Abstract
Adult Drosophila are decorated with several types of polarized cuticular structures, such as hairs and bristles. The morphogenesis of these takes place in pupal cells and is mediated by the actin and microtubule cytoskeletons. Mutations in flare (flr) result in grossly abnormal epidermal hairs. We report here that flr encodes the Drosophila actin interacting protein 1 (AIP1). In other systems this protein has been found to promote cofilin-mediated F-actin disassembly. In Drosophila cofilin is encoded by twinstar (tsr). We show that flr mutations result in increased levels of F-actin accumulation and increased F-actin stability in vivo. Further, flr is essential for cell proliferation and viability and for the function of the frizzled planar cell polarity system. All of these phenotypes are similar to those seen for tsr mutations. This differs from the situation in yeast where cofilin is essential while aip1 mutations result in only subtle defects in the actin cytoskeleton. Surprisingly, we found that mutations in flr and tsr also result in greatly increased tubulin staining, suggesting a tight linkage between the actin and microtubule cytoskeleton in these cells.
THE actin cytoskeleton plays conserved and key roles in a variety of cellular activities in eukaryotes, such as cell migration, endocytosis, cytokinesis, and cell shape changes (Baum 2002; Welch and Mullins 2002; Rodriguez et al. 2003; Etienne-Manneville 2004; Revenu et al. 2004; Schafer 2004; Pilot and Lecuit 2005; Dormann and Weijer 2006; Egea et al. 2006; Marston and Goldstein 2006; Mathur 2006). Among the model systems used to study actin and cellular morphogenesis are epidermal hairs, denticles, laterals, and bristles of insects such as Drosophila (Wong and Adler 1993; Petersen et al. 1994; Tilney et al. 1995; Dickinson and Thatcher 1997; Turner and Adler 1998; Baum 2002). These structures have dramatically polarized and reproducible morphologies and their development involves prominent actin filaments/bundles. These properties have facilitated the identification of mutations in genes that play important roles in regulating the actin cytoskeleton. Classic examples include singed (fascin) and forked that encode proteins that bundle actin filaments (Bryan et al. 1993; Cant et al. 1994; Petersen et al. 1994; Tilney et al. 1995). Mutations in these genes result in bent, twisted, and shortened bristles, arista laterals, and epidermal hairs. In addition to actin, growing hairs and bristles also contain centrally located microtubules (Petersen et al. 1994; Turner and Adler 1998; Tilney et al. 2000; He and Adler 2002; He et al. 2005).
The actin cytoskeleton is dynamic and its polymerization, depolymerization, and subcellular localization must be regulated. The conserved protein cofilin is one of the most important regulators of actin filament disassembly (Lappalainen et al. 1997; Welch and Mullins 2002; Balcer et al. 2003; Paavilainen et al. 2004). Despite extensive research some disagreement remains as to the biochemical mechanism by which cofilin promotes actin filament disassembly. The data suggest cofilin mediates both depolymerization at the pointed end of actin filaments and filament severing, but the relative importance of these two activities in vivo is not clear (Balcer et al. 2003; Paavilainen et al. 2004). Phosphorylation is a known conserved mechanism for cofilin regulation. Cofilin is inactivated by the phosphorylation of ser3, which is mediated by Lim kinase (Arber et al. 1998; Yang et al. 1998; Ohashi et al. 2000; Niwa et al. 2002; Paavilainen et al. 2004). In Drosophila this is reversed by Slingshot (Ssh)-mediated dephosphorylation (Niwa et al. 2002).
Consistent with Ssh activating cofilin and promoting actin depolymerization, mutations in ssh result in increased levels of F-actin accumulation in imaginal disc cells (Niwa et al. 2002). Such mutations also result in shortened, deformed, and split bristles, hairs, and arista laterals consistent with actin depolymerization being important for the cellular morphogenesis of these structures. The twinstar (tsr) gene encodes the Drosophila cofilin (Gunsalus et al. 1995). Mutations in this gene are also known to result in increased levels of F-actin in cells (Baum et al. 2000) and surviving tsr hypomorphs display abnormal bristles (Chen et al. 2001; Wahlstrom et al. 2001). tsr is essential at both the organismal and cell levels. The requirement for cell viability is thought to be due to cofilin being essential for cytokinesis (Gunsalus et al. 1995; Rogers et al. 2003). In contrast to the requirement for cofilin, ssh is not required for cell viability (Niwa et al. 2002; Rogers et al. 2003). This could be a consequence of enough cofilin activity remaining in an ssh mutant to allow cytokinesis or to a different phosphatase providing this function. Mutations in actin Capping Proteins also result in increased accumulation of F-actin and cell death in wing blade cells (Janody and Treisman 2006) and this is thought to be due to effects on epithelial integrity.
In yeast, animals, and plants the actin interacting protein 1 (AIP1) facilitates the ability of cofilin to promote actin depolymerization (Okada et al. 1999, 2006; Balcer et al. 2003; Mohri and Ono 2003; Ono 2003; Ketelaar et al. 2004; Ono et al. 2004; Paavilainen et al. 2004; Rodal et al. 2005; Clark et al. 2006). AIP1 has been found to bind directly to both actin and cofilin, but it is not thought that it has any actin depolymerizing activity on its own (Ono 2003; Ono et al. 2004; Rodal et al. 2005; Clark et al. 2006; Okada et al. 2006). In yeast cofilin is an essential gene, while AIP1 is not. However, a synthetic lethality is seen between cofilin and AIP1 consistent with AIP1 enhancing, but not being essential for, cofilin's depolymerization activity (Lappalainen et al. 1997; Rodal et al. 1999). In Caenorhabditis elegans there are two AIP1 homologs, UNC-78 and K08F9.2 (Ono 2001, 2003). UNC-78 is expressed in muscle and null mutations result in a severe disorganization of muscle actin and a defect in muscle contractility (Ono 2001). The second AIP1 gene has not been well studied and an RNAi screen did not detect a phenotype from knocking down the expression of this gene (Ono 2003). In Drosophila there is a single AIP1 homolog (CG10724), but no functional analysis of this gene has been reported. Reducing Arabidopsis AIP1 expression resulted in a wide range of developmental defects including reduced cell expansion (Ketelaar et al. 2004). The crystal structure of both the yeast and worm AIP1s have been solved (Voegtli et al. 2003; Mohri et al. 2004). The two structures are very similar and contain 14 WD40 domains arranged in two seven-blade β-propellers. The 14 WD40 domains are conserved in all AIP1 homologs and it is likely that the three-dimensional structure is as well.
We report here that CG10724 is the flare (flr) gene, which was discovered >30 years ago in a screen for mutations that produced wing hair phenotypes in mitotic clones (Garcia-Bellido and Dapena 1974). Indeed, flr has been used as a cell marker in many studies. We show that flr is essential for cell viability, that flr mutant cells show increased F-actin levels, and that F-actin in flr mutant cells is more stable than in neighboring wild-type wing cells. We further show that tsr mutations produce a mutant wing hair phenotype that is similar to flr. Our data argue that these two proteins function together in regulating the actin cytoskeleton in the fly epidermis. Somewhat unexpectedly we also found a marked increase in microtubule staining in flr and tsr mutant cells raising the possibility that some of the phenotypic consequences of flr and tsr mutations might be due to effects on the microtubule cytoskeleton. Consistent with flr functioning to promote F-actin depolymerization we found that FLR did not accumulate specifically in growing wing hairs, which are thought to require active actin polymerization.
Recently it was found that tsr function was required for the function of the fz pathway in the epidermis and the asymmetric accumulation of planar polarity proteins (Blair et al. 2006). We show that flr is also required for the development of normal wing cell planar polarity.
MATERIALS AND METHODS
Drosophila stocks:
Stocks that carried mutations required for generating and marking mitotic clones and stocks used for driving expression of transgenes were obtained from the Drosophila Stock Center in Bloomington. Mutant alleles of tsr (N121 and N96A), ssh (1-11 and 1-63), and CG10724 (EY05812) were also obtained from the stock center. Equivalent results were obtained with each of the tsr and ssh alleles, both of which are reported to be null alleles (Niwa et al. 2002; Ng and Luo 2004). The presented figures are of tsrN121 and ssh1-11. Most of the flr alleles were isolated in our laboratory after EMS mutagenesis. All of the alleles are homozygous lethal, with the exception of flr4, which is semilethal.
