has been shown that Dal81 is required for full induction of amino acid-induced SPS sensor-dependent AAP gene expression (Iraqui et al. 1999; Bernard and André 2001; Abdel-Sater et al. 2004b). In all of these pathways, Dal81 functions together with an inducer-specific transcription factor to activate target genes via inducer-specific sequences (van Vuuren et al. 1991; Talibi et al. 1995; Iraqui et al. 1999; Bernard and André 2001; Abdel-Sater et al. 2004b). However, while inducer-specific factors Uga3 (Noel and Turcotte 1998; Idicula et al. 2002), Dal82 (André and Jauniaux 1990; Olive et al. 1991; Dorrington and Cooper 1993), and Stp1 and Stp2 (De Boer et al. 2000; Nielsen et al. 2001; Abdel-Sater et al. 2004b) have been found to directly bind to specific upstream-activating sequences, direct binding of Dal81 to these elements has not been demonstrated. Consistently, the deletion of the putative Zn (II)2Cys6 DNA binding domain of Dal81 has no effect on the induction of allantoin/urea and GABA utilization pathways (Bricmont et al. 1991).
Our results demonstrate that Dal81 is important, but not absolutely required for SPS sensor-regulated gene expression under conditions when Stp1 and Stp2 accumulate in the nucleus. The SPS sensor-regulated promoters remained partially active in induced dal81Δ cells (Figure 2B) or dal81Δ cells carrying the dominant STP1-133 allele (Figure 3C). Conversely, in the absence of Stp1 and Stp2, Dal81 by itself was unable to induce sufficient expression of AGP1 or GNP1 to confer AzC sensitivity (Figure 3). Together these findings indicate that Dal81 enhances signaling mediated by Stp1 and Stp2. Similarly, the UASGABA element from the UGA1 promoter is capable of supporting low levels of GABA-induced reporter activation in dal81Δ cells (Talibi et al. 1995), which suggests that Uga3 is also able to independently activate transcription of UGA1. Thus, Dal81 appears to have an important and synergistic role in amplifying the induced expression of genes in several well-characterized nitrogen source utilization pathways, i.e., urea and allantoin, GABA, and SPS sensor pathways.
We found that Dal81 facilitates the binding of Stp1 to SPS sensor-regulated promoters (Figure 4). Notably, the decreased association of Stp1 with promoters in dal81Δ mutants was not due to impaired nuclear targeting or accumulation (Figure 5). Similarly, the nuclear localization of Dal82 is not changed in a dal81Δ mutant (Scott et al. 2000). In previous work by others, it was shown that the induction of a lacZ reporter construct via a 21-bp UASaa element from the AGP1 promoter is abolished by deleting either STP1 or DAL81 (Abdel-Sater et al. 2004b). Clearly, Stp1 and Dal81 exert their function via the same regulatory sequences. On the basis of our ChIP analysis (Figure 4), Dal81 functions to amplify the sensitivity of SPS sensor-mediated signaling by increasing the efficiency of Stp1 and Stp2 binding to target promoters.
The finding that RLS1 is identical to DAL81 provides novel insights into the functional significance of the Asi backup system. The inactivation of the Asi system results in constitutive gene activation owing to the amplifying function of Dal81, which enables robust transcription even in the presence of low levels of nuclear localized Stp1 and Stp2. In fact, the levels of immunoprecipitated Stp1 associated with AGP1 and GNP1 promoters in uninduced asi1Δ cells are indistinguishable to those observed in induced wild-type cells (Figure 4) in which all of the detectable Stp1 and Stp2 is in the nucleus (Andréasson and Ljungdahl 2002; Boban et al. 2006). Conversely, in the absence of Dal81, Stp1 promoter binding was barely detected in uninduced asi1Δ cells (Figure 4). This striking observation suggests that Dal81 ensures high affinity binding of Stp1 to target promoters. Consistently, in dal81Δ mutants, the repressing activity of the Asi proteins is dispensable, demonstrating that without Dal81-dependent amplification, the levels of precursor forms of Stp1 and Stp2 that escape cytoplasmic retention are insufficient to activate transcription.
