Abstract
The blaTEM-1 β-lactamase gene has become widespread due to the selective pressure of β-lactam use and its stable maintenance on transferable DNA elements. In contrast, blaSME-1 is rarely isolated and is confined to the chromosome of carbapenem-resistant Serratia marcescens strains. Dissemination of blaSME-1 via transfer to a mobile DNA element could hinder the use of carbapenems. In this study, blaSME-1 was determined to impart a fitness cost upon Escherichia coli in multiple genetic contexts and assays. Genetic screens and designed SME-1 mutants were utilized to identify the source of this fitness cost. These experiments established that the SME-1 protein was required for the fitness cost but also that the enzyme activity of SME-1 was not associated with the fitness cost. The genetic screens suggested that the SME-1 signal sequence was involved in the fitness cost. Consistent with these findings, exchange of the SME-1 signal sequence for the TEM-1 signal sequence alleviated the fitness cost while replacing the TEM-1 signal sequence with the SME-1 signal sequence imparted a fitness cost to TEM-1 β-lactamase. Taken together, these results suggest that fitness costs associated with some β-lactamases may limit their dissemination.
HORIZONTAL gene transfer must overcome several barriers before stable maintenance of the transferred gene in the recipient bacteria can occur. DNA entry, avoidance of restriction systems, and incorporation into the host replication machinery are necessary steps for transfer (Thomas and Nielsen 2005). Although fulfilling these criteria permits a gene to transfer, the gene must also confer a selective advantage to expand within the bacterial population (Berg and Kurland 2002). Such is the case with the rapid dissemination of β-lactamase genes in response to the selective pressure of β-lactam use. A better understanding of the barriers to horizontal gene transfer and possible costs associated with maintenance of the transferred gene may facilitate the development of strategies to reduce the spread of antibiotic resistance.
β-Lactamases provide bacterial resistance to β-lactam antibiotics by catalyzing the hydrolysis of these drugs. The widespread dissemination of β-lactamase genes represents an ongoing challenge to the efficacy of treatment with β-lactam antibiotics. A large collection of β-lactamases exists and can be classified into four groups (A–D) on the basis of primary amino acid sequence homologies (Ambler 1980). The most prevalent plasmid-borne β-lactamase in Enterobacteriaceae is the class A TEM-1 β-lactamase (Shah et al. 2004). The spread of this β-lactamase to >25 species of gram-negative bacteria has contributed to increased bacterial resistance to penicillins and early cephalosporins and thereby has reduced their efficacy as anti-bacterial agents. In response to increased bacterial resistance to these β-lactams, extended spectrum cephalosporins and β-lactamase inhibitors were introduced in the 1980s. The selective pressure resulting from the introduction of these antibiotics has resulted in the rapid evolution of TEM-1 β-lactamase. There are now >100 TEM-1 variants containing amino acid substitutions that allow the enzyme to hydrolyze extended-spectrum β-lactams and/or avoid inactivation by the β-lactamase inhibitors (http://www.lahey.org). To date, no TEM-1 variants capable of conferring resistance to a third class of β-lactams, the carbapenems, have surfaced.
Carbapenem resistance is often associated with decreased cell-wall permeability and the presence of class B or class D β-lactamases that are distantly related to the class A β-lactamase TEM-1 (Navon-Venezia et al. 2005). A few class A β-lactamases capable of hydrolyzing carbapenems (referred to as carbapenemases) have been discovered in clinical Enterobacteriaceae isolates. One such carbapenemase, SME-1, has been identified in several Serratia marcescens strains and is encoded by the chromosomal blaSME-1 gene (Yang et al. 1990; Naas et al. 1994). Genomic analysis of several carbapenem-resistant S. marcescens strains isolated from diverse geographical locations reveals a closer genetic relatedness among strains containing blaSME-1 than among S. marcescens strains lacking the blaSME-1 gene, and therefore they may represent a globally disseminated subtype of this species (Queenan et al. 2000). The blaSME-1 gene has not been identified on plasmids or other mobile genetic elements. This may have restricted the spread of the blaSME-1 gene.
Class A β-lactamases have no known function besides β-lactam hydrolysis and therefore incorporation of β-lactamase genes into a bacterial cell is not expected to alter metabolism (Jain et al. 1999). It has been hypothesized, however, that class C β-lactamases could participate in recycling of cell-wall components due to the structural similarity between β-lactam antibiotics and the DD-peptide bond found in the peptidoglycan layer (Bishop and Weiner 1992). A fitness cost associated with expression of the class C β-lactamase AmpC from Enterobacter cloacae in Salmonella enterica serotype Typhimurium has been documented (Morosini et al. 2000). However, no significant changes in the muropeptide composition of the peptidoglycan could be discerned. Regardless of the cause, the association with a fitness cost may explain why AmpC-producing Salmonella are seldom isolated (Morosini et al. 2000). Subsequent work by Hossain et al. (2004) found that another class C β-lactamase, CMY-7, impaired Salmonella typhimurium strain LT2 growth and cell invasion when encoded on a high-copy number plasmid.
In the course of working with the class A SME-1 carbapenemase, it was noted that cell lysis and occasional plasmid rearrangement events occurred in overnight cultures of Escherichia coli transformed with a plasmid encoding SME-1. These observations suggested a fitness cost may be associated with blaSME-1 in E. coli. The purpose of this investigation was to quantitatively establish the presence of a blaSME-1-associated fitness cost and then to determine the origins of the fitness cost. Genetic screens as well as designed mutant constructs were used to investigate specific features of the SME-1 gene as potential sources of the fitness cost. These methods revealed the requirement of the SME-1 enzyme in conferring a fitness cost to E. coli but suggest that enzymatic activity itself does not play a role.
MATERIALS AND METHODS
Bacterial strains and cloning:
Unless otherwise noted, all enzymes were obtained from New England Biolabs (Ipswich, MA). E. coli K12 XL1-Blue strain (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′proAB lacIqZΔM15 Tn10 (Tetr)]) was obtained from Stratagene (La Jolla, CA). E. coli B Ara+ and Ara− strains were a gift from R. E. Lenski (Bouma and Lenski 1988). pTP123 is a derivative of the pTrc99A vector encoding the LacI repressor and a multiple cloning site downstream of the Ptrc promoter (Petrosino et al. 1999). pTP123-TEM-1 and pTP123-SME-1 were constructed by inserting the blaTEM-1 and blaSME-1 genes, respectively, into the multiple cloning site of pTP123 (Majiduddin and Palzkill 2003a,b). pUC18-Kn was constructed by releasing the kanamycin resistance gene from the pKRP11 plasmid (Reece and Phillips 1995) with the HincII restriction enzyme and inserting the fragment into the end-filled pUC18 plasmid cut with AatII/BsaI. This resulted in a majority of the blaTEM-1 gene being replaced by the kanamycin resistance gene. pUC18-Kn-TEM-1 and pUC18-Kn-SME-1 were constructed by inserting the blaTEM-1 and blaSME-1 genes from pTP123-TEM-1 and pTP123-SME-1, respectively, into the multiple cloning site of pUC18-Kn using the SacI/XbaI and SacI/BamHI restriction enzymes, respectively. The SME-1 truncation mutant (pTP123-Trunc.SME-1) is a fortuitously isolated spontaneous mutant of pTP123-SME-1 that possesses a single base pair deletion of t363 (numbering according to Naas et al. 1994) resulting in an amber STOP codon at amino acid position 81 (numbering according to Ambler et al. 1991). The SME-1 ΔSTART mutant (pDCM104) was created by using Stratagene's QuickChange kit with pTP123-SME-1 as template and the SME-STARTdel primer that mutates the first and second codons to create a SpeI site. All primers used in this study are listed in Table 1. The pTP123-SME-1 C69V:C238V construct was created via use of the QuickChange kit and primers SME-C69V-Top and SME-C238V-Top simultaneously. Screening for positive clones was facilitated by the introduction of silent mutations creating a HindIII site by the SME-C69V-Top primer and both the creation of a SfoI site and destruction of a naturally occurring NdeI site by the SME-C238V-Top primer. The pTP123-SME-1 S70A:E166Q mutant was created stepwise using overlap PCR (Ho et al. 1989). First, the E166Q SME-1 mutant was created using internal primers SME-E166Q-Top and SME-E166Q-Bot and external primers SME-Sac and SME-BamHI with pTP123-SME-1 (Majiduddin and Palzkill 2003a) as template. In the first step, each half of the wild-type SME-1 gene was amplified separately using one internal and one external primer. This incorporated the desired mutation into the complementary ends of the two products, which were combined in a final PCR reaction also containing external primers. The full-length product with the desired mutation was then digested with the appropriate restriction enzymes along with DpnI to remove residual, host-derived plasmid DNA and ligated into the target plasmid treated with calf intestinal phosphatase (CIP). The S70A:E166Q SME-1 double mutant was created by overlap PCR using the internal primers SME-S70A-Top and SME-S70A-Bot and external primers SME-Sac and SME-BamHI with pTP123-SME-1 E166Q as template. To facilitate the creation of the TEM(SS)SME construct (pDCM102), a modified version of pTP123-TEM-1 was generated that contained a silent AgeI site in the signal sequence coding region of blaTEM-1. This modified pTP123-TEM-1, named pTP251, was created by overlap PCR using internal primers TEM-AgeI-Top and TEM-AgeI-Bot and external primers PDbla1 (Petrosino et al. 1999) and PDbla2. The pTP251 plasmid allows insertion of mature coding sequences behind the canonical blaTEM-1 signal sequence coding region. The SME-1 mature coding region was amplified by Pfu polymerase (Stratagene) using the primers TEM-SME and SME-BamHI and digested with AgeI/BamHI for ligation into similarly digested pTP251. To create the SME(SS)TEM construct (pDCM106-8), an overlap PCR reaction combining three products was used. Overlap PCR was used to create the following: product A was derived from pTP123-TEM-1 using primers PDbla1 and Pbla-ssSME-Bot, product B was derived from pTP123-SME-1 using primers Pbla-ssSME-Top and ssSME-mTEM-Bot, and product C was derived from pTP123-TEM-1 using primers ssSME-mTEM-Top and PDbla2. Full-length SME(SS)TEM insert was recovered by combining products A, B, and C with external primers PDbla1 and PDbla2, digested with SacI/XbaI, and ligated into similarly digested pTP123. The resulting SME(SS)TEM construct, pDCM106-8, has the 5′ UTR and ribosome binding site of blaTEM-1 followed by the encoded signal sequence of blaSME-1 fused to the mature blaTEM-1 sequence. All genes and their hybrids cloned into the pTP123 vector are under the transcriptional control of the Ptrc promoter. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was not used in the experiments because basal expression levels from the Ptrc promoter were sufficient to confer ampicillin resistance to all constructs expected to produce a functional β-lactamase. All PCR reactions utilized Pfu polymerase obtained from Stratagene; all primers were obtained from Integrated DNA Technologies (Coralville, IA). Silent mutations resulting in the introduction or deletion of restriction enzyme sites were engineered into primers by using the Primer Generator program found at http://www.med.jhu.edu/medcenter/ (Turchin and Lawler 1999). All constructs and several clones isolated from the genetic screens (see below) were sequenced using Applied Biosystems Instruments (Foster City, CA) Prism Big Dye DNA sequencing with an ABI 3100 automated sequencer. Prior to use in each experiment, each construct was transformed into E. coli K12 XL1-Blue or E. coli B Ara+/Ara− cells and spread on Luria broth (LB) plates (Difco Laboratories, Sparks, MD) containing the appropriate selective antibiotic. When possible, ampicillin was also included in LB plates to maintain functional (e.g., SME-1 expressing) clones.