Clonal analysis:
Somatic clones were generated using the FRT/FLP system. Pupal wing clones were marked by the loss of GFP. Unmarked clones were detected by mutant phenotypes.
Cytological techniques:
White prepupae were collected and aged until dissection. Immunostaining was done by standard techniques after fixation with paraformaldehyde (Adler et al. 2004). Fluorescent secondary antibodies and fluorescent phalloidin for staining the actin cytoskeleton were obtained from Molecular Probes. Anti-armadillo monoclonal antibody was obtained from the Developmental Studies Hybridoma Bank at the University of Iowa. The anti-tubulin and anti-acyl-tubulin antibodies were obtained from Sigma Chemical Company. Confocal images were obtained on an ATTO CARV confocal unit attached to a Nikon microscope. Images were obtained using Metamorph software and analyzed with Metamorph and Image J.
Latrunculin A experiments:
Fly tissues were dissected in Schneider's medium and then incubated in 50 μm latrunculin A (Lat-A), followed by fixation and staining by standard techniques.
Statistical analyses:
When possible groups were compared using either an analysis of variance or a t-test (if there were only two groups being compared). In some cases unequal variance led us to use the nonparametric Mann-Whitney Rank Sum test. The SigmaStat program was used.
RESULTS
Identification of the flare gene:
Epidermal cells mutant for flr elaborate highly abnormal hairs (Figures 1 and 2). Previous data indicated that flr mapped to the middle of 3L (Garcia-Bellido and Dapena 1974). This map position was refined to the 70AB region by the students of the Biology 411 class at the University of Virginia in the spring of 2005. In scanning the genes located in this region CG10724 stood out as an obvious candidate as it encodes the Drosophila AIP1 homolog. We obtained an allele of CG10724 (EY05812) associated with a P insertion into the first exon. This allele failed to complement a series of five independent flr alleles isolated in our laboratory. The P-insertion allele (henceforth called flr7) was not a null allele and when heterozygous with a weak allele (flr4) isolated in Charlottesville, a number of adult escapers were obtained. These showed an obvious flr phenotype in the distal region of the wing, with a progressively weaker phenotype as one moved proximally (Figure 1, A–C). We did not see a phenotype in other body regions, but it is likely that these alleles interfered with flr function in other tissues as flr4/flr7 flies had reduced viability (∼10% of expected). To determine if the P insertion in CG10724 was responsible for the recessive lethality of this chromosome as well as its failure to complement flr alleles we mobilized the P insert and obtained revertants based on the loss of the w+ gene contained in the transposon. Most of the putative revertants were homozygous viable and wild type at flare. In addition, we also isolated a number of partial revertants. These showed a very weak flr wing hair phenotype when homozygous or when heterozygous with flr4 (data not shown). Heteroallelic combinations that did not involve flr4 (or partial revertants of flr7) were all lethal.
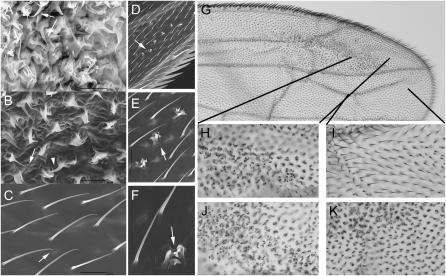
Figure 1.—
The flare mutant phenotype and rescue. A–C are scanning electron micrographs that show the distal, medial and proximal regions of flr7/flr4 hypomorphs. The panels show very strong, moderately strong, and very weak phenotypes, respectively. D–F are SEMs of a flr5 clone in the wing at several different magnifications. Arrows point to the mutant cells. Note the blebs and ridges. G is a low magnification image of a ptc-GAL4/UAS-flr∷GFP; flr7/flr4 fly. Wings from such flies are typically partly folded and do not lie flat. The flr mutant phenotype is rescued in the central region of the wing (the ptc domain) while the flr hypomorphic phenotype is obvious outside of the ptc expression domain. H and I are higher magnification images of parts of G that show mutant and rescued regions, respectively. The lines indicate where these regions are located on the low magnification image. Note the normal hair morphology of the rescued cells. J and K show the equivalent regions of an flr7/flr4 wing (no rescue control). Note that there is a strong flr phenotype in all distal regions in such wings.
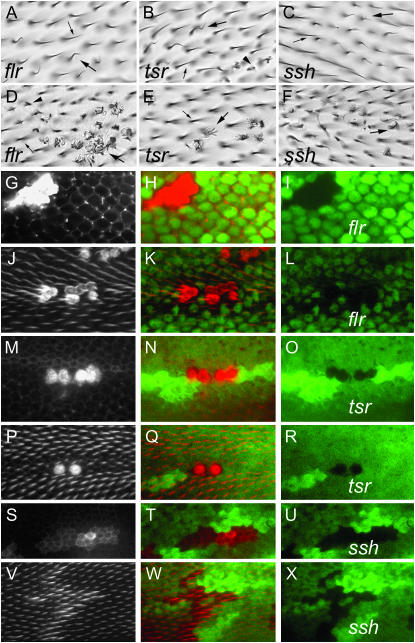
Figure 2.—
A comparison of the flr, tsr, and ssh mutant phenotypes. A–C are light micrographs that show respectively flr, tsr, and ssh clones that display weak phenotypes. Small arrows point to wild-type cells and large arrows to mutant cells displaying the weak phenotype. B also shows a few mutant cells with a strong phenotype (arrowhead). D–F show, respectively, flr, tsr, and ssh clones that display strong phenotypes for these genes. The large arrows point to mutant cells and small arrows to wild-type cells. Note the flr and tsr mutant cells show a similar phenotype and that this is stronger than the ssh phenotype. G–I show a flr clone marked by the loss of GFP and stained for F-actin using Alexa 568 phalloidin. These panels are of a pupal wing prior to hair morphogenesis (∼28 hr after white prepupa, awp). G shows the phalloidin staining, H the overlap, and I the GFP by itself. The phalloidin staining is shown in grayscale in G (this provides a bit more detail and contrast) and in red in the overlap (H). J–L are the equivalent for a clone in a pupal wing during hair morphogenesis (∼32 hr awp). M–O are the equivalent for a tsr clone prior to hair formation and P–R are the equivalent for a tsr clone during hair morphogenesis. S–U are the equivalent for an ssh clone prior to hair morphogenesis and V–X are the equivalent for an ssh clone during hair morphogenesis. For this figure the null alleles flr5, tsrN121, and ssh63-11 were used. There is substantial clone-to-clone variation in both the pupal and adult wings and this is visible in the set of clones displayed. For example, examine the two clones in B and the two cells on the right compared to the two cells on the left in M.
To confirm that we had correctly identified CG10724 as flr we constructed a UAS-flr∷GFP gene and obtained transgenic animals. When the expression of this transgene was driven in the center of the wing using ptc-Gal4 it rescued the wing hair phenotype seen in flr4/flr7 flies (i.e., w; ptc-GAL4/UAS-flr∷GFP; flr4/flr7 ) (see Figure 1, G–K). We tested the ability of the transgene to provide flr function in other cell types by constructing flies that carried strong flr alleles and ubiquitously expressed the transgene (i.e., act-Gal4/UAS-flr∷GFP; flr5/flr6). We obtained a small number of phenotypically normal flies. That these were recovered in smaller numbers than predicted suggested there were one or more tissues/developmental stages where flr function is essential and where the directed expression of the transgene did not provide optimal rescue. These rescue experiments confirmed that CG10724 was flare and that our GFP fusion protein was functional.
Genome annotation of CG10724 suggests that it encodes two isoforms that are transcribed from different promoters. The two mRNA isoforms contain different 5′-most exons followed by six common exons. The two flare proteins are predicted to contain 608 and 604 amino acids that differ at their 12 and 8 amino terminal residues. Our transgenic experiments utilized the 608 amino acid isoform.
