Abstract
French populations of the European corn borer consist of two sympatric and genetically differentiated host races. As such, they are well suited to study processes that could be involved in sympatric speciation, but the initial conditions of host-race divergence need to be elucidated. Gene genealogies can provide insight into the processes involved in speciation. We used DNA sequences of four nuclear genes to (1) document the genetic structure of the two French host races previously delineated with allozyme markers, (2) find genes directly or indirectly involved in reproductive isolation between host races, and (3) estimate the time since divergence of the two taxa and see whether this estimate is compatible with this divergence being the result of a host shift onto maize after its introduction into Europe ∼500 years ago. Gene genealogies revealed extensive shared polymorphism, but confirmed the previously observed genetic differentiation between the two host races. Significant departures from the predictions of neutral molecular evolution models were detected at three loci but were apparently unrelated to reproductive isolation between host races. Estimates of time since divergence between French host races varied from ∼75,000 to ∼150,000 years, suggesting that the two taxa diverged recently but probably long before the introduction of maize into Europe.
THE European corn borer (ECB), Ostrinia nubilalis Hübner (Lepidoptera: Crambidae), exists as a number of sympatric, genetically differentiated but partially interfertile taxa across its geographical range in northern Eurasia and northern America (Hudon et al. 1989). As such, it is a well-suited biological model to study processes that could be involved in the early steps of speciation, especially sympatric and ecological speciation (Schluter 2001; Via 2001; Berlocher and Feder 2002; Gavrilets 2003; Rundle and Nosil 2005).
The ECB's area of origin is thought to be central Asia (Hudon et al. 1989). Its larvae feed on a large number of wild and cultivated host plants (Caffrey and Worthley 1927; Hodgson 1928; Ponsard et al. 2004), but it is commonly found on maize (Zea mays L.). ECB populations collected as larvae on maize stalks in certain locations in Europe (Peña et al. 1988) and in North America (Klun 1975; Kochansky et al. 1975; Glover et al. 1990) are polymorphic with respect to sex pheromone communication. On the basis of this polymorphism, two pheromonal races can be distinguished: the Z-race using a 97:3 blend of Z:E isomers of the 11-tetradecenyl acetate (11-14:OAc), and the E-race using blends of Z11-14:OAc and E11-14:OAc in a 1:99 to 4:96 ratio (Klun 1975). The percentage of hybrid moths is lower than expected under random mating between E- and Z-race moths, suggesting partial reproductive isolation between the two pheromonal races (Glover et al. 1990).
In contrast to other locations in Europe and North America, ECB pheromonal races in France can be distinguished according to the host plant(s) they use (Bourguet et al. 2000; Martel et al. 2003). Indeed, in France, but not in the Balkans, Italy, and North America, maize is infested exclusively by moths using the Z sex pheromone blend (Pélozuelo et al. 2004), whereas hop (Humulus lupulus L.) and mugwort (Artemisia vulgaris L.) are infested exclusively by moths using the E pheromone blend (Thomas et al. 2003; Pélozuelo et al. 2004). These host races have thus been referred to as the “maize race” and the “mugwort–hop race,” respectively. The percentage of hybrids between the two host races is lower than those observed in the United States between the two pheromonal races (Malausa et al. 2005), suggesting that the pheromone difference is probably not the only factor serving to isolate the maize race from the mugwort–hop race.
Given that maize was introduced into Eurasia only ∼500 years ago and has become a major host plant of many ECB populations worldwide, it is reasonable to assume that populations of the maize race have adapted recently to this host plant (Thomas et al. 2003). It is also tempting to hypothesize that the substantial reproductive isolation and genetic divergence between the two French host races is the result of a host shift onto maize followed by host-plant specialization. Indeed, a set of ecological features that might constitute host-plant adaptations, such as emergence timing (Thomas et al. 2003) and oviposition preferences (Bethenod et al. 2005; Malausa et al. 2007), have been found to distinguish both host races in France. However, as with other well-documented host-race models [e.g., Rhagoletis pomonella (Feder et al. 2003), Zeiraphera diniana (Emelianov et al. 2004), Acyrthosiphon pisum (Via 1999), and Enchenopa binotata (Wood et al. 1999)] the exact circumstances of the divergence between ECB races remain elusive.
The genetic divergence between the two pheromonal races on the one hand and the two host races on the other hand was first investigated by means of allozyme markers. American surveys were performed on >30 loci (Harrison and Vawter 1977; Cardé et al. 1978; Cianchi et al. 1980) while 6 loci were analyzed in France (Bourguet et al. 2000; Martel et al. 2003; Thomas et al. 2003; Bontemps et al. 2004). Major differences in allelic frequencies were detected between the American pheromonal races at only 1 locus, Tpi (triose phosphate isomerase), and between French host races at 2 loci: Tpi and Mpi (mannose phosphate isomerase). More recently, the divergence between North American ECB pheromonal races was investigated using a gene genealogical approach based on DNA sequences at 4 nuclear loci, including Tpi, and at the mitochondrial COI locus (Dopman et al. 2005). Only Tpi showed clear differentiation of the two pheromonal strains: all haplotypes of the E-race, although sampled from two quite distant locations, formed a very homogeneous cluster that did not include any Z-race haplotype. For the mitochondrial COI gene, the results from Dopman et al. (2005) for American populations are in agreement with those of Martel et al. (2003), who did not find any differentiation within or between French ECB populations at this locus.
No other genetic data exist on ECB populations collected in France. As a consequence, the delineation of both host races remains incomplete and the hypothesis of divergence triggered by a host shift of some populations onto maize has never been tested. Moreover, no gene has been detected to be involved or located in a chromosomal region involved in host-plant adaptation and reproductive isolation of French host races. This work aims at examining nuclear DNA sequence polymorphism in French ECB populations collected on maize, mugwort, hop, pepper, cocklebur, sunflower, and sorghum. Specifically, our goal is to determine whether the distribution of polymorphisms (1) confirms the host-race delineation found with allozyme markers and possibly even reveals additional genetically differentiated taxa, (2) reveals patterns of monophyly in the host races or selective effects acting at some loci, and/or (3) provides an estimate of divergence time compatible with the scenario of host-race formation by host shift onto maize after its introduction into Europe ∼500 years ago.
MATERIALS AND METHODS
Insect samples:
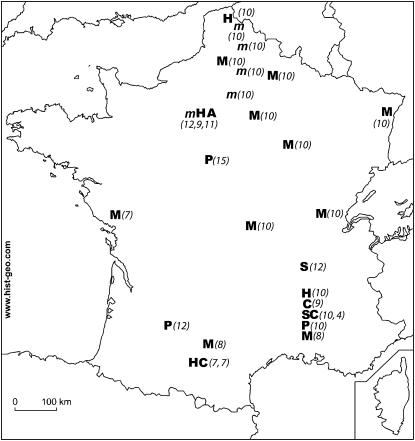
Samples of ECB populations were collected over the course of 2002 and 2003 as fifth larval instars from a total of seven plant species and 27 populations throughout France (Figure 1). In three cases (Muret, Pierrelatte, Grignon), populations from different plants were collected in the same sites. The sex of each individual was determined. Only females were used, as female Lepidoptera are heterogametic (ZW), i.e., haploid at all loci on the Z chromosome, while males are diploid (ZZ). We aimed at collecting ∼10 females for each population. Samples included populations collected from maize (N = 93, 10 populations), mugwort (N = 52, 5 populations), hop (N = 43, 4 populations), sorghum Sorghum sp. (N = 22, 2 populations), cocklebur Xanthium sp. (N = 20, 2 populations), pepper Capsicum frutescens L. (N = 37, 3 populations) and amaranth Amaranthus sp. (N = 11, 1 population). Populations collected from mugwort could be collected only in the northern part of France, as this plant species does not appear to be infested in the southern part of this country (Martel et al. 2003). The geographic distances between all pairs of sampling sites were estimated as linear distances, estimated in kilometers, between points plotted on a map of France (using Google Earth at http://earth.google.com/).
