Abstract
We report the construction of a linkage map for the moss Ceratodon purpureus (n = 13), based on a cross between geographically distant populations, and provide the first experimental confirmation of maternal chloroplast inheritance in bryophytes. From a mapping population of 288 recombinant haploid gametophytes, genotyped at 121 polymorphic AFLP loci, three gene-based nuclear loci, one chloroplast marker, and sex, we resolved 15 linkage groups resulting in a map length of ∼730 cM. We estimate that the map covers more than three-quarters of the C. purpureus genome. Approximately 35% of the loci were sex linked, not including those in recombining pseudoautosomal regions. Nearly 45% of the loci exhibited significant segregation distortion (α = 0.05). Several pairs of unlinked distorted loci showed significant deviations from multiplicative genotypic frequencies, suggesting that distortion arises from genetic interactions among loci. The distorted autosomal loci all exhibited an excess of the maternal allele, suggesting that these interactions may involve nuclear–cytoplasmic factors. The sex ratio of the progeny was significantly male biased, and the pattern of nonrandom associations among loci indicates that this results from interactions between the sex chromosomes. These results suggest that even in interpopulation crosses, multiple mechanisms act to influence segregation ratios.
THE moss Ceratodon purpureus (Hedw.) Brid. is used as a model system for the study of several biological processes (Shaw and Gaughan 1993; Shaw et al. 1997; Shaw and Beer 1999; McDaniel 2005; McDaniel and Shaw 2005; Thornton et al. 2005; Cove et al. 2006). However, progress toward identifying genes underlying traits of interest in C. purpureus is limited (Cove et al. 1996; Wagner et al. 1997; Esch et al. 1999; Kern and Sack 1999; Cove and Quatrano 2006; but see Brucker et al. 2005), largely because classical forward genetic approaches have not been developed. Here we describe the construction of a linkage map for C. purpureus (n = 13), derived from a cross between two geographically distant populations. Our immediate purpose was to document segregation patterns across the C. purpureus genome, particularly on the sex chromosomes. The linkage map additionally provides a foundation for subsequent mapping of loci underlying induced or natural phenotypic variants.
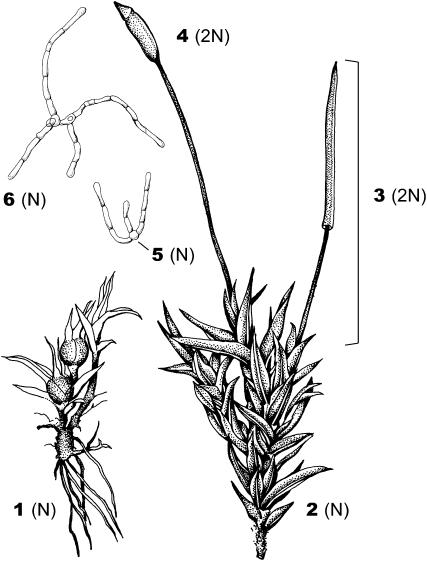
Like all land plants, the moss life cycle consists of a multicellular haploid gametophyte generation that alternates with a morphologically distinct diploid sporophyte generation (Figure 1). In angiosperms, the gametophyte generation consists of pollen and ovules, while the sporophyte generation is much larger and long lived. In contrast, the gametophyte is the dominant portion of the moss life cycle, and the sporophyte is ephemeral. C. purpureus is a dioecious species with a heteromorphic XY pair of chromosomes that presumably correlate with sex (Ramsay and Berrie 1982). Because sex determination happens at the haploid stage in bryophytes, the diploid sporophyte is always heterogametic (by convention labeled XY), and the X and Y chromosomes segregate to females and males, respectively, following meiosis (Figure 1). Stains of meiotic chromosomes indicate that recombination is suppressed in the middle of the heteromorphic chromosomes, although they pair in pseudoautosomal regions (PARs) at both distal ends of the chromosomes. Karyotypic analysis indicates that nearly one-third of the genome resides on this pair of chromosomes (Heitz 1932; Jachimsky 1935; J. Greilhuber, personal communication). Nevertheless, no physical or genetic evidence demonstrates that the heteromorphic pair of chromosomes in C. purpureus (or any species of moss) are in fact sex chromosomes.
Figure 1.—
Life cycle of C. purpureus. The mature haploid male (1) produces sperm, which fertilizes an egg on the haploid female (2), resulting in a diploid sporophyte (3). Meiosis occurs in the mature sporophyte (4), which produces recombinant haploid spores (two shown, 5). The spores germinate, forming a network of protonemal filaments (6). At maturity, male and female leafy gametophores (1, 2) develop on the protonema.
Experimental bryophyte crosses are achieved by growing a haploid male and female commingled under inductive conditions. The juvenile phase of the moss gametophyte consists of protonemal filaments that grow by serial extension and ultimately produce the mature and more conspicuous leafy stems. At sexual maturity, the haploid gametophyte of C. purpureus produces either sperm-producing antheridia or egg-producing archegonia. All gametes from a single haploid gametophyte are produced by mitosis and are therefore genetically identical (barring mutation). The sperm disperse through the environment to a female plant to fertilize an egg, forming an F1 diploid sporophyte that will remain attached to the female parent gametophyte throughout its development. Meiosis takes place in the mature sporophyte, which ultimately ruptures, releasing the (F2-like) recombinant male and female haploid spores. Maternal and paternal alleles should segregate at 1:1 ratios, but previous work indicates that natural, field-collected sporophytes may produce progeny with significant sex-ratio bias (Shaw and Gaughan 1993; Shaw and Beer 1999).
This study forms a component of a larger project aimed at understanding the genetic basis of population differentiation in C. purpureus. Previously we have documented genetic differentiation in morphological and life history traits among populations of the species (Shaw and Beer 1999; McDaniel 2005). As a first step toward understanding the genetic basis of these phenotypic differences, we generated recombinant progeny from a cross between isolates of C. purpureus from Ithaca, New York, and Otavalo, Ecuador. Taxonomically these populations belong to different subspecies (Burley and Pritchard 1990), although in previous molecular population genetic analyses we found no genealogical support for this arrangement (McDaniel and Shaw 2005). Here we describe widespread segregation distortion in the progeny of a cross between these two populations, which we suggest results from spore mortality. The patterns of nonrandom associations among unlinked distorted loci indicate that sex-specific lethality and complex nuclear–cytoplasmic interactions were the predominant forces generating genomewide patterns of distortion in this cross. We discuss these findings in the context of population divergence in the species.
MATERIALS AND METHODS
Generation of hybrid mapping population:
Diploid sporophytes from populations of C. purpureus near Otavalo, Ecuador, and Ithaca, New York, were chosen to represent extremes of the morphological distribution. To generate single spore isolates (SSIs) for study, the sporophytes were individually surface sterilized in a 0.6% sodium hypochlorite solution for 1 min, rinsed in sterile water, and placed in a test tube containing 1 ml of sterile water. The sporophyte was then ruptured, creating a suspension of spores. The spore suspension was plated on standard BCD media containing 920 mg/liter ammonium tartrate (Knight et al. 2002) and placed under 40 quanta/m2/sec continuous light at ∼20°. Spores were germinated and allowed to produce protonemal filaments several cells long. Spores from numerous sporophytes from both populations were cultivated, and SSIs were isolated before the protonema overgrew one another and transferred to a new plate where they were grown under the same light conditions as the spore suspension plates. When the plants had reached ∼1 cm in diameter, one SSI from each diploid sporophyte was transplanted on Turface MVP soil amendment (Hummert International, Saint Louis) in a Conviron growth chamber at 20° with 16-hr days. Male plants produce obvious macroscopic sperm-producing buds (perigonia); females were confirmed by observing the presence of archegonia using a light microscope.
A single male SSI from the New York (NY) population and a single female SSI from the Ecuador (Ec) population were chosen to create a mapping population (Figure 1). Tissue from the two individuals was ground in sterile water and pipetted onto Turface soil amendment in a Conviron growth chamber. The plants were watered from below twice a day for the first 2 weeks and once a day thereafter. The day length was set to 14 hr, with light intensity 120 quanta/m2/sec. The daytime temperature was set to 20° and the nighttime temperature to 16°. Male buds (perigonia) were evident after ∼4 weeks under this treatment, and young sporophytes appeared after 3 months. The temperature in the chamber was reduced to 14° daytime and 10° nighttime to promote sporophyte maturation. Maturation of the hybrid sporophyte, evident by swelling of the spore capsule, was complete after an additional 2 months. Spores from a single hybrid F1 sporophyte were germinated as described above, and nearly 500 SSIs were isolated. Due to fungal contamination over the course of cultivation, the final mapping population consisted of 288 individuals. These were grown for several weeks to generate tissue for DNA extraction.