The Flare protein:
BLAST searches showed that flr encoded the Drosophila AIP1 (actin interacting protein 1) homolog. Homologs of AIP1 are found in all animals, in plants, fungi, and other eukaryotes, and the protein is conserved throughout its length (supplemental Figure S1 at http://www.genetics.org/supplemental/). AIP1 has been extensively studied in yeast and worms (Fedorov et al. 1997; Rodal et al. 1999, 2005; Ono 2001; Mohri and Ono 2003; Voegtli et al. 2003; Mohri et al. 2004, 2006; Ono et al. 2004; Clark et al. 2006; Okada et al. 2006).
We isolated and sequenced DNA from three recently isolated flr alleles. The weak flr4 allele was associated with a change of ser158 to leu. This ser is conserved in all animal flr homologs (supplemental Figure S1 at http://www.genetics.org/supplemental/) and in homologs from more distantly related organisms such as Arabidopsis thaliana, Physarum polycephalum, and Trypanosoma cruzi. Consistent with this being a weak allele the residue is not absolutely conserved, with ser being replaced by ala in some fungi. The two strong alleles both encode partial FLR proteins. flr5 is associated with a small internal deletion that removes the sixth exon resulting in an internal deletion of 76 amino acids, from amino acid 482 to amino acid 558. This removes most of two WD40 motifs. flr6 contained a nonsense mutation that results in a truncated protein of 541 amino acids that is missing the carboxy terminal 67 amino acids. This region contains one entire and part of an additional WD40 motif.
To localize the FLR protein in fly cells we used the FLR∷GFP fusion protein. In pupal wing cells prior to and during hair formation FLR∷GFP was preferentially localized to the apical-lateral cell periphery (Figure 3, A–C) with at most only weak accumulation in the hair. When we expressed FLR∷GFP in growing bristles some accumulation was seen in the bristle but it did not preferentially accumulate there as does F-actin (Figure 2, D–F). The lack of accumulation of FLR in growing hairs and bristles is consistent with the evidence that actin polymerization is primarily distal and that actin filaments and bundles depolymerize in the cell body (Guild et al. 2002, 2005).
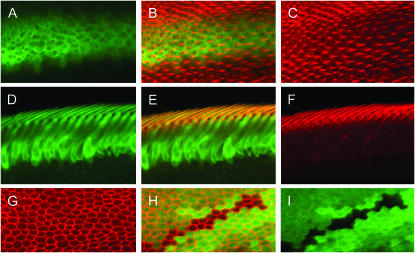
Figure 3.—
A–C are of 32-hr ptc-Gal4/UAS-flr∷GFP pupal wings stained with Alexa-568 phalloidin (red). B is the merged image. The ptc-Gal4 driver results in expression of flr-GFP in the central region of the wing. Note the lack of prominent hair accumulation of Flr∷GFP and that it is located peripherally. D–F are of 30-hr UAS-flr∷GFP/+; neur-Gal4/+ pupal wings stained with Alexa-568 phalloidin (red). The neur-Gal4 enhancer trap drives expression of flr∷GFP in the bristle sense organ cells. Note the intense F-actin staining of the developing bristles. Flr∷GFP is seen in the bristle but it does not specifically accumulate there compared to other parts of bristle sense organ cells. G–I are of 28-hr pupal wings containing flr4 clones marked by the loss of GFP that were stained with an anti-Armadillo monoclonal antibody. Note the lack of effect of the clone on Armadillo localization.
The flr mutant phenotype:
To better characterize the flr wing hair phenotype we examined both mutant clones and hypomorphic adult wings in the SEM as well as the light microscope (Figures 1, A–F, and 2, A and D). A range of phenotypes was seen. The most extreme cells showed multiple blebs and ridges. Less severely affected cells showed spikes that resembled small hairs in addition to the blebs and ridges. Still less severely affected cells showed hairs that were wider, often bent, and occasionally split. The weakest phenotype was a hair with slightly bent tip (Figures 1C and 2A).
Mutations in cofilin are thought to be cell lethal due to cofilin being required for cytokinesis (Gunsalus et al. 1995). The ability to recover clones of cells mutant for flr could be due to flr not being required for cytokinesis or to perdurance of wild-type protein. To distinguish between these possibilities we generated flr clones at various times after egg laying (ael) and compared clone size and frequency. As a control we used multiple wing hairs (mwh) as this gene is also located on 3L, is a good cuticular marker, and is not essential. As development proceeds the number of cells in the wing disc increases and this results in an increase in the frequency of clones induced at later stages. The resulting clones are smaller, however, due to a reduced time for proliferation. As expected when we examined mwh clones we found an increase in frequency and a decrease in size when clones were induced at later ages (Figure 4). flr mutant clones induced 4–5 days ael were recovered at a similar frequency to the mwh clones and they were of similar size. However, clones for strong flr alleles induced 3–4 days ael contained significantly fewer cells than those induced at 4–5 days ael. Thus, flr mutant cells were lost over time. For the hypmorphic flr4 allele the 3–4 day and 4–5 day clones were of similar size. This could be due to these cells stopping proliferating or to cell death balancing continued growth. Almost no flr clones were recovered from wings when clones were induced from 2–3 days ael. Consistent with the mechanism of cell death being due to a failure of cytokinesis occasional polyploid wing cells [identified by their size (Adler et al. 2000)] were seen in wings with flr clones (data not shown).
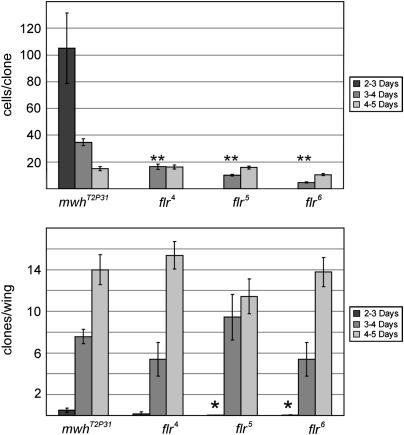
Figure 4.—
flr is required for cell proliferation and cell viability. (Top) The mean number of cells per clone as a function of the time of clone induction is shown. As a control we used mwh, as this is a nonessential gene that has long been used as a gratuitous cell marker. As expected, the size of mwh clones decreased dramatically with later times of induction. In contrast, for the two strong flr alleles mean clone size was smaller when clones were induced 3–4 days ael vs. 4–5 days ael. For the hypomorphic flr4 allele no difference in clone size was seen when clones were induced at 3–4 days ael vs. 4–5 days ael. flr clones induced at 3–4 days ael were significantly (P < 0.001) smaller than mwh clones induced at the same time (indicated by **) consistent with the flr clones showing decreased proliferation and/or increased cell death. Too few flr clones were recovered after induction at 2–3 days ael to allow us to compare their size. (Bottom) The frequency of clones per wing is compared for the four mutations as a function of time of induction. For both of the strong flr alleles the frequency of clones induced at 2–3 days ael was significantly (P < 0.05) lower than that of the control mwh clones indicating that the flr cells died.