Figure 1.—
Location of sampling sites. Host plants: M, maize; m, mugwort; P, pepper; H, hop; C, cocklebur; S, sorghum; A, amaranth. The number of females sampled in each population is indicated in parentheses. Letters grouped on the same line indicate sites where several plants, located <10 km from each other, were sampled.
Nuclear loci:
Sequence data were obtained from four nuclear loci: Mpi, Tpi, Pbp, and Ket. Mpi and Tpi are both enzymes of carbohydrate metabolism. Pbp encodes a pheromone binding protein, which displays affinity to certain pheromone components in insect species (Du and Prestwich 1995). Ket encodes kettin, a high molecular weight protein of the insect flight muscle (Kolmerer et al. 2000). Tpi and Ket loci are not thought to have relevance for host selection or pheromone production and response, but they are located on the Z sex chromosome that also carries genes affecting male behavioral response to female sex pheromones (Resp) and the major factor of postdiapause development time (Pdd) (Dopman et al. 2004, 2005).
DNA extraction, amplification, and sequencing:
DNA extraction followed the CTAB protocol (Doyle and Doyle 1987). An ∼1600-bp DNA fragment including the coding region (5 exons of a total length of 732 bp) and introns (∼870 bp) of Tpi was amplified using primers ECBtpi_for1A and ECBtpi_rev5 (5′-ATAGTTTACGAATTACGAGTT-3′) (Dopman et al. 2005) and PCR profile: 1.5 min at 95°, 35 cycles of 1 min at 95°, 30 sec at 55°, and 1 min at 72°, followed by 3 min at 72°. An ∼1400-bp fragment of Ket, including a 232-bp exon, was amplified using primers ECBketF and ECBketR (Dopman et al. 2005) and PCR profile: 1.5 min at 95°, 35 cycles of 1 min at 95°, 30 sec at 58°, and 1 min at 72°, followed by 3 min at 72°. An ∼400-bp fragment of Mpi, including two exons of 60 and 77 bp, was amplified with primers Mpi-15 and Mpi-16 (Leniaud et al. 2006) and PCR profile: 1.5 min at 95°, 35 cycles of 30 sec at 95°, 30 sec at 60°, and 30 sec at 72°, followed by 3 min at 72°. Finally, an ∼1650-bp DNA fragment, including Pbp and its three exons of 126, 181, and 182 bp, was amplified with primers ECEP5 and ECPA (Willett and Harrison 1999) and PCR profile: 1.5 min at 95°, 35 cycles of 30 sec at 95°, 30 sec at 60°, and 1 min at 72°, followed by 3 min at 72°. Raw sequences of all loci were submitted to GenBank under the accession nos. EF396347–EF396478.
Because Tpi and Ket are located on the Z chromosome and because we used only female ECB in our study, PCR products for these two genes were sequenced directly. Tpi PCR products were sequenced by Genome Express S.A. (Meylan, France). Ket PCR products were purified with 2 μl of Exosap (USB, Cleveland) solution for 5 μl of PCR product and sequenced with a General Electrics Healthcare automatic sequencer using the BigDye terminator kit 3.1 (General Electrics Healthcare, Little Chalfont, UK). Sequencing primers were ECBtpi_for1A and ECBtpi_rev5 for Tpi and ECBketF for Ket. Of 278 individual samples, we obtained 222 sequences unambiguously readable over the full length of the fragment for Tpi and 219 such sequences for Ket.
Because Mpi and Pbp are located on autosomes, PCR products containing parts of these two genes were cloned before sequencing. A total of 94 individuals were chosen among those sequenced for Tpi and Ket by randomly picking 2–3 individuals from each population. PCR products obtained from these individuals were sent to Genoscreen (Lille, France) for cloning using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA). PCR products were ligated in 96-well plates following the TA cloning kit protocol (2004) and transformed with the one-shot Top10 competent cells provided with the TA cloning kit, in 96-well plates (Millipore plasmid miniprepkit LSK P09624, Millipore, Billerica, MA). The transformation products were spread on an Luria Burani (LB)-ampicillin (100 μg · μl−1) solidified solution contained in petri dishes (AES–AEB521279). Individual colonies were then each transferred into a well of a 96-well plate (Millipore plasmid miniprepkit LSK P09624) and maintained in a liquid LB-ampicillin solution (100 μg · μl−1). Plasmids were extracted with a miniprepkit (Millipore) and sequenced with an Applied Biosystems 3730 XL automatic sequencer. Sequence reactions used the Bigdye 3.1 kit (Applied Biosystems, Foster City, CA). Mpi clones were sequenced using the two primers provided in the TOPO TA cloning kit (Invitrogen). Pbp clones were sequenced using the forward 5′-GACAAACTAAATTGTTAGGCA-3′ and the reverse 5′-GATTCCAACTCCATCGCAT-3′ primers, which were designed by Genoscreen to improve the quality of final sequences by increasing the length of the overlapping zone between forward and reverse sequences (giving a final complete sequence of ∼1300–1400 bp).
For both Mpi and Pbp, four clones were initially sequenced for each individual, with the hope of unambiguously determining at least one allele per locus. However, for ∼30% of the individuals, the sequencing of these four clones yielded more than two different haplotypes, a problem also reported for Pbp by Willett and Harrison (1999). In a diploid organism, this can be due to (1) multiple copies of the gene in the genome, (2) Taq DNA polymerase errors, or (3) in vitro recombination during the PCR. Taq errors and in vitro recombination are known to occur [“jumping PCR,” see Paabo et al. (1990); Bradley and Hillis (1997)]. We directly sequenced PCR products of individuals for which cloning had yielded more than two haplotypes and for which no length polymorphism among clone sequences was detected. We considered that Taq errors were the most likely explanation when substitutions observed in clone sequences were absent from direct PCR product sequences of the same individual. For all but five (when considering both Pbp and Mpi) of the remaining individuals, manual inspection of the sequences showed that the set of haplotype sequences obtained was compatible with a scenario of in vitro recombination, i.e., we found that the additional haplotypes found in PCR products at low frequencies consisted of a combination of successive fragments of the two sequences. In the five remaining individuals, we could not assess the compatibility of sequences with an in vitro recombination scenario, due to too large a number of Taq errors (including errors at polymorphic sites). For Pbp, we concluded that Taq errors occurred in ∼20% of the clones and in vitro recombination in ∼10% of the clones. For Mpi, these percentages were ∼10 and ∼5%, respectively.
To choose which haplotypes to keep for subsequent analyses, we proceeded as follows. When the initial four to eight clone sequences revealed only one or two haplotypes for a given individual, we used the most common sequence provided it was observed at least three times. When this number was not reached, we either sequenced additional clones when length polymorphism was observed or directly sequenced the PCR products when no length polymorphism was detected. For all individuals showing more than two haplotypes and no insertion/deletion (indel) in the region responsible for the existence of these apparently more than two haplotypes, we directly sequenced the PCR products and kept the clone sequence(s) that unambiguously matched the PCR product sequence. For individuals with fragment length polymorphisms for which direct sequencing did not help to distinguish true haplotype(s) from the artifact(s), we sequenced up to 12 additional clones and used only the sequence found in the majority of clones, provided this majority consisted of three or more clones. All other sequences were discarded.