TABLE 1.
Primers used for creating constructs
| Primer name | Sequencea |
|---|---|
| SME-STARTdel | 5′-CAATGACAGTTAATAGGTAAAGTTACTAGTAACAAAGTAAATTTTAAAACGGCTAGC-3′ |
| SME-Sac | 5′-GGGGCGGAGCTCAACTCATTCAACACTCGG-3′ |
| SME-BamHI | 5′-GGGGCGGGATCCGCGTCAAGGCCACAGTCAGCTCTAACGC-3′ |
| SME-C69V-Top | 5′-GCGGTTCCCTTTAGTCTCAAGCTTTAAAGGTTTTTTGGCGGCTGC-3′ |
| SME-C238V-Top | 5′-GGTGACAAAACTGGGAGCGTTGGCGCCTATGGTACTGCGAATG-3′ |
| SME-E166Q-Top | 5′-GGTTAGATCGCTGGCAGCTGGAACTTAACACTGC-3′ |
| SME-E166Q-Bot | 5′-GCAGTGTTAAGTTCCAGCTGCCAGCGATCTAACC-3′ |
| SME-S70A-Top | 5′-GCGGTTCCCTTTATGCGCTTCATTTAAAGGTTTTTTGGCGGCTGC-3′ |
| SME-S70A-Bot | 5′-GCAGCCGCCAAAAAACCTTTAAATGAAGCGCATAAAGGGAACCGC-3′ |
| PDbla1 | 5′-CGGGGAGCTCGTTTCTTAGACGTCAGGTGG-3′ |
| PDbla2 | 5′-CCCCGTCTAGACTTACCAATGCTTAATCAGT-3′ |
| TEM-AgeI-Top | 5′-CATTTTGCCTACCGGTTTTTGCTCAC-3′ |
| TEM-AgeI-Bot | 5′-GTGAGCAAAAACCGGTAGGCAAAATC-3′ |
| TEM-SME | 5′-GCCTACCGGTTTTTGCTAACAAAAGTGATGCTGC-3′ |
| Pbla-ssSME-Top | 5′-CAATAATATTGAAAAAGGAAGAGTATGTCAAACAAAGTAAATTTTAAAACGGCTAGC-3′ |
| Pbla-ssSME-Bot | 5′-GCTAGCCGTTTTAAAATTTACTTTGTTTGACATACTCTTCCTTTTTCAATATTATTG-3′ |
| ssSME-mTEM-Top | 5′-GCTTTGTCGGCATTTAATGCTCATGCTCACCCAGAAACGCTGG-3′ |
| ssSME-mTEM-Bot | 5′-CCAGCGTTTCTGGGTGAGCATGAGCATTAAATGCCGACAAAGC-3′ |
| pyrF-bla-top | 5′-CACGCGATTGTCGTCTGAAGGTCGGCAAAGAGATGGTTTCTTAGACGTCAGGTGGCACTTTT CGGGGAAATG-3′ |
| pyrF-bla-bot | 5′-GCGGTGCATCTTTGCCAAACGGAACCAGTGCCTCTTACCAATGCTTAATCAGTGAGGCACCTA TCTCAGCG-3′ |
| SME-1STOP-pyrF384bot | 5′-GGCTTCCATGCTGGTCAACACTGTCAATTAATCAATTGCCTGAATTGC-3′ |
Engineered restriction sites utilized for cloning and/or screening are underlined.
E. coli strains containing chromosomally encoded blaTEM-1 (TP112) or blaSME-1 (DCM105) inserted into the pyrF locus were created using λ Red homologous recombination in E. coli SW102 (Warming et al. 2005). In generating E. coli TP112, the blaTEM-1 gene along with its native promoter was PCR amplified from pBG66 (Huang et al. 1996) using primers pyrF-bla-top and pyrF-bla-bot to yield a PCR product with ∼33 nucleotides on each end capable of homologous recombination with the pyrF locus. Transformation of E. coli SW102 with this PCR product resulted in the deletion of codons F70–R131 of pyrF and insertion of blaTEM-1. Insertion of blaSME-1 into the pyrF locus was initiated by creating a plasmid, pDCM105, that encodes blaSME-1 downstream of the 5′ UTR of blaTEM-1. The plasmid pDCM105 was generated by an overlap PCR reaction using the pTP123-TEM-1 template with primers PDbla1 and Pbla-ssSME-bot combined with a PCR fragment from the pTP123-SME-1 template using primers Pbla-ssSME-top and SME-BamHI. Amplification of blaSME-1 from pDCM105 using the primers PDbla1 and SME-1STOP-pyrF384bot yielded a PCR product with 146 nucleotides of homology with the 5′ UTR of blaTEM-1 chromosomally encoded in TP112 and 26 nucleotides of homology with a region of pyrF downstream of the blaTEM-1 insertion. Transformation of E. coli TP112 with the pDCM105-derived PCR product replaced blaTEM-1 with blaSME-1 and further deleted pyrF codons E131–V147, thus creating strain DCM105 with blaSME-1 under the control of Pbla. P1 transduction was subsequently utilized to move the chromosomally encoded blaTEM-1 gene of E. coli TP112 into the E. coli B Ara+/Ara− strains (DCM106 and DCM107, respectively). Similarly, P1 transductants derived from DCM105 were utilized to move blaSME-1 into the E. coli B Ara+/Ara− strains (DCM108 and DCM109, respectively). Due to the deficiency of recA in the parental SW102 strain, the low-copy number plasmid pGE591 supplied the RecA protein in trans to enhance P1 phage production in the donor strains E. coli TP112 and DCM105 (Weisemann and Weinstock 1988). The P1 transduction protocol is based upon a protocol found in Miller (1992).
Genetic screen for spontaneous suppressors of a SME-1-mediated fitness cost:
Spontaneous suppressors of the SME-1-mediated fitness cost were selected by passaging E. coli B Ara+ containing pTP123-SME-1 for several days in LB media. Individual clones of pTP123-TEM-1 or pTP123-SME-1 were inoculated for growth overnight with shaking at 37°. Every 12 hr, 100 μl of culture was transferred to 10 ml of fresh LB broth containing chloramphenicol and grown again with shaking at 37°. Individual clones with a growth advantage are expected to overtake the culture. After 48 hr, a 1:106 dilution of the passaged culture was spread on LB plates containing 12.5 μg/ml chloramphenicol to obtain isolated colonies. DNA was isolated from seven pTP123-SME-1 clones and four pTP123-TEM-1 clones and subjected to restriction digest analysis. These clones were also patched onto LB plates containing 12.5 μg/ml chloramphenicol and plates containing both chloramphenicol and 100 μg/ml ampicillin to determine their resistance profile. As several of the pTP123-SME-1 clones possessed aberrant restriction digests and/or sensitivity to ampicillin, five of these clones were sequenced resulting in four unique clones being isolated (Cp.1–4).