Development and analysis of molecular markers:
Genomic DNA was extracted from sterile protonemal tissue from the two parents and all of progeny, using a cetyl trimethyl ammonium bromide extraction, described in McDaniel and Shaw (2005), modified for 1-ml extractions using a Genogrinder 2000 bead shaker (SPEX CertiPrep, Metuchen, NJ). The DNA was resuspended in 30 μl of TE. The concentration of DNA in each extraction was approximated by comparison to known quantities of λDNA on a 1% agarose gel stained with ethidium bromide. Portions of the adk and phy2 genes were sequenced from the two parents using primers and PCR amplification and sequencing conditions described in McDaniel and Shaw (2005) (Table 1). Under the same PCR conditions, an ∼1-kb portion of the des6 gene was amplified using the primers DES-F and DES-R (Table 1), designed from an mRNA sequence from C. purpureus (GenBank accession no. AF250734). The amplified fragment spans nucleotide positions 216–906 from the mRNA, including portions of exon 2 and 4, and all of exon 3, based on comparison to the Physcomitrella patens genomic sequence (GenBank accession no. AJ222981). The sequences of the parental alleles of all three loci were compared using Sequencher (GeneCodes, Ann Arbor, MI) to identify restriction site polymorphisms between the parental sequences. The adk sequences differed at two MseI sites, the phy2 sequences differed at a Bsp1286I site, and the des6 sequences differed at an XbaI site. For all progeny the loci were amplified in 8.3 μl PCR reactions and digested for 3 hr at 37° in a total volume of 10 μl. Differences in restriction banding pattern were resolved on a 2% agarose gel.
TABLE 1.
AFLP primer combinations
| AFLP primer pairs | No. of polymorphic bands |
|---|---|
| MseI-TCC/EcoRI-GTC | 29 |
| MseI-TCG/EcoRI-GTC | 21 |
| MseI-TGC/EcoRI-GTC | 28 |
| MseI-TGG/EcoRI-GTC | 43 |
Amplified fragment length polymorphisms (AFLPs) genotypes were generated for the two parents and all progeny using standard protocols (Vos et al. 1995). The restriction digest-ligation (RL) reactions were carried out simultaneously in 96-well plates containing ∼100 ng genomic DNA, the restriction enzymes EcoRI and MseI, adaptors, T4 ligase, and T4 ligase buffer, in a total reaction volume of 25 μl. The reaction was incubated at 37° overnight (∼16 hr). The RL product was then diluted with 50 μl water for use as the template for the preamplification reaction. For each individual, we carried out a single preamplification reaction, using standard EcoRI and MseI primers with a single selective base on each (G and T, respectively). In the selective amplification we used a single EcoRI primer with three selective bases (GTC), labeled with Hex, in combination with four MseI primers (TCC, TCG, TGC, TGG). The resulting selective amplification products were resolved on an ABI 3700 capillary sequencer (Applied Biosystems, Foster City, CA). An average of 30 polymorphic bands were scored for each primer combination (Table 2).
TABLE 2.
Gene-based primers
| Locus | Primers | Polymorphism |
|---|---|---|
| Adenosine kinase (adk) | 5′-GAAGAAGCCAGAAAACTGGGC-3′, 5′-GTCACCCCATCTTCAGCAAC-3′ | MseI site |
| Phytochrome 2 (phy2) | 5′-GGCATGGAAATGATGTGTTG-3′, 5′-CATCACTGTACCCATCTCG-3′ | Bsp1286 site |
| Δ6-fatty acid desaturase (des6) | 5′-GAGCACTTGGCAACGATG-3, 5′-CGTGAGACAACCATCCGC-3 | XbaI or MseI site |
| atpB-rbcL spacer | 5′-ACATCKARTACKGGACCAATAA-3, 5′-AACACCAGCTTTRAATCCAA-3 | TAAA repeat |
The ABI 3700 trace files were analyzed using the software Genographer (available at http://www.hordeum.montana.edu/genographer/). Bins 0.6 bp wide were used for scoring the presence or absence of AFLP bands. Lanes with low signal quality for particular primer combinations were deleted. Marker bands with few individuals showing polymorphism (i.e., the band present in <10% or >90% of individuals) and bands showing unusual peak-size heterogeneity among individuals were not scored. The presence/absence strings for each marker were concatenated and converted to the appropriate genotypes (either NY or Ec, depending upon which was the null allele).
Linkage map construction:
The full mapping population consisted of 288 individuals genotyped for 124 polymorphic markers, with a large number of individuals genotyped at each marker. We constructed a linkage map using MAPMAKER 3.0 software (Lander et al. 1987; Lincoln and Lander 1992). We used the GROUP command (two-point linkage criteria set to a LOD >6.0 and maximum distance between markers set to 37 cM) with the Kosambi mapping function to organize the markers into linkage groups. Where more than five markers formed a linkage group, the command ORDER, with error detection on, was used to find the most likely linear arrangement of markers. This function uses a starting subset of five markers, ordered with a threshold of LOD 3.0, and tries to place the remaining markers with a threshold of LOD 2.0. The error detection data and two-point distances were examined to identify potentially unreliable markers. These markers, as well as other markers that were linked but not placed, were evaluated using the TRY and MAP commands to determine whether their inclusion between other markers increased the map length by >5 cM. Such markers, as well as those with more than eight errors, were discarded. For linkage groups with five or fewer markers, marker orders were constructed by hand, using the LOD TABLE and MAP commands. The same strategy to remove unreliable markers from the larger linkage groups was also used for the smaller linkage groups. This process resulted in four classes of markers: (1) framework markers ordered into linkage groups in the final map; (2) markers that are tightly linked to framework markers, but cannot be placed into single intervals with certainty; (3) markers that link to one or more established group, but increase map length; and (4) unlinked markers.
We estimated the length L of the C. purpureus genome in two ways, following Fishman et al. (2001). First we added twice the average marker interval (the summed recombination fractions from all mapped intervals, divided by the number of intervals) to each linkage group. Second, we used method 4 of Chakravarti et al. (1991), which multiplies the length of each linkage group by a factor (m + 1)/(m – 1), where m is the number of markers in that linkage group. We estimated genome coverage by dividing the summed recombination fractions by our two estimates of L. The proportion c of the genome within d cM of a marker, assuming a random marker distribution, was estimated as c = 1 – e2dn/L, where n is the number of markers.
Identification of sex-limited markers:
To identify markers that cosegregated with sex, the recombinant haploid gametophytes were grown on Turface under the inductive conditions described above. Although not all recombinant progeny reached sexual maturity, we were able to confidently map the sex-determining region on the linkage map. To determine the inheritance pattern of the chloroplast, we identified a minisatellite polymorphism in the chloroplastic atpB–rbcL spacer between the two parents, and sequenced this locus in 10 recombinant progeny (5 males and 5 females), using primers and PCR conditions and primers described in McDaniel and Shaw (2003).
Analysis of segregation distortion:
Significant deviations from the expected 1:1 Mendelian ratios in haploid progeny were examined at each mapped marker using a χ2 test, at a significance level of α = 0.05, with 1 d.f. Since genotypes at linked loci are not independent, the meaning of statistical tests of segregation distortion is unclear. We therefore report only genomic regions of significant distortion, defined as three or more contiguous markers all showing significant distortion toward the same parent (at the α = 0.05 level). Segregation ratios also were calculated for males and females separately, and the significance of sex-specific distortion was evaluated with a χ2 test, using the complete mapping population allele frequency to generate an expected value.
