Abstract
Predicting the chromosomal location of mapped markers has been difficult because linkage maps do not reveal differences in crossover frequencies along the physical structure of chromosomes. Here we combine a physical crossover map based on the distribution of recombination nodules (RNs) on Solanum lycopersicum (tomato) synaptonemal complex 1 with a molecular genetic linkage map from the interspecific hybrid S. lycopersicum × S. pennellii to predict the physical locations of 17 mapped loci on tomato pachytene chromosome 1. Except for one marker located in heterochromatin, the predicted locations agree well with the observed locations determined by fluorescence in situ hybridization. One advantage of this approach is that once the RN distribution has been determined, the chromosomal location of any mapped locus (current or future) can be predicted with a high level of confidence.
WHILE linkage maps accurately describe gene order and the amount of crossing over between genes, they are less useful for predicting the physical locations of genes on chromosomes. This is because the frequency of crossing over along the length of chromosomes is not uniform, and as a result loci that are physically far apart on chromosomes can be nearby on linkage maps and vice versa (Sturtevant and Beadle 1939; Khush and Rick 1968; Sherman and Stack 1995; Zhong et al. 1999; Islam-Faridi et al. 2002; Budiman et al. 2004). Such discrepancies are impediments to applying linkage maps to guide genome sequence assembly or for gene discovery by chromosome walking (Budiman et al. 2004).
A variety of cytogenetic techniques have been used to determine the position of genes on chromosomes without relying on linkage maps. Widely used techniques include (1) cytogenetic analysis of chromosome duplications, deletions, and rearrangements (Khush and Rick 1968; Berger 2004); (2) genetic analysis of radiation hybrids whereby genes are assigned to particular chromosomal fragments on the basis of presence vs. absence tests (Hudson et al. 2001; Kynast et al. 2004); and (3) fluorescence in situ hybridization (FISH) involving hybridization of labeled DNA fragments to intact chromosomes to show the positions of complementary sequences (Fransz et al. 1996; De Jong et al. 1999; Jackson et al. 2000). While these techniques have been used to localize many genes on chromosomes, they are limited by special requirements, and they are not high throughput.
Recently, Anderson et al. (2004) reported a new approach for locating mapped loci on maize pachytene chromosomes. This technique depends on first mapping the frequency and location of recombination nodules (RNs) on synaptonemal complexes (SCs, pachytene bivalents). Assuming that each RN represents a crossover (Carpenter 1975; Herickhoff et al. 1993; Sherman and Stack 1995; Pigozzi and Solari 1999; Marcon and Moens 2003), variation in the frequency and distribution of RNs on SCs represents the variation in recombination rates along pachytene chromosomes. RN distributions can be converted to centimorgan (cM) distributions, thereby creating an RN–cM recombination map for each pachytene chromosome (Anderson et al. 2004). By relating crossover frequency to chromosome structure, the RN–cM map provides a means to predict the positions of mapped loci on pachytene chromosomes. While this approach also has special requirements, the advantage is that once the RN–cM map is made, the chromosomal positions of all mapped loci can be predicted easily, along with any additional markers placed on the genetic map in the future. The accuracy of this approach for maize was tested for markers on chromosome 9 by comparing the predicted chromosomal locations of loci from the UMC98 linkage map (Davis et al. 1999) with their observed physical locations determined independently by FISH. The correlation between the predicted and observed chromosomal locations was very strong (r2 = 0.996), indicating that most, if not all, loci on the linkage map can be positioned accurately on maize SCs using the RN–cM map [see the Morgan2McClintock Translator (http://www.lawrencelab.org/Morgan2McClintock) that uses maize and tomato RN–cM map data to predict the location of mapped loci on maize and tomato pachytene chromosomes, respectively (Lawrence et al. 2006)].
RN mapping data comparable to that in maize have been available for some time in tomato (Solanum lycopersicum var. Cherry; Sherman and Stack 1995), and in their study of the relationship of recombination rates and genome evolution, Stephan and Langley (1998) used these data to predict whether certain mapped loci are in areas of high or low recombination. However, the accuracy of positioning mapped loci on tomato pachytene chromosomes using the distribution of RNs has not been tested. One (potentially) complicating factor for tomato compared to maize is that cultivated tomato varieties are highly inbred with few polymorphisms, so the most complete molecular linkage map (EXPEN 2000) is based on an interspecific hybrid between tomato and S. pennellii (Tanksley et al. 1992; http://www.sgn.cornell.edu). Here we use the tomato RN–cM map in combination with the tomato EXPEN 2000 molecular linkage map to predict the locations of 17 mapped loci on pachytene chromosome 1 and compare these with the positions of the mapped loci determined independently by FISH.