A similar selection for SME-1 suppressors was performed in LB broth containing both 12.5 μg/ml chloramphenicol and 100 μg/ml ampicillin. Again, cultures containing pTP123-TEM-1 or pTP123-SME-1 were allowed to grow 12 hr before being transferred into fresh LB media containing both chloramphenicol and ampicillin. At 96 hr, individual clones were obtained by spreading a 1:106 dilution of the passaged culture on LB plates containing chloramphenicol and 100 μg/ml ampicillin. Seven pTP123-SME-1 clones were sequenced resulting in three unique clones being isolated (CpAp.1, 2, 3).
Library construction and selection:
Mutagenic PCR via inclusion of 0.0625 mm MnCl2 was performed with 5 units Taq polymerase (Promega, Madison, WI), 0.2 mm dNTPs (Bioline, Randolph, MA), 1.5 mm MgCl2, 1× Taq polymerase buffer (Promega), 100 ng of each primer SME-Sac and SME-BamHI, and 45 ng of pTP123-SME-1 template in a 100-μl reaction. A second series of 100-μl mutagenic PCR reactions were performed utilizing either 2.5 or 0.25 ng of the first reaction's product as template with either 0.0625 or 0.15 mm MnCl2 (a total of four reactions performed in duplicate). The PCR reaction parameters were as follows: 95° for 1 min; followed by 30 cycles of 95° for 1 min, 50° for 2 min, and 72° for 3 min; ending with 72° for 10 min. All eight mutagenic PCR reactions were pooled and column purified using a QIAGEN PCR purification kit (QIAGEN, Valencia, CA) before sequential digestion with SacI/BamHI/DpnI. Insert DNA was gel purified and ligated into CIP-treated pTP123 digested with SacI/BamHI. The ligation was phenol/chloroform extracted, transformed into electrocompetent E. coli K12 XL1-Blue, and spread onto LB plates containing chloramphenicol. A total of 1.72 × 105 CFUs were pooled. Plasmid DNA was isolated from two clones and sequencing showed six nucleotide changes in one clone and two nucleotide changes in the other. The library was subjected to two independent selections by passaging in LB broth with 12.5 μg/ml chloramphenicol or in LB broth with 12.5 μg/ml chloramphenicol and 100 μg/ml ampicillin as described above for the selection of spontaneous mutants. At 96 hr, DNA was isolated from individual clones and sequenced.
Growth curves:
Growth curve data were obtained by measuring cell density at OD600 on a Beckman-Coulter (Fullerton, CA) DU 800 spectrophotometer. Isolated clones of each construct, in either E. coli K12 XL1-Blue or E. coli B Ara+, were inoculated into 2 ml of LB broth with chloramphenicol and incubated overnight. Cultures were then diluted to an OD600 of 0.1 in 10 ml of LB with chloramphenicol and allowed to shake vigorously (295 rpm) at 37°. OD600 was monitored over a time course of 23 hr. The data represent an average of at least three independent experiments.
Culture viability:
Individual clones of E. coli K12 XL1-Blue transformed with pTP123, pTP123-TEM-1, pTP123-SME-1, pTP123-SME-1 S70A:E166Q, pDCM102, pDCM106-8, pUC18-Kn, pUC18-Kn-TEM-1, or pUC18-Kn-SME-1 were inoculated into 10 ml LB broth containing either chloramphenicol or kanamycin (according to plasmid resistance marker) and allowed to grow 24 hr with vigorous shaking (295 rpm) at 37°. Overnight cultures were diluted 1:106 and 100 μl was spread in duplicate onto LB plates and allowed to grow overnight at 37°. The number of colony-forming units per milliliter was determined for each culture and expressed relative to the number of colony-forming units per milliliter obtained from an overnight culture containing empty vector (pTP123 or pUC18-Kn). The data represent an average of at least six independent experiments. The t-test was utilized to assign statistical significance by comparison against data obtained with the relevant empty vector.
Competition experiments:
To directly compare fitness between two strains in a mixed culture, E. coli B strains differing by the ability to utilize arabinose (Ara+ and Ara−) were transformed with the desired plasmids and allowed to compete during overnight growth. Overnight starter cultures of the competing clones were mixed in a 50:50 ratio and 200 μl of the mixed culture was used to inoculate 20 ml of LB broth containing chloramphenicol. During the course of the experiment, dilutions of the mixed culture were spread onto tetrazolium arabinose (TA) indicator plates to determine the contribution of each strain to the total population. On TA plates, E. coli B Ara+ appears pinkish-white while the Ara− strain appears dark red (Levin et al. 1977; Bouma and Lenski 1988; Lenski 1988). Competitions were performed in both strain contexts (i.e., Ara+ pTP123-TEM-1 vs. Ara− pTP123-SME-1 and Ara− pTP123-TEM-1 vs. Ara+ pTP123-SME-1). Because no strain-specific effects were observed with the plasmid constructs, the competition data were averaged between the strain contexts. The competitions of plasmid-encoded β-lactamase genes were followed overnight. For strains possessing chromosomally encoded β-lactamase genes, competitions were conducted over 3 days during which time 200 μl of the culture was propagated into 20 ml of fresh LB media every 16 hr. Data obtained from these experiments represent an average of at least four independent experiments and were used to calculate a selection rate constant for each competition. The selection rate constant is the difference of the realized Malthusian parameters associated with the competing E. coli B strains and is dependent upon the relative growth rates of the competing strains (Lenski et al. 1991). It was determined by regression analysis using SigmaPlot with the following equation: loge R(t) = loge R(0) + st, where R is the ratio of the prevalence of the competing strains, t is time in days, and s is the selection rate constant having units of day−1 (Lenski et al. 1994; Travisano and Lenski 1996).
Minimum inhibitory concentration determination:
β-Lactam resistance levels were utilized to determine if the chimeric TEM(SS)SME (pDCM102) and SME(SS)TEM (pDCM106-8) clones conferred functional in vivo resistance levels equivalent to pTP123-SME-1 and pTP123-TEM-1, respectively. Both E-test strips (AB BIODISK, Piscatway, NJ) and broth dilution methods were utilized. E-test strips containing the β-lactams ampicillin or amoxicillin plus clavulanic acid were placed upon LB plates spread with a one-tenth dilution of an overnight culture of E. coli B transformed with pTP123-TEM-1, pTP123-SME-1, pDCM102, or pDCM106-8. For broth dilution minimum inhibitory concentration (MIC), twofold dilutions of ampicillin were tested in a 96-well format in which each well contained 1 × 104 bacteria in 100 μl. The 96-well plate was sealed and allowed to shake overnight at 37° before scoring for growth. Each assay was conducted in triplicate using independent overnight cultures. The generally accepted level of significance of plus or minus one twofold dilution was employed to determine if the chimeras displayed a significant effect on in vivo activity relative to the relevant wild-type construct.
Plasmid loss from strains grown in LB media:
The effect of wild type and mutant SME-1 genes on plasmid stability in E. coli B was determined by monitoring the rate of plasmid loss over time. Isolated colonies were inoculated into 10 ml of LB media lacking a selective antibiotic and grown at 37° with vigorous shaking for 24 hr. Every 24 hr, 5 μl of culture was transferred to 10 ml of fresh LB. Additionally, every 24 hr dilutions were spread in duplicate onto LB plates and LB plates containing a selective antibiotic (either chloramphenicol for the pTP123 series or kanamycin for the pUC18-Kn series). Because plasmid loss was evident in the pUC18-Kn-SME-1 construct after only 24 hr of growth, the possibility of plasmid loss on the original LB agar plate was investigated by resuspending colonies in LB broth and diluting and directly spreading onto LB plates and LB plates containing kanamycin. Colonies were counted to determine the percentage of bacteria that retained plasmid as ascertained by the ability to grow on plates containing antibiotic. Experiments were conducted at least in duplicate.
RESULTS
Establishing the existence of a blaSME-1-mediated fitness cost in E. coli:
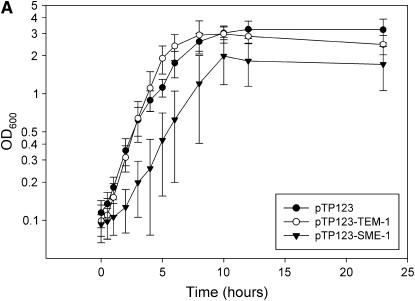
The observance of plasmid instability and cell lysis in overnight cultures containing plasmids encoding blaSME-1 suggested the possibility of a fitness cost associated with the SME-1 gene in E. coli. To quantify these observations, several independent assays were utilized in multiple genetic contexts to delineate a SME-1-mediated fitness cost in E. coli. Initially, the effect of the SME-1 gene on the growth of E. coli B and K12 XL1-Blue strain was determined by measuring the optical density of the culture over time. As seen in Figure 1A, the presence of blaTEM-1 encoded on the pTP123 plasmid (pTP123-TEM-1) had little impact on the growth rate of E. coli K12 XL1-Blue relative to strains transformed with empty vector (pTP123). However, E. coli K12 XL1-Blue transformed with plasmid-expressing SME-1 β-lactamase (pTP123-SME-1) exhibited an extended lag phase and an overall reduction of the final cell density in stationary phase. Similar results were obtained in E. coli B cells (data not shown).