To identify loci involved in lethal epistatic interactions in the NY–EC cross we tested for deviations from multiplicative genotypic frequencies for all pairwise combinations of markers (using a Python script available from S.F.M.) using the equation
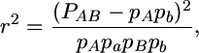 |
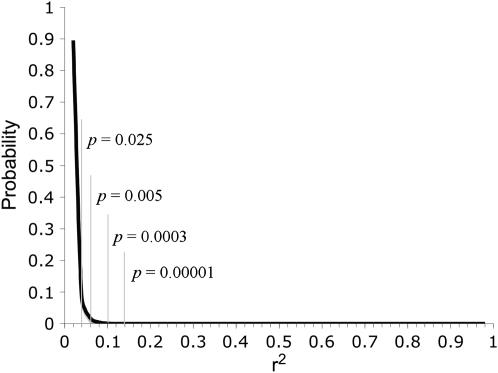
where pA, pa, pB, and pb are the allele frequencies at two loci, A and B, and P refers to one of the four possible two-locus haplotypes. To evaluate the statistical significance of associations among genotypes, we numerically generated a probability distribution for r2 using a mapping population size of 130 (the minimum number of individuals for any comparison, given the patterns of missing genotypes), and assuming no genotypes were absent (consistent with our data) and a multinomial probability function where each of the four genotypes was equally likely
 |
Establishing a threshold for statistical significance was complicated by the multiple tests and both the nonindependence of the tests of disequilibrium and the nonindependence of marker loci due to physical linkage. Our strategy, therefore, was to focus on interlocus interactions associated with the significantly distorted autosomal regions and the sex-chromosome distortion.
RESULTS
Construction of linkage map:
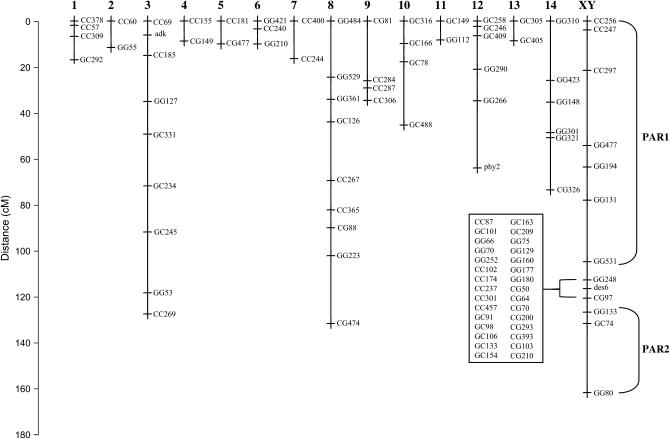
We generated 124 markers in total, 121 AFLPs (Table 2) and three gene-based markers (adk, phy2, and des6). We were able to assess the sex of more than half of the recombinant progeny (160 of 288), 59% of which were male. Using the three gene-based markers and 67 AFLPs, we resolved 15 linkage groups (Figure 2). The sex-determining locus colocalized nearly perfectly with approximately one-third of the markers generated for the map, including the des6 gene; most of these markers were not included in the framework map (see below). We used the allelic state at the des6 locus to determine the sex of the hybrids for which sex had not been determined anatomically (60% male; Table 3). All 10 surveyed individuals contained the chloroplast-linked Ec atpB-rbcL sequence, indicating that the chloroplast was maternally inherited.
Figure 2.—
Genetic map of C. purpureus showing 15 linkage groups, numbered above, including the XY pair of sex chromosomes. The y-axis shows centimorgan distances between pairs of linked markers. The recombining pseudoautosomal regions (PAR1 and 2) linked to the sex-correlated region of the XY chromosome are marked in brackets to the right of the linkage group; additional AFLP markers that cosegregated with sex are shown on the left.
TABLE 3.
Segregation ratios in male and female progeny
| Position (cM) | Frequency Ec allele
|
|||||
|---|---|---|---|---|---|---|
| Marker | n | χ2 | All progeny | Females | Males | |
| LG1 | ||||||
| 0 | CC378 | 275 | 3.36 | 0.58 | 0.57 | 0.58 |
| 1.9 | CC57 | 275 | 2.49 | 0.57 | 0.52 | 0.60 |
| 4.8 | CC309 | 275 | 0.96 | 0.54 | 0.55 | 0.53 |
| 9.7 | GC292 | 281 | 0.22 | 0.52 | 0.54 | 0.54 |
| LG2 | ||||||
| 0 | CC60 | 275 | 0.22 | 0.52 | 0.57 | 0.49 |
| 11.6 | GG55 | 282 | 2.30 | 0.56 | 0.62 | 0.53 |
| LG3 | ||||||
| 0 | CC69 | 275 | 14.40 | 0.66 | 0.59 | 0.71 |
| 6 | adk | 263 | 9.05 | 0.63 | 0.62 | 0.64 |
| 8.9 | CC185 | 275 | 21.60 | 0.70 | 0.66 | 0.72 |
| 20.2 | GG127 | 186 | 1.55 | 0.56 | 0.59 | 0.55 |
| 14.2 | GC331 | 281 | 2.44 | 0.57 | 0.64 | 0.50 |
| 22.6 | GC234 | 281 | 3.60 | 0.58 | 0.56 | 0.58 |
| 20.1 | GC246 | 281 | 22.72 | 0.70 | 0.74 | 0.68 |
| 26.4 | GG53 | 282 | 6.38 | 0.61 | 0.62 | 0.62 |
| 9.2 | CC269 | 275 | 29.33 | 0.73 | 0.73 | 0.74 |
| LG4 | ||||||
| 0 | CC155 | 275 | 2.23 | 0.56 | 0.53 | 0.58 |
| 8.7 | CG249 | 270 | 0.74 | 0.54 | 0.54 | 0.55 |
| LG5 | ||||||
| 0 | CC181 | 275 | 6.77 | 0.61 | 0.59 | 0.63 |
| 9.9 | CG477 | 131 | 6.42 | 0.66 | 0.58 | 0.69 |
| LG6 | ||||||
| 0 | GG421 | 282 | 5.17 | 0.60 | 0.59 | 0.60 |
| 3.5 | CC240 | 275 | 1.75 | 0.56 | 0.54 | 0.57 |
| 6.5 | GG210 | 282 | 1.20 | 0.55 | 0.53 | 0.57 |
| LG7 | ||||||
| 0 | CC400 | 275 | 3.36 | 0.58 | 0.52 | 0.60 |
| 16.3 | CC244 | 275 | 11.35 | 0.64 | 0.59 | 0.64 |
| LG8 | ||||||
| 0 | GG484 | 186 | 0.17 | 0.52 | 0.53 | 0.53 |
| 24.5 | GG529 | 282 | 0.57 | 0.47 | 0.48 | 0.47 |
| 9.6 | GG361 | 282 | 9.19 | 0.63 | 0.59 | 0.67 |
| 9.9 | GC126 | 281 | 21.92 | 0.70 | 0.67 | 0.69 |
| 25.5 | CC267 | 275 | 24.89 | 0.71 | 0.74 | 0.70 |
| 12.8 | CC365 | 275 | 17.82 | 0.68 | 0.68 | 0.68 |
| 7.7 | CG88 | 270 | 5.40 | 0.60 | 0.59 | 0.62 |
| 12.3 | GG223 | 282 | 30.89 | 0.73 | 0.74 | 0.73 |
| 29.6 | CG474 | 131 | 0.46 | 0.46 | 0.42 | 0.51 |
| LG9 | ||||||
| 0 | GC81 | 281 | 22.72 | 0.70 | 0.68 | 0.71 |
| 26 | CC284 | 275 | 1.98 | 0.44 | 0.43 | 0.46 |
| 3.1 | CC287 | 275 | 0.31 | 0.48 | 0.44 | 0.51 |
| 5.5 | CC306 | 275 | 0.09 | 0.51 | 0.47 | 0.54 |
| LG10 | ||||||
| 0 | GC316 | 281 | 0.22 | 0.52 | 0.50 | 0.54 |
| 9.8 | GC166 | 281 | 0.09 | 0.51 | 0.56 | 0.46 |
| 7.9 | GC78 | 281 | 0.09 | 0.51 | 0.52 | 0.50 |
| 27.7 | GC488 | 281 | 0.14 | 0.48 | 0.44 | 0.52 |
| LG11 | ||||||
| 0 | GC149 | 281 | 0.30 | 0.52 | 0.53 | 0.50 |
| 8.3 | GG112 | 282 | 0.86 | 0.54 | 0.57 | 0.52 |
| LG12 | ||||||
| 0 | GC258 | 281 | 6.62 | 0.61 | 0.58 | 0.65 |
| 2.3 | GC24B | 281 | 10.01 | 0.63 | 0.62 | 0.67 |
| 4.1 | GC409 | 281 | 12.86 | 0.65 | 0.68 | 0.65 |
| 14.6 | GG290 | 282 | 4.79 | 0.59 | 0.59 | 0.59 |
| 13.8 | GG266 | 282 | 1.39 | 0.55 | 0.57 | 0.54 |
| 29.2 | phy2 | 200 | 0.64 | 0.54 | 0.53 | 0.52 |
| LG13 | ||||||
| 0 | GC305 | 281 | 2.71 | 0.57 | 0.55 | 0.57 |
| 8.6 | GC406 | 281 | 0.94 | 0.46 | 0.41 | 0.48 |
| LG14 | ||||||
| 0 | GG310 | 282 | 0.26 | 0.52 | 0.64 | 0.43 |
| 25.9 | GG423 | 282 | 2.84 | 0.57 | 0.58 | 0.55 |
| 9.5 | GG148 | 282 | 5.56 | 0.60 | 0.62 | 0.59 |
| 13.2 | GG301 | 282 | 9.71 | 0.63 | 0.62 | 0.64 |
| 2.2 | GG321 | 138 | 12.19 | 0.71 | 0.69 | 0.71 |
| 22.7 | CG326 | 140 | 1.73 | 0.58 | 0.49 | 0.63 |
| XY | ||||||
| 0 | CC256 | 275 | 5.11 | 0.40 | 0.42 | 0.40 |
| 3.9 | CC247 | 275 | 2.23 | 0.44 | 0.42 | 0.45 |
| 17.6 | CC297 | 275 | 2.23 | 0.44 | 0.44 | 0.44 |
| 32.8 | GG477 | 282 | 2.05 | 0.44 | 0.39 | 0.48 |
| 9.3 | GG194 | 282 | 0.01 | 0.50 | 0.50 | 0.49 |
| 14.5 | GG131 | 282 | 19.92 | 0.69 | 0.97 | 0.50 |
| 26.8 | GG531 | 282 | 0.45 | 0.53 | 0.91 | 0.27 |
| 8 | GG248 | 282 | 0.71 | 0.46 | 0.97 | 0.12 |
| 3.7 | des6 | 266 | 5.48 | 0.40 | 1.00 | 0.00 |
| 4.2 | CG97 | 270 | 5.81 | 0.40 | 0.90 | 0.07 |
| 6.1 | GG133 | 282 | 11.35 | 0.36 | 0.88 | 0.01 |
| 5 | GC74 | 281 | 12.86 | 0.35 | 0.65 | 0.16 |
| 30.1 | GG80 | 144 | 18.00 | 0.25 | 0.59 | 0.05 |
Average marker spacing, 10.41 cM. Overall frequency Ec allele, 0.56.