MATERIALS AND METHODS
Recombination map for tomato SC 1 based on the frequency and distribution of RNs:
See Sherman and Stack (1995) for a more complete description of map construction. The average absolute length (26.8 μm) and arm ratio (3.00) for SC 1 were taken from Sherman and Stack (1992), as adjusted by Peterson et al. (1996). Of the original 457 SC 1's analyzed by Sherman and Stack (1995), the data for 51 were corrupted and the data from an additional 141 SCs were eliminated because the arm ratios were outside the range allowed (2.7–3.3) for SC 1, leaving a total of 265 SC 1's for use in producing the RN–cM map used in this study. The new RN–cM map for SC 1 is slightly shorter than the original (122.9 cM vs. 124 cM), but otherwise hardly changed in the distribution of recombination events. These data as well as RN–cM data for all tomato SCs and SC arms are available as supplemental data at http://www.genetics.org/supplemental/. To make the RN–cM map, SC 1 was divided into 0.2-μm bins (except around the centromere where the two bins on either side were 0.3 μm). The number of RNs in each bin was multiplied by 50 (50 cM/RN, Sherman and Stack 1995), and this product was divided by the total number of SCs observed (265) to obtain the centimorgan (map unit) value for each 0.2-μm bin. The bins were summed along the length of the SC starting from the tip of the short arm to obtain a cumulative RN–cM map that correlates centimorgan distances with physical positions on SC 1.
Adjusting the EXPEN 2000 molecular linkage map to the size of the RN–cM map:
Seventeen loci on SC 1 were selected from the EXPEN 2000 molecular linkage map [that was downloaded from the SOL Genomics Network web site (http://www.sgn.cornell.edu) on January 9, 2007]. Because the total EXPEN 2000 map length for chromosome 1 differs from the corresponding RN–cM map length (Table 1), the EXPEN 2000 map positions were adjusted proportionally for each arm to fit the length of the RN–cM map following the procedures described by Anderson et al. (2004). These adjusted values are now termed the RN–cM values for each marker (Table 1).
TABLE 1.
Genetic and cytological characteristics for chromosome (SC) 1
| Genetic length (cM)
|
|||
|---|---|---|---|
| Arm | Length (μm) | RN–cM | EXPEN 2000a |
| Short | 6.7 | 26.9 | 32.7 |
| Long | 20.1 | 96.0 | 132.3 |
| Total | 26.8 | 122.9 | 165 |
Based on SSR266 FISH localization near centromere.
Predicting the chromosomal positions of genetically mapped loci:
The chromosomal position of each genetically mapped locus was predicted with and without reference to the RN–cM map. For the RN–cM predictions, the RN–cM value for each locus was matched to the corresponding chromosomal location on the RN–cM map. For EXPEN 2000 predictions in short arms, the linkage (centimorgan) position of each marker was divided by the centimorgan length of the arm to obtain a percentage value, and that percentage value was multiplied by the average length of the SC 1 short arm in micrometers to yield the physical location of the marker measured from the tip of the short arm. EXPEN 2000 predictions in long arms were determined similarly except the centimorgan length of the short arm was subtracted from the centimorgan positions of the marker and measurements were made from the kinetochore (centromere).
Growing plants for SC spreads:
The same line of tomatoes (Solanum lycopersicum var. Cherry) that was used to prepare the RN–cM map (Sherman and Stack 1995) was grown from seed and maintained as before in a controlled temperature greenhouse with supplemental lighting.
Preparation of SC spreads for FISH:
Spreads of tomato SCs were prepared for FISH generally as described by Peterson et al. (1999) with some modifications. Briefly, when a tomato anther was confirmed to contain primary microsporocytes in pachytene by an aceto-orcein squash, the remaining anthers in the bud were transferred into a depression slide containing 200 μl of aqueous digestion medium [0.56 mm monobasic potassium phosphate, 0.2% potassium dextran sulfate, 1 mm CaCl2, 0.1 mm (acid) PIPES, 0.7 m mannitol, 1% polyvinyl pyrrolidone, pH 5.1] and 3 mg of desalted cytohelicase. Anthers were bisected and their contents squeezed out with dissecting needles. After a 10 min digestion, rods of protoplasts were drawn into a micropipet for a total volume of no more than 0.5 μl of protoplast suspension. The protoplast suspension was expelled into 10 μl of aqueous bursting medium (0.05% Nonidet P-40, 0.1% BSA, 0.3% formaldehyde (CHOH), 0.001% potassium dextran sulfate) at the end of a plastic pipet tip, and the droplet was placed on a cleaned, glow-discharged glass slide. Ten microliters of bursting medium was added, and then the slide was taken to a hood where it was sprayed with 30 sweeps from a nebulizer containing aqueous 4% CHOH, pH 8.5. The slides were allowed to air dry, briefly washed in water, air dried again, and stored at −80°. Slides were scanned by phase microscopy to record the location of good SC spreads before FISH. These SC spreads are essentially the same as those made on plastic-coated slides for RN mapping by electron microscopy (Sherman and Stack 1995)
Preparation of Cot 100 nuclear DNA for use in chromosomal in situ suppression hybridization:
Nuclear DNA was isolated from etiolated tomato seedlings (Peterson and Stack 1997). The DNA was sheared to ∼500 bp by sonication, denatured, and renatured to Cot 100 (Zwick et al. 1997; Chang 2004). Cot 100 DNA was selected because it should include most of the repetitive sequences in the tomato genome (Peterson et al. 1999). The remaining single-stranded DNA was digested with S1 nuclease. Double-stranded Cot 100 DNA was isolated from this digest using chloroform–isoamyl alcohol and then precipitated with ethanol. After brief air drying, the DNA was resuspended in distilled water.