This procedure resulted in a much smaller data set than we were initially hoping to gather: of the 94 individuals (i.e., 188 alleles) initially included in the cloning protocol, 79 and 83 individuals finally yielded one usable sequence each for Pbp and Mpi, respectively.
Data analysis:
Sequences were aligned using CLUSTALW 1.83 (Thompson et al. 1994) and adjusted manually using BIOEDIT 7.01 (Hall 1999). The correspondence between sequences and chromatograms was checked manually using CHROMAS 2.3 (Technelysium Pty, Tewantin, Queensland, Australia). All sequences were blasted (March 6, 2006) using the BlastN and BlastX functions of the NCBI–BLAST website (http://www.ncbi.nlm.nih.gov/BLAST/), which confirmed that they all corresponded to the expected locus.
For Tpi, Ket, and Pbp, several complex sets of indels were observed, corresponding to substitutions and length polymorphism within the indels. As the sequences were long and the number of informative sites was high, we excluded indels from our analysis. For Mpi, we failed to align the entire data set because a region of ∼200 bp (of 300–400 bp, depending on the length of the indels) proved extremely variable. Hence, we restricted our analysis to the ∼160-bp region for which the sequences could be aligned. Delineation of coding and noncoding regions as well as positions of variable sites in the final alignments is shown for each gene in the supplemental online material (http://www.genetics.org/supplemental/).
We used DNASP 4.01 (Rozas et al. 2003) to calculate the following summary statistics describing our data set: number of haplotypes (H), haplotype diversity (Hd), nucleotide diversity estimated by π (Nei 1987) and θ (Watterson 1975), and minimum number of recombination events (Rm). These descriptive statistics were estimated for the entire data set and for data sets restricted to populations collected on the same host-plant species.
Mutation models (Jukes and Cantor, Kimura 2P, Tajima and Nei, Tamura, Tamura and Nei) available in ARLEQUIN 3.01 (Excoffier et al. 2005) were fitted to the data and their likelihood was tested for each locus using the MODELTEST 3.7 program (Posada and Crandall 1998). Tamura and Nei's model was retained for Tpi (uniform substitution rates among sites), Ket (uniform substitution rates), and Pbp (different substitution rates, γ = 0.77), and Kimura's 2P was retained for Mpi (different substitution rates, γ = 0.253). To look for possible clustering by host plant, neighbor joining (NJ) trees (inferences tested by a bootstrap procedure of 1000 replications) were constructed with MEGA 3.1 software (Kumar et al. 2004) for each locus, using the most appropriate mutation model as determined by MODELTEST 3.7.
The level of genetic differentiation between groups of populations collected on the same host-plant species was assessed by calculating among- and within-group fixation indices (FCT and FSC) in an analysis of molecular variance (AMOVA) using ARLEQUIN 3.01. Again, the most appropriate mutation model to be used in the AMOVA was chosen for each locus according to prior tests using MODELTEST 3.7. The P-values associated with among-group fixation indices were obtained by permutation tests performed by ARLEQUIN 3.01 (10,000 permutations). As the number of available sequences varied among loci, the mean FCT value across all four loci could not be obtained using the standard procedure of ARLEQUIN 3.01. Instead, we calculated the mean FCT value over the four loci and combined the corresponding P-values using Fisher's method (Fisher 1932).
We also tested for a correlation between the matrices of pairwise genetic and geographic distances with Mantel tests performed with GENEPOP 3.4 (Raymond and Rousset 1995). Pairwise genetic distances were calculated for each locus by computing pairwise FST in the “Population Comparisons” section of ARLEQUIN 3.01 (the haplotype distance matrices were computed using mutational models chosen according to the goodness of fit assessed with MODELTEST 3.7). The average of this pairwise genetic distance was calculated as the average of the four distance values estimated at the four loci for the pair of populations considered. This analysis was conducted using data on populations sampled from (1) maize, (2) mugwort, or (3) all host-plant species. Populations collected from plants other than maize and mugwort were not numerous enough to test for isolation by distance within each data subset.
Possible departures from the standard neutral model of molecular evolution—potentially revealing demographic events or the existence of selective effects at certain loci—were tested for each locus by Tajima's D-test, Fu and Li's D*-test, and Fu's Fs-test using DNASP 4.01. Neutrality tests were performed on the entire data set and on data subsets each consisting of populations collected on the same host-plant species, provided at least three populations collected on this host plant were available. Thus, tests on separate host-related groups were conducted only for maize, pepper, mugwort, and hop. P-values associated with these tests were obtained from coalescent simulations performed by DNASP 4.01 taking into account estimates of the intragenic recombination parameter R. Following Fu (1997), the P-value limit for significance in Fu's Fs tests was set to P = 0.02 instead of 0.05. The signature of selective constraints on coding regions was tested using the CODEML program of the PAML package (YANG, 1997) by comparing likelihood of models where the ratio of synonymous and nonsynonymous substitutions (dN/dS) was either fixed to 1 (neutral) or let as a free parameter.
The time elapsed since the initial divergence between populations was estimated using the updated version of the Isolation Model (IM) program released July 31, 2006 (see Hey and Nielsen 2004 for details on the initial release) on the subset of sequences obtained for larvae collected on maize and mugwort. Again, only sequences obtained for individuals collected on one of these two host plants were used because the individuals collected on the other host plants could not be assigned to one of the host races with complete certainty. Tpi and Mpi were excluded from this analysis as neutrality tests revealed a significant departure from neutral evolution at these loci in at least one population (see results). Since intragenic recombination was detected, we followed Hey and Nielsen's (2004) advice and used, for any given locus, only the largest region of the sequence that was not subject to recombination. The presence of recombination between sites was checked using the Hudson and Kaplan algorithm (1985) included in DNASP 4.01. We analyzed two data sets: both included the longest portion (511 bp) of Ket (where intragenic recombination was rare), but they differed in the region of Pbp that was used (158 bp in one data set and 194 bp in the other). Summary descriptive statistics for these two data sets are detailed in supplemental online material (http://www.genetics.org/supplemental/). Although IM did not reject the infinite site mutation model for the two loci, results of MODELTEST 3.7 clearly indicated that the HKY model (Hasegawa–Kishino–Yano: Hasegawa et al. 1985) provided a better fit to the data for both Ket and Pbp. Thus, we decided to use this latter mutation model for IM runs. First runs used parameter values recommended by Hey and Nielsen (2004) for priors of upper bounds of population size theta, migration rate m, and divergence time t. Following the advice of the IM manual (version of July 31, 2006), convergence and mixing of runs were evaluated by examining (1) the level of autocorrelation between final and initial parameter values, (2) the estimated value of the effective sample size (ESS), (3) the parameter trends plots, and (4) the consistency of results over different runs using the same model (three runs were performed for each data set). Data were not informative enough to estimate all parameters of complex models including different values of theta, m, and population size fluctuations. Runs with such models did not converge and/or resulted in flat posterior distribution for m and t. We thus used the simplest possible model, consisting of one ancestral population splitting into two populations of same size and experiencing symmetric gene flow. In final runs, priors were set to 8 for theta, 10 for t, and 75 for m. Optimal chain swapping rates and ESS numbers were obtained with 20 chains and the following heating scheme: “-f g -g1 0.7 -g2 0.9.” The burn-in period was set to 500,000 iterations and convergence of runs was reached after a duration composed of between ∼7 and ∼12 million iterations. Finally, the “−p5” option was used to have a closer look at gene flow between populations by estimating the maximum likelihood number of migration events and the mean time at which they occurred in genealogies that had at least one migration event. Estimates of the mean mutation rate per gene per generation and of the number of generations per year are required to convert results into demographic units. Mutation rates per site in nuclear genes are generally expected to be ∼10−9 per nucleotide site per generation, or 10−6 per (entire) gene per year (Futuyma 1998; Ridley 2004). To be conservative we used a mutation rate of 10−8 per site per generation, which corresponded to a value of ∼2.8 × 10−6 and ∼3.1 × 10−6 per gene per generation for data set A and data set B, respectively. Again, to be conservative, as the ECB can achieve two generations per year, we chose to use a generation time (g) of 0.5 year. This resulted in the mutation rate per gene per year (μ) being ∼5.6 × 10−6 and ∼6.2 × 10−6 for data sets A and B, respectively.