Figure 1.—
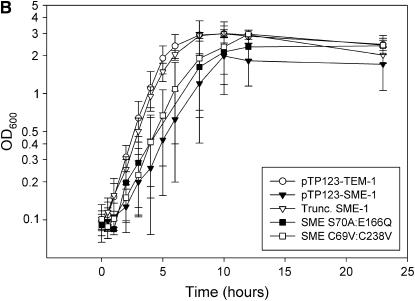
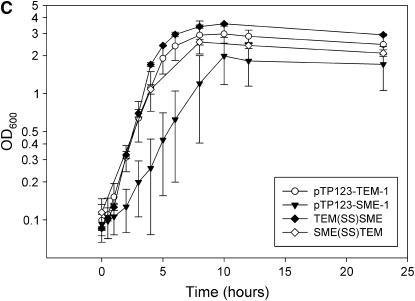
Effect of β-lactamase genes on E. coli growth rates. (A) Growth curves in LB broth of E. coli K12 XL1-Blue containing pTP123, pTP123-TEM-1, or pTP123-SME-1 were obtained by monitoring OD600 over time. (B) Growth curves of a frameshifted SME-1 clone and point mutants that eliminate enzymatic activity (S70A:E166Q) or disulfide bond formation (C69V:C238V). (C) Growth curves of chimeric constructs in which signal sequences of TEM-1 and SME-1 have been exchanged. Error bars represent standard deviation of at least three independent experiments.
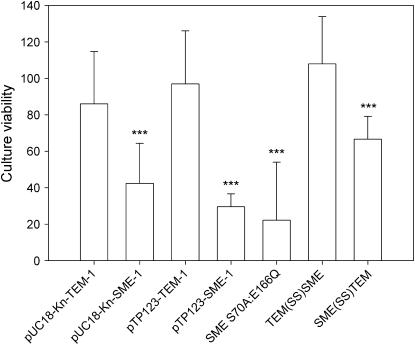
Culture viability experiments were utilized to determine the effect of the blaSME-1 gene on the number of colony-forming cells present in overnight cultures of E. coli K12 XL1-Blue. Overnight cultures of E. coli K12 XL1-Blue transformed with plasmid-encoding blaSME-1 were diluted and spread onto LB plates. The number of colony-forming units per milliliter was determined and is presented in Figure 2 relative to the number of colony-forming units per milliliter obtained from an overnight culture containing empty vector. Relative to empty vector, the blaSME-1 gene confers reduced culture viability when encoded by either a pTrc99A-based vector (pTP123 series, P < 0.001) or a pUC18-based vector (pUC18-Kn series, P < 0.001) (Figure 2). The gene encoding TEM-1, blaTEM-1, had no statistically significant effect on the number of colony-forming cells in either plasmid context as determined by this assay (pTP123-TEM-1, P = 0.62; pUC18-Kn-TEM-1, P = 0.07). These results are consistent with a blaSME-1-mediated fitness cost that is present in multiple genetic contexts.
Figure 2.—
Effect of β-lactamase genes on culture viability. The number of colony-forming units per milliliter is indicative of culture viability. Data for E. coli K12 XL1-Blue containing the indicated plasmids were normalized to E. coli K12 XL1-Blue harboring isogenic plasmid without a β-lactamase gene. Two genetic contexts (pTP123 parent and pUC18-Kn parent plasmids) were examined. Error bars represent the standard deviation of at least six independent experiments. Significance is based on t-test against empty vector: ***P < 0.001.
To quantitate the fitness cost associated with the SME-1 gene, competition experiments were performed (Bouma and Lenski 1988). Initially, Ara+ or Ara− E. coli B strains were transformed with either pTP123-SME-1 or empty vector pTP123, mixed in a 50:50 ratio, and competed over a 24-hr time course. The relative contribution of each construct to the total culture population was tracked by spreading dilutions onto TA indicator plates. E. coli B Ara+ is capable of utilizing arabinose and appears white or light pink on TA indicator plates while an E. coli B Ara− strain is unable to utilize arabinose and appears dark red on TA indicator plates (Levin et al. 1977; Bouma and Lenski 1988; Lenski 1988). Within 5 hr, <10% of the mixed culture contained the strain harboring the pTP123-SME-1 plasmid. As a more stringent test of fitness, the Ara+ or Ara− E. coli B strain containing the pTP123-SME-1 plasmid was competed against the reciprocal Ara+/− E. coli B strain containing isogenic pTP123-TEM-1. After an overnight competition, 98 ± 3% of the culture was composed of the pTP123-TEM-1 containing strain. To facilitate comparisons between experiments, the selection rate constant of each competition was determined (Table 2). The negative selection rate constant, −7.0 ± 2.4 day−1, represents a significant fitness cost associated with carriage of the pTP123-SME-1 plasmid relative to pTP123-TEM-1. This provides direct evidence of a blaSME-1-mediated fitness cost in E. coli B.
TABLE 2.
Effect of the SME-1 gene and its derivatives on E. coli B fitness
| Competition experiment | Selection rate constant (day−1) |
|---|---|
| pTP123-SME-1 vs. pTP123 | −10.7 ± 0.7*** |
| pTP123-SME-1 vs. pTP123-TEM-1 | −7.0 ± 2.4*** |
| chSME-1 Ara+vs. chTEM-1 Ara− | −1.3 ± 0.5*** |
| chSME-1 Ara−vs. chTEM-1 Ara+ | −0.2 ± 0.1** |
| pTP123-SME-1 vs. Trunc. SME-1 | −2.2 ± 1.3*** |
| pTP123-SME-1 vs. SMEΔSTART | −3.8 ± 2.8*** |
| SMEΔSTART vs. pTP123-TEM-1 | −0.3 ± 0.7 |
| pTP123-SME-1 vs. SME S70A:E166Q | −0.9 ± 1.4 |
| pTP123-SME-1 vs. SME C69V:C238V | 1.4 ± 1.0* |
| pTP123-SME-1 vs. TEM(SS)SME | −3.9 ± 1.3*** |
| TEM(SS)SME vs. pTP123-TEM-1 | −0.9 ± 1.2 |
| SME(SS)TEM vs. pTP123-TEM-1 | −1.4 ± 1.1** |
| pTP123-SME-1 vs. SME(SS)TEM | −4.4 ± 1.2*** |
A negative selection rate constant is indicative of a fitness cost associated with the initial construct (e.g., SME-1) relative to the construct it is competing against (e.g., TEM-1). Significance is based on paired t-test: *0.05 < P < 0.01; **0.01 < P < 0.001; ***P < 0.001.
To determine if the fitness cost due to blaSME-1 was dependent upon copy number, the blaSME-1 was transferred to the chromosome of the E. coli Ara+/Ara− strains and competitions against E. coli Ara+/Ara− strains encoding a chromosomal blaTEM-1 gene were performed. Although a strain-dependent effect is apparent, in all competitions the E. coli B strain encoding a chromosomal copy of blaTEM-1 outcompeted the reciprocal strain encoding blaSME-1 (Table 2). It should be noted that the chromosomally encoded blaSME-1 differs from the vector-encoded blaSME-1 in that it possesses the 5′ UTR of blaTEM-1. Although this establishes the presence of a fitness cost associated with the blaSME-1 coding region, the difference of selection coefficients observed between chromosomally encoded and plasmid-encoded blaSME-1 could be copy number or 5′ UTR dependent. However, the data are consistent with the blaSME-1 gene conferring a fitness cost to E. coli B even when in a chromosomal context.