FISH:
Bacterial artificial chromosomes (BACs) containing each of the 17 mapped loci from the tomato HindIII BAC library (http://www.sgn.cornell.edu) were isolated by standard protocols and labeled by nick translation with either biotin or digoxygenin (DIG) using the manufacturer's instructions (Roche).
FISH was performed as described (Zhong et al. 1996; Chang 2004) with the following modifications. Glass microscope slides with SC spreads were incubated for 1 min each in 45% acetic acid, freshly prepared 1:3 acetic ethanol, and 100% ethanol before air drying for 30 min at 65°. Slides were then treated for 1 hr at 37° with 100 μg/ml RNase A in 2× SSC, washed in 2× SSC, washed in 0.01 n HCl, incubated for 6 min in 5 μg/ml of pepsin in 0.01 n HCl, washed in water, and fixed for 10 min in 1% formalin in PBS plus 50 mm MgCl2. Subsequently, slides were washed in 2× SSC and dehydrated through an alcohol series before drying in warm air.
The hybridization mixture (20 μl/slide), consisting of 1–50 ng of one or more labeled (probe) DNAs per slide, Cot 100 DNA at 50–100× probe DNA amount, 50% formamide, 10% sodium dextran sulfate, 0.25% SDS, and 2× SSC, was placed on slides with SC spreads, and cover glasses were added. The slides were heated for 2.5 min on a hot block at 80° to denature the DNA and then incubated horizontally in a moist chamber for at least 8 hr at 37°. (All steps where temperature is not specified were at room temperature.) After removal of cover glasses in 2× SSC and two 5-min washes with 2× SSC, the slides were washed three times for 5 min each in 50% formamide in 2× SSC at 42° (80% stringency). Blocking and antibody incubations were performed in 1-hr increments at 37° as described (Zhong et al. 1996; Chang 2004). The antibodies differed depending on the probe label and included (in this order but not necessarily in each experiment) mouse anti-biotin 1:100 (Roche), biotinylated donkey anti-mouse 1:250 (Jackson ImmunoResearch) and/or sheep anti-DIG tetramethyl rhodamine (TRITC) 1:125 (Roche), and streptavidin–FITC 1:250 (Roche) and/or donkey anti-sheep–TRITC 1:100 (Jackson ImmunoResearch). Slides were dehydrated through an ethanol series, dried, and cover glasses were mounted using 15 μl of Vectashield containing 5 μg/ml of DAPI.
Microscopy:
Microscopy and photography were performed with a Olympus Provis microscope equipped for phase and fluorescence microscopy using DAPI, FITC, and TRITC filter cubes with zero pixel shift. A cooled Optronics black and white camera was used for photography. Images (8-bit) were artificially colored and merged using Picture Frame 2.3.
Determining the positions of BACs on SC 1:
Before slides were used for FISH, good spreads of SCs were photographed with phase contrast illumination, and stage coordinates were recorded. After FISH, the same SC spreads were rephotographed, and the phase and fluorescent images were merged. SC lengths and kinetochore positions were measured from the phase images using the computer program MicroMeasure (http://www.biology.colostate.edu/MicroMeasure). SC 1 as well as most of the other 11 tomato SCs can be readily identified on the basis of relative length and arm ratios (Sherman and Stack 1992). Positions of fluorescent foci, indicating BAC locations, were measured from kinetochores on the merged images and converted to percentages of arm lengths. The average percentage location of each BAC was multiplied by the average SC 1 arm length (in micrometers) to position the BAC on the model (average) SC 1 (Sherman and Stack 1992; Peterson et al. 1996). The positions and euchromatin/heterochromatin boundaries were taken from Sherman and Stack (1992; also see http://www.sgn.cornell.edu/cview/map.pl?map_id=13). Statistics were performed with Minitab version 4.
RESULTS
Marker selection:
Seventeen BACs that contain markers spanning the length of the EXPEN 2000 linkage map for chromosome 1 were selected for FISH localization (Table 2, Figure 1, http://www.sgn.cornell.edu). Thirteen markers had been mapped with high confidence (LOD = 3), while the other 4 markers were mapped with lower confidence (LOD ≤ 2). The latter markers were chosen in spite of their low LOD scores because there are only a few markers in the regions of the linkage map where they are found.
TABLE 2.