RESULTS
All loci were polymorphic, but the extent of polymorphism varied among loci (Table 1). Haplotype diversity was comparable at Ket, Pbp, and Mpi (∼0.9, Table 1) and markedly lower at Tpi (∼0.7, Table 1). Nucleotide diversity, whether estimated by π or θ, was an order of magnitude lower at Tpi and Ket (10−3) than at Pbp and Mpi (10−2). Intragenic recombination (Rm, Hudson and Kaplan 1985) was detected at all four loci, although at quite different frequencies (Table 1). Descriptive statistics within populations collected from each plant are available as supplemental online material (http://www.genetics.org/supplemental/).
TABLE 1.
Descriptive statistics of polymorphism found at each of the four nuclear loci, for the entire data set
| Locus | N | H | Hd | π | θ | Ps | Rm |
|---|---|---|---|---|---|---|---|
| Tpi | 222 | 28 | 0.740 (0.023) | 0.00166 (0.00017) | 0.00358 (0.00062) | 33 | 2 |
| Ket | 219 | 23 | 0.893 (0.009) | 0.00407 (0.00017) | 0.00650 (0.00130) | 25 | 4 |
| Pbp | 79 | 38 | 0.940 (0.016) | 0.02101 (0.00086) | 0.02164 (0.00184) | 139 | 14 |
| Mpi | 83 | 17 | 0.902 (0.013) | 0.04935 (0.00110) | 0.03151 (0.00630) | 25 | 2 |
Standard deviation of estimates is in parentheses. N, number of sequences; H, number of haplotypes; Hd, haplotype diversity; π, pi nucleotide diversity; θ, theta nucleotide diversity; Ps, number of polymorphic sites; Rm, minimum number of recombination events.
Differentiation between populations:
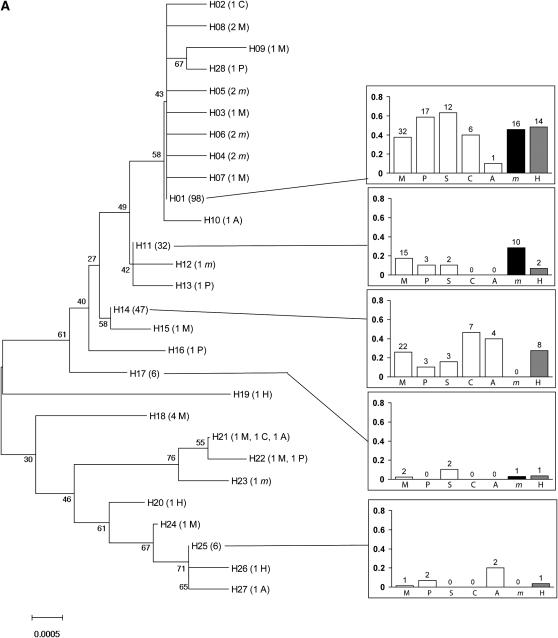
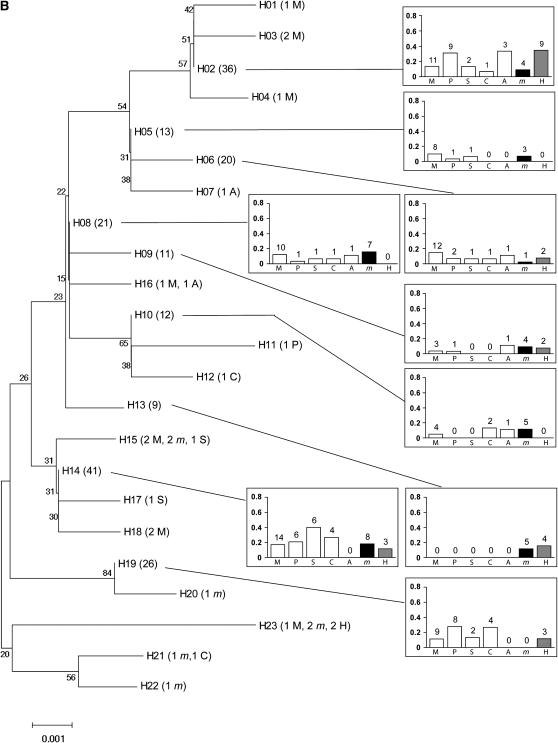
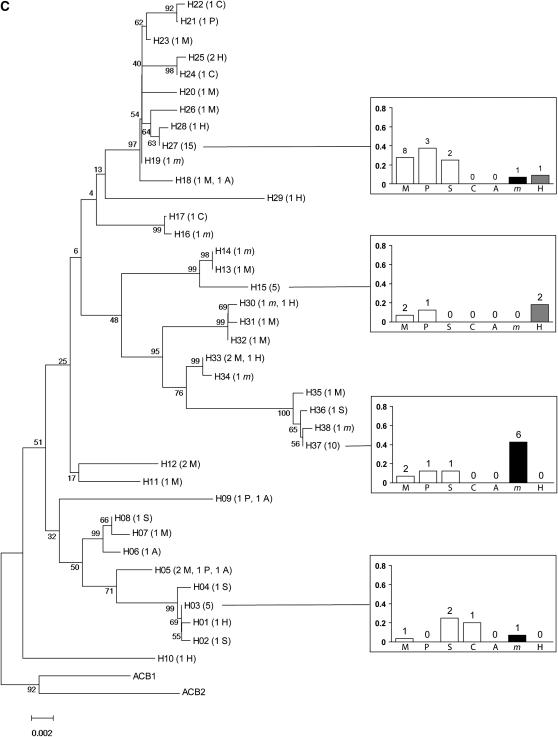
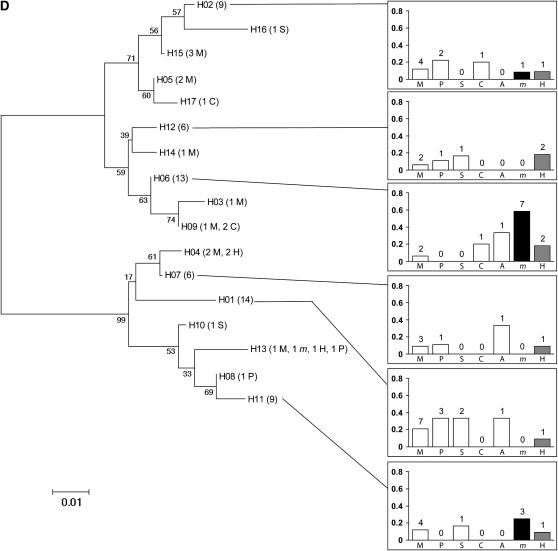
Over all populations and all loci, individuals did not clearly cluster according to their host plant in NJ trees (Figure 2). Rather, populations collected from any pair of plants typically shared several haplotypes. The only exception was one Pbp haplotype that was shared by 43% of the individuals collected from mugwort but occurred much less frequently, if at all, in populations collected from other host-plant species.