The persistence of antibiotic resistance genes in the absence of selection suggests a neutral fitness effect. A lack of a fitness cost may contribute to the failure of antibiotic cessation to remove reservoirs of β-lactam resistance in a natural environment (Liebana et al. 2006). To address the persistence of blaSME-1, the effect of the gene on the rate of plasmid loss in the absence of selection was ascertained. Cultures were inoculated with isolated colonies and then propagated into fresh LB media every 24 hr for 5 days. The proportion of the culture harboring plasmid was determined by comparing the number of colony-forming units obtained on LB plates to the number of colony-forming units obtained on plates with the appropriate selective antibiotic (chloramphenicol for the pTP123 series and kanamycin for the pUC18-Kn series). By day 3, a majority of viable cells in the culture had lost the pTP123-SME-1 plasmid (Figure 3). In contrast, neither the pTP123 nor the pTP123-TEM-1 vectors displayed any appreciable levels of plasmid loss during the course of the experiment. In the context of the pUC18-Kn vector, the blaSME-1 gene yielded a 57 ± 22% loss of the SME-1 construct within 24 hr while the pUC18-Kn-TEM-1 culture displayed no plasmid loss within 24 hr (data not shown). Resuspension and dilution of pUC18-Kn-SME-1 colonies directly taken from the agar plate showed no plasmid loss occurring on the plate itself (109 ± 21%). The simplest interpretation of these results is that plasmid-free cells arise in the culture by passive loss of the SME-1-encoding plasmids and subsequently expand in the population to overtake the culture. However, these data do not distinguish between plasmid segregation, plasmid stability, or fitness cost as being the root cause of plasmid loss mediated by blaSME-1 (Lenski and Bouma 1987). Taken together, the results of these experiments indicate that the presence of the SME-1 gene results in reduced growth, lower culture viability, impaired fitness, and plasmid loss in E. coli and that these effects are not specific to a plasmid or strain context.
Figure 3.—
Plasmid loss in the absence of antibiotic selection. Overnight cultures were spread onto LB agar plates and LB agar plates containing chloramphenicol to determine the percentage of cells harboring plasmid. Errors bars represent the standard deviation for at least two independent experiments.
Genetic analysis of the blaSME-1-mediated fitness cost:
To determine the source of the blaSME-1-mediated fitness cost, mutants of blaSME-1 that ameliorate the fitness cost were isolated under several selection schemes. An overnight culture of E. coli B containing the pTP123-SME-1 plasmid was passaged into fresh LB media with chloramphenicol for 1 week. Clones with mutations that confer a fitness advantage are expected to expand in number and dominate the culture. The presence of chloramphenicol in the culture maintains the chloramphenicol-resistant plasmid backbone but does not select for a functional SME-1 gene. As a control, E. coli B containing pTP123-TEM-1 were passaged in parallel. To determine the effect of passaging on β-lactamase expression, each culture was diluted and spread onto plates containing the β-lactam ampicillin at the end of the experiment. Also, culture supernatant was immunoblotted with anti-β-lactamase sera to determine if β-lactamase was being produced and secreted into the media. As determined by these experiments, expression of TEM-1 β-lactamase was not affected by passaging. However, the SME-1 culture lost β-lactamase expression as determined by a lack of ampicillin-resistant clones and immunoblotting results (data not shown). DNA sequencing of the SME-1 gene from passaged clones revealed that an insertion sequence (IS1) transposon had been inserted into the SME-1 gene. This insertion event occurred at least three independent times (Table 3, Cp.2–4) as evident by differences between clones in the exact insertion point of the IS1 element (nucleotides 453, 457, and two at 460; numbering according to Naas et al. 1994). Also, one passaged pTP123-SME-1 clone was found to have an 11-bp deletion in the N-terminus of the blaSME-1 coding region that resulted in a frameshift and truncation at Phe19 (Table 3, Cp.1). Although such genetic events are expected to occur with low frequency in E. coli cultures, the emergence of these mutants to replace the original wild-type construct suggests that these clones must have a selective advantage in the absence of a β-lactam antibiotic. No DNA rearrangements or mutations were detected in clones isolated from the pTP123-TEM-1 culture passaged in parallel under the same conditions. These data further support the existence of a blaSME-1-mediated fitness cost and provide evidence for the requirement of the SME-1 enzyme in conferring the fitness cost.
TABLE 3.
Mutant SME-1 genes selected in the presence or absence of ampicillin
| 5′UTR | Signal sequence | Mature sequence | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spontaneous | ||||||||||||
| Cp.1a | F19b | |||||||||||
| Cp.2 (2)c | IS1d | |||||||||||
| Cp.3 | IS1 | |||||||||||
| Cp.4 | IS1 | |||||||||||
| CpAp.1e (4) | N20Y | |||||||||||
| CpAp.2 (2) | IS5 | |||||||||||
| CpAp.3 | IS10 | |||||||||||
| Library | ||||||||||||
| Cp.5 | S2P | K273E | ||||||||||
| Cp.6 | L14b | |||||||||||
| CpAp.4 | Δa73 | S2A | Q128R | |||||||||
| CpAp.5 (3) | N20S | |||||||||||
| CpAp.6 | K4R | F19L | S62P | |||||||||
| CpAp.7 | F19L | |||||||||||
| CpAp.8 (2) | t121c | a124g | F19L | |||||||||
Cp clones isolated under chloramphenicol selection.
STOP codon.
Clones isolated multiple times are indicated by number in parenthesis.
Occurance of insertion sequence as described in text.
CpAp clones isolated under chloramphenicol plus 100 μg/ml ampicillin selection.
In a similar experiment, a strain containing the pTP123-SME-1 plasmid was passaged in LB media containing both chloramphenicol and 100 μg/ml ampicillin. Inclusion of ampicillin prevents the emergence of nonfunctional, insertion or deletion mutants in the population and thereby requires alternative means to suppress the blaSME-1-mediated fitness cost. The pTP123-TEM-1 construct was passaged in parallel as a control. After 4 days of passaging, DNA was isolated and sequenced from individual clones. Again, clones from the pTP123-TEM-1 culture displayed the expected DNA migration pattern and did not have any mutations as determined by DNA sequencing. However, clones from the pTP123-SME-1 culture clones with aberrant DNA migration had either IS5 or IS10 elements inserted downstream of the blaSME-1 gene (Table 3, CpAp.2 and CpAp.3). IS elements mediating polar effects on gene transcription have been described and may be altering SME-1 protein levels in this case (Schneider and Lenski 2004). In several clones with the expected DNA mobility, a nonsynonymous point mutation occurred causing a single N20Y amino acid substitution in the SME-1 signal sequence near the signal cleavage site.
To obtain more information concerning the source of the blaSME-1-associated fitness cost, a library of SME-1 mutant clones was created by error-prone PCR. As with the selection for spontaneous suppressors of the fitness cost, this library was subjected to selection by passaging in either the presence or absence of ampicillin for several days with the expectation that clones with a fitness advantage would expand in number and dominate the culture. Signal sequence mutations were present in all of the clones isolated from the library under both passaging conditions. In some cases, mutations outside of the signal sequence were found in combination with signal sequence mutations (Table 3, library clones).
The prevalence of signal sequence mutations may reflect a direct role of the SME-1 signal sequence in the fitness effect or could indicate an indirect suppression of this phenomenon by preventing normal delivery of the SME-1 protein to the periplasmic space. To address this issue, periplasmic extracts were obtained from the most prevalent mutants, F19L and N20S, and were analyzed by immunoblot. As with wild-type SME-1, both of these signal sequence mutants delivered >95% of the SME-1 protein to the soluble fraction of the periplasmic extract and did not have any discernable effect on expression levels (data not shown). The signal sequence substitutions F19L and N20S therefore do not appear to suppress the fitness effect by preventing delivery of the mature SME-1 to the periplasmic space.
Defining structural components of blaSME-1 involved in imparting a fitness cost to E. coli:
The repeated isolation of nonfunctional SME-1 clones with deletions or insertions from the genetic screens suggests a requirement for the SME-1 protein in conferring a fitness effect. A frameshift mutant of blaSME-1 (pTP123-Trunc.SME-1) was fortuitously isolated and found to exhibit a growth curve similar to that of E. coli K12 XL1-Blue transformed with empty vector pTP123 or pTP123-TEM-1 (Figure 1B). Overnight competition of the wild-type pTP123-SME-1 construct vs. the truncation mutant yielded a selection rate constant of −2.2 ± 1.3 day−1, indicating a competitive advantage conferred by the frameshift mutation (Table 2). This confirms that some component of the SME-1 protein is necessary to mediate the fitness cost associated with blaSME-1. To further confirm that the SME-1 protein was necessary to confer a fitness cost, the ATG START codon of the SME-1 gene was mutated to ACT for the purpose of preventing translation. Competition between E. coli B Ara+/Ara− strains transformed with either the wild-type SME-1 construct or the START codon mutation construct (pTP123-SMEΔSTART) resulted in a selection rate constant similar to that obtained from the competition of wild-type SME-1 against the SME truncation mutant (Table 2). This again supports the hypothesis that the SME-1 protein is involved in the fitness cost. To further confirm the absence of a fitness cost associated with the pTP123-SME-1ΔSTART mutant, a competition of E. coli B Ara+/Ara− strains containing either pTP123-SME-1ΔSTART or the pTP123-TEM-1 construct was conducted. Competition of wild-type TEM-1 against the START codon mutant of SME-1 resulted in a selection rate constant of 0.34 ± 0.71 day−1 (Table 2). These data indicated no statistically significant difference in fitness between these constructs. The START codon mutant is therefore a suppressor of the SME-1 mediated fitness effect as it outcompetes wild-type pTP123-SME-1 and lacks any detectable fitness cost as determined by competition against pTP123-TEM-1. This confirms that at least some portion of the fitness cost is associated with an intrinsic characteristic of the SME-1 protein.