Chromosome 1 markers from the EXPEN 2000 map
| Marker position (cM)
|
Location on SC 1 (μm) from tip of short arm and as % of arm length from centromereb
|
||||||
|---|---|---|---|---|---|---|---|
| Marker | BACa | LOD | EXPEN 2000 | Adjusted to RN–cM map | Observed (FISH) | Predicted (RN–cM) | Predicted (EXPEN 2000) |
| C2_At5g06370 | 130I12 | 2 | 18.5 | 15.1 | 0.9 | 2.0 | 3.8 |
| 87S | 70S | 43S | |||||
| CT87 | 069E17 | 2 | 21.7 | 17.7 | 1.1 | 2.2 | 4.4 |
| 84S | 67S | 34S | |||||
| T1650 | 262O22 | 3 | 22 | 17.9 | 1.3 | 2.2 | 4.5 |
| 81S | 67S | 33S | |||||
| T1619 | 003D15 | 3 | 29 | 23.6 | 2.4 | 2.8 | 5.9 |
| 64S | 58S | 12S | |||||
| TG378c | 252G05 | 2 | 31.9 | 26 | 9.1 | 4.4 | 6.5 |
| 12L | 34S | 3S | |||||
| SSR266 | 054N01 | 2 | 32.7 | 26.9 | 6.8 | 6.7 | 6.7 |
| 0.5L | 0C | 0C | |||||
| T1957 | 095K03 | 3 | 34 | 27.6 | 11.5 | 10 | 6.9 |
| 24L | 16L | 1L | |||||
| T1704 | 305F14 | 3 | 39 | 31.3 | 11.9 | 11.4 | 7.7 |
| 26L | 23L | 5L | |||||
| T0825 | 155M04 | 3 | 46 | 36.4 | 14.3 | 12.4 | 8.7 |
| 38L | 28L | 10L | |||||
| cLET-5-J13 | 329A12 | 3 | 52 | 40.7 | 16.5 | 13.2 | 9.6 |
| 49L | 32L | 14L | |||||
| TG460 | 208M24 | 3 | 70 | 53.8 | 17.8 | 14.8 | 12.4 |
| 55L | 40L | 28L | |||||
| cLET-7-E12 | 108J06 | 3 | 88 | 66.9 | 18.4 | 17.0 | 15.1 |
| 58L | 51L | 42L | |||||
| C2_At2g3870 | 309D12 | 3 | 92.5 | 70.2 | 19.2 | 17.4 | 15.8 |
| 62L | 53L | 45L | |||||
| T1488 | 051C15 | 3 | 109 | 82.2 | 20.4 | 19.8 | 18.3 |
| 68L | 65L | 58L | |||||
| T1109 | 245N21 | 3 | 140 | 104.7 | 23.6 | 24.2 | 23.0 |
| 84L | 87L | 81L | |||||
| C2_At2g15890 | 008L19 | 3 | 150 | 112.0 | 25.0 | 25.0 | 24.5 |
| 91L | 91L | 89L | |||||
| T1306 | 088L02 | 3 | 165 | 122.9 | 26.7 | 26.4 | 26.8 |
| 100L | 98L | 100L | |||||
Each BAC number is prefaced by Le_HBa for the HindIII library number assignment.
Marker position as fractional length of short (S) or long (L) arm from centromere (C) equals centiMcClintocks.
Marker mapped to wrong arm.
Figure 1.—
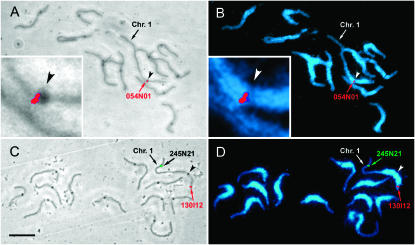
Tomato pachytene SC spreads viewed by phase contrast with superimposed FISH (A and C) and by UV illumination after DAPI staining and superimposed FISH (B and D). The dark spots observed on each SC by phase contrast are kinetochores, and the kinetochore of chromosome 1 is indicated in each image by an arrowhead. Pericentromeric heterochromatin stains more brightly with DAPI than distal euchromatic portions of the chromosomes. The locations of three BACs—054N01 (A and B, red signal), 0245N21 (C and D, green signal), and 130I12 (C and D, red signal) on chromosome 1—are indicated by arrows and text. The centromeric region of chromosome 1 has been enlarged (insets, A and B) to show that the FISH signal for 054N01 (marker SSR266) is located close to the centromere. Bar (A–D), 10 μm.
Predicting the cytological positions of genetically mapped loci on SC 1:
The first step in predicting the cytological positions of genetically mapped loci on SC 1 was to adjust the length of the EXPEN 2000 map to match that of the RN–cM map (see materials and methods and Tables 1 and 2). This adjustment is optimal when the position of the centromere is known on the linkage map and the adjustments can be made for each arm rather than for the whole SC (Anderson et al. 2004). One BAC (P054N01), containing the SSR266 marker at 32.7 cM on the EXPEN 2000 map, was observed by FISH to be located very near the centromere (Figure 1, A and B). Because there is essentially no crossing over in the heterochromatin nearest the centromere (Sherman and Stack 1995; Figure 2), we used 32.7 cM as the linkage map position of the centromere.
Figure 2.—
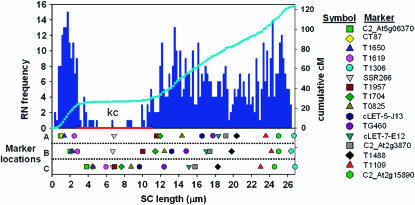
Comparing predicted and observed marker positions on chromosome 1. (Top) The bar graph shows the distribution of RNs in 0.2-μm intervals along SC 1 that is represented on the x-axis. The short arm is to the left, pericentric heterochromatin is marked with a red bar, and the position of the kinetochore (equals centromere) is marked by “kc.” The light blue line superimposed on the RN distribution represents the cumulative cM value for each interval along the chromosome. (Bottom) The chromosomal positions of markers as observed by FISH (row A), predicted from the RN–cM map (row B), and predicted from the EXPEN 2000 map (row C) are shown. The same markers are shown in each row with a symbol with the same color and shape. The gray inverted triangle represents the SSR266 marker that is located near or at the centromere (see key on the right). The clustering of markers near the centromere in row C is typical of genetic maps in areas of low recombination.