Figure 2.—
Neighbor joining trees for the four loci: (A) Tpi, (B) Ket, (C) Pbp, (D) Mpi. H01–H30: haplotype number, followed by the number of individuals carrying each haplotype (in parentheses) and the host plant(s) from which these individuals were collected (names are coded as in Figure 1). For haplotypes shared by more than four individuals sampled from more than four different plants, a diagram (right) represents (i) among all individuals collected from a given plant, the proportion sharing the haplotype (bars) and (ii) the corresponding number of individuals from this plant (above bars). When sequences were available, Ostrinia furnacalis (the Asian corn borer, ACB) was used as an outgroup.
Fixation indices (Table 2) essentially showed that the population structure detected in French populations with allozyme markers (Bourguet et al. 2000; Martel et al. 2003; Leniaud et al. 2006) was also detectable at DNA sequence level. Significant genetic divergence was found between populations collected on mugwort and populations collected on any of the other host plants except hop. Conversely, populations collected on maize did not show any significant divergence from populations collected on any other host plant except mugwort. Moreover, divergences from populations collected on mugwort (estimated by FCT over all genes) were of similar magnitude for populations collected on maize, cocklebur, pepper, and sorghum. No significant divergence was detected in the present study between populations collected on hop and those collected on maize. Restricting the AMOVA to northern France (Table 2) did not change the conclusion about populations feeding on maize and mugwort being significantly divergent and about populations feeding on mugwort and hop not being significantly divergent. However, in this region, significant divergence between populations from maize and hop was detected.
TABLE 2.
Fixation indices between European corn borer populations collected from maize, mugwort, and hop, and between maize, mugwort, and other plants
| Comparison
|
Tpi
|
Ket
|
Pbp
|
Mpi
|
All genes
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Plant 1 | Plant 2 | FCT | FSC | FCT | FSC | FCT | FSC | FCT | FSC | FCT | FSC |
| Overall France | |||||||||||
| Maize | Mugwort | 0.05** | −0.01 | 0.03* | 0.06** | 0.14** | −0.03 | 0.00 | 0.12 | 0.06*** | 0.05* |
| Hop | 0.00 | 0.04 | 0.01 | 0.09** | −0.05 | 0.04 | −0.06 | 0.17* | 0.00 | 0.08** | |
| Pepper | 0.01 | −0.01 | −0.01 | 0.12*** | −0.06 | 0.01 | −0.07 | 0.07 | 0.00 | 0.05** | |
| Sorghum | 0.01 | −0.01 | 0.01 | 0.05 | 0.05 | −0.03 | −0.09 | 0.09 | 0.02 | 0.03 | |
| Xanthium | −0.02 | −0.01 | 0.05 | 0.05* | 0.04 | −0.01 | 0.18 | 0.13 | 0.07 | 0.05 | |
| Mugwort | Hop | 0.04 | 0.11* | −0.02 | 0.13** | 0.12* | −0.08 | −0.06 | 0.16 | 0.04 | 0.10** |
| Pepper | 0.01 | 0.01 | 0.05 | 0.16*** | 0.15* | −0.14 | 0.06 | −0.13 | 0.07* | 0.04** | |
| Sorghum | 0.00 | 0.03 | 0.05 | 0.06* | 0.17*** | −0.23 | 0.03 | −0.09 | 0.06*** | 0.02 | |
| Xanthium | 0.09 | 0.04 | 0.04 | 0.06* | 0.31** | −0.23 | 0.08 | 0.03 | 0.13*** | 0.03 | |
| Northern France only | |||||||||||
| Maize | Mugwort | 0.05 | 0.01 | 0.06** | 0.03 | 0.25*** | −0.16 | −0.10 | 0.31* | 0.09*** | 0.09 |
| Hop | 0.00 | 0.10 | 0.11*** | 0.08 | 0.05 | −0.14 | −0.14 | 0.42** | 0.04*** | 0.15** | |
| Mugwort | Hop | −0.05 | 0.13 | −0.05 | 0.14*** | 0.12* | −0.20 | −0.23 | 0.21 | 0.03 | 0.12** |
FCT is relative to among-group variance and FSC to within-group variance. Within-population variance is not shown. P-values testing whether indices are significantly >0 were obtained by permutation tests with 10,000 permutations (Schneider et al. 2000). *P < 0.05, **P < 0.01, ***P < 0.001.
Mantel tests revealed no correlation between matrices of pairwise genetic and geographic distances, be it with populations collected from mugwort (P = 0.141), from maize (P = 0.438), or from all host-plant species (P = 0.477).
Selective effects:
Neutrality tests performed on the entire data set showed that Tpi and Mpi were not at mutation-drift equilibrium. Indeed, except for the Fu and Li's D* for Mpi, values of the three statistics were all significantly different from zero for Tpi and Mpi (Table 3). Tpi was characterized by negative values, whereas Mpi displayed positive values. For Ket, the values of the three statistics were negative but none of them were significantly different from zero. For Pbp, D*, D, and Fs values were very close to zero with no particular trend. Results of the analyses performed on each host-plant-specific data set were consistent with these global trends, except in hop populations for Ket (Table 3). Finally and noteworthy, the three different test statistics used in this analysis lead to consistent results (Tajima's D revealed five while the two other tests revealed four significant departures from neutral evolution)(Table 3). dN/dS was significantly different from one in the coding regions of Tpi (dN/dS = 0.22) and Pbp (dN/dS = 0.11) but not in those of Mpi (dN/dS = 0.44) and Ket (dN/dS = 0; only three sites were variable).
TABLE 3.
Tests for neutral evolution of molecular polymorphism
|
Tpi
|
Ket
|
Pbp
|
Mpi
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | D | D* | Fs | N | D | D* | Fs | N | D | D* | Fs | N | D | D* | Fs | |
| All populations | 222 | −1.64* | −3.86** | −12.66** | 219 | −1.03 | −1.25 | −7.04 | 79 | −0.19 | −0.38 | −0.01 | 83 | 1.23* | 0.23 | 1.52* |
| Maize | 86 | −1.24 | −0.74 | −2.90 | 81 | −0.72 | −0.37 | −3.93 | 29 | −0.12 | 0.04 | 1.67 | 35 | 1.03 | 0.62 | 0.76 |
| Pepper | 29 | −1.03 | −0.41 | −0.40 | 29 | 0.19 | −0.73 | −0.35 | 8 | −0.46 | −0.25 | 3.10 | 10 | 0.83 | 0.57 | 1.64 |
| Mugwort | 35 | −1.88** | −2.45* | −1.90 | 44 | −1.11 | −0.10 | −4.07 | 14 | 0.14 | −0.25 | 3.67 | 12 | 1.07 | 1.06 | 5.29* |
| Hop | 28 | −0.72 | −0.27 | 0.20 | 26 | 0.21 | 1.47* | 0.94 | 11 | −0.15 | −0.02 | 1.46 | 11 | 0.28 | 0.05 | −0.28 |
N, number of sequences; D, Tajima's D; D*, Fu and Li's D*; Fs, Fu's Fs. *P < 0.05 (P < 0.02 for Fu's Fs); **P < 0.01.