Using a variety of measures, we estimate that our map covers ∼70% of the C. purpureus genome. The total map length was 728.8 cM, with an average distance of 10.4 cM between markers (range: 1.9–32.8 cM). Adding twice the average marker spacing to each linkage group (to account for chromosome ends beyond terminal markers) we calculate the total genome to be 1040 cM. Using the Chakravarti et al. (1991) method 4, we arrive at a slightly larger estimate of 1076 cM. Using the formula c = 1 – e2dn/L (see materials and methods) we estimated that 74 and 93% of the genome lay within 10 and 20 cM of a mapped marker, respectively.
Of the markers not included in our framework map, 38 were placed unambiguously on intervals in the map with LOD scores greater than 6.0: one of these was on LG2; five were on LG3; two were on LG8; and 30 were in the sex-determining region. Eight additional markers were ambiguously placed on the ends of two chromosomes, although one of the placements had a LOD >6, and was greater than 10 times the LOD score of alternative placement. Seven of these markers were placed on the sex chromosome, but conflicted with LG14, and one was placed on LG8, but conflicted with LG3. Three additional markers placed near a single chromosome with LOD >3.0, one each on LG4, LG8, and LG12. Five markers (∼4%) were unplaced on the map.
Patterns of segregation distortion:
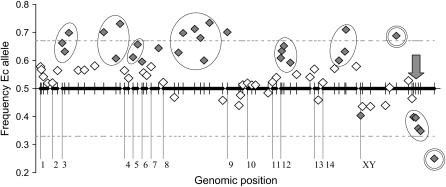
Approximately 45% of the markers on our linkage map (32 of 70 markers), as well as sex, deviated significantly from the expected 1:1 ratio (Table 3; Figure 3). Removing the three markers that were tightly linked to the nonrecombining sex-correlated region reduced this number slightly (43.8%). The most distorted marker (CGG80, on the sex chromosome) had a frequency of pEc = 0.25. The majority of the significantly distorted markers (27 of 32 markers) grouped into seven chromosomal regions each consisting of two or more markers distorted in the same direction: six were located in autosomal regions (the distal portions of LG3, both loci on LG5, the middle of LG8, the end of LG12, and the middle of LG14) and were all distorted toward the Ec (maternal) parent. The last encompassed the sex-determining region and the single marker on PAR2 and was distorted toward the NY parent. Single loci on LG6, LG7, LG9, and PAR1 showed an excess of Ec alleles, and a single locus in PAR1 showed a significant Ec deficit. We emphasize that this is a minimum estimate of the number of distorted regions; highly distorted AFLP markers could have been misinterpreted as monomorphic and not scored, and in a higher density map the multilocus regions may resolve into multiple distorted loci.
Figure 3.—
Genomic segregation rates in the NY–Ec cross. The y-axis represents the frequency of the Ec allele in the segregating progeny. The thick horizontal lines represent linkage groups (numbered below), with the hashes show marker locations, and the dashed lines indicate allele frequencies of 0.33 and 0.67, consistent with a two-locus lethal interaction (see discussion). The gray vertical arrow shows the position of the sex-determining region. The diamonds represent the frequency of the Ec allele at each marker locus. The expected frequency of the Ec allele is 0.5. Solid diamonds indicate loci showing significant deviation from 1:1 segregation (α = 0.05); the allele frequency threshold for significance varied due to marker sample size (see Table 3). The ovals inscribe chromosomal regions where linked markers exhibited significant distortion in the same direction. The double circles indicate markers showing sex-specific segregation distortion.
The sex chromosomes and autosomes exhibited distinct patterns of distortion. No autosomal markers were significantly distorted toward the NY parent, and only 6 of 57 autosomal markers exhibited frequencies of pEc < 0.5. In contrast, 10 of the 13 markers on the sex chromosome had an excess of the NY allele, 6 of them (including the 4 tightly linked to the nonrecombining sex-correlated region) exhibiting significant distortion.
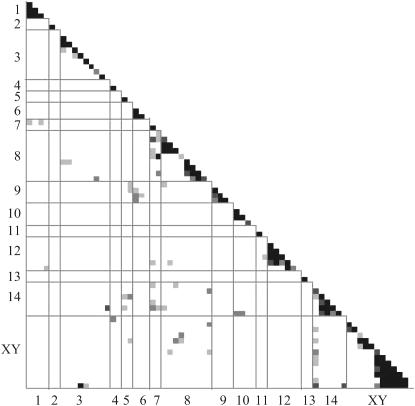
The difference between autosomes and sex chromosomes was also true of deviations from multiplicative genotypic frequencies. Overall, autosomal loci showed significantly nonrandom associations with other autosomal loci, and nonrandom associations involving the sex chromosome were largely independent of autosomal variation (, Figures 4 and 5). Nonrandom associations between markers with an Ec deficit and those with an Ec excess, or complementary distortion, more often involved sex-linked markers or those in the PARs. The four marker pairs showing complementary distortion with the highest r2 values all involved sex chromosome–sex chromosome interactions, while only 2 of the top 11 involved only autosomal markers. Two interactions were between the PAR2 and LG14; seven of the markers that remained unmapped because they placed on two chromosomes conflicted between these two genomic regions, consistent with a strong interaction. We found only a single pair of loci with NY excesses exhibiting strong nonrandom associations (r2 > 0.06), with both loci on the sex chromosome. In contrast, nonrandom pairwise associations between markers both showing an excess of the Ec allele generally did not involve the sex chromosomes; 2 of the top 9 interactions were between an autosomal marker and a PAR marker.
Figure 4.—
Numerically generated probability distribution of r2, assuming a sample size of 130, equal allele frequencies, and multiplicative genotypic frequencies.
Figure 5.—
Genomewide map of deviations from pairwise multiplicative genotypic frequencies, measured by r2. White, r2 < 0.04; light gray, r2 > 0.04 (P < 0.025); gray, r2 > 0.06 (P < 0.005); dark gray, r2 > 0.10 (P < 0.0003); black, r2 > 0.14 (P < 0.00001).