The physical (micrometers) positions of markers on SC 1 were predicted in two ways (Figure 2, Table 2). One method (EXPEN 2000 predictions) assumed that crossing over was even along the length of the chromosome. Here, markers from the EXPEN 2000 linkage map were superimposed directly on SC 1 on the basis of corresponding percentage values of genetic and physical arm length. The other method (RN–cM predictions) used the RN–cM map to adjust for variation in crossover frequency along the SC. Here, markers were positioned on the chromosomes by matching the RN–cM value of the marker to the location of the centimorgan value on the RN–cM map (Figure 2).
Comparing predicted loci positions to those observed by FISH:
Of the 17 BACs localized on SC 1 by FISH, the BAC containing the TG378 marker was located by FISH to the long arm although the marker had been genetically mapped to the short arm. TG378 was mapped with rather low confidence (LOD = 2), and the discrepancy could have been due to its presence in pericentric heterochromatin where recombination is suppressed and mapping is difficult (Figure 2). In any case, the TG378 marker was not included in Figure 2 and was not included in the data used for regression analysis.
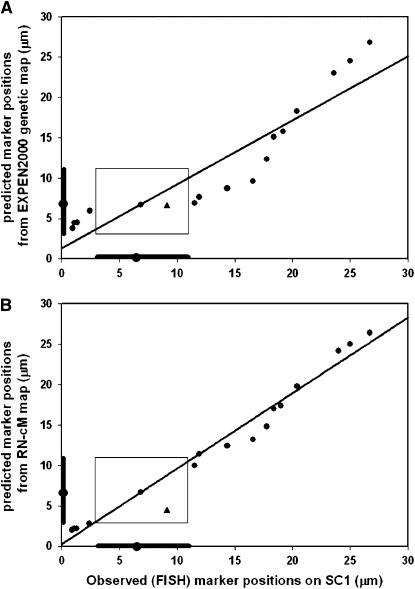
Regression analysis revealed that positions of loci determined by FISH and positions of loci predicted by using the EXPEN 2000 linkage map, i.e., without regard to the RN–cM map, agree well (Figure 3A; y = 0.792x + 1.35; r2 = 0.849). This is at least partially due to the fact that predicted and observed loci are in the same order within the confines of the pachytene chromosome. When the positions of loci determined by FISH were compared with their predicted positions using the RN–cM map, the fit is significantly better and close to the theoretical expectation of perfect fit with a slope of 1 and an intercept of 0 (Figure 3B; y = 0.931x + 0.29; r2 = 0.978). Another approach to judging the accuracy of the two methods of prediction is a comparison of the physical distance between the predicted locations and the observed (FISH) locations on the pachytene chromosome. The average distance between observed positions of loci and EXPEN 2000 predictions is 3.1 ± 2.0 μm compared to 1.1 ± 1.0 μm for RN–cM predictions. Converting these SC distances to DNA amounts on the basis of an estimate of 1.55 Mb/μm of SC per chromatid in euchromatin (Peterson et al. 1996) gives average differences between observed and predicted positions of 4.8 Mb (3.1 μm × 1.55 Mb/μm) for EXPEN 2000 predictions and 1.7 Mb (1.1 μm × 1.55 Mb/μm) for RN–cM predictions. For both predictions, discrepancies between the predicted and observed locations were particularly obvious for markers in low recombination regions near the centromere (kinetochore, Figure 2).
Figure 3.—
Graphs comparing the observed marker locations by FISH with those predicted from the genetic map (A) and those predicted from the RN–cM map (B). The locations of pericentric heterochromatin and the centromere are indicated by darker bars and a circle, respectively, along the x- and y-axes of each graph. The rectangle in each graph indicates the physical location of pericentric heterochromatin. The TG378 marker is indicated by a triangle and was not included in the regression analyses because it was mapped to the wrong arm. Regression equations are (A) y = 0.792x + 1.35, r2 = 0.849; (B) y = 0.931x + 0.29, r2 = 0.978.
DISCUSSION
Using the EXPEN 2000 molecular linkage map in combination with the RN–cM map for tomato, we have predicted the location of 17 mapped loci on pachytene chromosome 1 and found a close correspondence to locations demonstrated by BAC–FISH. This is similar to the result obtained in maize (Anderson et al. 2004), and together the two studies indicate that the location of mapped loci can be accurately predicted on pachytene chromosomes of angiosperms and probably any organism if the following conditions are met: (1) pachytene chromosomes are individually recognizable, (2) there is a saturated linkage map, and (3) there is an RN–cM map. The last requirement is the most problematic since few RN–cM maps have been prepared. However, maps of Mlh1 fluorescent foci on pachytene chromosomes (Froenicke et al. 2002) are a reasonable substitute in mammals and birds because in these animals Mlh1 foci correspond to RNs (Barlow and Hultén 1998; Anderson et al. 1999; Pigozzi 2001; Marcon and Moens 2003). On the other hand, this does not seem to be case in tomato where Lhuissier et al. (2007) report 30% fewer Mlh1 foci than RNs on pachytene chromosomes.
