Abstract
Branching morphogenesis of the Drosophila tracheal system relies on the fibroblast growth factor receptor (FGFR) signaling pathway. The Drosophila FGF ligand Branchless (Bnl) and the FGFR Breathless (Btl/FGFR) are required for cell migration during the establishment of the interconnected network of tracheal tubes. However, due to an important maternal contribution of members of the FGFR pathway in the oocyte, a thorough genetic dissection of the role of components of the FGFR signaling cascade in tracheal cell migration is impossible in the embryo. To bypass this shortcoming, we studied tracheal cell migration in the dorsal air sac primordium, a structure that forms during late larval development. Using a mosaic analysis with a repressible cell marker (MARCM) clone approach in mosaic animals, combined with an ethyl methanesulfonate (EMS)-mutagenesis screen of the left arm of the second chromosome, we identified novel genes implicated in cell migration. We screened 1123 mutagenized lines and identified 47 lines displaying tracheal cell migration defects in the air sac primordium. Using complementation analyses based on lethality, mutations in 20 of these lines were genetically mapped to specific genomic areas. Three of the mutants were mapped to either the Mhc or the stam complementation groups. Further experiments confirmed that these genes are required for cell migration in the tracheal air sac primordium.
REGULATION of gas and fluid exchanges at the level of barrier epithelia is a key feature common to all organisms of the animal kingdom. To achieve this function, epithelia often acquire a tubular architecture where functional units occur repetitively, form in many cases an interconnected network, and create a large interface of interaction with their environment. This organization is achieved during embryogenesis via a process called branching morphogenesis, which relies on distinct cellular behavior often including cell division, cell migration, cell rearrangements, cell shape changes, and cell death (Hogan and Kolodziej 2002; Affolter et al. 2003; Lubarsky and Krasnow 2003). Growth factors, including fibroblast growth factor (FGF) molecules, are known to be crucial for the regulation of these processes (Warburton et al. 2000; Ghabrial et al. 2003).
In Drosophila melanogaster, Breathless/fibroblast growth factor receptor (Btl/FGFR) is implicated in the Branchless/FGF (Bnl/FGF)-dependent migration of tracheal cells during the development of the embryonic tracheal system (Klambt et al. 1992; Sutherland et al. 1996; Affolter et al. 2003; Ghabrial et al. 2003; Uv et al. 2003). Btl/FGFR is expressed in migrating tracheal cells, whereas the Bnl/FGF ligand is expressed in single or groups of ectodermal and mesodermal cells, in a highly dynamic pattern that prefigures tracheal branch outgrowth. In the absence of either the receptor or the ligand, tracheal cells do not migrate. Additional factors including the cytoplasmic adaptor Downstream-of-FGFR (Dof) and the FGFR coreceptors Sulfateless (Slf) and Sugarless (Sgl) have been shown to be required for FGFR signaling (Michelson et al. 1998; Vincent et al. 1998; Imam et al. 1999; Lin et al. 1999; Pellegrini 2001). However, the important maternal contribution of proteins of the Ras/MAP kinase pathway to the egg makes it difficult to link the function of this signaling cassette to FGFR-dependent cell migration, as the homozygous mutant offspring of heterozygous parents show no phenotype (Affolter and Weijer 2005). In addition, eggs deriving from homozygous female germ-line clone-bearing mutations affecting genes of the Ras/MAP kinase pathway display extremely severe developmental defects, hindering any detailed analysis in the tracheal system. This shortcoming can be circumvented to a large extent by analyzing clones of mutant cells in mosaic Drosophila larvae, where much of the maternal contribution has been consumed. Interestingly, during late larval development, the tracheal system is extensively remodeled to give rise to the adult respiratory organs (Whitten 1980; Manning and Krasnow 1993). During this period, a structure referred to as the dorsal air sac primordium buds from a tracheal branch called the transverse connective, in the second thoracic segment, and undergoes a morphogenetic process that relies both on cell division and on cell migration (Sato and Kornberg 2002; Guha and Kornberg 2005). The mosaic analysis with a repressible cell marker (MARCM) clone technique (Lee and Luo 1999, 2001) has been adapted to genetically dissect tracheal cell migration and proliferation in the developing dorsal air sac primordium (Cabernard and Affolter 2005). It was shown that while cell proliferation and survival in the primordium require the epidermal growth factor receptor (EGFR) signaling pathway, FGFR signaling is strictly required for tracheal cell migration at the tip of the primordium (Sato and Kornberg 2002; Cabernard and Affolter 2005). The effects of both EGFR and FGFR require the Ras/MAP kinase cassette. Interestingly, the ETS transcription factor Pointed (Brunner et al. 1994; O'Neill et al. 1994) is required exclusively for FGF-dependent cell migration, but dispensable for EGF-regulated cell division/survival (Cabernard and Affolter 2005). Although these studies provide evidences that the Ras/MAP kinase pathway is involved in tracheoblast migration, additional factors involved in interpreting either the FGF or the EGF signal remain to be identified.
To gain further insight into the regulation of FGF-dependent tracheal cell migration, we took advantage of the MARCM clone technique and carried out a large-scale screen of a collection of fly lines carrying randomly induced mutations. In this article, we describe a screening approach that allowed us to successfully isolate mutant fly strains displaying tracheal cell migration defects during the morphogenesis of the dorsal air sac primordium. We also present the identification of two complementation groups required for cell migration during air sac morphogenesis.
MATERIALS AND METHODS
Drosophila stocks:
D. melanogaster lines were raised at 25° using standard conditions. Ethyl methanesulfonate (EMS) mutant lines were generated according to standard mutagenesis procedures (see accompanying article by Baer et al. 2007, this issue). Isogenic FRT40A males were fed on 30 mm EMS to generate random mutations in the genome. The following MARCM strain (Cabernard and Affolter 2005) was used during the screen: 70hsFLP/70hsFLP; tubGal80, FRT40A/CyO; btlenhancer-mRFP1moe, btlGal4-UAS-CD8-GFP/TM6C. Deficiency lines generated by Exelixis (Parks et al. 2004) were used for complementation tests. The following mutant lines were used to map lethal hits and/or to recover MARCM mutant clones: btlH82delta3 (Reichman-Fried et al. 1994), gluk08819, Mhc1, Mhc3, Mhc4 (Mogami et al. 1986), and Mhc3, FRT40A (kindly provided by P. Rorth) (Borghese et al. 2006). The Mhc1 mutant allele was recombined with FRT40A using standard genetic methods.
Generation of MARCM clones in the developing air sac primordium:
MARCM clones were generated following the procedure described previously in Cabernard and Affolter (2005). MARCM virgin females were crossed en masse to the mutant FRT40A lines of interest. Embryos of the progeny were submitted to a heat shock 4–6 hr after egg laying for 1 hr at 38° in a circulating water bath and kept at 25° until larvae reached third instar. Third instar larvae bearing GFP-positive clones were collected using a Leica MZFLIII GFP stereomicroscope. Larval wing discs were dissected in PBS and mounted in Schneider Cell Medium (GIBCO, Grand Island, NY). Pictures of air sac primordia were taken using a Leica TCS SP2 confocal system with the Leica Confocal Software and deconvoluted with Huygens Essential (Version 2.3.0) and subsequently processed with the Imaris 4.0.4 software (Bitplane).