The tsr (twinstar) gene encodes Drosophila cofilin, which is required for cell viability (Gunsalus et al. 1995). Several papers have reported that tsr clones have increased F-actin accumulation, thus mutant cells must be viable under some circumstances (Baum et al. 2000; Baum and Perrimon 2001). A likely explanation is that like flr, tsr mutant cells might survive for some time due to perdurance of wild-type gene product. If so would they also display a flr like epidermal hair phenotype? To test this we induced tsr mutant clones in larvae of various ages and examined adult wings. When clones were induced from 4–5 days ael, small clones that displayed a strong flr-like hair phenotype were seen (Figure 2, B and E). These clones were rarely larger than 8 cells (the largest clone we found was 20 cells) and inducing clones earlier in development did not result in larger clones (data not shown). Indeed, few clones were recovered when they were induced 3–4 days ael. The mutant hair phenotypes and observations on clone recovery suggested the tsr phenotype was slightly stronger than flr. Consistent with the hypothesis that flr and tsr functioned together we found that reducing tsr gene dose from two to one enhanced the phenotype of flr hypomorphs resulting in lethality (i.e., tsr−/+; flr4/ flr7 flies died as young larvae).
slingshot (ssh) encodes a phosphatase that reverses the phosphorylation-mediated inactivation of cofilin (Niwa et al. 2002). ssh mutant clones produce epidermal hairs that are often split, multipled, and deformed (Niwa et al. 2002). We examined ssh clones on the wing found a range of phenotypes that were similar to weak to moderate flr clones (Figure 2). Large ssh clones were obtained and we saw no evidence for ssh being required for cell viability. This was consistent with results reported previously by others (Niwa et al. 2002).
flr and actin stability:
We examined the flr mutant phenotype in pupal wings in both marked clones and in hypomorphic wings and found a dramatic cell-autonomous increase in the level of F-actin (Figure 2, G–L). Similar increases in F-actin accumulation have been reported for both tsr and ssh mutant clones and we confirmed these results (Figure 2, M–X) (Baum et al. 2000; Baum and Perrimon 2001; Niwa et al. 2002). The increase in F-actin was seen both prior to and during hair morphogenesis. In Z sections increased F-actin could be detected both apically and basally (supplemental Figure S2 at http://www.genetics.org/supplemental/). To compare the relative phenotypes of flr, tsr, and ssh we examined F-actin accumulation in clones mutant for each of these in parallel experiments (Figure 2). Dramatic increases were seen in all three although our qualitative evaluation of the phenotypes was that tsr ≥ flr > ssh. This was similar to the ranking with respect to cell lethality and hair morphology and suggested all of these phenotypes had a similar basis. It is interesting that in many ssh clones we did not see a dramatic pupal hair phenotype, as might be expected from the adult hair phenotype. Growing ssh mutant hairs often appeared longer, thicker, and brighter staining than their wild-type neighbors but were not notably distorted (Figure 2). This suggested that the ssh hair phenotype might be due in part to an inability to depolymerize F-actin late in hair morphogenesis.
Biochemical studies done on cofilin and AIP1 from a variety of sources argue that these proteins serve to promote actin filament disassembly (Mohri and Ono 2003; Mohri et al. 2004, 2006; Ono et al. 2004; Rodal et al. 2005; Brieher et al. 2006; Clark et al. 2006; Okada et al. 2006). In vivo evidence for this has been obtained in yeast (Okada et al. 2006). To determine if FLR promoted actin filament disassembly in fly cells we cultured pupal wings and wing discs that contained marked flr clones for varying periods of time in the presence of Lat-A. These tissues were then fixed and stained for F-actin with a fluorescent phalloidin. Lat-A binds to actin monomers and prevents them from being incorporated into new actin filaments. Thus, in cells cultured with Lat-A no new actin filaments are formed (nor old ones extended) and we can follow the disassembly of preexisting actin filaments (Okada et al. 2006). In experiments on both pupal wings and wing discs (and in control experiments on other tissues) we observed that different cellular actin filaments disappeared at different rates. For example, muscle actin filaments were quite stable. Even after 90 min in Lat-A actin filaments in muscle (body wall or gut) did not appear to be changed (supplemental Figure S3 at http://www.genetics.org/supplemental/). In contrast, dispersed cytoplasmic actin filaments in a variety of cells types (e.g., pupal wing cells, wing disc cells, and salivary gland cells) were rapidly lost during culture in Lat-A (Figure 5, A–J, Figure S3). From casual observation it appeared that flr mutant clone cells did not lose phalloidin staining as rapidly as their wild-type neighbors. To quantify this we compared the level of phalloidin staining of mutant cells with their wild-type neighbors (see materials and methods for details). Prior to culture in Lat-A, flr mutant cells had a four- to fivefold higher level of phalloidin staining than neighboring wild-type cells. This increased during culture in the presence of Lat-A to 10-fold or greater (Figure 5K). Such changes were not seen when cells were cultured in the absence of Lat-A (Figure 5K). Thus, F-actin was more stable in the mutant than wild-type cells. The kinetics of loss of phalloidin staining varied somewhat from experiment to experiment. We suspect that this is due to variation in the rate of uptake of the Lat-A in the cultured pupal wings. This tissue is very fragile and we were only able to obtain good viability if the tissue was left within its sac of pupal cuticle, which likely provided a partial barrier to uptake of the drug. The presence of the pupal cuticle results in wings tending to float, which further increases the possibility of altered kinetics of drug uptake.
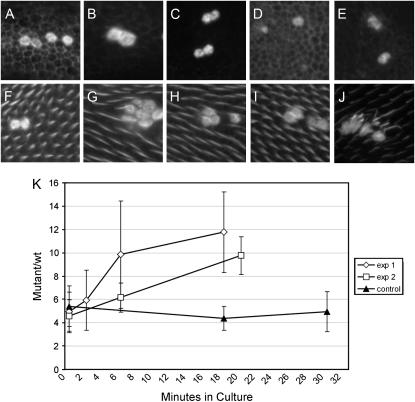
Figure 5.—
Mutations in flr stabilize wing cell F-actin. (A–J) Confocal micrographs of wing tissue containing flr5 clones stained with Alexa-568 phalloidin. A–C are of 24–28-hr pupal wings after 0, 6, and 18 min of culture in the presence of Lat-A. The flr mutant cells have greatly elevated F-actin staining levels. Note the loss of most of the F-actin staining in the wild-type cells after 6 min in the presence of Lat-A. D and E show third instar wing discs containing flr clones after 0 and 2 min, respectively, in Lat-A. F–J show 32–34-hr pupal wings containing flr clones after 0, 2, 6, 18, and 40 min of culture in Lat-A. Note the quick loss of general F-actin staining during culture in Lat-A and that the wing hair staining is relatively stable. Note that by 40 min in Lat-A wild-type wing hair F-actin staining is substantially reduced. The time required to see such an effect varied from experiment to experiment. K shows a plot of relative F-actin staining in flr clones compared to neighboring wild-type cells for pupal wings cultured for various periods of time in Lat-A. Two independent experiments are shown. A control experiment where pupal wings were cultured without any Lat-A is also shown. Note there is no change in relative actin staining in mutant vs. wild-type cells in the control experiment even after 30 min of culture.
The experiments described above were done on pupal wings prior to hair formation, which occurred ∼32 hr after white prepupae. In several experiments we also examined flr clone-bearing pupal wings that were in the process of hair formation. Wild-type cell F-actin hair staining was relatively resistant to Lat-A treatment (Figure 5, F–J), although with long incubations (40–90 min) we did often see a reduction in staining. Dispersed F-actin staining remained quite sensitive to Lat-A in these wings; thus the stability of hair staining appears to be due to greater F-actin stability in the hair subcellular compartment and not to slower uptake of drug.
flr mutations disrupt the microtubule cytoskeleton:
The dramatic hair and increased F-actin phenotypes of flr, tsr, and ssh mutant cells suggested the possibility that other aspects of cell structure might be disrupted. To test this possibility we examined mutant clones by immunostaining with anti-tubulin and anti-acyl-tubulin antibodies. Clones of flr (Figure 6) and tsr (data not shown) cells showed highly increased staining using both antibodies. This was seen both prior to and during the process of hair morphogenesis. The increase and alteration in tubulin immunostaining appeared quite similar to that for the actin cytoskeleton with substantial colocalization of the staining. This suggested the possibility that the increased microtubule staining was a consequence of the increased levels of F-actin. Only a weak and variable increase in anti-tubulin staining was seen in ssh clone cells (data not shown). This could be due to the increase in F-actin often being less dramatic in ssh clone cells than in flr or tsr cells.
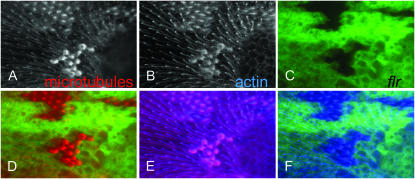
Figure 6.—
Mutations in flr and tsr alter the distribution of microtubules. A–F show a region of a 34-hr pupal wing with several flr4 clones that are marked by the loss of GFP (C). A shows tubulin immunostaining and B shows F-actin staining with Alexa 647 phalloidin. A and B are shown in grayscale to maximize contrast. This was particularly important for B as phalloidin staining with Alexa 647 phalloidin after immunostaining often results in a relatively weak signal. A is in red and B in blue in the merged images (D–F). D is a merge of A and C, E a merge of A and B, and F a merge of B and C. Note the accumulation of both F-actin and tubulin staining in the flr clone.