In principle, the predicted locations of mapped sequences on tomato pachytene chromosomes should exactly match observed locations by FISH. However, some markers, especially those located more proximally in the euchromatin of the long arm of tomato chromosome 1, differ more from their predicted locations (Figures 2 and 3B). This must be due to differences between the molecular map and the RN–cM map. Indeed, the tomato EXPEN 2000 molecular map is longer than the RN–cM map for the whole genome (1460 cM vs. 1086.5 cM, respectively) as well as for chromosome 1 (165 cM vs. 122.9 cM, respectively). This was compensated for by proportionally reducing arm lengths on the molecular map to make them the same length as arms on the RN–cM map. However, this mathematical reduction was applied uniformly along both arms of SC 1, and it is unlikely that the discrepancies between the two maps would be consistently distributed proportionally along the lengths of the chromosome arms. In addition, there are obvious differences in the techniques used to create the EXPEN 2000 and the RN–cM maps, and Sherman and Stack (1995) pointed out other factors that could contribute to differences in crossover patterns, including dissimilar crossover patterns in primary microsporocytes vs. megasporocytes, misclassification of phenotypes, problems interpreting map distances involving two or more crossovers between markers, incorrect marker order, unknown rates of gene conversion, and missing RNs. We do not know to what extent any of these factors contribute to differences between the tomato molecular and RN–cM maps, but it is revealing that many of these factors could apply equally well to locating mapped sequences on maize pachytene chromosomes where the match between predicted and observed marker locations is even better than in tomato (r2 = 0.996 vs. 0.978; Anderson et al. 2004). However, a prominent difference in the maize and tomato molecular linkage maps is that the maize map is based on intraspecific crosses (http://www.maizegdb.org) while the tomato map is based on an interspecific hybrid (http://www.sgn.cornell.edu), and we think this is likely to be the most important factor contributing to differences between the EXPEN 2000 linkage map and RN–cM map in tomato.
Why is the best tomato linkage map based on an interspecific hybrid? Tomato has a low level of polymorphism that is probably due to its being a self-compatible species that passed through a genetic bottleneck during domestication (Miller and Tanksley 1990; Stephan and Langley 1998; Baudry et al. 2001). Therefore, a hybrid between tomato and S. pennellii was used to make the molecular map to take advantage of the numerous molecular polymorphisms between the two species (Tanksley and Rick 1980). The use of the hybrid was further justified because (1) the two species are closely related diploids with the same chromosome number (2n = 2x = 24) and similar chromosome morphology; (2) homeologous chromosomes in the hybrid synapse completely; (3) hybrids form homeologous bivalents with only slightly fewer chiasmata at diakinesis and metaphase I than either S. pennellii or tomato; (4) homeologous segregation in hybrids is normal (no univalents); (5) hybrids are moderately fertile (25%) with fertility reduction probably due to genetic, not chromosomal, incompatibility; and (6) the S. pennellii and tomato genomes appear to be largely homosequential (Khush and Rick 1963; Tanksley and Rick 1980). While it was realized that there might be differences in crossover patterns between tomato and the hybrid, it was assumed that such differences are minor enough to be ignored for mapping purposes (Tanksley et al. 1992). Our FISH localization of mapped sequences in conjunction with the RN–cM map indicates that this assumption is generally justified for chromosome 1. However, some divergence was observed, particularly in the more proximal euchromatin of the long arm, and this could be due to differences in crossover patterns between tomato and the hybrid (Figures 2 and 3B).
Certain structural differences in the tomato and S. pennellii genomes lend support to the suggestion that the pattern of crossing over is somewhat different in the hybrid compared to tomato. For example, even though the morphology of the chromosomes is similar in the two species (Khush and Rick 1963), the genome of S. pennellii is 20% larger than the genome of tomato (http://www.rbgkew.org.uk/cval/database1.html). Because genome size and SC length are closely correlated in plants (Anderson et al. 1985), some mismatch is expected between the shorter tomato chromosomes and the longer S. pennellii chromosomes. In this regard, Khush and Rick (1963) reported examples of mismatched borders of pericentric heterochromatin and mismatched chromomeres at pachytene in the hybrid, and we have observed unilateral buckles, presumably of the S. pennellii lateral elements, in hybrid SCs (our unpublished observations). It is possible that such mispairing and synapsis proximally could obstruct the spread of crossover interference and thereby change crossover patterns and frequencies in the hybrid compared to tomato (Sybenga 1996).
On the other hand, markers in the euchromatic part of the short arm and the distal region of the long arm of chromosome 1 are very close to their predicted positions (Figures 2 and 3B, Table 2). This may be related to the tendency for chromosomes to initiate homologous pairing and synapsis distally (Stack and Anderson 1986; Zickler and Kleckner 1998). Assuming this to be the case in the hybrid, better homeologous matching and more crossing over may occur distally, while the progression of synapsis proximally may involve more mismatch and suppression of proximal crossing over (Figure 3B).