Interactions involving PAR-linked markers were stronger than those involving the sex-determining region. Only the two markers directly adjacent to the nonrecombining sex-correlated region, GG131 and GG80, exhibited significant sex-specific segregation distortion (P < 0.05, χ2 = 4.52, 4.48, respectively, d.f. = 1). Marker GG131 was unbiased in males but nearly completely fixed for the Ec allele in females and distorted in the opposite direction from the sex-determining region. Marker GG80 was unbiased in females but the NY allele was at high frequency in males, more strongly distorted than sex (Table 3).
DISCUSSION
Here we report the construction of a linkage map in the moss C. purpureus, derived from a cross between two geographically distant populations. This represents the first linkage map in a bryophyte and the first empirical demonstration of maternal inheritance of the chloroplast in this lineage of plants. Three important observations result from our analysis of segregation patterns in this cross. First, we provide genetic evidence suggesting that the large heteromorphic chromosomes of C. purpureus are likely to be sex chromosomes. Second, we report that autosomal segregation distortion was widespread and almost exclusively toward the maternal parent, which we suggest may result from nuclear–cytoplasmic interactions. Third, we found significant sex-ratio distortion, similar to natural crosses in the species.
Genetic confirmation of genome structure:
The haploid genome of C. purpureus contains 13 chromosomes (Crum and Anderson 1981), with a heterogeneous distribution of chromosome sizes (J. Greilhuber, personal communication). We found 15 linkage groups, suggesting that at least two pairs of linkage groups should coalesce with the addition of more markers. A variety of estimates suggest that our map, while preliminary, covers approximately three-quarters of the C. purpureus genome. We estimate that ∼93% of the genome lies within 20 cM of a marker, and only 4% of the markers were unlinked to any other marker. We should point out that the length of our map is shorter than the 1300 cM expected from two crossovers per chromosome. Whether this reflects the biological reality, for example, because the genome contains acrosomic chromosomes that have only one crossover, or an artificially shortened map due to incomplete coverage or hybrid lethality, is uncertain.
The most striking feature of the karyotype of C. purpureus is the large heteromorphic pair of chromosomes, comprising approximately one-third of the genome (Heitz 1932; Jachimsky 1935; J. Greilhuber, personal communication). In the liverwort Sphaerocarpus texana similarly large chromosomes were correlated with sex using tetrad analysis (Allen 1917). Here we found that more than 35% of the AFLP markers and the des6 gene cosegregated with sex (Figure 2). Assuming the AFLP markers are randomly distributed throughout the genome, the heteromorphic pair of chromosomes should contain approximately this number of loci (although Y chromosomes may be particularly AFLP dense; see Liu et al. 2004). In meiotic chromosome stains in C. purpureus, the heteromorphic pair of chromosomes paired at their distal ends at metaphase, but a large region of heterochromatin was evident in the center of both chromatids (Ramsay and Berrie 1982). In our linkage map, recombining markers were linked to either end of the sex-correlated markers, similar to other sex chromosomes where a nonrecombining sex-determining region lies between two recombining PARs. Ultimately, however, we need physical evidence in addition to these genetic data to conclusively demonstrate that the large, heteromorphic chromosomes are indeed sex chromosomes.
Approximately 100 cM of the total linkage map (∼1000 cM) were found on the recombining portions of the sex chromosome. Given that the genome size of C. purpureus is ∼300 Mb (Lamparter et al. 1998; Voglmayr 2000) and assuming that one-third of the physical genome is located on the sex chromosome, we estimate that there are ∼4.5 cM/Mb on the autosomes (900 cM / 200 Mb). Assuming this rate also applies to the recombining regions of the sex chromosomes, we estimate the PARs together occupy ∼20 Mb and the nonrecombining portion comprises almost 80 Mb. Increasing the map length only would reduce our estimate of the physical size of the PARs and increase the size of the nonrecombining sex-specific region.
In contrast to sex chromosomes in mammals, the Y chromosome in plants is generally larger than autosomes (Charlesworth et al. 2005). In dioecious mosses, genes on both the X and Y chromosomes are expressed in the haploid state. This likely precludes the loss of gametophyte-essential genes on both sex chromosomes, unlike on the mammalian Y chromosome, where degenerating alleles may be complemented by homologs on the X chromosome. However, since the moss sex chromosomes presumably derive from an ancestral pair of autosomes, the retention of genes by itself cannot explain their larger size. Hood et al. (2004) suggested that transposons may not be efficiently purged on haploid sex chromosomes and may comprise a large portion of the extra genetic material. Alternatively, selection may favor the translocation of genes with sex-specific function to the sex chromosomes because they can specialize on either male or female-specific function at the DNA sequence level. However, beyond the sex-linked des6 protein, which does not have an obviously sex-specific function (incorporating a double bond into a hydrocarbon chain; Sperling et al. 2000), we do not know what genes are located there. We are now screening an expression library for additional markers for the linkage map as well as to study sex chromosome gene content.
Segregation distortion due to population divergence:
In a cross between two haploids, the maternal and paternal alleles are each expected at a frequency of 0.5 among the recombinant haploid progeny. Here we report that 45% of the loci in our mapping population deviated significantly from the 1:1 expectation (Table 3; Figure 3). This estimate is similar to that found in intraspecific crosses in angiosperms (Zamir and Tadmor 1986; Jenczewski et al. 1997; Lu et al. 2000; Fishman et al. 2001; Harushima et al. 2001; Schwarz-Sommer et al. 2003; Kuittinen et al. 2004; Hall and Willis 2005; Bratteler et al. 2006). The distorted loci tended to be clustered along the linkage groups; 6 of the 12 regions showing significant distortion contained at least three markers, suggesting that the distorted markers were nonrandomly distributed throughout the genome (Figure 3). Autosomal distorted markers were almost exclusively distorted toward the maternal (Ec) parent. All progeny carried the Ec chloroplast type, suggesting that the cytoplasm as a whole was maternally inherited.
Because of the nature of controlled crosses in bryophytes we do not believe that the segregation distortion in our map resulted from any of the several prezygotic mechanisms that act in diploids (e.g., Rieseberg and Carney 1998). The moss life cycle allows the elimination of gamete competition, because each haploid gametophyte produces only a single genotype of (mitotic) gametes. Additionally, the production of haploid moss spores is akin to male meiosis, in that all four products go on to form viable spores or gametophytes. Thus, meiotic drive mechanisms that exploit the inherent asymmetries of female meiosis, where three products form nonreproductive polar bodies (Henikoff et al. 2001; Pardo-Manuel de Villena and Sapienza 2001; Fishman and Willis 2005), cannot function in C. purpureus. Unconditionally deleterious alleles also are unlikely to explain the genomewide segregation patterns, particularly because we crossed vigorous haploid isolates. While it is possible that spontaneous deleterious mutations arose, such mutations must have arisen early in the development of the sporophyte used to generate our mapping population.
Our results, therefore, are most consistent with segregation distortion resulting from impaired development of certain genotypes. Since we isolated haploid gametophytes for our mapping population early in development, it is likely that spores of some (mostly NY) genotypes failed to germinate or stopped developing shortly thereafter. We do not have estimates of germination rate for this cross, although germination rates are highly variable in natural crosses in C. purpureus. SD in this artificial cross could be due to a population-specific response to some aspect of our culture conditions (growth medium, light, temperature, or fungal contamination); Ec genotypes could have been better adapted to the conditions encountered in our growth chambers and therefore were overrepresented in the progeny. However, on the basis of our experience with cultivating C. purpureus and other species of moss, this possibility is remote. In this experiment and others, some clones of a genotype have succumbed to fungal contamination while others have not. Moreover, the experimental conditions we used are standard for routine cultivation of several distantly related mosses. We have had success cultivating C. purpureus spores from a wide variety of localities under these conditions, and wild collected individuals from the NY population grew slightly but significantly faster than Ec individuals under our conditions (S. F. McDaniel, J. H. Willis, A. J. Shaw, unpublished data).
Alternatively, germination failure could result from deleterious epistatic effects among loci. The most general epistatic model to explain autosomal SD in a wide cross, independently proposed by Dobzhansky (1937) and Muller (1942), is based on improper interactions between heterospecific alleles at multiple loci. Such alleles function properly in their native genetic background, but are incompatible when brought together in a hybrid. Considerable evidence indicates that Dobzhansky–Muller (DM) incompatibilities play an important role in post-zygotic reproductive isolation in diverse species pairs (Coyne and Orr 2004). If the phenotype of one of the two recombinant two-locus genotypes (i.e., ANY, BEc) is sufficiently deleterious such that we would not have isolated such spores, as predicted by the DM model, the complementary (AEc, BNY) alleles will be overrepresented in a hybrid population of gametophytes. The two-locus haploid model predicts that the maximum allele frequency should be 0.67, consistent with most of the distorted loci in the NY–Ec mapping population (Table 3; Figure 3).