We also stained flr, tsr, and ssh clones using anti-armadillo antibody and did not see any consistent differences between mutant and wild-type neighbors indicating that the adherens junctions were normal (Figure 3, G–I). Similar results were also obtained using anti-DE-cadherin antibodies on flr and ssh mutant cells. Thus, basic apical basal polarity did not appear to be altered. Some mutant cells in flr clones showed a substantially increased level of anti-DE-cadherin immunostaining compared to their wild-type neighbors but this was not a consistent result and its significance is unclear.
flr and wing planar polarity:
Recently it was found that a viable hypomorphic tsr genotype resulted in planar polarity phenotypes and that the asymmetric accumulation of planar polarity proteins was disrupted (Blair et al. 2006). The conclusion of this study was that the actin cytoskeleton was not only the target of the planar cell polarity pathway but also was required for the function of the pathway. In examining flr clones in adult wings we found occasional clones where a neighboring wild-type cell produced either multiple hairs or a hair of obviously abnormal polarity (Figure 7, C and D). The frequency of detecting such clones varied from 9% to 3% (total n = 599 clones) in experiments that involved three different alleles (flr4, flr6, and flr5) and two ways of generating clones (e.g., vg-Gal4 UAS-flp vs. hs-flp). We also found this to be the case for tsr clones [8% of clones (n = 710)] but it was seen much less frequently for ssh clones (<1% of 365 clones). This behavior is also seen for clones mutant for essentially any of the planar polarity genes suggesting that in the PCP pathway was not functioning normally in flr cells (Park et al. 1996; Adler 2002). To test this we immunostained pupal wings bearing marked flr clones using an anti-Stan/Fmi monclonal antibody (Chae et al. 1999; Usui et al. 1999). We found that the normal zig-zag staining of the Stan protein was disrupted (Figure 7, A and B). In some cells the protein accumulated on an anterior/posterior boundary and in some cases the disruption extended into neighboring wild-type cells.
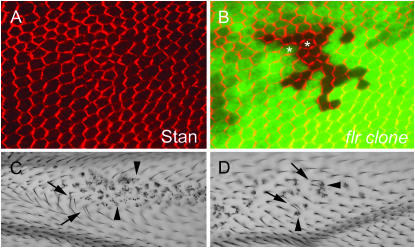
Figure 7.—
flr functions in planar cell polarity. A and B show an flr5 clone in a pupal wing marked by the loss of GFP (B). Both panels show immunostaining of Stan protein in red. Note the disruption of the Stan zig-zag staining pattern in some clone and neighboring cells (several examples are marked by asterisks). C and D are examples of flr5 adult wing clones that show weak domineering nonautonomy for planar polarity. Arrows point to wild-type cells that have produced more than one hair (a common planar polarity phenotype). In some cases hairs no longer point distally. Arrowheads point to flr mutant cells.
DISCUSSION
cofilin and AIP1:
At first glance, the relationship between cofilin and AIP1 appears to be simple; however, it shows some complexity and differences between organisms. In yeast it is clear the two proteins function together to promote actin filament disassembly, but the data argue strongly that cofilin is the more important of the two. The cofilin gene is essential, while AIP1 null mutant cells are viable and show only subtle actin cytoskeleton abnormalities (Rodal et al. 1999; Clark et al. 2006; Okada et al. 2006). Biochemical experiments also argued that AIP1 has no actin depolymerizing activity on its own (Okada et al. 1999; Rodal et al. 1999; Ono 2003). Hence, AIP1 can be considered to be a cofactor that regulates/stimulates cofilin activity. Previous data had suggested that at AIP1 might be functionally more important in organisms other than yeast. For example, AIP1 had been found to have a role in cytokinesis in both Dictyostelium and mammalian cells (Konzok et al. 1999; Gerisch et al. 2004; Fujibuchi et al. 2005). The data in this study show that in Drosophila at the genetic level cofilin (tsr) and AIP1 (flr) are functionally of similar importance. Data previously published (Gunsalus et al. 1995; Blair et al. 2006) or reported in this study established that both genes were essential for organismal and cell viability, and that mutations in either gene resulted in grossly deformed epidermal hairs, an abnormal accumulation of F-actin and microtubules, and interfered with the functioning of the frizzled-based planar cell polarity system. The cellular phenotypes of tsr clones appeared on average slightly stronger than those of flr, but the end point for both types of clones was the same (death) and the strongest cell viable phenotypes were equivalent. Hence, we suggest the differences were due to greater perdurance of flr protein and/or mRNA. It remains unclear why cofilin appeared to be of substantially greater importance than aip1 in yeast, while that was not the case for fly cells. Part of the explanation could be due to both cofilin and AIP1 being required for cytokinesis in fly (and presumably other animal cells). This is essential for animal cells, but budding yeast does not carryout cytokinesis. A complete explanation, however, will require the identification of an essential process in yeast that requires cofilin, but not AIP1. Once such a process is identified it will be interesting to examine this in fly cells. The phenotypes of ssh clones stand in contrast to those of tsr and flr. While there was substantial overlap, the ssh clones phenotypes were less severe and as reported previously (Niwa et al. 2002) ssh does not appear to be required for cell viability as are tsr and flr.
How do mutations in these genes alter the MT cytoskeleton?
Extensive genetic and biochemical experiments with cofilin and AIP1 from other systems have established these proteins as being key players in the turnover of actin filaments (Ono 2003; Paavilainen et al. 2004; Clark et al. 2006). Thus, it was not surprising that mutations in both of these genes led to dramatic over-accumulation of F-actin in fly epidermal cells. However, none of the published data suggested that there would also be a dramatic increase in tubulin immunostaining, which we detected in wing cells. What is the basis for this? We suspect that the increase in tubulin immunostaining was due to a localized accumulation of polymerized microtubules and not to an overall increase in cell tubulin abundance, although we could not test this biochemically due to our inability to obtain a pure population of mutant cells. If we assume this is the case there are two possible mechanisms to explain the increase in tubulin immunostaining. One is that cofilin and AIP1 directly function to promote microtubule depolymerization, much as they do to promote actin filament disassembly. We think this hypothesis is unlikely. Since both flr and tsr mutations resulted in a similar increase in tubulin immunostaining both genes/proteins would have to share this unknown function. Both cofilin and AIP1 have been extensively studied biochemically and we are not aware of any report that these proteins can interact with microtubules or tubulin. In addition, extensive genetic experiments, particularly in yeast, have failed to show any evidence for a genetic interaction between either cof1 or aip1 and tubulin or microtubule components. An alternative hypothesis is that flr and tsr mutations result in an increased concentration of F-actin and this in turn results in increased tubulin staining. There is a substantial literature on proteins that mediate connections between the actin and microtubule cytoskeletons (e.g., Roper et al. 2002; Miller et al. 2004) although none of the interactions are as dramatic as the one we found. One example of a gene that encodes a protein that links the actin and microtubule cytoskeletons and that function in the morphogenesis of the fly wing is short stop (shot), which encodes a spectraplakin family member. However, it is unlikely that this protein plays an important role in linking the actin and microtubule cytoskeletons during hair morphogenesis as shot mutant wing cells produce normal-looking hairs (Roper et al. 2002).
Planar polarity and the actin cytoskeleton:
We found that clones of flr mutant cells resulted in disorganized Stan/Fmi accumulation in clone cells and in neighboring wild-type cells. We also found by examining adult wings that flr and tsr clones sometimes produced planar polarity-like phenotypes in neighboring wild-type cells. These observations are consistent with the previous demonstration that tsr function is essential for the development of normal planar polarity (Blair et al. 2006). In that study a hypomorphic tsr genotype produced relatively strong planar polarity phenotypes, without producing any notable hair morphology phenotype. One difference between our results and those of Blair et al. (2006) is that we only saw planar polarity phenotypes in or next to flr clones that produced abnormal hairs. We did not see such phenotypes in regions of wings from viable hypomorphs where hair morphology was normal. The observations that planar polarity phenotypes can arise from mutations in tsr, flr, and Lim kinase (Blair et al. 2006) argue strongly that this is a consequence of defects in actin dynamics. We cannot rule out the hypothesis that the planar polarity phenotypes involve the indirect disruption of the microtubule cytoskeleton.