Mapping of lethal mutations:
Lethal mutations induced on the left arm of the second chromosome were genetically mapped by screening for noncomplementation of lethality, using deficiencies generated by Exelixis, which uncover 80% of the left arm of the second chromosome (Thibault et al. 2004). In a further candidate gene approach, known lethal mutations affecting genes located in the genomic regions determined by deficiency mapping were tested for lethality in trans to mutant candidate lines. Other mutant lines were obtained from the Bloomington Stock Center.
Rescue constructs:
To generate a UAS-stam rescue construct, a full-length stam cDNA (LD02639) was subcloned into the pUAST vector. Transgenic flies were generated according to standard transformation protocols. Only insertions in the third chromosome were kept for the rescue experiments performed in combination with MARCM analysis.
Sequencing experiments:
Identification of the affected gene for the 2L2896 and 2L3297 lines was achieved by DNA sequencing. The 2L2896 and 2L3297 lines were balanced over a CyO-YFP balancer chromosome. YFP-negative homozygous mutant embryos were sorted using a Leica MZFLIII GFP stereomicroscope. Genomic DNA from these embryos was extracted and used as a template for PCR amplification of the Stam, Ial, and Dnz1 coding regions. Primers were designed along these DNA regions to sequence the entire open reading frames. The primer pairs that yielded the point mutations for the 2L2896 line have the following sequences: 5′-GGTCTACGCAGGAGGAAGTACACC-3′ and 5′-CTCAATCGGGGGATCGGG-3′ for the C16-to-T substitution and 5′-CGGGTGGATTCCCACCGG-3′ for the G1513-to-A substitution. The following primers allowed the identification of the mutations in the 2L3297 lines: 5′-CCGAGCTGGAACGCGTCG-3′ and 5′-GTGGCACCTGCCCCTGCGG-3′ for the T1283-to-C substitution and 5′-CGGGTGGATTCCCACCGG-3′ and 5′-CCCTGTGGTGGCGGTGCC-3′ for the T1583-to-C substitution.
RESULTS
Screen procedure overview:
To identify genes involved in FGF-dependent migration of tracheal cells during morphogenesis of the dorsal air sac primordium in Drosophila, we carried out a large-scale mosaic MARCM clone screen (Lee and Luo 1999, 2001) for fly lines displaying cell migration defects. We designed a F3 mutagenesis scheme to establish mutant fly stocks carrying random EMS-induced mutations. Since our analysis was focused on genes located on the left arm of the second chromosome, we used a FRT40A chromosome in the EMS-treated stock (Figure 1A and accompanying article by Baer et al. 2007).
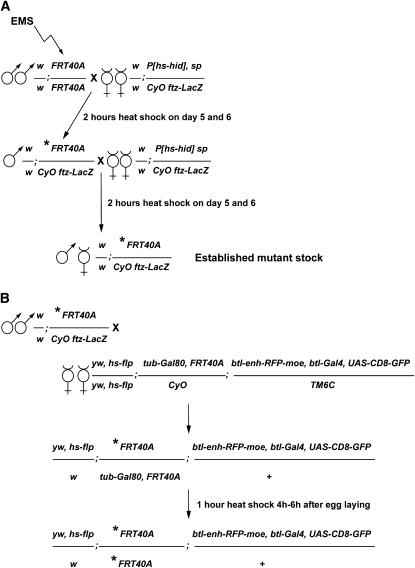
Figure 1.—
Mutagenesis and crossing scheme to generate MARCM clones of cells homozygous for mutations on chromosome 2L. (A) Scheme for the establishment of Drosophila stocks carrying mutations on the second chromosome. Ethyl methanesulfonate (EMS)-induced mutations were randomly generated in the genome of males bearing a FRT40A chromosome. EMS-treated males were subsequently crossed to females carrying an hs-hid construct and a balancer chromosome. The asterisk represents the induced mutation. Balanced mutant stocks were established in two generations. A heat-shock regime applied to the progeny of the F0 and F1 generation induced the expression of the hs-hid construct and the death of animals due to ectopic apoptosis. Therefore, establishment of the heterozygous mutant stocks did not require virgin female collection. (B) Crossing scheme to induce MARCM clones in the Drosophila larval tracheal system. F2 heterozygous mutant males were crossed to so-called MARCM females carrying a heat-shock-flipase (hs-flp) source, a FRT40A chromosome recombined to a tubulin-Gal80 (tub-Gal80) construct, and a third chromosome bearing the breathless-Gal4 (btl-Gal4), UAS-CD8-green fluorescent protein (UAS-CD8-GFP), and breathless enhancer-red fluorescent protein-moesin (btl-enh-RFP-moe) constructs. Heat-shock treatment of generation F3 induced the Flp-driven recombination at FRT sequences, which segregate the tub-Gal80 construct away from the induced mutation. Therefore, the btl-Gal4-dependent expression of CD8-GFP was possible only in clones of cells homozygous for the induced mutation (see also Figure 3A).
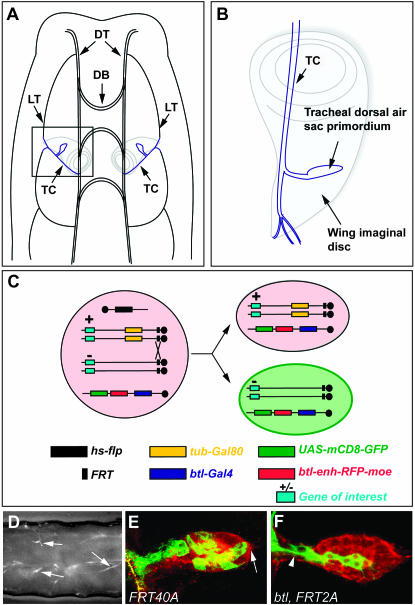
To induce MARCM mutant clones, ∼10 males of each of these putative heterozygous mutant lines were crossed en masse to ∼30 so-called FRT40A MARCM females; these females carry a heat-shock-flipase (hs-flp) source, a FRT40A chromosome recombined to a tubulin-Gal80 (tub-Gal80) construct, and a third chromosome bearing a breathless-Gal4 (btl-Gal4), a UAS-CD8-green fluorescent protein (UAS-CD8-GFP), and a breathless enhancer-red fluorescent protein-moesin (btl-enh-RFP-moe) construct (Cabernard and Affolter 2005) (Figure 1B). Using this genetic setup, mutant clones can be induced via FLP-mediated recombination at FRT40A sites in early embryonic stages, and visualized as GFP-positive groups of cells following the loss of Gal80. The Gal80-independent action of the btl enhancer enables the visualization of the entire tracheal system by expression of the RFP-moe fusion construct (Figure 2, C, E, and F). The dorsal air sac primordium buds from a tracheal branch called the transverse connective (TC) in the second thoracic segment (Figure 2A) and grows on the underlying wing imaginal disc during the third larval instar period (Sato and Kornberg 2002; Guha and Kornberg 2005) (Figure 2B). FLP-driven recombination was induced in the early embryo according to the procedure described (Cabernard and Affolter 2005). Embryos were subsequently allowed to develop and third instar larvae displaying GFP-labeled patches of cells in the tracheal system were collected (Figure 2D). Wing discs were dissected and air sac primordia bearing MARCM clones were analyzed using laser confocal microscopy in live tissues (Figure 2, E and F), without any fixation or staining requirement.