Although a fitness effect of the SME-1 protein related to translation, processing, or export was not excluded by the data, we chose to investigate the role of several unique structural features of SME-1 in mediating a fitness cost. Specifically, the role of SME-1's carbapenemase activity in conferring a growth defect upon E. coli was tested by creating a double mutant that removes residues critical for hydrolysis of β-lactam antibiotics. In agreement with studies of structurally related class A β-lactamases (Strynadka et al. 1992; Matagne and Frere 1995; Minasov et al. 2002; Nukaga et al. 2003; Chen et al. 2005a,b; Meroueh et al. 2005), the S70A:E166Q mutant of SME-1 is unable to confer detectable resistance to the β-lactam ampicillin. Also, the disulfide bridge near the SME-1 active site (Sougakoff et al. 2002) was tested for its role in conferring a fitness cost via a SME-1 double mutant, C69V:C238V. Neither the ablation of enzyme activity in pTP123-SME-1 S70A:E166Q nor the removal of the unique disulfide bridge in pTP123-SME-1 C69V:C238V alleviates the SME-1-mediated toxicity as determined by the effect of these mutants on E. coli K12 XL1-Blue growth curves (Figure 1B). E. coli K12 XL1-Blue transformed with the pTP123-SME-1 S70A:E166Q construct also exhibits decreased culture viability, similar to wild-type SME-1, as measured by the number of colony-forming cells per milliliter relative to empty vector (Figure 2). A competition between E. coli B strains containing either the wild-type pTP123-SME-1 construct or the catalytically deficient S70A:E166Q SME-1 mutant resulted in a stable population with neither construct predominating in the culture as indicated by a selection rate constant of −0.9 ± 1.4 (Table 2). The SME-1 disulfide bond mutant was also used in competition experiments against E. coli B containing wild-type SME-1. The C69V:C238V double mutant did not suppress the SME-1-mediated fitness cost and may possibly exacerbate the fitness cost phenotype (Table 2). Although some component of the SME-1 protein is associated with a fitness cost, neither enzyme activity nor disulfide bridge formation is requisite for the fitness cost of the SME-1 β-lactamase in E. coli.
In light of the repeated isolation of signal sequence mutants when selecting for enhanced fitness of pTP123-SME-1-containing E. coli, a construct was created that replaced the SME-1 signal sequence with the TEM-1 signal sequence and is designated as TEM(SS)SME. As measured by the MIC of ampicillin and amoxicillin plus clavulanic acid, the TEM(SS)SME and pTP123-SME-1 clones conferred similar levels of β-lactam resistance (ampicillin MIC = 512 μg/ml for each clone and amoxicillin plus clavulanic acid MIC = 32 μg/ml and 8–12 μg/ml for pTP123-SME-1 and TEM(SS)SME, respectively). E. coli K12 XL1-Blue transformed with the TEM(SS)SME construct displays a growth curve similar to that of E. coli K12 XL1-Blue transformed with pTP123-TEM-1 (Figure 1C). Additionally, TEM(SS)SME-expressing E. coli K12 XL1-Blue cells no longer display the reduced culture viability associated with expression of wild-type SME-1 (Figure 2). The importance of the SME-1 signal sequence in conferring a fitness cost upon E. coli was further investigated by competing strains containing the TEM(SS)SME construct against wild-type SME-1 and wild-type TEM-1-containing constructs, separately. As seen in Table 2, the E. coli B strain carrying pTP123-SME-1 suffers a significant competitive disadvantage relative to the strain containing the TEM(SS)SME plasmid. Also, in competition experiments of the TEM(SS)SME construct vs. wild-type TEM-1 there is little evidence of any residual fitness cost associated with the TEM(SS)SME construct (Table 2). The TEM(SS)SME chimera consistently abrogates the SME-1-mediated effects upon E. coli growth, viability, and fitness.
The ability of the TEM-1 signal sequence to suppress the aforementioned effects of SME-1 may be due a direct role of the SME-1 signal sequence in the fitness cost. To determine if the SME-1 signal sequence was sufficient to confer a fitness cost, a construct was created by fusing the SME-1 signal sequence coding region to the mature coding region of the TEM-1 gene and is designated SME(SS)TEM. As seen with the TEM(SS)SME construct, the SME(SS)TEM construct affords equivalent levels of β-lactam resistance as seen in E. coli B transformed with pTP123-SME-1 (ampicillin MIC = >4096 μg/ml for each clone; amoxicillin plus clavulanic acid MIC = 12 μg/ml for each clone). Although the growth curve of the SME(SS)TEM construct is similar to the pTP123-TEM-1 growth curve (Figure 1C), the resolution of this assay may not be sensitive enough to a detect subtle differences in growth rates. Culture viability is reduced for E. coli K12 XL1-Blue transformed with the SME(SS)TEM compared to pTP123-TEM-1 (Figure 2). The reduction in culture viability, however, is not as pronounced as that observed with wild-type pTP123-SME-1. Competition of the SME(SS)TEM construct against wild-type pTP123-TEM-1 shows that the presence of the SME-1 signal sequence imparts a fitness cost to the blaTEM-1 gene. The selection rate constant of −1.4 ± 1.1 obtained from this competition is also lower than that obtained when competing full-length SME-1 against TEM-1 (Table 2). Additionally, competition of pTP123-SME-1 against the SME(SS)TEM construct results in the fusion construct overtaking the wild-type SME-1 construct. In this case, a selection rate constant of zero would be expected if the entirety of the SME-1 fitness cost lies within the SME-1 signal sequence coding region. This result may indicate that the SME-1 signal sequence requires the context of the SME-1 mature protein to exert its entire fitness effect. Nevertheless, although the SME(SS)TEM construct cannot conclusively account for the entirety of the blaSME-1-mediated fitness cost, these results reveal a significant contribution of the SME-1 signal sequence in conferring a fitness cost to E. coli.
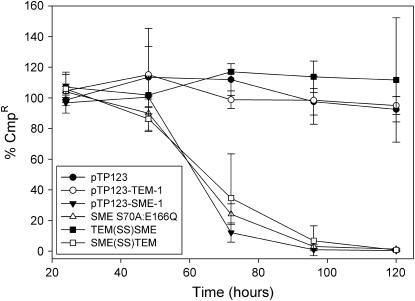
To confirm the results of competition experiments, spontaneous plasmid loss was also monitored for several of the SME-1 mutant constructs. In agreement with previous assays, no difference could be discerned between strains containing the pTP123-SME-1 construct and the isogenic catalytic knock-out, S70A:E166Q (Figure 3). Also consistent with previous data, replacement of the SME-1 signal sequence with the TEM-1 signal sequence (TEM(SS)SME) suppressed any fitness cost as evident by maintenance of this construct until the terminus of the experiment. Fusing the SME-1 signal sequence to the mature TEM-1 coding region [SME(SS)TEM] is sufficient to confer rapid plasmid loss at a rate similar to what is observed with full-length SME-1. Taken together, these results indicate that the SME-1 protein results in a negative fitness effect upon E. coli and that the SME-1 signal sequence contributes to this effect.
DISCUSSION
Antibiotic resistance that occurs via mutation of an antibiotic target often results in a fitness cost to the bacteria under permissive conditions. This suggests that the removal of antibiotic pressure will reduce the prevalence of resistant bacteria. However, the effectiveness of this strategy is dependent upon a fitness cost that can be overcome or reduced in several ways (Bouma and Lenski 1988; Lenski 1988; Andersson 2006). First, antibiotic resistance genes are often genetically linked in the form of multi-resistant mobile DNA elements and selection of one resistant determinate can result in the maintenance of other resistance genes by linkage (Enne et al. 2001; Weldhagen 2004; Bean et al. 2005). Second, fitness costs are typically negated by the appearance of compensatory mutations that alleviate the fitness cost while preserving the resistance phenotype (Bouma and Lenski 1988; Schrag et al. 1997; Björkman et al. 1998, 2000; Levin et al. 2000; Besier et al. 2005; Kugelberg et al. 2005; Luo et al. 2005; Nilsson et al. 2006; Zhang et al. 2006). Without a significant fitness cost, there is no selective pressure to drive a loss of the resistance determinant. Finally, multiple routes of resistance can exist and be highly variable with regard to the fitness costs they engender (Björkman et al. 1998; Gagneux et al. 2006; Kassen and Bataillon 2006). Therefore, a spectrum of resistant clones can exist; some with no fitness costs or even enhanced fitness under permissive conditions.
In the case of SME-1 β-lactamase expression in E. coli B and E. coli K12 XL1-Blue, the burden on the host bacterium is significant as determined by growth curves, culture viability, plasmid instability, and competition against E. coli B expressing TEM-1 β-lactamase. The fitness cost of blaSME-1 in E. coli appears to be present when encoded on either a pTrc99A-derived (pTP123) or a pUC18-derived (pUC18-Kn) plasmid as evident by a decrease in culture viability and an increase in the rate of plasmid loss relative to isogenic plasmids encoding blaTEM-1. In using high-copy number plasmids, gene dosage effects may amplify the SME-1 mediated fitness costs observed. However, even when chromosomally encoded in the E. coli Ara+/Ara− strains, a blaSME-1-mediated fitness cost is detectable when competing against E. coli Ara+/Ara− strains encoding blaTEM-1.