Divergence time between populations:
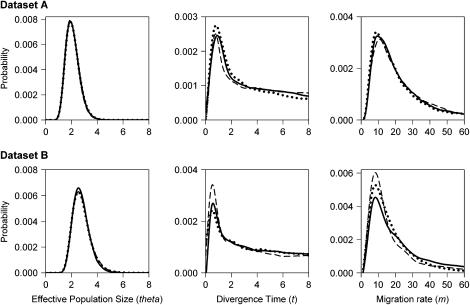
Overall, results were consistent over data sets and replicates (Figure 3, Table 4), albeit quite imprecise, as posterior distributions were long-tailed for t and m. The effective population size theta estimate with the highest maximum likelihood was consistently ∼150,000–200,000 individuals (see Table 4 for estimates of the 90% highest posterior density interval, the HPD90% interval, resulting from the different runs). The estimated divergence time t varied between ∼75,000 and ∼150,000 years. Despite a large HPD90% interval, in no instance did its lower bound include values compatible with a divergence time of 500 years. In all cases, the likelihood values attributed to this divergence time were extremely low. An unrealistically high (>10−4 and >5 × 10−5 for data sets A and B, respectively) mutation rate per gene per year would be necessary for the lower bound of the HPD90% interval of the divergence time to include the value of 500 years. Estimates for the migration rate m ranged from 7 to 11 and HPD90% intervals were large (Table 4). Finally, estimates for 2Nm between the two taxa were relatively high, i.e., ∼10.
Figure 3.—
Posterior distribution produced by the IM program for the effective population sizes theta of maize and mugwort populations, their divergence times t, and the migration rates m for each of the three runs using data set A and data set B (containing a common set of Ket sequences and two different parts of Pbp sequences; see supplemental material for details at http://www.genetics.org/supplemental/) .
TABLE 4.
Results of isolation model simulations
| Data set | Replicate no. | Effective population size (theta)
|
Divergence time (t)
|
Migration rate (m)
|
|||
|---|---|---|---|---|---|---|---|
| HiPt | HPD90% | HiPt | HPD90% | HiPt | HPD90% | ||
| A | 1 | 1.835 | 1.235–2.925 | 0.955 | 0.195–8.795 | 9.863 | 2.888–45.038 |
| 2 | 1.875 | 1.205–2.885 | 0.835 | 0.265–9.995a | 10.613 | 3.038–48.188 | |
| 3 | 1.785 | 1.225–2.935 | 0.765 | 0.155–9.085 | 9.338 | 2.663–47.963 | |
| Parameters converted into demographical units (individuals for theta or years for t) | |||||||
| 1 | 161,450 | 108,660–257,352 | 168,049 | 34,314–1,547,629 | 2Nm = | 9.05 | |
| 2 | 164,969 | 106,020–253,833 | 146,933 | 46,632–1,758,790a | 9.95 | ||
| 3 | 157,051 | 107,780–258,232 | 134,615 | 27,275–1,598,659 | 8.33 | ||
| B | 1 | 2.495 | 1.705–3.745 | 0.455 | 0.205–9.965 | 7.750 | 2.250–56.450 |
| 2 | 2.585 | 1.705–3.895 | 0.545 | 0.025–8.945 | 7.750 | 1.750–47.250 | |
| 3 | 2.535 | 1.655–3.845 | 0.495 | 0.025–9.355 | 7.650 | 2.050–38.050 | |
| Parameters converted into demographical units (individuals for theta or years for t) | |||||||
| 1 | 198,107 | 135,380–274,859 | 72,256 | 32,555–1,582,472 | 2Nm = | 9.67 | |
| 2 | 205,253 | 135,380–309,269 | 86,548 | 3,970–1,420,493 | 10.02 | ||
| 3 | 201,283 | 131,410–305,299 | 78,607 | 3,970–1,485,602 | 9.70 | ||
Model parameters estimates (theta, t, m) for data set A and B. Parameters were converted into demographic units using the following formulas: theta (individuals) = theta/(4 × μ); T(years) = t/μ. The migration parameter 2Nm was calculated by multiplying (0.5 × theta) and m. HiPt, bin with the maximum residence time; HPD90%, 90% highest posterior density interval.
Due to the long-tailed posterior distribution, the HPD90% upper bound was higher than the upper bound of the prior distribution and could not be estimated.
DISCUSSION
The genetic structure at the four nuclear loci we studied (i.e., Tpi, Ket, Mpi, and Pbp) was significantly influenced by the host plants on which the ECB populations were collected (Table 2). ECB populations collected on mugwort were significantly differentiated from those collected on maize. In addition, populations collected on pepper, sorghum, and cocklebur were genetically more similar to those feeding on maize than to those feeding on mugwort. Hence, our former conclusion based exclusively on allozyme markers also holds when investigating polymorphism at the DNA sequence level: in France, ECB populations are divided into at least two host races (Bourguet et al. 2000; Martel et al. 2003; Thomas et al. 2003). One, referred to as the maize race, is feeding mostly on maize but occasionally on pepper, sorghum, cocklebur, and sunflower (Leniaud et al. 2006). The second, referred to as the mugwort–hop race, feeds on mugwort and hop.
Although in general our results fit quite well with the host-race delineation based on allozyme data, we did find one discrepancy. When populations from all over France were included in the analysis, we failed to detect any differentiation between populations collected on maize and on hop. Such differentiation could be detected only when our analysis was restricted to populations from northern France. In southern France, mugwort stands are apparently free of ECB (Martel et al. 2003), whereas maize fields are often substantially infested by this pest (A. Weissenberger, unpublished data), and hop is restricted to wild stands along rivers (T. Malausa, D. Bourguet and S. Ponsard, personal observations). Hence, in this geographical area, adult moths of the mugwort–hop race are probably at low density and strongly outnumbered by adults of the maize race. As the two host races still hybridize in field conditions (Malausa et al. 2005), populations of the mugwort–hop race may thus receive higher gene flow from populations feeding on maize in southern than in northern France.
Overall, the level of divergence between populations was low, even when comparing populations collected on maize with populations collected from mugwort. These two groups of populations displayed extensive shared polymorphisms (Table 1), and consequently none of the trees based on any of the four markers exhibits any clustering of the two ECB host races (Figure 2). This suggests that reproductive isolation between the maize-Z and mugwort–hop-E races is recent and/or incomplete. More generally, reproductive barriers between diverging or recently diverged species are often semipermeable, which leads to patterns of shared alleles at most loci. Shared polymorphism between closely related species has been observed in other animals. For example, Machado et al. (2002) observed shared polymorphism at 12 out of 14 nuclear loci between Drosophila pseudoobscura and D. persimilis. In field crickets, Broughton and Harrison (2003) did not find any fixed differences at 4 nuclear loci between Gryllus firmus and G. pennsylvanicus, two species that are estimated to have diverged ∼103–104 generations ago. In R. pomonella, a model species thought to have experienced a recent host shift, the sequences of three nuclear loci did not cluster according to the host plant from which the individuals were collected (Feder et al. 2003). Similarly, recently diverged cichlid fish species also exhibit a pattern of shared polymorphisms (Hey et al. 2004). Our results thus tend to confirm that the two French Ostrinia taxa constitute two recently diverged host races or sibling species, rather than two distant species in which divergence would be strong at most loci, including nuclear genes (Blum et al. 2003; Barr and McPheron 2006; see review in Hare 2001).