Figure 2.—
Localization of the dorsal air sac primordium in a Drosophila larva and generation and visualization of MARCM clones in the air sac primordium. (A) Schematic representation of the dorsal view of a Drosophila larva. The dorsal air sac primordium (blue) buds from a tracheal branch called the transverse connective (TC) and grows on the underlying wing imaginal disc (gray). DT, dorsal trunk; DB, dorsal branch; LT, lateral trunk; TC, transverse connective. (B) Schematic representation of the localization of a dorsal air sac primordium on the wing imaginal disc: magnification of the inset shown in A. (C) Schematic representation of the induction of MARCM clones. In all cells of animals heterozygous for the mutation of interest (+/−), the expression of the Gal80 repressor, which is under the control of a tubulin promoter, inhibits the Gal4-dependent expression of CD8-GFP. The expression of RFP-moesin is driven in all tracheal cells by the btl enhancer (red background). Upon heat-shock treatment of heterozygous animals, Flp expression is induced and drives the site-directed recombination at FRT sites, which ultimately leads, after mitosis, to the segregation of Gal80 and Gal4. The expression of CD8-GFP (green background) is therefore induced in cells homozygous for the mutation of interest (−/−). (D) GFP stereomicroscope view of a Drosophila larva of the F3 generation (see Figure 1B), after heat-shock treatment. Arrows indicate GFP-positive MARCM clones scattered in the tracheal system. Anterior is to the left, dorsal is up. (E and F) Confocal micrographs of a Drosophila third instar larval dorsal air sac primordium. All tracheal cells are labeled in red (RFP-moesin). Cells belonging to the MARCM clones are labeled in green (CD8-GFP). Clones are homozygous for a FRT40A (E) or btl, FRT42A chromosome (F). Arrows indicate the distal tip of the air sac primordium. Arrowheads indicate the proximal region of the air sac primordium.
It has previously been reported that MARCM clones of wild-type cells contribute to the growing tip of the air sac primordium (arrow in Figure 2E) in ∼70% of the cases (Cabernard and Affolter 2005). It was also shown that the FGFR signaling pathway is crucial for tracheal cell migration, as MARCM clones mutant for the Drosophila btl/FGFR or for certain downstream effectors of the FGFR signaling pathway remain in the proximal region of the air sac primordium (arrowhead in Figure 2F) and never colonize the migrating tip of the primordium (Cabernard et al. 2004). In an attempt to isolate genes involved in tracheal cell migration, mutant lines displaying a migration defect with <40% of the MARCM clones present at the air sac distal tip were kept for further analysis. Putative mutant lines were systematically retested a second and eventually a third time.
Screen result overview:
A total of 1123 lines were screened. Statistical distribution of the different phenotypes with respect to the presence/absence and localization of MARCM clones is summarized in Table 1. In 11% (N = 122) of the lines, we did not recover any individuals displaying MARCM clones in the larval tracheal system. In 8% (N = 90) of the lines, clones were observed in the tracheal system, but not recovered in the air sac primordium. In 77% (N = 864) of the lines tested, MARCM clones showed wild-type behavior, as GFP-labeled clones colonized the air sac primordium distal tip according to wild-type frequencies.
TABLE 1.
Overview of the screen
| Tested mutant lines | 1123 |
| Lines with larvae displaying no MARCM clones | 122 (11%) |
| Lines with air sacs displaying no MARCM clones | 90 (8%) |
| Lines showing no tracheal cell migration and proliferation phenotype | 864 (77%) |
| Lines showing a tracheal cell migration phenotype (class I) | 38 |
| Class I lethal lines | 34 |
| Class I not lethal lines | 4 |
| Lines showing a tracheal cell proliferation phenotype (class II) | 9 |
| Class II lethal lines | 8 |
| Class II not lethal lines | 1 |
A total of 1123 lines were tested. Numbers refer to the amount of lines for which F3 third instar larvae displayed a given phenotype after heat-shock treatment and induction of MARCM clones. Percentage values shown in parentheses refer to the total number of strains that were tested. Lines displaying a tracheal cell migration phenotype were classified in two categories: class I (strict migration phenotype) and class II (migration and proliferation phenotype). In both categories, lethal and viable alleles were recovered.
In the remaining lines (4%; N = 47), a consistent phenotype was observed with respect to two criteria: the position of MARCM clones within the air sac primordium and the size distribution of the MARCM clones. Indeed, it has been shown previously that MARCM clones mutant for components of the Ras/MAPK pathway display both a migration phenotype and a proliferation defect, since mutant clone sizes are rather small when compared to marked wild-type clones (Cabernard and Affolter 2005). In contrast, MARCM clones homozygous for btl/FGFR grow normally, but never populate the air sac primordium distal tip (Cabernard and Affolter 2005). Taking these two aspects into consideration, we assigned the 47 lines displaying a migration phenotype to two distinct classes (Table 1): mutant lines displaying a strict migration phenotype (showing wild-type-like clone sizes) were assigned to class I, whereas lines showing both a migration phenotype and a reduced clone size distribution were assigned to class II.
Class I mutants:
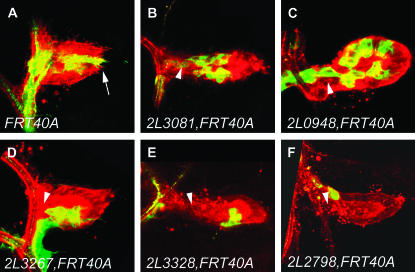
The class I category consisted of 38 fly lines (compare Figure 3, B–D, to Figure 3A). Thirty-four class I mutant lines were homozygous lethal, and 4 lines were homozygous viable (Tables 1 and 2). Two of the 34 homozygous lethal mutant class I lines (2L3267 and 2L2870) displayed a phenotype similar to the complete loss of FGF signaling; i.e., no clones were observed at the distal tip of the air sac primordium (Table 2 and Figure 3D). In the other class I mutants, the statistical distribution of MARCM clones at the distal tip ranked from 3.3 to 38.1% (Table 2).