In summary, we have addressed how the SPS-sensing pathway provides the proper balance between two opposing parameters that influence accurate transcriptional responses, i.e., the ability to promote gene expression in a highly specific and sensitive manner vs. the need to prevent gene activation in the absence of inducing signals. Dal81 contributes greatly to the sensitivity and responsiveness of amino acid-induced gene activation by amplifying signals, whereas, the inner nuclear membrane Asi proteins ensure that SPS sensor-regulated genes are expressed only after amino acid-dependent processing of Stp1 and Stp2. The fact that, in the presence of Dal81, only low levels of Stp1 and Stp2 can mediate what amounts to a fully induced state underscores the importance of regulatory mechanisms that negatively modulate gene expression.
Acknowledgments
We thank the other members of Ljungdahl laboratory for constructive comments throughout the course of this work. This research was supported by the Ludwig Institute for Cancer Research and the Swedish Research Council.
References
- Abdel-Sater, F., M. El Bakkoury, A. Urrestarazu, S. Vissers and B. André, 2004. a Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol. Cell Biol. 24: 9771–9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Sater, F., I. Iraqui, A. Urrestarazu and B. André, 2004. b The external amino acid signaling pathway promotes activation of Stp1 and Uga35/Dal81 transcription factors for induction of the AGP1 gene in Saccharomyces cerevisiae. Genetics 166: 1727–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André, B., and J. C. Jauniaux, 1990. Nucleotide sequence of the DURM gene coding for a positive regulator of allophanate-inducible genes in Saccharomyces cerevisiae. Nucleic Acids Res. 18: 7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson, C., and P. O. Ljungdahl, 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16: 3158–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson, C., and P. O. Ljungdahl, 2004. The N-terminal regulatory domain of Stp1p is modular and fused to an artificial transcription factor confers full SPS-sensor control. Mol. Cell. Biol. 24: 7503–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson, C., E. P. A. Neve and P. O. Ljungdahl, 2004. Four permeases import proline and the toxic proline analogue azetidine-2-carboxylate into yeast. Yeast 21: 193–199. [DOI] [PubMed] [Google Scholar]
- Andréasson, C., S. Heessen and P. O. Ljungdahl, 2006. Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 20: 1563–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi, A., and G. R. Fink, 1992. The yeast Ca(2+)-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell 3: 633–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, F., and B. André, 2001. Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. Mol. Microbiol. 41: 489–502. [DOI] [PubMed] [Google Scholar]
- Boban, M., A. Zargari, C. Andréasson, S. Heessen, J. Thyberg et al., 2006. Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors. J. Cell Biol. 173: 695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricmont, P. A., J. R. Daugherty and T. G. Cooper, 1991. The DAL81 gene product is required for induced expression of two differently regulated nitrogen catabolic genes in Saccharomyces cerevisiae. Mol. Cell Biol. 11: 1161–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivanlou, A. H., and J. E. Darnell, Jr., 2002. Signal transduction and the control of gene expression. Science 295: 813–818. [DOI] [PubMed] [Google Scholar]
- Burke, B., and C. L. Stewart, 2002. Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell Biol. 3: 575–585. [DOI] [PubMed] [Google Scholar]
- Burke, D., D. Dawson and T. Stearns, 2000. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Sping Harbor, NY.
- Connelly, C., and P. Hieter, 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86: 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert, D., S. Vissers and B. André, 1991. The pleiotropic UGA35(DURL) regulatory gene of Saccharomyces cerevisiae: cloning, sequence and identity with the DAL81 gene. Gene 97: 163–171. [DOI] [PubMed] [Google Scholar]
- De Boer, M., P. S. Nielsen, J. P. Bebelman, H. Heerikhuizen, H. A. Andersen et al., 2000. Stp1p, Stp2p and Abf1p are involved in regulation of expression of the amino acid transporter gene BAP3 of Saccharomyces cerevisiae. Nucleic Acids Res. 28: 974–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrington, R. A., and T. G. Cooper, 1993. The DAL82 protein of Saccharomyces cerevisiae binds to the DAL upstream induction sequence (UIS). Nucleic Acids Res. 21: 3777–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr, S. D., A. Vassarotti, J. Garrett, B. L. Geller, M. Takeda et al., 1986. The amino terminus of the yeast F1-ATPase beta-subunit precursor functions as a mitochondrial import signal. J. Cell Biol. 102: 523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg, H., and P. O. Ljungdahl, 2001. a Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21: 814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg, H., and P. O. Ljungdahl, 2001. b Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40: 91–109. [DOI] [PubMed] [Google Scholar]