An important reason for preparing the tomato molecular linkage map was to facilitate chromosome walking to genes (Tanksley et al. 1992). This approach assumes that tight linkage between a molecular locus and a gene locus indicates close physical proximity. While this can be true in some cases, we and others have shown that the validity of this assumption differs along the length of tomato pachytene chromosomes (Zhong et al. 1999; Budiman et al. 2004). The RN–cM map can help overcome this limitation by defining the relation between map distances and physical distances on pachytene chromosomes so that more accurate estimates of the physical distance between two loci can be made. The importance of this improvement in accuracy is illustrated by considering that the average distance between observed and predicted positions of loci based on the linkage map alone is almost 5 Mb, while the average distance between observed and predicted distances based on the combined linkage and RN–cM maps is <2 Mb. Such an increase in the accuracy of estimating the position of loci can be a significant aid in the initial efforts to find genes by chromosome walking as well as in genome sequence assembly. Even after the tomato genome sequence is completed (http://www.sgn.cornell.edu), the tomato RN–cM map will continue to be useful for defining the location of genes on chromosome structure.
Acknowledgments
This work was supported in part by National Science Foundation grants DBI-0116076 (S.M.S.), DBI-0421634 (S.M.S.), and MCB-0314644 (L.K.A.).
References
- Anderson, L. K., S. M. Stack, M. H. Fox and C. Zhang, 1985. The relationship between genome size and synaptonemal complex length in higher plants. Exp. Cell Res. 156: 367–378. [DOI] [PubMed] [Google Scholar]
- Anderson, L. K., A. Reeves, L. M. Webb and T. Ashley, 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151: 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L. K., N. Salameh, H. W. Bass, L. C. Harper, W. Z. Cande et al., 2004. Integrating genetic linkage maps with pachytene chromosome structure in maize. Genetics 166: 1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow, A. L., and M. A. Hultén, 1998. Crossing over analysis at pachytene in man. European Journal of Human Genetics 6: 350–358. [DOI] [PubMed] [Google Scholar]
- Baudry, E., C. Kerdelhué, H. Innan and W. Stephan, 2001. Species and recombination effects on DNA variability in the tomato genus. Genetics 158: 1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, F., 2004. Imprinting—a green variation. Science 303: 483–485. [DOI] [PubMed] [Google Scholar]
- Budiman, M. A., S.-B. Chang, S. Lee, T. J. Yang, H.-B. Zhang et al., 2004. Localization of jointless-2 gene in the centromeric region of tomato chromosome 12 based on high resolution genetic and physical mapping. Theor. Appl. Genet. 108: 190–196. [DOI] [PubMed] [Google Scholar]
- Carpenter, A. T. C., 1975. Electron microscopy of meiosis in Drosophila melanogaster females: II: the recombination nodule—a recombination-associated structure at pachytene? Proc. Natl. Acad. Sci. USA 72: 3186–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S.-B., 2004. Cytogenetic and molecular studies on tomato chromosomes using diploid tomato and tomato monosomic additions in tetraploid potato. Ph.D. Dissertation, Wageningen University, Wageningen, The Netherlands.
- Davis, G. L., M. D. Mcmullen, C. Baysdorfer, T. Musket, D. Grant et al., 1999. A maize map standard with sequenced core markers, grass genome reference points and 932 expressed sequence tagged sites (ESTs) in a 1736-locus map. Genetics 152: 1137–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong, J. H., P. F. Fransz and P. Zabel, 1999. High resolution FISH in plants—techniques and applications. Trends Plant Sci. 4: 258–263. [DOI] [PubMed] [Google Scholar]
- Fransz, P. F., C. Alonso-Blanco, T. B. Liharska, A. J. M. Peeters, P. Zabel et al., 1996. High resolution physical mapping in Arabidopsis thaliana and tomato by fluorescence in situ hybridization to extended DNA fibres. Plant J. 9: 421–430. [DOI] [PubMed] [Google Scholar]
- Froenicke, L., L. K. Anderson, J. Weinberg and T. Ashley, 2002. Male mouse recombination maps for each autosome identified by chromosome painting. Am. J. Hum. Genet. 71: 1353–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herickhoff, L., S. Stack and J. Sherman, 1993. The relationship between synapsis, recombination nodules and chiasmata in tomato translocation heterozygotes. Heredity 71: 373–385. [Google Scholar]
- Hudson, T. J., D. M. Church, S. Greenaway, H. Nguyen, A. Cook et al., 2001. A radiation hybrid map of mouse genes. Nat. Genet. 29: 201–205. [DOI] [PubMed] [Google Scholar]