However, our estimates of divergence time between taxa suggest that the abundant shared polymorphism between both Ostrinia host races is likely due to a high level of gene flow having occurred since the divergence of taxa, rather than to a short time since divergence. Using mutation rates usually observed at nuclear genes (Futuyma 1998; Ridley 2004) in IM simulations, we estimated the divergence time to be between ∼75,000 and 150,000 years and 2Nm to have values ∼10. Although these estimates are rough, they seem compatible either with an allopatric divergence followed by secondary contact between both taxa or with a sympatric divergence, but they do not support a scenario of recent divergence triggered by a shift onto maize ∼500 years ago. Indeed, one would have to assume a mutation rate >5 × 10−5 substitutions per gene per year to obtain divergence time estimates whose confidence intervals include 500 years. Although little is known about rates of evolution at the two loci Ket and Pbp, such a value seems unrealistically high. An earlier divergence may thus have occurred due to previous host shift (e.g., onto Sorghum sp. or Panicum sp.). This pattern of slow divergence under a regime of high gene flow strengthens the conclusions of Emelianov et al. (2004), who concluded from several studies on the larch budmoth Z. diniana that a long and relatively stable sympatric phase of genetic divergence in the presence of gene flow is often a feature of speciation.
Finally, we did not find evidence that any of the genes studied marked a genome region involved in reproductive isolation between the two Ostrinia host races. This result contrasts with the findings of Dopman et al. (2005) who documented a clear clustering of the North American E pheromonal race at Tpi and a departure of its polymorphism from neutral evolution restricted to this same E-race. In France, differentiation at Tpi is significant but low (Table 4) and there is only little evidence that its departure from a neutral molecular evolution was restricted to populations of the E-race (Table 3). Further, this departure can be explained, as in the United States, by one or several selective sweep(s), but it remains unclear whether these sweeps were caused by the selective constraints detected on the coding region of Tpi or by selection acting on a gene located near Tpi. This latter hypothesis was supported by Dopman et al. (2005) because Pdd, a locus involved in the determinism of the postdiapause-development duration (a trait probably under strong selection in most populations), maps at the same chromosomal location as Tpi. However, neither Tpi nor Pdd seems to contribute to reproductive isolation between French host races: divergence at Tpi was not greater than that observed at other loci and was negligible when compared to that estimated between Ostrinia American pheromonal races (Dopman et al. 2005) or between host races of Z. diniana in genomic regions thought to be involved in reproductive isolation (Emelianov et al. 2004).
Conclusion:
Although French host races of the ECB have been found to be reproductively isolated (Malausa et al. 2005), analysis of molecular polymorphisms at four nuclear loci did not reveal a clear pattern of monophyly for each race. Instead, extensive haplotype sharing was observed for all genes, even at the Tpi locus in which a clear separation of E and Z populations was previously observed in North America (Dopman et al. 2005). However, the results of this study confirmed the significant genetic differentiation observed in previous work on the French ECB populations using allozyme markers (Bourguet et al. 2000; Martel et al. 2003; Thomas et al. 2003; Malausa et al. 2005; Leniaud et al. 2006). IM coalescent-based simulations allowed estimation of the time since divergence of the two host races between 75,000 and 150,000 years. Although additional data from mitochondrial or microsatellite loci will be needed to confirm this first estimation, this suggests that a scenario of speciation by host shift onto maize, after the introduction of this plant into Europe ∼500 years ago, is not likely. Finally, although neutrality tests revealed several departures from the mutation-drift equilibrium for the molecular polymorphism of two genes, we did not identify any gene involved in the reproductive isolation between the two French host races.
Acknowledgments
We thank all those who helped with sampling [N. Eychenne, L. Folcher, M. Delos, the staff of the different Services Régionaux de Protection des Végétaux, several farmer families, H. Clerc (AIREL), and L. Pélozuelo]. We are grateful to M. Blanchi, P. Ceotto, and J.C. Malausa, who kindly accepted to handle long IM runs on their computers, and to M. Fontaine, S. Piry, and M. Ciosi for their advice and technical help. Financial support for this study was provided by the Institut Français de la Biodiversité (AO Biodiversité et Changement Global), the Centre National de la Recherche Scientifique (AO Impact des Biotechnologies sur les Agrosystèmes), the Action Concertée Incitative–Fonds National de la Science Ecco (Ecosphère Continentale: Processus et Modélisation), and the European Union project ProBenBt. We also thank the Service des relations internationales of University P. Sabatier-Toulouse III that funded T.M.'s visit to Cornell University for 5 weeks in 2005. Development of the molecular markers was funded by National Research Initiative CSREES grant 2001-35391-1123 and National Science Foundation grant DEB-0415343 to R.G.H. We finally thank the Comité de Biovigilance and the Direction Générale de l'Alimentation of the French Ministry of Agriculture for funding most of the sampling performed for this study.
References
- Barr, N. B., and B. A. McPheron, 2006. Molecular phylogenetics of the genus Ceratitis (Diptera: Tephritidae). Mol. Phylogenet. Evol. 38: 216–230. [DOI] [PubMed] [Google Scholar]
- Berlocher, S., and J. Feder, 2002. Sympatric speciation in phytophagous insects: Moving beyond controversy? Ann. Rev. Entomol. 47: 773–815. [DOI] [PubMed] [Google Scholar]
- Bethenod, M.-T., Y. Thomas, F. Rousset, B. Frérot, L. Pélozuelo et al., 2005. Genetic isolation between two sympatric host plant races of the European corn borer, Ostrinia nubilalis Hübner. II. Assortative mating and host plant preferences for oviposition. Heredity 94: 264–270. [DOI] [PubMed] [Google Scholar]
- Blum, M. J., E. Bermingham and K. Dasmahapatra, 2003. A molecular phylogeny of the neotropical butterfly genus Anartia (Lepidoptera: Nymphalidae). Mol. Phylogenet. Evol. 26: 46–55. [DOI] [PubMed] [Google Scholar]
- Bontemps, A., D. Bourguet, L. Pélozuelo, M.-T. Bethenod and S. Ponsard, 2004. Managing the evolution of Bacillus thuringiensis resistance in natural populations of the European corn borer, Ostrinia nubilalis: host plant, host race and pherotype of adult males at aggregation sites. Proc. R. Soc. London B 271: 2179–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguet, D., M.-T. Bethenod, C. Trouvé and F. Viard, 2000. Host-plant diversity of the European corn borer Ostrinia nubilalis: What value for sustainable transgenic insecticidal Bt maize? Proc. R. Soc. London B 267: 1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, R. D., and D. M. Hillis, 1997. Recombinant DNA sequences generated by PCR amplification. Mol. Biol. Evol. 14: 592–597. [DOI] [PubMed] [Google Scholar]
- Broughton, R. E., and R. G. Harrison, 2003. Nuclear gene genealogies reveal historical, demographic and selective factors associated with speciation in field crickets. Genetics 163: 1389–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey, D., and L. Worthley, 1927. A progress report on the investigations of the European corn borer. United States Department of Agriculture Bulletin no. 1476. United States Department of Agriculture, Washington, DC.
- Cardé, R., W. Roelofs, R. Harrison, A. Vawter, P. Brussard et al., 1978. European corn borer: Pheromone polymorphism or sibling species? Science 199: 555–556. [DOI] [PubMed] [Google Scholar]
- Cianchi, R., S. Maini and L. Bullini, 1980. Genetic distance between pheromone strains of the European corn borer, Ostrinia nubilalis: different contribution of variable substrate, regulatory and non regulatory enzymes. Heredity 45: 383–388. [Google Scholar]
- Dopman, E. B., S. M. Bogdanowicz and R. G. Harrison, 2004. Genetic mapping of sexual isolation between E and Z pheromone strains of the European corn borer (Ostrinia nubilalis). Genetics 167: 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopman, E. B., L. Perez, S. M. Bogdanowicz and R. G. Harrison, 2005. Consequences of reproductive barriers for genealogical discordance in the European corn borer. Proc. Natl. Acad. Sci. USA 102: 14706–14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J., and J. Doyle, 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem. Bull. 19: 11–15. [Google Scholar]
- Du, G. H., and G. D. Prestwich, 1995. Protein-structure encodes the ligand-binding specificity in pheromone binding-proteins. Biochemistry 34: 8726–8732. [DOI] [PubMed] [Google Scholar]
- Emelianov, I., F. Marec and J. Mallet, 2004. Genomic evidence for divergence with gene flow in host races of the larch budmoth. Proc. R. Soc. London B 271: 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier, L., L. Laval and S. Schneider, 2005. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- Feder, J. L., S. H. Berlocher, J. B. Roethele, H. Dambroski, J. J. Smith et al., 2003. Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl. Acad. Sci. USA. 100: 10314–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. A., 1932. Statistical Methods For Research Workers, Ed. 4. Oliver & Boyd, London.