Why are flr mutant wing hairs so abnormal?
The hairs produced by both flr and tsr wing cells are far more abnormal than those seen due to mutations in genes that encode actin bundling proteins (Bryan et al. 1993; Petersen et al. 1994), capping protein (Hopmann et al. 1996; Hopmann and Miller 2003; Frank et al. 2006), cytoplasmic myosins such as myosin VII (Turner and Adler 1998; Kiehart et al. 2004), or other proteins thought to modulate actin polymerization, such as profilin (Verheyen and Cooley 1994) or Arp2/3 (Hudson and Cooley 2002) . The reason for this is uncertain and leads to the question as to what is the basis for the extreme flr phenotype? One possibility is that the disassembly of F-actin in growing hairs is essential for morphogenesis. If this were the case we predict that early hairs would look normal but that later ones would get progressively more abnormal. Some evidence supporting this possibility was obtained. Occasional ssh pupal wing clone cells had hairs that appeared relatively normal except for being longer, thicker, and brighter staining than their wild-type neighbors. We expect that these cells went on to produce abnormal adult cuticular hairs due to a defect in late stages of morphogenesis. We did not see such hairs in flr and tsr mutant cells as for these mutations hairs were abnormal from the start of hair morphogenesis. This could be due to these mutations producing on average much more severe cellular phenotypes. Even for ssh a late function is unlikely to be the whole story as early ssh hairs were often abnormal. Indeed, cells mutant for any of these genes showed increased levels of F-actin at all times observed from third instar wing discs to pupal wings before, during, and after hair morphogenesis. It is also worth noting that other experiments showed that hair F-actin is much more stable than general cellular F-actin. Thus, we expect that hair morphogenesis would be relatively insensitive to a decrease in F-actin disassembly. Further evidence against the hair phenotype being due to a direct effect on hair F-actin disassembly is that the Flr protein did not accumulate in growing hairs.
A second possibility is that the hair defect in flr and tsr mutant cells is a consequence of the buildup of stable F-actin in the cell leading to a lack of G-actin required for hair growth. We cannot rule out this hypothesis; however, in those cases where we could see a discrete mutant hair it did not stain less strongly for F-actin as would be predicted by this hypothesis. Further, the inhibition of actin polymerization with drugs such as latrunculin A does not phenocopy the blebs and ridges so prominent in flr and tsr mutant hairs (Turner and Adler 1998; Geng et al. 2000; He and Adler 2002).
A third possibility to explain the grossly abnormal hair morphology phenotypes of flr, tsr, and ssh is that the actin cytoskeleton is involved in multiple aspects of hair morphogenesis and that the phenotype is due to additive effects (Figure 8). The morphogenesis of the hair is known to be under at least two types of control—spatial and temporal. The activity of the fz-based planar/tissue polarity system restricts hair initiation to the distal-most part of the apical surface of cells (Wong and Adler 1993). Temporal control of hair initiation is likely to involve the turn on of expression of shavenoid/kojak (He and Adler 2002; Ren et al. 2006), perhaps due to its expression being regulated by the shavenbaby and/or grainy head transcription factors (Lee and Adler 2004; Chanut-Delalande et al. 2006). The planar cell polarity (PCP) proteins are known to become restricted to the proximal and distal sides of cells and it is generally believed that feedback interactions between these proteins lead to the formation of these two protein domains (Tree et al. 2002; Amonlirdviman et al. 2005). There is evidence that both the microtubule and actin cytoskeletons serve to organize intracellular transport to help establish these domains (Shimada et al. 2005; Blair et al. 2006). Defects in actin disassembly could interfere with the formation of these domains (Blair et al. 2006). The initial formation of the PCP domains could polarize the microtubule and/or actin cytoskeletons to direct intracellular transport of PCP proteins to the correct domains providing the system with positive feedback. It is likely that the PCP proteins function by localizing the activity of downstream effector proteins. One of these, the Inturned protein, has been found to be localized to the proximal side of wing cells under the direction of the PCP proteins (Adler et al. 2004). This localization is also likely to require the activity of the actin and microtubule cytoskeletons and disruption of these can at least partially phenocopy inturned mutations (Turner and Adler 1998). The activity of the PCP and PCP effector proteins likely results in one or more proteins localizing at and marking the site for hair initiation. Once this site is selected the growth of the hair will presumably involve the extension of the membrane and the transport of hair-building proteins into the growing hair. This is another cellular function that is likely to involve the cytoskeleton and once again we anticipate this will involve a positive feedback system. For example, a mark at the distal tip of a growing hair could recruit and polarize the cytoskeleton to direct intracellular transport into the hair to drive outgrowth as well as to further stabilize the mark and maintain cytoskeleton organization. This could be the basis for the cytoskeleton-based refinement process that is needed for insuring a single hair is formed (Adler 2002). This would be analogous to what is seen in budding yeast (Pruyne et al. 2004) and migrating cells (Etienne-Manneville 2004; Kodama et al. 2004; Dormann and Weijer 2006). The dramatic phenotype of flr and tsr mutant epidermal cells could be a consequence of their profound effect on actin function and the additive effects of disrupting multiple actin-dependent processes. It is also important to remember that mutations in flr and tsr also resulted in an abnormal distribution of microtubules. Thus, some of the effects of flr and tsr mutations could be due to an indirect disruption of microtubule function.
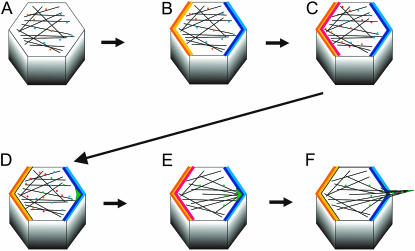
Figure 8.—
A model for the multiple roles of the cytoskeleton in hair morphogenesis. Early in pupal life (A) domains of planar polarity proteins (PPP) have not yet been established. A slight proximal-distal bias to the microtubule cytoskeleton results in a bias in the intracellular transport of PPP (Shimada et al. 2005). This combined with an intraplanar polarity protein feedback inhibition leads to the establishment of proximal and distal domains (Tree et al. 2002) (B). These domains could enhance the bias to the microtubule cytoskeleton providing a further feedback enhancement. The establishment of PPP domains leads to the formation of domains of planar polarity effector proteins (PPEP) (C). At this time this has only been documented for Inturned at the proximal side of wing cells (Adler et al. 2004). The localization of PPEP leads to the accumulation of hair-promoting proteins (HPP) in the vicinity of the distal-most vertex of the cell (D). This leads to the capture of microtubules and/or actin filaments (E), which promotes the further accumulation of HPP at the distal tip of the growing hair which in turn leads to the further capture of cytoskeletal elements and hair outgrowth (F). At the present there is no direct evidence for any early developmental stage proximal distal bias to the actin cytoskeleton.
A final model to explain the grossly abnormal morphology of flr and tsr hairs is that the large accumulation of F-actin and microtubules below the apical surface of the epidermal cells acts as a mechanical block to hair elongation and outgrowth. These abnormal cytoskeletal structures could be linked to the plasma membrane cytoskeleton and inhibit its normal outgrowth. This could act as another component of the additive model suggested above.
Acknowledgments
Core facilities used in carrying out the research were supported by the University of Virginia Cancer Center. Fly stocks were obtained from the Drosophila stock center at Indiana University in Bloomington, Tadashi Uemura, and Frank Laski. This study was supported by grants from the National Institute of General Medical Science to P.N.A.