- Forsberg, H., M. Hammar, C. Andréasson, A. Moliner and P. O. Ljungdahl, 2001. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics 158: 973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Idicula, A. M., G. L. Blatch, T. G. Cooper and R. A. Dorrington, 2002. Binding and activation by the zinc cluster transcription factors of Saccharomyces cerevisiae. Redefining the UASGABA and its interaction with Uga3p. J. Biol. Chem. 277: 45977–45983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqui, I., S. Vissers, F. Bernard, J. O. de Craene, E. Boles et al., 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell Biol. 19: 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, E., E. Dubois, C. Hennaut and J. M. Wiame, 1981. Positive regulatory elements involved in urea amidolyase and urea uptake induction in Saccharomyces cerevisiae. Curr. Genet. 4: 13–18. [DOI] [PubMed] [Google Scholar]
- Jørgensen, M. U., M. B. Bruun, T. Didion and M. C. Kielland-Brandt, 1998. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast 14: 103–114. [DOI] [PubMed] [Google Scholar]
- Kippert, F., 1995. A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol. Lett. 128: 201–206. [DOI] [PubMed] [Google Scholar]
- Klasson, H., G. R. Fink and P. O. Ljungdahl, 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell Biol. 19: 5405–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, P. S., B. van den Hazel, T. Didion, M. de Boer, M. Jørgensen et al., 2001. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol. Gen. Genet. 264: 613–622. [DOI] [PubMed] [Google Scholar]
- Noel, J., and B. Turcotte, 1998. Zinc cluster proteins Leu3p and Uga3p recognize highly related but distinct DNA targets. J. Biol. Chem. 273: 17463–17468. [DOI] [PubMed] [Google Scholar]
- Olive, M. G., J. R. Daugherty and T. G. Cooper, 1991. DAL82, a second gene required for induction of allantoin system gene transcription in Saccharomyces cerevisiae. J. Bacteriol. 173: 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, S., R. Dorrington, V. Svetlov, A. E. Beeser, M. Distler et al., 2000. Functional domain mapping and subcellular distribution of Dal82p in Saccharomyces cerevisiae. J. Biol. Chem. 275: 7198–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger, S., A. Hecht, K. Luo and M. Grunstein, 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11: 83–93. [DOI] [PubMed] [Google Scholar]
- Talibi, D., M. Grenson and B. André, 1995. Cis- and trans-acting elements determining induction of the genes of the gamma-aminobutyrate (GABA) utilization pathway in Saccharomyces cerevisiae. Nucleic Acids Res. 23: 550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, C. M., A. J. Koleske, D. M. Chao and R. A. Young, 1993. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73: 1361–1375. [DOI] [PubMed] [Google Scholar]
- Turoscy, V., and T. G. Cooper, 1982. Pleiotropic control of five eucaryotic genes by multiple regulatory elements. J. Bacteriol. 151: 1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vuuren, H. J., J. R. Daugherty, R. Rai and T. G. Cooper, 1991. Upstream induction sequence, the cis-acting element required for response to the allantoin pathway inducer and enhancement of operation of the nitrogen-regulated upstream activation sequence in Saccharomyces cerevisiae. J. Bacteriol. 173: 7186–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers, S., B. André, F. Muyldermans and M. Grenson, 1989. Positive and negative regulatory elements control the expression of the UGA4 gene coding for the inducible 4-aminobutyric-acid-specific permease in Saccharomyces cerevisiae. Eur. J. Biochem. 181:357–361. [DOI] [PubMed] [Google Scholar]
- Vissers, S., B. André, F. Muyldermans and M. Grenson, 1990. Induction of the 4-aminobutyrate and urea-catabolic pathways in Saccharomyces cerevisiae. Specific and common transcriptional regulators. Eur. J. Biochem. 187: 611–616. [DOI] [PubMed] [Google Scholar]
- Wu, B., K. Ottow, P. Poulsen, R. F. Gaber, E. Albers et al., 2006. Competitive intra- and extracellular nutrient sensing by the transporter homologue Ssy1p. J. Cell Biol. 173: 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zargari, A., M. Boban, S. Heessen, C. Andréasson, J. Thyberg et al., 2007. Inner nuclear membrane proteins Asi1, Asi2 and Asi3 function in concert to maintain the latent properties of transcription factors Stp1 and Stp2. J. Biol. Chem. 282: 594–605. [DOI] [PubMed] [Google Scholar]