Figure 3.—
Migration defects of various mutants isolated during the screen. Confocal micrographs of the dorsal air sac primordium of a Drosophila third instar larva are shown. All tracheal cells are labeled in red (RFP-moesin) and MARCM clones are labeled in green (CD8-GFP). The FRT40A chromosome (A) was used as a wild-type control. Isolated mutants were classified in two categories (see also Table 1): class I mutants, characterized by a strict migration phenotype and displaying clones of normal size, such as observed in the lines 2L3081, FRT40A (B), 2L0948, FRT40A (C), and 2L3267, FRT40A (D), and class II mutants, showing a migration defect and additionally a reduced size of MARCM clones, such as observed in lines 2L3328, FRT40A (E) and 2L2798, FRT40A (F). Arrows indicate the distal tip of the air sac primordium. Arrowheads indicate the proximal region of the air sac primordium.
TABLE 2.
Overview of the phenotype and mapping of the isolated class I mutant lines
| Clones at the distal tip | Clones at the proximal region | Analyzed clones | % at the distal tip | Lethal line | Mapping to | Does not complement to | Identified gene | |
|---|---|---|---|---|---|---|---|---|
| 2L 3267 | 0 | 26 | 26 | 0.0 | Yes | Df(2L)exel6039, 8036 | ||
| 2L 2870 | 0 | 39 | 39 | 0.0 | Yes | |||
| 2L 2677 | 1 | 29 | 30 | 3.3 | Yes | |||
| 2L 2436 | 1 | 15 | 16 | 6.3 | Yes | Df(2L)exel8012, 7021 | ||
| 2L 3186 | 1 | 13 | 14 | 7.1 | Yes | Df(2L)exel7038 | ||
| 2L 3189 | 1 | 12 | 13 | 7.7 | Yes | |||
| 2L 3081 | 4 | 31 | 35 | 11.4 | Yes | Df(2L)exel8038 | ||
| 2L 2881 | 2 | 14 | 16 | 12.5 | Yes | Df(2L)exel6027, 7067 | mhc1 | mhc |
| 2L 3298 | 1 | 6 | 7 | 14.3 | Yes | Df(2L)exel7034 | ||
| 2L 2475 | 1 | 6 | 7 | 14.3 | Yes | Df(2L)exel6042 | 2L1665 | |
| 2L 2653 | 2 | 11 | 13 | 15.4 | Yes | Df(2L)exel6047, 7055 | ||
| 2L 1683 | 2 | 11 | 13 | 15.4 | Yes | Df(2L)exel7038 | ||
| 2L 3297 | 4 | 21 | 25 | 16.0 | Yes | Df(2L)exel8026, 7049 | 2L2896 | stam |
| 2L 2896 | 5 | 24 | 29 | 17.2 | Yes | Df(2L)exel7038, 8026, 7049 | 2L3297 | stam |
| 2L 2828 | 6 | 26 | 32 | 18.8 | Yes | |||
| 2L 2985 | 4 | 15 | 19 | 21.1 | Yes | |||
| 2L 3073 | 3 | 10 | 13 | 23.1 | Yes | |||
| 2L 2845 | 10 | 32 | 42 | 23.8 | Yes | |||
| 2L 2853 | 8 | 25 | 33 | 24.2 | Yes | |||
| 2L 2718 | 2 | 5 | 7 | 28.6 | Yes | |||
| 2L 2775 | 7 | 17 | 24 | 29.2 | Yes | Df(2L)exel6028 | ||
| 2L 1710 | 4 | 9 | 13 | 30.8 | Yes | |||
| 2L 1685 | 7 | 15 | 22 | 31.8 | Yes | Df(2L)exel8003 | ||
| 2L 948 | 9 | 18 | 27 | 33.3 | Yes | |||
| 2L 2921 | 6 | 12 | 18 | 33.3 | Yes | |||
| 2L 1561 | 3 | 6 | 9 | 33.3 | Yes | Df(2L)exel6042 | ||
| 2L 1540 | 12 | 22 | 34 | 35.3 | Yes | Df(2L)exel6044 | ||
| 2L 1809 | 10 | 18 | 28 | 35.7 | Yes | |||
| 2L 2938 | 9 | 16 | 25 | 36.0 | Yes | Df(2L)exel6277 | ||
| 2L 1665 | 9 | 16 | 25 | 36.0 | Yes | Df(2L)exel6042 | 2L2475 | |
| 2L 3179 | 8 | 14 | 22 | 36.4 | Yes | Df(2L)exel7022 | ||
| 2L 2615 | 8 | 14 | 22 | 36.4 | Yes | Df(2L)exel6031 | ||
| 2L 3011 | 13 | 22 | 35 | 37.1 | Yes | Df(2L)exel6042, 8040 | ||
| 2L 1794 | 8 | 13 | 21 | 38.1 | Yes | |||
| 2L 2468 | 1 | 7 | 8 | 12.5 | No | |||
| 2L 2366 | 1 | 7 | 8 | 12.5 | No | |||
| 2L 883 | 4 | 12 | 16 | 25.0 | No | |||
| 2L 3064 | 7 | 15 | 22 | 31.8 | No |
Lines displaying a strict tracheal cell migration phenotype, characterized by the observation of <40% of MARCM clones reaching the distal tip of the air sac primordium (see also Figure 3, B–D), were retained for further analysis (class I mutants). We recovered 38 strains meeting this criterion (see also Table 1). For each line, numbers refer to the amount of MARCM clones observed at the distal tip of the air sac primordium (column 2) and in the proximal region (column 3), to the total number of observed clones (column 4), and to the percentage of MARCM clones localized at the air sac primordium distal tip (column 5; Figure 3). We recovered 34 homozygous lethal lines and 4 homozygous viable lines. Exelixis deficiencies, other independent class I mutants or previously characterized alleles, and names of mutants belonging to the same complementation group as other class I mutants we isolated are indicated in columns 7, 8, and 9, respectively.
Class II mutants:
Class II contains nine mutant lines, MARCM clones of which displayed both a migration and a proliferation phenotype (compare Figure 3, E and F, to Figure 3A). Eight class II mutants were homozygous lethal, and one was homozygous viable (Tables 1 and 3).
TABLE 3.
Overview of the phenotype of the isolated class II mutant lines
| Clones at the distal tip | Clones at the proximal region | Analyzed clones | % at the distal tip | Lethal line | |
|---|---|---|---|---|---|
| 2L 3194 | 0 | 7 | 7 | 0.0 | Yes |
| 2L 1748 | 0 | 3 | 3 | 0.0 | Yes |
| 2L 1682 | 0 | 5 | 5 | 0.0 | Yes |
| 2L 2983 | 0 | 1 | 1 | 0.0 | Yes |
| 2L 2834 | 0 | 3 | 3 | 0.0 | Yes |
| 2L 2798 | 0 | 7 | 7 | 0.0 | Yes |
| 2L 2686 | 3 | 23 | 26 | 11.5 | Yes |
| 2L 3328 | 3 | 21 | 24 | 12.5 | Yes |
| 2L 1738 | 4 | 15 | 19 | 21.1 | No |
Class II mutant lines displayed migration phenotypes meeting our screening criteria (<40% of MARCM clones reaching the air sac primordium distal tip). However, class II mutant lines also displayed MARCM clones of reduced size (see also Figure 3, E and F). We recovered nine strains meeting this criterion (see also Table 1). For each line, numbers refer to the amount of MARCM clones observed at the distal tip of the air sac primordium (column 2) and in the proximal region (column 3), to the total number of observed clones (column 4), and to the percentage of MARCM clones localized at the distal tip of the air sac primordium (column 5; Figure 3, E and F). We recovered eight homozygous lethal lines and one homozygous viable line.