- Fu, Y. X., 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma, D., 1998. Evolutionary Biology. Sinauer Associates, Sunderland, Massachussetts.
- Gavrilets, S., 2003. Perspective: models of speciation: What have we learned in 40 years? Evolution 57: 2197–2215. [DOI] [PubMed] [Google Scholar]
- Glover, T., M. Campbell, P. Robbins and W. Roelofs, 1990. Sex-linked control of sex pheromone behavioral responses in European corn borer moths (Ostrinia nubilalis) confirmed with TPI marker gene. Arch. Insect Biochem. Physiol. 15: 67–77. [Google Scholar]
- Hall, T., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids. Symp. Ser. 41: 95–98. [Google Scholar]
- Hare, M. P., 2001. Prospects for nuclear gene phylogeography. Trends Ecol. Evol. 16: 700–706. [Google Scholar]
- Harrison, R. G., and A. T. Vawter, 1977. Allozyme differentiation between pheromone strains of the European corn borer, Ostrinia nubilalis. Ann. Entomol. Soc. Am. 70: 717–720. [Google Scholar]
- Hasegawa, M., H. Kishino and T. Yano, 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160–174. [DOI] [PubMed] [Google Scholar]
- Hey, J., and R. Nielsen, 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey, J., Y. J. Won, A. Sivasundar, R. Nielsen and J. A. Markert, 2004. Using nuclear haplotypes with microsatellites to study gene flow between recently separated Cichlid species. Mol. Ecol. 13: 909–919. [DOI] [PubMed] [Google Scholar]
- Hodgson, B., 1928. The host plants of the European corn borer in New England. Tech. Bull. 77: 1–63. [Google Scholar]
- Hudon, M., E. LeRoux and D. Harcourt, 1989. Seventy years of European corn borer (Ostrinia nubilalis) research in North America. Agric. Zool. Rev. 3: 53–96. [Google Scholar]
- Hudson, R., and N. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klun, J. A., 1975. Insect sex pheromones: intraspecific pheromonal variability of Ostrinia nubilalis in North America and Europe. Env. Entomol. 4: 891–894. [Google Scholar]
- Kochansky, J., R. Cardé, J. Liebherr and W. Roelofs, 1975. Sex pheromone of the European corn borer, Ostrinia nubilalis (Lepidoptera: Pyralidae), in New York. J. Chem. Ecol. 1: 225–231. [Google Scholar]
- Kolmerer, B., J. Clayton, V. Benes, T. Allen, C. Ferguson et al., 2000. Sequence and expression of the kettin gene in Drosophila melanogaster and Caenorhabditis elegans. J. Mol. Biol. 296: 435–448. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinformatics 5: 150–163. [DOI] [PubMed] [Google Scholar]
- Leniaud, L., P. Audiot, D. Bourguet, B. Frérot, G. Genestier et al., 2006. Genetic structure of European and Mediterranean maize borer populations on several wild and cultivated host plants. Entomol. Exp. Appl. 120: 51–62. [Google Scholar]
- Machado, C. A., R. M. Kliman, J. A. Markert and J. Hey, 2002. Inferring the history of speciation from multilocus DNA sequence data: The case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 19: 472–488. [DOI] [PubMed] [Google Scholar]
- Malausa, T., M. T. Bethenod, A. Bontemps, D. Bourguet, J. M. Cornuet et al., 2005. Assortative mating in sympatric host races of the European corn borer. Science 308: 258–260. [DOI] [PubMed] [Google Scholar]
- Malausa, T., B. Pélissié, V. Piveteau, C. Pélissier, B. Bourguet et al., 2007. Differences in oviposition behaviour of two sympatric sibling species of the Ostrinia genus. Bull. Entomol. Res. (in press). [DOI] [PubMed]
- Martel, C., A. Réjasse, F. Rousset, M.-T. Bethenod and D. Bourguet, 2003. Host-plant-associated genetic differentiation in northern french populations of the European corn borer. Heredity 90: 141–149. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Paabo, S., D. M. Irwin and A. C. Wilson, 1990. DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem. 265: 4718–4721. [PubMed] [Google Scholar]
- Pélozuelo, L., C. Malosse, G. Genestier, H. Guenego and B. Frérot, 2004. Host-plant specialization in pheromone strains of the European corn borer Ostrinia nubilalis in France. J. Chem. Ecol. 30: 335–352. [DOI] [PubMed] [Google Scholar]
- Peña, A., H. Arn, H. Buser, S. Rauscher, F. Bigler et al., 1988. Sex pheromone of European corn borer, Ostrinia nubilalis: polymorphism in various laboratory and field strains. J. Chem. Ecol. 14: 1359–1366. [DOI] [PubMed] [Google Scholar]
- Ponsard, S., M. Bethenod, A. Bontemps, L. Pélozuelo, M. Souqual et al., 2004. Carbon stable isotopes: a tool for studying the mating, oviposition, and spatial distribution of races of European corn borer, Ostrinia nubilalis, among host plants in the field. Can. J. Zool. 82: 1177–1185. [Google Scholar]
- Posada, D., and K. Crandall, 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Raymond, M., and F. Rousset, 1995. GENEPOP (version 1.2): populations genetics software for exact tests and ecumenicism. J. Heredity 86: 248–249. [Google Scholar]
- Ridley, M., 2004. Evolution. Blackwell Scientific, Oxford.
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Rundle, H., and P. Nosil, 2005. Ecological speciation. Ecol. Lett. 8: 336–352. [Google Scholar]
- Schluter, D., 2001. Ecology and the origin of species. Trends Ecol. Evol. 16: 372–380. [DOI] [PubMed] [Google Scholar]
- Schneider, S., D. Roessli and L. Excoffier, 2000. Arlequin ver 2.00: a software for population genetics data analysis. University of Geneva, Geneva.
- Thomas, Y., M.-T. Bethenod, L. Pélozuelo, B. Frérot and D. Bourguet, 2003. Genetics isolation between two sympatric host-plant races of the European corn borer, Ostrinia nubilalis Hübner. I. Sex pheromone, moth emergence timing, and parasitism. Evolution 57: 261–273. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via, S., 1999. Reproductive isolation between sympatric races of pea aphids. I. Gene flow restriction and habitat choice. Evolution 53: 1446–1457. [DOI] [PubMed] [Google Scholar]
- Via, S., 2001. Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol. Evol. 16: 381–390. [DOI] [PubMed] [Google Scholar]
- Watterson, G., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Willett, C. S., and R. G. Harrison, 1999. Insights into genome differentiation: Pheromone-binding protein variation and population history in the European corn borer (Ostrinia nubilalis). Genetics 153: 1743–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, T. K., K. J. Tilmon, A. B. Shantz, C. K. Harris and J. Pesek, 1999. The role of host-plant fidelity in initiating insect race formation. Evol. Ecol. Res. 1: 317–332. [Google Scholar]
- Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]