Lethal DM interactions also result in a correlation between allelic states at the interacting loci (Jiang et al. 2000; Payseur and Hoekstra 2005). This is because the ANY and BEc alleles in the preceding example are found only with the homospecific complementary allele, but never together (e.g, the ANY allele is found only with the BNY, and the BEc allele is found only with the AEc). If SD results from heterospecific interactions between autosomal loci, we expect to find nonrandom associations between loci distorted in opposite directions (e.g., the NY allele overrepresented at one locus, and the Ec allele overrepresented at the interacting partner). The autosomal SD in the NY–Ec cross, however, was overwhelmingly toward the Ec (maternal) parent, with only sex chromosome-linked loci (both the sex-determining region and the two PARs) distorted toward the NY parent (except marker GG131—see below). We calculated the correlations between allelic states for all possible pairwise combinations of loci and found that almost none of the Ec-distorted loci exhibited deviations from multiplicative genotypic frequencies with loci on the sex chromosome (the only exception being two distorted loci on LG8). This indicated that the distortion at these Ec-biased autosomal loci did not result from sex-related heterospecific interactions.
Instead, we found significant nonrandom associations between loci that both had an excess of the Ec allele (Table 4; Figure 4). This observation suggests that some NY two-locus genotypes may have had deleterious interactions with an additional Ec factor that was not linked to any segregating marker in our mapping population; such factors may have been absent either due to chance or, more likely, because they were maternally inherited cytoplasmic elements (Burke and Arnold 2001; Willett and Burton 2001; Levin 2003; Fishman and Willis 2006; Harrison and Burton 2006; Willett 2006; Chase 2007). This model implies that these genetic interactions involve multiple complementary loci, where the presence of a homospecific allele at either autosomal locus is sufficient for proper function. (If only the NY two-locus genotype were lost, the two Ec alleles would be found at 67% in the hybrid population, roughly equivalent to the genomic distortion toward the Ec parent; Figure 3.) It is possible that some of the interlocus associations we report had no causative relationship and that the distortion was driven by single nuclear loci that failed to interact with the cytoplasm. In this case, however, lethal single-locus interactions would result in the fixation of the nuclear Ec allele in the progeny, and we would not have detected such loci in our screen for polymorphic loci. Strongly distorted linked alleles would have been detected, but we found only one locus with PEc = 0.75 − 0.9, while most of the distorted autosomal loci segregated at PEc = 0.6 – 0.7. Nevertheless, we cannot eliminate that either single locus or more complex nuclear–cytoplasmic (or other autosome–autosome) interactions contributed to spore lethality.
TABLE 4.
Patterns of nonrandom association among pairs of unlinked markers
| Locus 1 | Locus 2 | R2 | n | P(Ec)1 | χ2 | P(Ec)2 | χ2 | |
|---|---|---|---|---|---|---|---|---|
| Ec excess at both loci | ||||||||
| LG7-LG8 | CC244 | CC267 | 0.15 | 275 | 0.64 | 11.35 | 0.71 | 24.89 |
| LG3-LG14 | CC269 | GG321 | 0.12 | 135 | 0.71 | 12.03 | 0.71 | 12.03 |
| PAR1-LG14 | CG477 | GG148 | 0.07 | 131 | 0.66 | 6.42 | 0.60 | 2.39 |
| LG8-PAR1 | GG361 | GG194 | 0.06 | 282 | 0.63 | 9.19 | 0.50 | 0.01 |
| LG7-LG8 | CC244 | GG223 | 0.06 | 269 | 0.65 | 11.60 | 0.74 | 29.98 |
| LG6-LG9 | GG421 | CC306 | 0.06 | 269 | 0.59 | 4.11 | 0.52 | 0.15 |
| LG7-LG14 | CC400 | GG321 | 0.06 | 135 | 0.53 | 0.18 | 0.71 | 12.03 |
| LG5-LG14 | CC181 | GG321 | 0.06 | 135 | 0.61 | 3.11 | 0.71 | 12.03 |
| LG5-LG14 | CC181 | GG148 | 0.06 | 269 | 0.61 | 6.47 | 0.61 | 6.04 |
| Ec excess at one locus | ||||||||
| PAR1-XY | GG131 | des6 | 0.24 | 260 | 0.69 | 18.47 | 0.40 | 5.61 |
| PAR1-XY | GG131 | GG133 | 0.24 | 282 | 0.69 | 19.92 | 0.36 | 11.35 |
| XY-PAR2 | GG531 | GG80 | 0.16 | 144 | 0.54 | 0.50 | 0.25 | 18.00 |
| PAR1-XY | GG131 | GG248 | 0.16 | 282 | 0.69 | 19.92 | 0.46 | 0.71 |
| LG14-PAR2 | GG310 | GG80 | 0.12 | 144 | 0.53 | 0.22 | 0.25 | 18.00 |
| PAR1-PAR2 | GG131 | GG80 | 0.09 | 144 | 0.67 | 8.00 | 0.25 | 18.00 |
| LG3-LG8 | GC246 | CG474 | 0.07 | 129 | 0.68 | 8.56 | 0.47 | 0.31 |
| LG6-LG9 | GG421 | CC287 | 0.07 | 269 | 0.59 | 4.11 | 0.48 | 0.15 |
| LG8-PAR1 | GC126 | GG477 | 0.07 | 275 | 0.69 | 20.82 | 0.45 | 1.53 |
| LG4-XY | CC155 | CC256 | 0.06 | 275 | 0.56 | 2.23 | 0.40 | 5.11 |
| PAR1-PAR1 | CC297 | GG131 | 0.06 | 269 | 0.43 | 2.28 | 0.69 | 18.96 |
| Ec deficit at both loci | ||||||||
| XY-PAR2 | des6 | GG80 | 0.36 | 129 | 0.40 | 2.83 | 0.26 | 14.42 |
| LG3-PAR2 | GG127 | GG80 | 0.24 | 48 | 0.50 | 0 | 0.31 | 3.38 |
Genomics of sex-ratio distortion:
The significant excess of males in the mapping population indicates that carriers of the Ec X chromosome suffered greater mortality than carriers of the NY Y chromosome. Sex-ratio distortion is reported from interpopulation crosses in a number of experimental systems (Mercot et al. 1995; Taylor 1999; Dermitzakis et al. 2000; Tao et al. 2001; Orr and Irving 2005). The level of sex-ratio distortion in this artificial cross was qualitatively indistinguishable from that found in field-collected sporophytes of C. purpureus, where both male- and female-biased families were frequent (Shaw and Gaughan 1993; Shaw and Beer 1999). A wide variety of dioecious bryophytes show strong population-level sex-ratio variation (reviewed in Bisang and Hedenas 2005), suggesting that this may be a common feature of bryophyte biology (although because most of these estimates were based on sex ratios evident in field-collected herbarium specimens, it is uncertain whether this reflects the primary sex ratio, differential attrition of one sex, or simply differences in sex expression).
No sex-linked loci showed significant deviations from multiplicative genotypic frequencies with any autosomal loci, suggesting that sex-ratio distortion in the NY–Ec cross does not result from autosomal allelic variants that differentially affect the fitness of males and females. Therefore, we suspect that the biased sex ratio resulted from epistatic interactions between loci within the sex-correlated region of the sex chromosome (i.e., sex chromosome meiotic drive; Lyttle 1993; Jaenike 2001; Taylor and Ingvarsson 2003) or as a result of linkage to interacting loci in the PARs. In the first case, we would not be able to detect complementary distortion at the interacting loci because recombination is suppressed on both the male and female chromosomes. This hypothesis is additionally complicated by the fact that sex-ratio distortion in dioecious mosses is associated not with a loss of gametes (as in male animals) but with a reduction in absolute numbers of offspring spores (Nauta and Hoekstra 1993). This fitness cost of course could be offset by selection on sex ratio itself (Godfray and Werren 1996) or a pleiotropic selective advantage of distorter alleles to the diploid sporophyte (Dalstra et al. 2003; van der Gaag et al. 2003).