As our aim was to identify genes implicated in FGF-driven tracheal cell migration, we started our analysis with class I mutants, which show a strict migration phenotype. Since the FGFR signaling pathway is crucial for numerous steps throughout development, mutations affecting factors playing important roles in this pathway are likely to be homozygous lethal. Thus, we have chosen to focus our analysis on those 34 class I mutants that were homozygous lethal. Our strategy to identify genes involved in distal tip cell migration during dorsal air sac development was the following. First, we identified the lethal hit in the mutant line. Second, we investigated whether the gene responsible for lethality was indeed involved in distal tip migration, either by performing rescue experiments in MARCM clones or via the identification of additional mutations in the same gene and testing whether these mutations caused the same MARCM mutant phenotype (Tables 1 and 2).
Mapping of class I mutants and complementation analysis:
Lethal hits in candidate lines were mapped using complementation analysis. We took advantage of the Exelixis targeted deficiency kit, as each deficiency uncovers only ∼25 genes (Parks et al. 2004). Exelixis deficiencies cover ∼80% of transcription units of chromosomal arm 2L described by the FlyBase Consortium. All the EMS mutant lines were crossed to homozygous lethal Exelixis deficiency lines and the progeny of these crosses was scored for the absence of viable trans-heterozygotes. This approach allowed us to map lethal hits in 20 mutant strains (Table 2). On the basis of these lethality tests, we found that 18 lines carried at least one lethal hit (no complementation between the lethal hit and either one deficiency or a group of overlapping deficiencies) and that 2 lines carried at least two lethal hits (no complementation between the lethal hit and two nonoverlapping deficiencies) on the left arm of the second chromosome.
Mutant lines mapping to the same genomic area were crossed inter se to determine whether they belonged to the same complementation group. Using this procedure, we found that 2L1665 and 2L2475, both carrying a lethal hit mapping to Df(2L)exel6042, as well as 2L2896 and 2L3297, carrying a lethal hit mapping to Df(2L)exel7049 and Df(2L)exel8026, did not complement each other's lethality (Table 2), suggesting that these mutations represent two independent alleles of the same gene.
To identify the affected loci in the 20 mapped mutant lines, we used available lethal mutations (previously isolated mutations or transposon insertions) in the region uncovered by the corresponding Exelixis deficiencies and tested whether these mutations complemented the lethality of the corresponding EMS-induced mutants. Alternatively, when no lethal mutation was available in the region of interest, a sequencing approach was used. These approaches led to the identification of two complementation groups responsible for lethality (see below).
Gene identification:
Mutant line 2L2881:
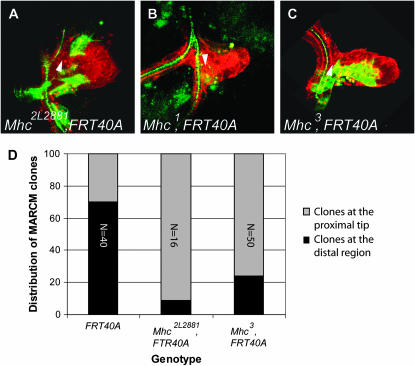
In 2L2881, only 12.5% of the homozygous mutant MARCM clones were capable of reaching the distal tip of the air sac primordium (Table 2 and Figure 4, A and D).
Figure 4.—
Homozygous Mhc mutant MARCM clones display a migration phenotype. (A–C) Confocal micrographs of a Drosophila third instar larval dorsal air sac primordium. All tracheal cells are labeled in red (RFP-moesin). Cells belonging to the MARCM clones are labeled in green (CD8-GFP). MARCM clones were induced in individuals heterozygous for Mhc2L2881 (A), Mhc1 (B), and Mhc3 (C). Arrowheads indicate the proximal region of the air sac primordium. (D) Graphical representation of the statistical distribution of MARCM clones in the dorsal air sac primordium (gray, localization at the proximal region; black, localization at the distal growing tip). The FRT40A chromosome was used as a wild-type control. Wild-type clones colonize the distal growing air sac tip in 70% of the cases. This proportion is dramatically reduced in the Mhc2L2881 and Mhc3 mutants. The numbers refer to the total number of observed clones.
Genetic mapping showed that the lethal mutation present in 2L2881 did not complement two overlapping Exelixis deficiencies, Df(2L)Exel6027 and Df(2L)Exel7067 (Table 2). This narrowed the region of interest down to 12 genes, including gluon (glu) and Myosin heavy chain (Mhc), two essential genes. Lethal alleles of glu (gluk08819) and of Mhc [Mhc1, Mhc3, and Mhc4 (Mogami et al. 1986)] were then tested by complementation analysis; we found that the Mhc1, Mhc3, and Mhc4 alleles did not complement the lethality observed in 2L2881, whereas gluk08819 did. These results suggested that 2L2881 carried a lethal hit in the Mhc gene.
We next tested whether mutations in Mhc displayed a phenotype similar to 2L2881 by analyzing the behavior of MARCM clones homozygous for the amorphic Mhc1 or the hypomorphic Mhc3 allele in the air sac primordium. To do so, we recombined the Mhc1 allele onto the FRT40A chromosome and used a previously described Mhc3, FTR40A recombinant allele (Borghese et al. 2006), respectively. Although many air sac primordia were screened, only one tiny MARCM Mhc1mutant clone, consisting of one single labeled cell, was recovered at the back of the air sac primordium (Figure 4B). However, we did observe a migration defect for the Mhc3 allele, on the basis of the finding that only 24% of MARCM mutant Mhc3 clones reached the distal air sac tip (Figure 4, C and D). These results suggest that Mhc is indeed required for tracheal cell migration in the air sac primordium.
These observations also suggest that the cell migration process we analyze is more strongly affected by the Mhc1 allele than by the 2L2881 allele. Mhc1 is an embryonic lethal allele: late embryos are devoid of muscles and are incapable of moving and hatching (Mogami et al. 1986). In contrast, we did recover viable homozygous 2L2881 first instar larvae (data not shown). Furthermore, staining of F-actin in muscles using TRITC-phalloïdin showed that muscle fibers can still be observed in homozygous 2L2881 embryos, whereas they are absent in Mhc1 homozygotes (data not shown). Altogether, these results suggest that 2L2881 is a hypomorphic Mhc allele.