- Islam-Faridi, M. N., K. L. Childs, P. E. Klein, G. Hodnett, M. A. Menz et al., 2002. A molecular cytogenetic map of sorghum chromosome 1: fluorescence in situ hybridization analysis with mapped bacterial artificial chromosomes. Genetics 161: 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S. A., Z. Cheng, M. L. Wang, H. M. Goodman and J. Jiang, 2000. Comparative fluorescence in situ hybridization mapping of a 431-kb Arabidopsis thaliana bacterial artificial chromosome contig reveals the role of chromosomal duplications in the expansion of the Brassica rapa genome. Genetics 156: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush, G. S., and C. M. Rick, 1963. Meiosis in hybrids between Lycopersicon esculentum and Solanum pennellii. Genetica 33: 167–183. [Google Scholar]
- Khush, G. S., and C. M. Rick, 1968. Cytogenetic analysis of the tomato genome by means of induced deficiencies. Chromosoma 23: 452–484. [Google Scholar]
- Kynast, R. G., R. J. Okagaki, M. W. Galatowitsch, S. R. Granath, M. S. Jacobs et al., 2004. Dissecting the maize genome by using chromosome addition and radiation hybrid lines. Proc. Natl. Acad. Sci. USA 101: 9921–9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C. J., T. E. Seigfried, H. W. Bass and L. K. Anderson, 2006. Predicting chromosomal locations of genetically mapped loci in maize using the Morgan2McClintock translator. Genetics 172: 2007–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhuissier, F. G. P., H. H. Offenberg, P. E. Wittich, N. O. E. Vischer and C. Heyting, 2007. The mismatch repair protein MLH1 marks a subset of strongly interfering crossovers in tomato. Plant Cell 19: 862–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon, E., and P. Moens, 2003. Mlh1p and Mlh3p localize to precociously induced chiasmata of okadaic acid treated mouse spermatocytes. Genetics 165: 2283–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. C., and S. D. Tanksley, 1990. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor. Appl. Genet. 80: 437–448. [DOI] [PubMed] [Google Scholar]
- Peterson, D. G., and S. Stack, 1997. A method of isolating milligram quantities of “polyphenol-free” nuclear DNA from tomato. Rept. Tomato Genet. Coop. 47: 18–20. [Google Scholar]
- Peterson, D. G., H. J. Price, J. S. Johnston and S. M. Stack, 1996. DNA content of heterochromatin and euchromatin in tomato (Lycopersicon esculentum) pachytene chromosomes. Genome 39: 77–82. [DOI] [PubMed] [Google Scholar]
- Peterson, D. G., N. Lapitan and S. M. Stack, 1999. Localization of single- and low-copy sequences on tomato synaptonemal complex spreads using fluorescence in situ hybridization (FISH). Genetics 152: 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigozzi, M. I., 2001. Distribution of MLH1 foci on the synaptonemal complexes of chicken oocytes. Cytogenet. Cell Genet. 95: 129–133. [DOI] [PubMed] [Google Scholar]
- Pigozzi, M. I., and A. J. Solari, 1999. Recombination nodule mapping and chiasma distribution in spermatocytes of the pigeon, Columba livia. Genome 42: 308–314. [DOI] [PubMed] [Google Scholar]
- Sherman, J. D., and S. M. Stack, 1992. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. V. Tomato (Lycopersicon esculentum) karyotype and idiogram. Genome 35: 354–359. [Google Scholar]
- Sherman, J. D., and S. M. Stack, 1995. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141: 683–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack, S. M., and L. K. Anderson, 1986. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. II. Synapsis in Lycopersicon esculentum. Am. J. Bot. 73: 264–281. [Google Scholar]
- Stephan, W., and C. H. Langley, 1998. DNA polymorphism in Lycopersicon and crossing-over per physical length. Genetics 150: 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A., and G. Beadle, 1939. An Introduction to Genetics. W. B. Saunders, Philadelphia.
- Sybenga, J., 1996. Recombination and chiasmata: few but intriguing discrepancies. Genome 39: 473–484. [DOI] [PubMed] [Google Scholar]
- Tanksley, S. D., and C. M. Rick, 1980. Isozymic gene linkage map of the tomato: Applications in genetics and breeding. Theor. Appl. Genet. 57: 161–170. [DOI] [PubMed] [Google Scholar]
- Tanksley, S. D., M. W. Ganal, M. C. Prince, M. C. De Vicente, M. W. Bonierbale et al., 1992. High density molecular linkage maps of the tomato and potato genomes. Genetics 132: 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, X., P. F. Fransz, J. Wennekes-Van Eden, P. Zabel, A. Van Kammen et al., 1996. High-resolution mapping on pachytene chromosomes and extended DNA fibres by fluorescence in-situ hybridization. Plant Mol. Biol. Rep. 14: 232–242. [Google Scholar]
- Zhong, X.-B., J. Bodeau, P. F. Fransz, V. M. Williamson, A. Van Kammen et al., 1999. FISH to meiotic pachytene chromosomes of tomato locates the root-knot nematode resistance gene Mi-1 and the acid phosphatase gene Aps-1 near the junction of euchromatin and pericentromeric heterochromatin of chromosome arms 6S and 6L, respectively. Theor. Appl. Genet. 98: 365–370. [Google Scholar]
- Zickler, D., and N. Kleckner, 1998. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32: 619–697. [DOI] [PubMed] [Google Scholar]
- Zwick, M. S., R. E. Hanson, T. D. Mcknight, M. N. Islam-Faridi, D. M. Stelly et al., 1997. A rapid procedure for the isolation of Cot-1 DNA from plants. Genome 40: 138–142. [DOI] [PubMed] [Google Scholar]