References
- Adler, P. N., 2002. Planar signaling and morphogenesis in Drosophila. Dev. Cell 2: 525–535. [DOI] [PubMed] [Google Scholar]
- Adler, P. N., J. Liu and J. Charlton, 2000. Cell size and the morphogenesis of wing hairs in Drosophila. Genesis 28: 82–91. [DOI] [PubMed] [Google Scholar]
- Adler, P. N., C. Zhu and D. Stone, 2004. Inturned localizes to the proximal side of wing cells under the instruction of upstream planar polarity proteins. Curr. Biol. 14: 2046–2051. [DOI] [PubMed] [Google Scholar]
- Amonlirdviman, K., N. A. Khare, D. R. Tree, W. S. Chen, J. D. Axelrod et al., 2005. Mathematical modeling of planar cell polarity to understand domineering nonautonomy. Science 307: 423–426. [DOI] [PubMed] [Google Scholar]
- Arber, S., F. A. Barbayannis, H. Hanser, C. Schneider, C. A. Stanyon et al., 1998. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393: 805–809. [DOI] [PubMed] [Google Scholar]
- Balcer, H. I., A. L. Goodman, A. A. Rodal, E. Smith, J. Kugler et al., 2003. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13: 2159–2169. [DOI] [PubMed] [Google Scholar]
- Baum, B., 2002. Winging it—actin on the fly. Dev. Cell 2: 125–126. [DOI] [PubMed] [Google Scholar]
- Baum, B., and N. Perrimon, 2001. Spatial control of the actin cytoskeleton in Drosophila epithelial cells. Nat. Cell Biol. 3: 883–890. [DOI] [PubMed] [Google Scholar]
- Baum, B., W. Li and N. Perrimon, 2000. A cyclase-associated protein regulates actin and cell polarity during Drosophila oogenesis and in yeast. Curr. Biol. 10: 964–973. [DOI] [PubMed] [Google Scholar]
- Blair, A., A. Tomlinson, H. Pham, K. C. Gunsalus, M. L. Goldberg et al., 2006. Twinstar, the Drosophila homolog of cofilin/ADF, is required for planar cell polarity patterning. Development 133: 1789–1797. [DOI] [PubMed] [Google Scholar]
- Brieher, W. M., H. Y. Kueh, B. A. Ballif and T. J. Mitchison, 2006. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J. Cell Biol. 175: 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, J., R. Edwards, P. Matsudaira, J. Otto and J. Wulfkuhle, 1993. Fascin, an echinoid actin-bundling protein, is a homolog of the Drosophila singed gene product. Proc. Natl. Acad. Sci. USA 90: 9115–9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant, K., B. A. Knowles, M. S. Mooseker and L. Cooley, 1994. Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J. Cell Biol. 125: 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, J., M. J. Kim, J. H. Goo, S. Collier, D. Gubb et al., 1999. The Drosophila tissue polarity gene starry night encodes a member of the protocadherin family. Development 126: 5421–5429. [DOI] [PubMed] [Google Scholar]
- Chanut-Delalande, H., I. Fernandes, F. Roch, F. Payre and S. Plaza, 2006. Shavenbaby couples patterning to epidermal cell shape control. PLoS Biol. 4: e290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., D. Godt, K. Gunsalus, I. Kiss, M. Goldberg et al., 2001. Cofilin/ADF is required for cell motility during Drosophila ovary development and oogenesis. Nat. Cell Biol. 3: 204–209. [DOI] [PubMed] [Google Scholar]
- Clark, M. G., J. Teply, B. K. Haarer, S. C. Viggiano, D. Sept et al., 2006. A genetic dissection of Aip1p's interactions leads to a model for Aip1p-cofilin cooperative activities. Mol Biol Cell 17: 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, W. J., and J. W. Thatcher, 1997. Morphogenesis of denticles and hairs in Drosophila embryos: involvement of actin-associated proteins that also affect adult structures. Cell Motil. Cytoskeleton 38: 9–21. [DOI] [PubMed] [Google Scholar]
- Dormann, D., and C. J. Weijer, 2006. Chemotactic cell movement during Dictyostelium development and gastrulation. Curr. Opin. Genet. Dev. 16: 367–373. [DOI] [PubMed] [Google Scholar]
- Egea, G., F. Lazaro-Dieguez and M. Vilella, 2006. Actin dynamics at the Golgi complex in mammalian cells. Curr. Opin. Cell Biol. 18: 168–178. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S., 2004. Actin and microtubules in cell motility: Which one is in control? Traffic 5: 470–477. [DOI] [PubMed] [Google Scholar]
- Fedorov, A. A., P. Lappalainen, E. V. Fedorov, D. G. Drubin and S. C. Almo, 1997. Structure determination of yeast cofilin. Nat. Struct. Biol. 4: 366–369. [DOI] [PubMed] [Google Scholar]
- Frank, D. J., R. Hopmann, M. Lenartowska and K. G. Miller, 2006. Capping protein and the Arp2/3 complex regulate nonbundle actin filament assembly to indirectly control actin bundle positioning during Drosophila melanogaster bristle development. Mol. Biol. Cell 17: 3930–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujibuchi, T., Y. Abe, T. Takeuchi, Y. Imai, Y. Kamei et al., 2005. AIP1/WDR1 supports mitotic cell rounding. Biochem. Biophys. Res. Commun. 327: 268–275. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido, A., and J. Dapena, 1974. Induction, detection and characterization of cell differentiation mutants in Drosophila. Molec. Gen. Genet. 128: 117–130. [DOI] [PubMed] [Google Scholar]
- Geng, W., B. He, M. Wang and P. N. Adler, 2000. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics 156: 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch, G., J. Faix, J. Kohler and A. Muller-Taubenberger, 2004. Actin-binding proteins required for reliable chromosome segregation in mitosis. Cell Motil. Cytoskeleton 57: 18–25. [DOI] [PubMed] [Google Scholar]
- Guild, G. M., P. S. Connelly, K. A. Vranich, M. K. Shaw and L. G. Tilney, 2002. Actin filament turnover removes bundles from Drosophila bristle cells. J. Cell Sci. 115: 641–653. [DOI] [PubMed] [Google Scholar]
- Guild, G. M., P. S. Connelly, L. Ruggiero, K. A. Vranich and L. G. Tilney, 2005. Actin filament bundles in Drosophila wing hairs: hairs and bristles use different strategies for assembly. Mol. Biol. Cell 16: 3620–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus, K. C., S. Bonaccorsi, E. Williams, F. Verni, M. Gatti et al., 1995. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J. Cell Biol. 131: 1243–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, B., and P. N. Adler, 2002. The genetic control of arista lateral morphogenesis in Drosophila. Dev. Genes Evol. 212: 218–229. [DOI] [PubMed] [Google Scholar]
- He, Y., X. Fang, K. Emoto, Y. N. Jan and P. N. Adler, 2005. The tricornered Ser/Thr protein kinase is regulated by phosphorylation and interacts with furry during Drosophila wing hair development. Mol. Biol. Cell 16: 689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann, R., and K. G. Miller, 2003. A balance of capping protein and profilin functions is required to regulate actin polymerization in Drosophila bristle. Mol. Biol. Cell 14: 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann, R., J. A. Cooper and K. G. Miller, 1996. Actin organization, bristle morphology, and viability are affected by actin capping protein mutations in Drosophila. J. Cell Biol. 133: 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, A. M., and L. Cooley, 2002. A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J. Cell Biol. 156: 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janody, F., and J. E. Treisman, 2006. Actin capping protein alpha maintains vestigial-expressing cells within the Drosophila wing disc epithelium. Development 133: 3349–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar, T., E. G. Allwood, R. Anthony, B. Voigt, D. Menzel et al., 2004. The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr. Biol. 14: 145–149. [DOI] [PubMed] [Google Scholar]
- Kiehart, D. P., J. D. Franke, M. K. Chee, R. A. Montague, T. L. Chen et al., 2004. Drosophila crinkled, mutations of which disrupt morphogenesis and cause lethality, encodes fly myosin VIIA. Genetics 168: 1337–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama, A., T. Lechler and E. Fuchs, 2004. Coordinating cytoskeletal tracks to polarize cellular movements. J. Cell Biol. 167: 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzok, A., I. Weber, E. Simmeth, U. Hacker, M. Maniak et al., 1999. DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility. J. Cell Biol 146: 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen, P., E. V. Fedorov, A. A. Fedorov, S. C. Almo and D. G. Drubin, 1997. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 16: 5520–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., and P. N. Adler, 2004. The grainy head transcription factor is essential for the function of the frizzled pathway in the Drosophila wing. Mech. Dev. 121: 37–49. [DOI] [PubMed] [Google Scholar]