Mutant lines 2L2896 and 2L3297:
2L2896 and 2L3297 showed a migration phenotype of equivalent strength: in 2L2896, only 17.2% of MARCM clones were found at the distal tip of the air sac primordium and in 2L3297, 16% of the clones reached the tip (Table 2 and Figure 5, A, B, and E).
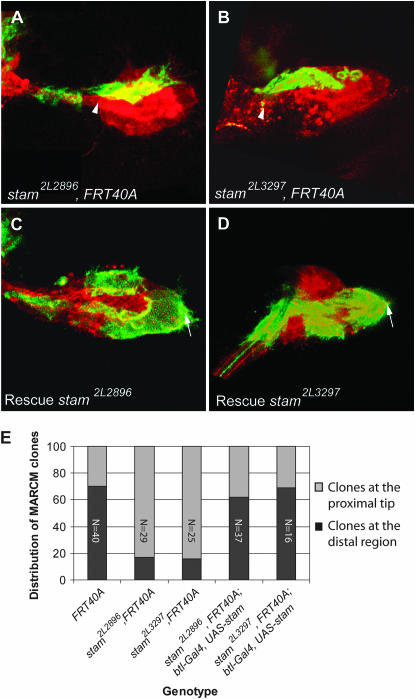
Figure 5.—
Homozygous stam mutant MARCM clones display a migration phenotype. (A–D) Confocal micrographs of a Drosophila third instar larval dorsal air sac primordium. All tracheal cells are labeled in red (RFP-moesin). Cells belonging to the MARCM clones are labeled in green (CD8-GFP). MARCM clones were induced for the stam2L2896, FRT40A (A) and for the stam2L3297, FRT40A (B) mutant. Overexpression of a wild-type stam cDNA in tracheal cells of stam2L2896, FRT40A (C) and stam2L3297, FRT40A (D) mutants rescued the migration phenotype. Arrows indicate the distal tip of the air sac primordium. Arrowheads indicate the proximal region of the air sac primordium. (E) Graphical representation of the statistical distribution of MARCM clones in the dorsal air sac primordium (gray, localization at the proximal region; black, localization at the distal growing tip). The FRT40A chromosome was used as a wild-type control. Wild-type clones colonized the distal tip of the growing air sac primordium in 70% of the cases. This proportion is dramatically reduced in the stam2L2896, FRT40A and stam2L3297, FRT40A mutants. Overexpression of stam in mutant MARCM clones restored a wild-type migration profile. The numbers refer to the total number of observed clones.
2L2896 and 2L3297 did not complement each other's associated lethality, and both mutations mapped to the overlapping region of Df(2L)Exel8026 and Df(2L)Exel7049 (Table 2). This region contains three genes: DNZDHHC/NEW1 zinc finger protein 11 (Dnz1), IplI-aurora-like kinase (ial), and signal transducing adaptor molecule (stam). Since no lethal mutations affecting any of these genes were available, we chose to identify the mutated gene by a systematic sequencing approach. We sequenced the open reading frames of Dnz1 (828 bp), ial (987 bp), and stam (2067 bp) in genomic DNA preparations from FRT40A wild-type embryos as well as from homozygous 2L2896 and 2L3297 mutant embryos (see materials and methods).
We did not find any nucleotide change in the Dnz1 and the ial coding region, suggesting that theses genes are not responsible for the lethality in these two mutant lines. However, nucleotide changes were found when comparing the stam gene sequence between the parental, nonmutagenized FRT40A chromosome, used as a wild-type control, and both mutant alleles. In 2L2896, we found a C16-to-T transition and a G1513-to-A transition. The first substitution causes a nonsense mutation from Gln6 to a stop codon, and the second mutation induces a transition of an Ala505 to a Thr. The presence of a stop codon at amino acid position 6 suggests that 2L2896 does not produce a functional Stam protein and represents a null allele. In the case of 2L3297, we also detected two nucleotide substitutions: a T1283 to C, and a T1583 to C. These modifications induce a Phe428-to-Ser and a Tyr528-to-His transition, respectively. It is worth mentioning that an in silico analysis revealed that Tyr528 is in an environment favorable to tyrosine phosphorylation (data not shown). If Tyr528 were indeed phosphorylated in vivo, this post-translational modification would no longer be possible in the stam2L3297 mutant allele. Further biochemical analyses of the Stam protein are required to test this hypothesis.
To investigate whether the cell migration defects observed in 2L2896 and 2L3297 were indeed due to the mutations we identified in the stam coding region, we overexpressed a UAS-stam transgene in homozygous mutant MARCM clones using a btl-Gal4 driver and observed a full rescue of the migration defects (Figure 5, C and D): 62 and 69% of the rescued MARCM clones reached the distal tip of the air sac primordium in 2L2896 and 2L3297 animals, respectively (Figure 5E). Altogether, these results strongly suggest that the tracheal cell migration phenotypes observed in the 2L2896 and 2L3297 alleles are due to the disruption of stam.
DISCUSSION
A MARCM clones screen for tracheal cell migration defects:
To better understand the molecular basis of the important role of FGF signaling in the regulation of cell migration, we conducted an EMS-genetic screen of the left arm of the second chromosome and analyzed the migration of mutant mitotic MARCM clones generated in the dorsal air sac primordium.
Since the analysis of MARCM clones in the air sac requires a labor-intensive dissection step, we restricted our initial screen to 1123 EMS mutant lines. The number of genes present on chromosomal arm 2L is estimated to be between 2700 and 2800. Therefore, the screen we performed was far away from being saturating. Indeed, most of the lethal complementation groups we isolated are represented by a single allele; two individual alleles were recovered only for two complementation groups. In addition, we did not isolate novel alleles of pten, which is the only locus located on chromosomal arm 2L known to be required for tracheal cell migration in the air sac primordium (Cabernard and Affolter 2005). However, as this screen indeed allowed us to recover mutations affecting tracheal cell migration, a larger screen on 2L and similar approaches involving the other chromosomal arms should be performed to identify additional factors involved in FGF-dependent tracheal cell migration.
Screen result summary and isolated mutants:
Of the 1123 lines we have tested, 122 (11%) displayed no MARCM clones in the tracheal system (Table 1). The corresponding mutations may affect genes implicated in cell viability. Alternatively, the FRT site could be mutated, making recombination no longer possible. Interestingly, in the case of 90 lines (8%), no clones were observed in the air sac primordium although MARCM clones were detected in the mature larval tracheal system (Table 1). During third instar larval development, the second thoracic tracheal metamere is repopulated by tracheoblasts; a small subset of the latter subsequently gives rise to the dorsal air sac primordium (Guha and Kornberg 2005). We postulate that genes required for the process of tracheoblast repopulation would fall into this phenotypic category if mutated.