Although we cannot eliminate that sex-chromosome meiotic drive contributes to sex-ratio bias, hypotheses involving only the sex-determining region do not explain the SD we found on both PARs. Indeed, the two markers most closely linked to the sex-determining region are either more distorted than the sex-linked loci (GG80) or significantly distorted in the other direction (GG131; Figure 3). This observation suggests that the sex-ratio distortion may have been a by-product of interactions between linked loci in the PARs and other loci. Loci in both PARs showed nonrandom genotypic frequencies with several autosomal loci (Figure 5), as well as sex-specific distortion (Tables 3 and 4; Figures 3 and 5). We expected complementary allele frequencies for PAR loci in the two sexes (e.g., 0.80 in males, 0.20 in females, for a paternal allele) due to linkage to the sex-determining locus, but GG80 was slightly distorted (PEc = 0.59) in the Ec X chromosome background, but strongly distorted (PEc = 0.05) in the NY Y chromosome background. This observation suggested a lethal interaction involving the Ec GG80 and a NY Y-linked factor. The converse was found at marker GG131 where the NY allele was unbiased in males (PEc = 0.50) but at a frequency of 0.03 in the Ec X chromosome background.
Without a physical map it is difficult to distinguish whether the placement of GG80 and GG131 adjacent to the sex-determining region reflects true linkage or strong genetic interactions, which could cause us to misplace such markers in our map. Indeed, seven unmapped markers exhibited strong alternative placements between PAR2 and LG14, consistent with either of these hypotheses. However, Rice (1987) demonstrated that alleles causing sex-specific lethality can more easily fix in populations if they arise linked to the sex chromosome of the sex they benefit (i.e., male lethals must be linked to the X chromosome). The fixation of such alleles is predicted to lead to selection for suppressed recombination between the sex-determining region and the sexually antagonistic locus (Rice 1987). While this theory was developed for diploids, where only one sex has a sex-specific chromosome (i.e., the male-specific Y), it applies equally well to male or female moss sex chromosomes. Sex-ratio distortion could be evident in the progeny of a cross between populations that had fixed different PAR-linked sex lethals and recombination modifiers. Given our current data, however, we know of no way to test whether loci that appear to interact with GG80 or GG131 (those in the middle of LG3 and on the distal ends of LG14; Figure 5) are recombination modifiers, as the sexual antagonism hypothesis would suggest.
Conclusions:
The results from our interpopulation cross provide genetic confirmation of cytological observations regarding the size and meiotic pairing patterns of the sex chromosomes of C. purpureus. Our analysis of segregation patterns suggests that SD results from multiple phenomena that likely evolved within populations as well as in the process of population divergence. For example, we cannot exclude that the sex-ratio variation in this cross results from heterospecific epistatic interactions, although previous studies (Shaw and Gaughan 1993; Shaw and Beer 1999) show that similar sex-ratio distorters (potentially meiotic drivers or sexually antagonistic alleles) segregate at appreciable frequencies within natural populations of C. purpureus. We have little information regarding the genetic basis of adaptive differences among populations in the species, but the distortion toward the maternal parent suggests that nuclear–cytoplasmic interactions may be a major component of divergence. These results indicate that the raw material for reproductive isolation may be segregating within species with distributions spanning ecological clines, but identifying the relative contributions of these various incompatibilities to the speciation process is an ongoing challenge.
Acknowledgments
Members of the Willis lab, in particular A. Bouck, A. L. Case, L. Fishman, and M. C. Hall, provided helpful guidance at various stages of this research. C. J. Cox, E. Y. Walker, and S. Wooley helped generate the r2 values and graphics. D. J. Cove, L. Jesson, and L. C. Moyle made critical comments on earlier drafts of this manuscript. P. Eckel made the drawing in Figure 1. S.F.M. thanks his dissertation committee at Duke University, P. S. Manos, M. D. Rausher, A. J. Shaw, R. Vilgalys, and J. H. Willis. A collecting trip to Ecuador was supported by a National Geographic Society grant to A.J.S., and the labwork was supported by a National Science Foundation–Doctoral Dissertation Improvement Grant to S.F.M.
References
- Allen, C. E., 1917. A chromosome difference correlated with sex differences in Sphaerocarpos. Science 46: 466–467. [DOI] [PubMed] [Google Scholar]
- Bisang, I., and L. Hedenas, 2005. Sex ratio patterns in dioicous bryophytes re-visited. J. Bryol. 27: 207–219. [Google Scholar]
- Bratteler, M., C. Lexer and A. Widmer, 2006. A genetic linkage map of Silene vulgaris based on AFLP markers. Genome 49: 320–327. [DOI] [PubMed] [Google Scholar]
- Brucker, G., F. Mittmann, E. Hartmann and T. Lamparter, 2005. Targeted site-directed mutagenesis of a heme oxygenase locus by gene replacement in the moss Ceratodon purpureus. Planta 220: 864–874. [DOI] [PubMed] [Google Scholar]
- Burke, J. M., and M. L. Arnold, 2001. Genetics and the fitness of hybrids. Ann. Rev. Genet. 35: 31–52. [DOI] [PubMed] [Google Scholar]
- Burley, J. S., and N. M. Pritchard, 1990. Revision of the genus Ceratodon (Bryophyta). Harv. Pap. Bot. 2: 1–76. [Google Scholar]
- Chakravarti, A., L. K. Lasher and J. E. Reefer, 1991. A maximum-likelihood method for estimating genome length using genetic-linkage data. Genetics 128: 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., B. Charlesworth and G. Marais, 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. [DOI] [PubMed] [Google Scholar]
- Chase, C. D., 2007. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genetc. 23: 81–90. [DOI] [PubMed]
- Cove, D. J., and R. S. Quatrano, 2006. Agravitropic mutants of the moss Ceratodon purpureus do not complement mutants having a reversed gravitropic response. Plant Cell Environ. 29: 1379–1387. [DOI] [PubMed] [Google Scholar]
- Cove, D. J., R. S. Quatrano and E. Hartmann, 1996. The alignment of the axis of asymmetry in regenerating protoplasts of the moss, Ceratodon purpureus, is determined independently of axis polarity. Development 122: 371–379. [DOI] [PubMed] [Google Scholar]
- Cove, D., M. Bezanilla, P. Harries and R. Quatrano, 2006. Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 57: 497–520. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Crum, H., and L. Anderson, 1981. Mosses of Eastern North America. Columbia University Press, New York.
- Dalstra, H. J. P., K. Swart, A. J. M. Debets, S. J. Saupe and R. F. Hoekstra, 2003. Sexual transmission of the [Het-s] prion leads to meiotic drive in Poldospora anserina. Proc. Natl. Acad. Sci. USA 100: 6616–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis, E. T., J. P. Masly, H. M. Waldrip and A. G. Clark, 2000. Non-Mendelian segregation of sex chromosomes in heterospecific Drosophila males. Genetics 154: 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, T. H., 1937. Genetics and the Origin of Species. Columbia University Press, New York.