SME-1 β-lactamase is normally encoded on the chromosome of rare S. marcescens strains (Naas et al. 1994) and is positively regulated by a LysR family protein, SmeR (Naas et al. 1995). Cloning the blaSME-1 and blaTEM-1 genes into constructs with identical promoters (Ptrc in pTP123, Plac in pUC18-Kn, and Pbla in the chromosomal constructs) allows direct comparisons of fitness costs mediated by the gene products but excludes the contribution of transcriptional regulation under native conditions. Although beyond the scope of this work, it would be interesting to investigate the impact of blaSME-1 on S. marcescens S6 fitness. The carbapenemase most closely related to SME-1, IMI-2, is to our knowledge the only plasmid-encoded carbapenemase that is inducible by a cis-encoded LysR-type regulator (Aubron et al. 2005). Dissemination of the blaSME-1 gene to other bacterial species may be contingent upon gene copy number and the presence of transcriptional regulatory factors.
The widespread dissemination of the TEM-1 β-lactamase among gram-negative bacteria exemplifies the robust nature of the blaTEM-1 gene and, by extension, has led to the assumption that β-lactamases do not carry fitness costs in general. However, evidence for a fitness cost associated with expression of the E. cloacae AmpC gene in S. enterica serotype Typhimurium (Morosini et al. 2000), the CMY-7 gene in S. typhimurium strain LT2 (Hossain et al. 2004), and, here, expression of the S. marcescens S6 SME-1 gene in E. coli suggests that general statements cannot be made concerning compatibility of β-lactamase genes among even closely related bacterial species. A closer examination of the causes for these fitness costs may offer better insight into and perhaps aid in the prevention of β-lactamase dissemination.
The SME-1 enzyme is relatively isolated among class A β-lactamases with respect to its ability to hydrolyze carbapenems. It has been proposed that structural similarities between both substrate (the β-lactam antibiotics vs. D-Ala-D-Ala peptidoglycan side chain) and enzyme (class A and C β-lactamases vs. DD-peptidases) could indicate an in vivo interaction of β-lactamases with cell-wall components (Bishop and Weiner 1992; Morosini et al. 2000). Marginal peptidase activity of β-lactamases as well as β-lactamase activity of DD-peptidases has been described in vitro (Rhazi et al. 1999). The potential in vivo association of SME-1 with cell-wall components provided the impetus for an investigation of the effect of the SME-1 carbapenemase activity upon E. coli fitness. As determined by the inability of the catalytically inactivated S70A:E166Q SME-1 mutant to suppress the observed fitness cost, enzymatic activity does not contribute significantly to the fitness cost of SME-1.
The repeated isolation of signal sequence mutations from independent genetic screens suggested a role of the signal sequence in mediating the SME-1 fitness cost. To investigate the role of the SME-1 signal sequence in conferring a fitness cost, hybrid TEM-SME β-lactamases were constructed. Exchange of SME-1's native signal sequence for the TEM-1 signal sequence ameliorated the fitness cost of SME-1. Inversely, attaching the SME-1 signal sequence to the mature TEM-1 coding sequence imparted significant toxicity to this normally benign β-lactamase in the form of reduced culture viability, impaired fitness, and plasmid loss in E. coli. This implicates the SME-1 signal sequence as a source of the SME-1 mediated fitness cost.
Previous studies have described the ability of the signal peptides derived from colicin lysis proteins to reduce growth and viability in E. coli, even in the absence of the mature coding region of the lysis protein (Kanoh et al. 1991; van Der Wal et al. 1992, 1994). The enhanced stability associated with the colicin lysis protein's signal sequences may sequester some component of the protein translocation machinery. Alternatively, the accumulation of these signal sequences may result in pore formation in the cytoplasmic membrane leading to decreased cell viability. Biochemical analysis of the EJh holin, utilized by EJ-1 phage to escape the host cell, revealed that oligomerization of a single transmembrane α-helix is sufficient to generate transmembrane pores (Haro et al. 2003). The SME-1 signal sequence may be affecting E. coli in a similar manner. Further mutagenic analysis of the SME-1 β-lactamase may offer insight into how the signal sequence influences the deleterious effect of SME-1 in E. coli.
The continuing erosion of available antibiotic treatment options has spurred interest in seeking novel antibiotic targets and compounds. Maintenance of existing and future therapies may be facilitated by defining mechanisms of resistance and barriers to horizontal transfer of these resistance elements. The identification of a SME-1-mediated fitness cost allows the direct application of genetic techniques that have been utilized to understand structural features of β-lactamase function and evolution. The ability to utilize β-lactamase as a model system offers a means to investigate structural determinants of fitness costs and may lead to a broader understanding of how fitness costs affect the horizontal transfer of resistance genes across genera of bacteria.
Acknowledgments
We thank Fahd Majiduddin for creating the pTP123-SME-1 E166Q clone from which the pTP123-SME-1 S70A:E166Q construct was derived. We also thank Ji Yuan and Tulin Ayvaz for technical assistance. The E. coli B Ara+ and Ara− strains were provided by Richard Lenski. This work was supported by National Institutes of Health grant AI32956 to T.P.
References
- Ambler, R. P., 1980. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 289: 321–331. [DOI] [PubMed] [Google Scholar]
- Ambler, R. P., A. F. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris et al., 1991. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 276(Pt 1): 269–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, D. I., 2006. The biological cost of mutational antibiotic resistance: Any practical conclusions? Curr. Opin. Microbiol. 9: 461–465. [DOI] [PubMed] [Google Scholar]
- Aubron, C., L. Poirel, R. J. Ash and P. Nordmann, 2005. Carbapenemase-producing Enterobacteriaceae, U.S. rivers. Emerg. Infect. Dis. 11: 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean, D. C., D. M. Livermore, I. Papa and L. M. C. Hall, 2005. Resistance among Escherichia coli to sulphonamides and other antimicrobials now little used in man. J. Antimicrob. Chemother. 56: 962–964. [DOI] [PubMed] [Google Scholar]
- Berg, O. G., and C. G. Kurland, 2002. Evolution of microbial genomes: sequence acquisition and loss. Mol. Biol. Evol. 19: 2265–2276. [DOI] [PubMed] [Google Scholar]
- Besier, S., A. Ludwig, V. Brade and T. A. Wichelhaus, 2005. Compensatory adaptation to the loss of biological fitness associated with acquisition of fusidic acid resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49: 1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, R. E., and J. H. Weiner, 1992. Coordinate regulation of murein peptidase activity and AmpC beta-lactamase synthesis in Escherichia coli. FEBS Lett. 304: 103–108. [DOI] [PubMed] [Google Scholar]
- Björkman, J., D. Hughes and D. I. Andersson, 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95: 3949–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkman, J., I. Nagaev, O. G. Berg, D. Hughes and D. I. Andersson, 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287: 1479–1482. [DOI] [PubMed] [Google Scholar]
- Bouma, J. E., and R. E. Lenski, 1988. Evolution of a bacteria/plasmid association. Nature 335: 351–352. [DOI] [PubMed] [Google Scholar]
- Chen, Y., J. Delmas, J. Sirot, B. Shoichet and R. Bonnet, 2005. a Atomic resolution structures of CTX-M beta-lactamases: extended spectrum activities from increased mobility and decreased stability. J. Mol. Biol. 348: 349–362. [DOI] [PubMed] [Google Scholar]
- Chen, Y., B. Shoichet and R. Bonnet, 2005. b Structure, function, and inhibition along the reaction coordinate of CTX-M beta-lactamases. J. Am. Chem. Soc. 127: 5423–5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enne, V. I., D. M. Livermore, P. Stephens and L. M. C. Hall, 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357: 1325–1328. [DOI] [PubMed] [Google Scholar]
- Gagneux, S., C. D. Long, P. M. Small, T. Van, G. K. Schoolnik et al., 2006. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312: 1944–1946. [DOI] [PubMed] [Google Scholar]
- Haro, A., M. Velez, E. Goormaghtigh, S. Lago, J. Vazquez et al., 2003. Reconstitution of holin activity with a synthetic peptide containing the 1–32 sequence region of EJh, the EJ-1 phage holin. J. Biol. Chem. 278: 3929–3936. [DOI] [PubMed] [Google Scholar]
- Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen and L. R. Pease, 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59. [DOI] [PubMed] [Google Scholar]
- Hossain, A., M. D. Reisbig and N. D. Hanson, 2004. Plasmid-encoded functions compensate for the biological cost of AmpC overexpression in a clinical isolate of Salmonella typhimurium. J. Antimicrob. Chemother. 53: 964–970. [DOI] [PubMed] [Google Scholar]