Forty-seven lines (4%) exhibited a phenotype corresponding to our screening criteria, i.e., <40% of MARCM clones reaching the air sac primordium distal tip. We isolated 34 class I lines bearing a strict migration defect (Table 1). Two of these lines, 2L3267 and 2L2870, displayed a strict FGFR-like phenotype, as all MARCM clones were observed at the proximal air sac region (Table 2 and Figure 3D). This strong phenotype has so far been observed exclusively for mutants affecting components of the FGFR pathway: btl/FGFR, dof, slf, and pnt (Cabernard and Affolter 2005). The 2L3267 and 2L2870 lines do not map to any of these loci and therefore most likely carry mutations affecting essential and novel genes specifically linked to FGF-dependant cell migration. Identification and characterization of these two interesting loci are currently under investigation.
Of the selected candidate lines, nine belong to the class II, since they showed both a tracheal cell migration and a proliferation phenotype. Interestingly, this behavior resembles the phenotype observed for mutations in genes implicated in the Ras/MAPK pathway (Cabernard and Affolter 2005). Therefore, class II mutants could affect factors of this pathway, although it is obviously possible that some EMS-mutated genes identified in the screen code for components unrelated to the regulation of Ras or of its downstream targets. Identification and characterization of these loci will provide more insight into their precise function in the regulation of tracheal cell migration and proliferation.
Mapping strategy of the mutant loci:
Using the Exelixis deficiency kit we mapped lethal hits to specific genomic areas on chromosomal arm 2L in 20 candidate lines. For the remaining candidate lines, which are not mapped, we are currently testing the alternative Bloomington 2L deficiency kit, which covers 94.8% of the 2L chromosomal arm. However, we cannot exclude that in some lines the hit responsible for lethality is located on chromosomal arm 2R and is different from the mutation responsible for the migration phenotype located on chromosomal arm 2L.
Identification of the Mhc gene:
We identified a lethal hit in the Mhc gene. Mhc encodes a Myosin heavy chain protein and belongs to the Myosin II class of molecules also termed conventional myosins (Yamashita et al. 2000). Although Mhc is known as the muscle Mhc isoform, its function is not restricted to muscles. Indeed, expression of the Mhc gene has been shown to be regulated in border cells by the transcriptional regulator Slow border cells (Slbo) (Borghese et al. 2006; Wang et al. 2006), a key regulator of border cell migration in the egg chamber during oogenesis. Moreover, border cell migration is impaired in the Mhc3 allele (Borghese et al. 2006). Several genes required for border cell migration, like slbo, pvr (necessary for Drosophila VEGF-dependent signaling), and blistered/DSRF, were previously tested for their possible involvement in tracheal cell migration in the air sac. However, MARCM clones homozygous for mutations in these genes displayed a wild-type migration behavior (C. Cabernard, unpublished results). These results suggest different mechanisms of cell migration depending on cell and tissue context. However, Mhc is required in both cases, suggesting that it constitutes a more general actor in cell migration.
Identification of the stam gene:
In our screen, we identified 2L2896 and 2L3297 as carriers of independent and distinct alleles of the stam locus. Two human Stam homologs have been described (Takeshita et al. 1996; Endo et al. 2000; Pandey et al. 2000; Takata et al. 2000). Stam proteins are constitutive binding partners of the Hepatocyte growth factor-regulated substrate (Hrs) (Asao et al. 1997; Takata et al. 2000). Vertebrate Stam and Hrs colocalize at the level of early endosomes and are involved in vesicle trafficking and sorting of ubiquitinated membrane proteins into multivesicular bodies (MVB) (Bache et al. 2003; Kanazawa et al. 2003). In mammals, both proteins are known to be involved in the regulation of receptor tyrosine kinase (RTK) signaling via stimulation of the degradation of activated EGFR (for review, see Clague and Urbe 2006 and references therein). In Drosophila, Hrs is also required for MVB maturation and downregulation of several signaling receptors, including RTKs (Lloyd et al. 2002; Jekely and Rorth 2003). However, no stam loss-of-function mutants have been described so far in Drosophila, and its function has not been investigated yet. It will be of interest to test whether Drosophila stam regulates tracheal cell migration by regulating protein levels of activated Btl/FGFR. A study investigating this hypothesis will be presented elsewhere.
Acknowledgments
We thank K. Basler, M. Yoshihara, P. Rorth, the Blomington Stock Center, and the Drosophila Genetic Resource Center (KIT Tokyo) for sending flies; C. Dossenbach for participating in initial steps of the screen; and B. Bruno, G. Evora, and K. Mauro for technical support. We are grateful to B. Bello for invaluable comments on the manuscript. Studies in the laboratory of M. A. are supported by research grants from the Swiss National Science Foundation, the cantons Basel-Stadt and Basel-Land, and the European Commission via the FP6 Network of Excellence “Cells into organs.” Studies in the laboratory of M. L. are supported by the International Graduate School in Genetics and Functional Genomics and the Deutsche Forschungsgemeinschaft (grants LE546/3-1 and LE546/3-2).
References
- Affolter, M., and C. J. Weijer, 2005. Signaling to cytoskeletal dynamics during chemotaxis. Dev. Cell 9: 19–34. [DOI] [PubMed] [Google Scholar]
- Affolter, M., S. Bellusci, N. Itoh, B. Shilo, J. P. Thiery et al., 2003. Tube or not tube: remodeling epithelial tissues by branching morphogenesis. Dev. Cell 4: 11–18. [DOI] [PubMed] [Google Scholar]
- Asao, H., Y. Sasaki, T. Arita, N. Tanaka, K. Endo et al., 1997. Hrs is associated with STAM, a signal-transducing adaptor molecule. Its suppressive effect on cytokine-induced cell growth. J. Biol. Chem. 272: 32785–32791. [DOI] [PubMed] [Google Scholar]
- Bache, K. G., C. Raiborg, A. Mehlum and H. Stenmark, 2003. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278: 12513–12521. [DOI] [PubMed] [Google Scholar]
- Baer, M. M., A. Bilstein and M. Leptin, 2007. A clonal genetic screen for mutants causing defects in larval tracheal morphogenesis in Drosophila. Genetics 176: 2279–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese, L., G. Fletcher, J. Mathieu, A. Atzberger, W. C. Eades et al., 2006. Systematic analysis of the transcriptional switch inducing migration of border cells. Dev. Cell 10: 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, D., K. Ducker, N. Oellers, E. Hafen, H. Scholz et al., 1994. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature 370: 386–389. [DOI] [PubMed] [Google Scholar]