- Marston, D. J., and B. Goldstein, 2006. Actin-based forces driving embryonic morphogenesis in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 16: 392–398. [DOI] [PubMed] [Google Scholar]
- Mathur, J., 2006. Local interactions shape plant cells. Curr. Opin. Cell Biol. 18: 40–46. [DOI] [PubMed] [Google Scholar]
- Miller, A. L., Y. Wang, M. S. Mooseker and A. J. Koleske, 2004. The Abl-related gene (Arg) requires its F-actin-microtubule cross-linking activity to regulate lamellipodial dynamics during fibroblast adhesion. J. Cell Biol. 165: 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri, K., and S. Ono, 2003. Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J. Cell Sci. 116: 4107–4118. [DOI] [PubMed] [Google Scholar]
- Mohri, K., S. Vorobiev, A. A. Fedorov, S. C. Almo and S. Ono, 2004. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 279: 31697–31707. [DOI] [PubMed] [Google Scholar]
- Mohri, K., K. Ono, R. Yu, S. Yamashiro and S. Ono, 2006. Enhancement of actin-depolymerizing factor/cofilin-dependent actin disassembly by actin-interacting protein 1 is required for organized actin filament assembly in the Caenorhabditis elegans body wall muscle. Mol. Biol. Cell 17: 2190–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, J., and L. Luo, 2004. Rho GTPases regulate axon growth through convergent and divergent signaling pathways. Neuron 44: 779–793. [DOI] [PubMed] [Google Scholar]
- Niwa, R., K. Nagata-Ohashi, M. Takeichi, K. Mizuno and T. Uemura, 2002. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 108: 233–246. [DOI] [PubMed] [Google Scholar]
- Ohashi, K., T. Hosoya, K. Takahashi, H. Hing and K. Mizuno, 2000. A Drosophila homolog of LIM-kinase phosphorylates cofilin and induces actin cytoskeletal reorganization. Biochem. Biophys. Res. Commun. 276: 1178–1185. [DOI] [PubMed] [Google Scholar]
- Okada, K., T. Obinata and H. Abe, 1999. XAIP1: a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J. Cell Sci. 112(Pt 10): 1553–1565. [DOI] [PubMed] [Google Scholar]
- Okada, K., H. Ravi, E. M. Smith and B. L. Goode, 2006. Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments. Mol. Biol. Cell 17: 2855–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, S., 2001. The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 152: 1313–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, S., 2003. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: new blades for twisted filaments. Biochemistry 42: 13363–13370. [DOI] [PubMed] [Google Scholar]
- Ono, S., K. Mohri and K. Ono, 2004. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/Cofilin-bound actin filaments. J. Biol. Chem. 279: 14207–14212. [DOI] [PubMed] [Google Scholar]
- Paavilainen, V. O., E. Bertling, S. Falck and P. Lappalainen, 2004. Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol. 14: 386–394. [DOI] [PubMed] [Google Scholar]
- Park, W. J., J. Liu, E. J. Sharp and P. N. Adler, 1996. The Drosophila tissue polarity gene inturned acts cell autonomously and encodes a novel protein. Development 122: 961–969. [DOI] [PubMed] [Google Scholar]
- Petersen, N. S., D. H. Lankenau, H. K. Mitchell, P. Young and V. G. Corces, 1994. forked proteins are components of fiber bundles present in developing bristles of Drosophila melanogaster. Genetics 136: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot, F., and T. Lecuit, 2005. Compartmentalized morphogenesis in epithelia: from cell to tissue shape. Dev. Dyn. 232: 685–694. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., A. Legesse-Miller, L. Gao, Y. Dong and A. Bretscher, 2004. Mechanisms of polarized growth and organelle segregation in yeast. Ann. Rev. Cell Dev. Biol. 20: 559–591. [DOI] [PubMed] [Google Scholar]
- Ren, N., B. He, D. Stone, S. Kirakodu and P. N. Adler, 2006. The shavenoid gene of Drosophila encodes a novel actin cytoskeleton interacting protein that promotes wing hair morphogenesis. Genetics 172: 1643–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenu, C., R. Athman, S. Robine and D. Louvard, 2004. The co-workers of actin filaments: from cell structures to signals. Nat. Rev. Mol. Cell Biol. 5: 635–646. [DOI] [PubMed] [Google Scholar]
- Rodal, A. A., J. W. Tetreault, P. Lappalainen, D. G. Drubin and D. C. Amberg, 1999. Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 145: 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, O. C., A. W. Schaefer, C. A. Mandato, P. Forscher, W. M. Bement et al., 2003. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5: 599–609. [DOI] [PubMed] [Google Scholar]
- Rogers, S. L., U. Wiedemann, N. Stuurman and R. D. Vale, 2003. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162: 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper, K., S. L. Gregory and N. H. Brown, 2002. The ‘spectraplakins’: cytoskeletal giants with characteristics of both spectrin and plakin families. J. Cell Sci. 115: 4215–4225. [DOI] [PubMed] [Google Scholar]
- Schafer, D. A., 2004. Regulating actin dynamics at membranes: a focus on dynamin. Traffic 5: 463–469. [DOI] [PubMed] [Google Scholar]
- Shimada, Y., S. Yonemura, H. Ohkura, D. Strutt and T. Uemura, 2006. Polarized transport of Frizzled along the planar microtubule arrays in Drosophila wing epithelium. Dev. Cell 10: 209–222. [DOI] [PubMed] [Google Scholar]
- Tilney, L. G., M. S. Tilney and G. M. Guild, 1995. F actin bundles in Drosophila bristles. I. Two filament cross-links are involved in bundling. J. Cell Biol. 130: 629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney, L. G., P. S. Connelly, K. A. Vranich, M. K. Shaw and G. M. Guild, 2000. Actin filaments and microtubules play different roles during bristle elongation in Drosophila. J. Cell Sci. 113(Pt 7): 1255–1265. [DOI] [PubMed] [Google Scholar]
- Tree, D. R. P., J. M. Shulman, R. Rousset, M. P. Scott, D. Gubb et al., 2002. Prickle mediates feedback amplification to generate asymmetric planar cell polarity signaling. Cell 109: 1–11. [DOI] [PubMed] [Google Scholar]
- Turner, C. M., and P. N. Adler, 1998. Distinct roles for the actin and microtubule cytoskeletons in the morphogenesis of epidermal hairs during wing development in Drosophila. Mech. Dev. 70: 181–192. [DOI] [PubMed] [Google Scholar]
- Usui, T., Y. Shima, Y. Shimada, S. Hirano, R. W. Burgess et al., 1999. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 98: 585–595. [DOI] [PubMed] [Google Scholar]
- Verheyen, E. M., and L. Cooley, 1994. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development 120: 717–728. [DOI] [PubMed] [Google Scholar]
- Voegtli, W. C., A. Y. Madrona and D. K. Wilson, 2003. The structure of Aip1p, a WD repeat protein that regulates Cofilin-mediated actin depolymerization. J. Biol. Chem. 278: 34373–34379. [DOI] [PubMed] [Google Scholar]
- Wahlstrom, G., M. Vartiainen, L. Yamamoto, P. K. Mattila, P. Lappalainen et al., 2001. Twinfilin is required for actin-dependent developmental processes in Drosophila. J. Cell Biol. 155: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, M. D., and R. D. Mullins, 2002. Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 18: 247–288. [DOI] [PubMed] [Google Scholar]
- Wong, L. L., and P. N. Adler, 1993. Tissue polarity genes of Drosophila regulate the subcellular location for prehair initiation in pupal wing cells. J. Cell Biol. 123: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, N., O. Higuchi, K. Ohashi, K. Nagata, A. Wada et al., 1998. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393: 809–812. [DOI] [PubMed] [Google Scholar]