- Huang, W., J. Petrosino, M. Hirsch, P. S. Shenkin and T. Palzkill, 1996. Amino acid sequence determinates of β-lactamase structure and activity. J. Mol. Biol. 258: 688–703. [DOI] [PubMed] [Google Scholar]
- Jain, R., M. C. Rivera and J. A. Lake, 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. USA 96: 3801–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh, S., H. Masaki, S. Yajima, T. Ohta and T. Uozumi, 1991. Signal peptide of the colicin E2 lysis protein causes host cell death. Agric. Biol. Chem. 55: 1607–1614. [PubMed] [Google Scholar]
- Kassen, R., and T. Bataillon, 2006. Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nat. Genet. 38: 484–488. [DOI] [PubMed] [Google Scholar]
- Kugelberg, E., S. Lofmark, B. Wretlind and D. I. Andersson, 2005. Reduction of the fitness burden of quinolone resistance in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 55: 22–30. [DOI] [PubMed] [Google Scholar]
- Lenski, R. E., 1988. Experimental studies of pleiotropy and epistasis in Escherichia coli. I. Variation in competitive fitness among mutants resistant to virus T4. Evolution 42: 425–432. [DOI] [PubMed] [Google Scholar]
- Lenski, R. E., and J. E. Bouma, 1987. Effects of segregation and selection on instability of plasmid pACYC184 in Escherichia coli B. J. Bacteriol. 169: 5314–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski, R. E., M. R. Rose, S. C. Simpson and S. C. Tadler, 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138: 1315–1341. [Google Scholar]
- Lenski, R. E., S. C. Simpson and T. T. Nguyen, 1994. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 176: 3140–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, B. R., F. M. Stewart and L. Chao, 1977. Resource-limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. Am. Nat. 111: 3–24. [Google Scholar]
- Levin, B. R., V. Perrot and N. Walker, 2000. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154: 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebana, E., M. Batchelor, K. L. Hopkins, F. A. Clifton-Hadley, C. J. Teale et al., 2006. Longitudinal farm study of extended-spectrum beta-lactamase-mediated resistance. J. Clin. Microbiol. 44: 1630–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, N., S. Pereira, O. Sahin, J. Lin, S. Huang et al., 2005. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. USA 102: 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majiduddin, F. K., and T. Palzkill, 2003. a Amino acid sequence requirements at residues 69 and 238 for the SME-1 beta-lactamase to confer resistance to beta-lactam antibiotics. Antimicrob. Agents Chemother. 47: 1062–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majiduddin, F. K., and T. Palzkill, 2003. b An analysis of why highly similar enzymes evolve differently. Genetics 163: 457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matagne, A., and J. M. Frere, 1995. Contribution of mutant analysis to the understanding of enzyme catalysis: the case of class A beta-lactamases. Biochim. Biophys. Acta 1246: 109–127. [DOI] [PubMed] [Google Scholar]
- Meroueh, S. O., J. F. Fisher, H. B. Schlegel and S. Mobashery, 2005. Ab initio QM/MM study of class A beta-lactamase acylation: dual participation of Glu166 and Lys73 in a concerted base promotion of Ser70. J. Am. Chem. Soc. 127: 15397–15407. [DOI] [PubMed] [Google Scholar]
- Miller, J. H., 1992. A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Minasov, G., X. Wang and B. K. Shoichet, 2002. An ultrahigh resolution structure of TEM-1 beta-lactamase suggests a role for Glu166 as the general base in acylation. J. Am. Chem. Soc. 124: 5333–5340. [DOI] [PubMed] [Google Scholar]
- Morosini, M. I., J. A. Ayala, F. Baquero, J. L. Martinez and J. Blazquez, 2000. Biological cost of AmpC production for Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 44: 3137–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas, T., L. Vandel, W. Sougakoff, D. M. Livermore and P. Nordmann, 1994. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A beta-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob. Agents Chemother. 38: 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas, T., D. M. Livermore and P. Nordmann, 1995. Characterization of an LysR family protein, SmeR from Serratia marcescens S6, its effect on expression of the carbapenem-hydrolyzing beta-lactamase Sme-1, and comparison of this regulator with other beta-lactamase regulators. Antimicrob. Agents Chemother. 39: 629–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon-Venezia, S., R. Ben-Ami and Y. Carmeli, 2005. Update on Pseudomonas aeruginosa and Acinetobacter baumannii infections in the healthcare setting. Curr. Opin. Infect. Dis. 18: 306–313. [DOI] [PubMed] [Google Scholar]
- Nilsson, A. I., A. Zorzet, A. Kanth, S. Dahlstrom, O. G. Berg et al., 2006. Reducing the fitness cost of antibiotic resistance by amplification of initiator tRNA genes. Proc. Natl. Acad. Sci. USA 103: 6976–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukaga, M., K. Mayama, A. M. Hujer, R. A. Bonomo and J. R. Knox, 2003. Ultrahigh resolution structure of a class A beta-lactamase: on the mechanism and specificity of the extended-spectrum SHV-2 enzyme. J. Mol. Biol. 328: 289–301. [DOI] [PubMed] [Google Scholar]
- Petrosino, J., G. Rudgers, H. Gilbert and T. Palzkill, 1999. Contributions of aspartate 49 and phenylalanine 142 residues of a tight binding inhibitory protein of beta-lactamases. J. Biol. Chem. 274: 2394–2400. [DOI] [PubMed] [Google Scholar]
- Queenan, A. M., C. Torres-Viera, H. S. Gold, Y. Carmeli, G. M. Eliopoulos et al., 2000. SME-type carbapenem-hydrolyzing class A beta-lactamases from geographically diverse Serratia marcescens strains. Antimicrob. Agents Chemother. 44: 3035–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece, K.S., and G. J. Phillips, 1995. New plasmids carrying antibiotic-resistance cassettes. Gene 165: 141–142. [DOI] [PubMed] [Google Scholar]
- Rhazi, N., M. Galleni, M. I. Page and J. M. Frere, 1999. Peptidase activity of beta-lactamases. Biochem. J. 341(Pt 2): 409–413. [PMC free article] [PubMed] [Google Scholar]
- Schneider, D., and R. E. Lenski, 2004. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res. Microbiol. 155: 319–327. [DOI] [PubMed] [Google Scholar]
- Schrag, S. J., V. Perrot and B. R. Levin, 1997. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc. R. Soc. Lond. 264: 1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, A. A., F. Hasan, S. Ahmed and A. Hameed, 2004. Characteristics, epidemiology and clinical importance of emerging strains of Gram-negative bacilli producing extended-spectrum beta-lactamases. Res. Microbiol. 155: 409–421. [DOI] [PubMed] [Google Scholar]
- Sougakoff, W., G. L'Hermite, L. Pernot, T. Naas, V. Guillet et al., 2002. Structure of the imipenem-hydrolyzing class A beta-lactamase SME-1 from Serratia marcescens. Acta Crystallogr. D Biol. Crystallogr. 58: 267–274. [DOI] [PubMed] [Google Scholar]
- Strynadka, N. C., H. Adachi, S. E. Jensen, K. Johns, A. Sielecki et al., 1992. Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 A resolution. Nature 359: 700–705. [DOI] [PubMed] [Google Scholar]
- Thomas, C. M., and K. M. Nielsen, 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3: 711–721. [DOI] [PubMed] [Google Scholar]
- Travisano, M., and R. E. Lenski, 1996. Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics 143: 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchin, A., and J. F. Lawler, Jr., 1999. The primer generator: a program that facilitates the selection of oligonucleotides for site-directed mutagenesis. Biotechniques 26: 672–676. [DOI] [PubMed] [Google Scholar]
- van der Wal, F. J., B. Oudega, M. M. Kater, C. M. Ten Hangen-Jongman, F. K. de Graaf et al., 1992. The stable BRP signal peptide causes lethality but is unable to provoke the translocation of cloacin DF13 across the cytoplasmic membrane of Escherichia coli. Mol. Microbiol. 6: 2309–2318. [DOI] [PubMed] [Google Scholar]
- van der Wal, F. J., Q. A. Valent, C. M. Ten Hangen-Jongman, F. K. de Graaf, B. Oudega et al., 1994. Stability and function of the signal peptide of the pCloDF13-derived bacteriocin release protein. Microbiology 140: 369–378. [DOI] [PubMed] [Google Scholar]
- Warming, S., N. Costantino, D. L. Court, N. A. Jenkins and N. G. Copeland, 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisemann, J. M., and G. M. Weinstock, 1988. Mutations at the cysteine codons of the recA gene of Escherichia coli. DNA 7: 389–398. [DOI] [PubMed] [Google Scholar]
- Weldhagen, G. F., 2004. Integrons and beta-lactamases–a novel perspective on resistance. Int. J. Antimicrob. Agents 23: 556–562. [DOI] [PubMed] [Google Scholar]
- Yang, Y. J., P. J. Wu and D. M. Livermore, 1990. Biochemical characterization of a beta-lactamase that hydrolyzes penems and carbapenems from two Serratia marcescens isolates. Antimicrob. Agents Chemother. 34: 755–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., O. Sahin, P. F. Mcdermott and S. Payot, 2006. Fitness of antimicrobial-resistant Campylobacter and Salmonella. Microbes Infect. 8: 1972–1978. [DOI] [PubMed] [Google Scholar]