- Cabernard, C., and M. Affolter, 2005. Distinct roles for two receptor tyrosine kinases in epithelial branching morphogenesis in Drosophila. Dev. Cell 9: 831–842. [DOI] [PubMed] [Google Scholar]
- Cabernard, C., M. Neumann and M. Affolter, 2004. Cellular and molecular mechanisms involved in branching morphogenesis of the Drosophila tracheal system. J. Appl. Physiol. 97: 2347–2353. [DOI] [PubMed] [Google Scholar]
- Clague, M. J., and S. Urbe, 2006. Endocytosis: the DUB version. Trends Cell Biol. 16: 551–559. [DOI] [PubMed] [Google Scholar]
- Endo, K., T. Takeshita, H. Kasai, Y. Sasaki, N. Tanaka et al., 2000. STAM2, a new member of the STAM family, binding to the Janus kinases. FEBS Lett. 477: 55–61. [DOI] [PubMed] [Google Scholar]
- Ghabrial, A., S. Luschnig, M. M. Metzstein and M. A. Krasnow, 2003. Branching morphogenesis of the Drosophila tracheal system. Annu. Rev. Cell Dev. Biol. 19: 623–647. [DOI] [PubMed] [Google Scholar]
- Guha, A., and T. B. Kornberg, 2005. Tracheal branch repopulation precedes induction of the Drosophila dorsal air sac primordium. Dev. Biol. 287: 192–200. [DOI] [PubMed] [Google Scholar]
- Hogan, B. L., and P. A. Kolodziej, 2002. Organogenesis: molecular mechanisms of tubulogenesis. Nat. Rev. Genet. 3: 513–523. [DOI] [PubMed] [Google Scholar]
- Imam, F., D. Sutherland, W. Huang and M. A. Krasnow, 1999. stumps, a Drosophila gene required for fibroblast growth factor (FGF)-directed migrations of tracheal and mesodermal cells. Genetics 152: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jekely, G., and P. Rorth, 2003. Hrs mediates downregulation of multiple signalling receptors in Drosophila. EMBO Rep. 4: 1163–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa, C., E. Morita, M. Yamada, N. Ishii, S. Miura et al., 2003. Effects of deficiencies of STAMs and Hrs, mammalian class E Vps proteins, on receptor downregulation. Biochem. Biophys. Res. Commun. 309: 848–856. [DOI] [PubMed] [Google Scholar]
- Klambt, C., L. Glazer and B. Z. Shilo, 1992. breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline glial cells. Genes Dev. 6: 1668–1678. [DOI] [PubMed] [Google Scholar]
- Lee, T., and L. Luo, 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. [DOI] [PubMed] [Google Scholar]
- Lee, T., and L. Luo, 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24: 251–254. [DOI] [PubMed] [Google Scholar]
- Lin, X., E. M. Buff, N. Perrimon and A. M. Michelson, 1999. Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development 126: 3715–3723. [DOI] [PubMed] [Google Scholar]
- Lloyd, T. E., R. Atkinson, M. N. Wu, Y. Zhou, G. Pennetta et al., 2002. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell 108: 261–269. [DOI] [PubMed] [Google Scholar]
- Lubarsky, B., and M. A. Krasnow, 2003. Tube morphogenesis: making and shaping biological tubes. Cell 112: 19–28. [DOI] [PubMed] [Google Scholar]
- Manning, G., and M. Krasnow, 1993. Development of the Drosophila tracheal system, pp. 609–685 in The Development of Drosophila, edited by A. Martinez-Arias and M. Bate. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Michelson, A. M., S. Gisselbrecht, E. Buff and J. B. Skeath, 1998. Heartbroken is a specific downstream mediator of FGF receptor signalling in Drosophila. Development 125: 4379–4389. [DOI] [PubMed] [Google Scholar]
- Mogami, K., P. T. O'Donnell, S. I. Bernstein, T. R. Wright and C. P. Emerson, Jr., 1986. Mutations of the Drosophila myosin heavy-chain gene: effects on transcription, myosin accumulation, and muscle function. Proc. Natl. Acad. Sci. USA 83: 1393–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, E. M., I. Rebay, R. Tjian and G. M. Rubin, 1994. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78: 137–147. [DOI] [PubMed] [Google Scholar]
- Pandey, A., M. M. Fernandez, H. Steen, B. Blagoev, M. M. Nielsen et al., 2000. Identification of a novel immunoreceptor tyrosine-based activation motif-containing molecule, STAM2, by mass spectrometry and its involvement in growth factor and cytokine receptor signaling pathways. J. Biol. Chem. 275: 38633–38639. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., K. R. Cook, M. Belvin, N. A. Dompe, R. Fawcett et al., 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- Pellegrini, L., 2001. Role of heparan sulfate in fibroblast growth factor signalling: a structural view. Curr. Opin. Struct. Biol. 11: 629–634. [DOI] [PubMed] [Google Scholar]
- Reichman-Fried, M., B. Dickson, E. Hafen and B. Z. Shilo, 1994. Elucidation of the role of breathless, a Drosophila FGF receptor homolog, in tracheal cell migration. Genes Dev. 8: 428–439. [DOI] [PubMed] [Google Scholar]
- Sato, M., and T. B. Kornberg, 2002. FGF is an essential mitogen and chemoattractant for the air sacs of the drosophila tracheal system. Dev. Cell 3: 195–207. [DOI] [PubMed] [Google Scholar]
- Sutherland, D., C. Samakovlis and M. A. Krasnow, 1996. branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell 87: 1091–1101. [DOI] [PubMed] [Google Scholar]
- Takata, H., M. Kato, K. Denda and N. Kitamura, 2000. A hrs binding protein having a Src homology 3 domain is involved in intracellular degradation of growth factors and their receptors. Genes Cells 5: 57–69. [DOI] [PubMed] [Google Scholar]
- Takeshita, T., T. Arita, H. Asao, N. Tanaka, M. Higuchi et al., 1996. Cloning of a novel signal-transducing adaptor molecule containing an SH3 domain and ITAM. Biochem. Biophys. Res. Commun. 225: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. [DOI] [PubMed] [Google Scholar]
- Uv, A., R. Cantera and C. Samakovlis, 2003. Drosophila tracheal morphogenesis: intricate cellular solutions to basic plumbing problems. Trends Cell Biol. 13: 301–309. [DOI] [PubMed] [Google Scholar]
- Vincent, S., R. Wilson, C. Coelho, M. Affolter and M. Leptin, 1998. The Drosophila protein Dof is specifically required for FGF signaling. Mol. Cell 2: 515–525. [DOI] [PubMed] [Google Scholar]
- Wang, X., J. Bo, T. Bridges, K. D. Dugan, T. C. Pan et al., 2006. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev. Cell 10: 483–495. [DOI] [PubMed] [Google Scholar]
- Warburton, D., M. Schwarz, D. Tefft, G. Flores-Delgado, K. D. Anderson et al., 2000. The molecular basis of lung morphogenesis. Mech. Dev. 92: 55–81. [DOI] [PubMed] [Google Scholar]
- Whitten, J., 1980. The tracheal system, pp. 499–540 in The Genetics and Biology of Drosophila, edited by M. Ashburner and T. R. F. Wright. Academic Press, New York.
- Yamashita, R. A., J. R. Sellers and J. B. Anderson, 2000. Identification and analysis of the myosin superfamily in Drosophila: a database approach. J. Muscle Res. Cell Motil. 21: 491–505. [DOI] [PubMed] [Google Scholar]