- Esch, H., E. Hartmann, D. Cove, M. Wada and T. Lamparter, 1999. Phytochrome-controlled phototropism of protonemata of the moss Ceratodon purpureus: physiology of the wild type and class 2 ptr-mutants. Planta 209: 290–298. [DOI] [PubMed] [Google Scholar]
- Fishman, L., and J. H. Willis, 2005. A novel meiotic drive locus almost completely distorts segregation in mimulus (monkeyflower) hybrids. Genetics 169: 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman, L., and J. H. Willis, 2006. A cytonuclear incompatibility causes anther sterility in Mimulus hybrids. Evol. Int. J. Org. Evol. 60: 1372–1381. [DOI] [PubMed] [Google Scholar]
- Fishman, L., A. J. Kelly, E. Morgan and J. H. Willis, 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159: 1701–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfray, H. C. J., and J. H. Werren, 1996. Recent developments in sex ratio studies. Trends Ecol. Evol. 11: 59–63. [DOI] [PubMed] [Google Scholar]
- Hall, M. C., and J. H. Willis, 2005. Transmission ratio distortion in intraspecific hybrids of Mimulus guttatus: implications for genomic divergence. Genetics 170: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, J. S., and R. S. Burton, 2006. Tracing hybrid incompatibilities to single amino acid substitutions. Mol. Biol. Evol. 23: 559–564. [DOI] [PubMed] [Google Scholar]
- Harushima, Y., M. Nakagahra, M. Yano, T. Sasaki and N. Kurata, 2001. A genome-wide survey of reproductive barriers in an intraspecific hybrid. Genetics 159: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz, E., 1932. Geschlectschromosomen bei einem Laubmoos. Ber. Dtsh. Bot. Ges. 30: 204–206. [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Hood, M. E., J. Antonovics and B. Koskella, 2004. Shared forces of sex chromosome evolution in haploid-mating and diploid-mating organisms: Microbotryum violaceum and other model organisms. Genetics 168: 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachimsky, H., 1935. Beitrag zur Kenntnis von Geschlectschromosomen und Heterochromatin bei Mooses. Jahr. Wiss. Botani 81: 203–238. [Google Scholar]
- Jaenike, J., 2001. Sex chromosome meiotic drive. Ann. Rev. Ecol. Syst. 32: 25–49. [Google Scholar]
- Jenczewski, E., M. Gherardi, I. Bonnin, J. M. Prosperi, I. Olivieri et al., 1997. Insight on segregation distortions in two intraspecific crosses between annual species of Medicago (Leguminosae). Theor. Appl. Genet. 94: 682–691. [Google Scholar]
- Jiang, C.-x., P. W. Chee, X. Draye, P. L. Morrell, C. W. Smith et al., 2000. Multilocus interactions restrict gene introgression in interspecific populations of polyploid Gossypium (cotton). Evolution 54: 798–814. [DOI] [PubMed] [Google Scholar]
- Kern, V. D., and F. D. Sack, 1999. Irradiance-dependent regulation of gravitropism by red light in protonemata of the moss Ceratodon purpureus. Planta 209: 299–307. [DOI] [PubMed] [Google Scholar]
- Knight, C. D., D. J. Cove, A. C. Cuming and R. S. Quatrano, 2002. Moss gene technology, pp. 285–301 in Molecular Plant Biology, edited by P. M. Gilmartin and C. Bowler. Oxford University Press, Oxford.
- Kuittinen, H., A. A. de Haan, C. Vogl, S. Oikarinen, J. Leppala et al., 2004. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamparter, T., G. Brucker, H. Esch, J. Hughes, A. Meister et al., 1998. Somatic hybridisation with aphototropic mutants of the moss Ceratodon purpureus: genome size, phytochrome photoreversibility, tip-cell phototropism and chlorophyll regulation. J. Plant Physiol. 153: 394–400. [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Levin, D. A., 2003. The cytoplasmic factor in plant speciation. Syst. Bot. 28: 5–11. [Google Scholar]
- Lincoln, S. E., and E. S. Lander, 1992. Systematic detection of errors in genetic linkage data. Genomics 14: 604–610. [DOI] [PubMed] [Google Scholar]
- Liu, Z. Y., P. H. Moore, H. Ma, C. M. Ackerman, M. Ragiba et al., 2004. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427: 348–352. [DOI] [PubMed] [Google Scholar]
- Lu, C. G., J. S. Zou and H. Ikehashi, 2000. Gamete abortion locus detected by segregation distortion of isozyme locus Pgi1 in indica-japonica hybrids of rice (Oryza sativa L.). Breed. Sci. 50: 235–239. [Google Scholar]
- Lyttle, T. W., 1993. Cheaters sometimes prosper: distortion of Mendelian segregation by meiotic drive. Trends Genet. 9: 205–210. [DOI] [PubMed] [Google Scholar]
- McDaniel, S. F., 2005. Genetic correlations do not constrain the evolution of sexual dimorphism in the moss Ceratodon purpureus. Evol. Int. J. Org. Evol. 59: 2353–2361. [PubMed] [Google Scholar]
- McDaniel, S. F., and A. J. Shaw, 2003. Phylogeographic structure and cryptic speciation in the trans-Antarctic moss Pyrrhobryum mnioides. Evol. Int. J. Org. Evol. 57: 205–215. [DOI] [PubMed] [Google Scholar]
- McDaniel, S. F., and A. J. Shaw, 2005. Selective sweeps and intercontinental migration in the cosmopolitan moss Ceratodon purpureus (Hedw.) Brid. Mol. Ecol. 14: 1121–1132. [DOI] [PubMed] [Google Scholar]
- Mercot, H., B. Llorente, M. Jacques, A. Atlan and C. Montchamp-Moreau, 1995. Variability within the Seychelles cytoplasmic incompatibility system in Drosophila simulans. Genetics 141: 1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Nauta, M. J., and R. F. Hoekstra, 1993. Evolutionary dynamics of spore killers. Genetics 135: 923–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., and S. Irving, 2005. Segregation distortion in hybrids between the Bogota and USA subspecies of Drosophila pseudoobscura. Genetics 169: 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., and C. Sapienza, 2001. Nonrandom segregation during meiosis: the unfairness of females. Mamm. Genome V12: 331–339. [DOI] [PubMed] [Google Scholar]
- Payseur, B. A., and H. E. Hoekstra, 2005. Signatures of reproductive isolation in patterns of single nucleotide diversity across inbred strains of mice. Genetics 171: 1905–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay, H., and G. Berrie, 1982. Sex determination in bryophytes. J. Hattori Bot. Lab. 52: 255–274. [Google Scholar]
- Rice, W. R., 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41: 911–914. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., and S. E. Carney, 1998. Plant hybridization. New Phytol. 140: 599–624. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer, Z., E. D. Silva, R. Berndtgen, W. E. Lonnig, A. Muller et al., 2003. A linkage map of an F-2 hybrid population of Antirrhinum majus and A. molle. Genetics 163: 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, A. J., and J. F. Gaughan, 1993. Control of sex-ratios in haploid populations of the moss, Ceratodon purpureus. Am. J. Bot. 80: 584–591. [DOI] [PubMed] [Google Scholar]
- Shaw, A. J., B. S. Weir and F. H. Shaw, 1997. The occurrence and significance of epistatic variance for quantitative characters and its measurement in haploids. Evolution 51: 348–353. [DOI] [PubMed] [Google Scholar]
- Shaw, J., and S. C. Beer, 1999. Life history variation in gametophyte populations of the moss Ceratodon purpureus (Ditrichaceae). Am. J. Bot. 86: 512. [PubMed] [Google Scholar]
- Sperling, P., M. Lee, G. Thomas, U. Zahringer, S. Stymne et al., 2000. A bifunctional Delta(6)-fatty acyl acetylenase/desaturase from the moss Ceratodon purpureus: a new member of the cytochrome b(5) superfamily. Eur. J. Biochem. 267: 3801–3811. [DOI] [PubMed] [Google Scholar]
- Tao, Y., D. L. Hartl and C. C. Laurie, 2001. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc. Natl. Acad. Sci. USA 98: 13183–13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, D. R., 1999. Genetics of sex ratio variation among natural populations of a dioecious plant. Evolution 53: 55–62. [DOI] [PubMed] [Google Scholar]
- Taylor, D. R., and P. K. Ingvarsson, 2003. Common features of segregation distortion in plants and animals. Genetica 117: 27–35. [DOI] [PubMed] [Google Scholar]
- Thornton, L. E., N. Keren, I. Ohad and H. B. Pakrasi, 2005. Physcomitrella patens and Ceratodon purpureus, mosses as model organisms in photosynthesis studies. Photosynth. Res. 83: 87–96. [DOI] [PubMed] [Google Scholar]
- van der Gaag, M., A. J. M. Debets and R. F. Hoekstra, 2003. Spore killing in the fungus Podospora anserina: A connection between meiotic drive and vegetative incompatibility? Genetica 117: 59–65. [DOI] [PubMed] [Google Scholar]
- Voglmayr, H., 2000. Nuclear DNA Amounts in Mosses (Musci). Ann. Bot. 85: 531–546. [Google Scholar]
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Vandelee et al., 1995. AFLP: a new technique for DNA-fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, T. A., D. J. Cove and F. D. Sack, 1997. A positively gravitropic mutant mirrors the wild-type protonemal response in the moss Ceratodon purpureus. Planta 202: 149–154. [DOI] [PubMed] [Google Scholar]
- Willett, C. S., 2006. Deleterious epistatic interactions between electron transport system protein-coding loci in the copepod Tigriopus californicus. Genetics 173: 1465–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett, C. S., and R. S. Burton, 2001. Viability of cytochrome C genotypes depends on cytoplasmic backgrounds in Tigriopus californicus. Evolution 55: 1592–1599. [DOI] [PubMed] [Google Scholar]
- Zamir, D., and Y. Tadmor, 1986. Unequal segregation of nuclear genes in plants. Bot. Gaz. 147: 355–358. [Google Scholar]