Abstract
It is often suggested that heterozygosity at major histocompatibility complex (MHC) loci confers enhanced resistance to infectious diseases (heterozygote advantage, HA, hypothesis), and overdominant selection should contribute to the evolution of these highly polymorphic genes. The evidence for the HA hypothesis is mixed and mainly from laboratory studies on inbred congenic mice, leaving the importance of MHC heterozygosity for natural populations unclear. We tested the HA hypothesis by infecting mice, produced by crossbreeding congenic C57BL/10 with wild ones, with different strains of Salmonella, both in laboratory and in large population enclosures. In the laboratory, we found that MHC influenced resistance, despite interacting wild-derived background loci. Surprisingly, resistance was mostly recessive rather than dominant, unlike in most inbred mouse strains, and it was never overdominant. In the enclosures, heterozygotes did not show better resistance, survival, or reproductive success compared to homozygotes. On the contrary, infected heterozygous females produced significantly fewer pups than homozygotes. Our results show that MHC effects are not masked on an outbred genetic background, and that MHC heterozygosity provides no immunological benefits when resistance is recessive, and can actually reduce fitness. These findings challenge the HA hypothesis and emphasize the need for studies on wild, genetically diverse species.
THE extremely polymorphic genes of the major histocompatibility complex (MHC) encode the class I and II molecules, which present pathogen-derived peptide antigens to T-cells and thus initiate specific immune responses against parasites and pathogens (Klein 1986; Janeway et al. 2005). The crucial role of MHC genes in disease resistance suggests that pathogen-driven balancing selection could be the ultimate evolutionary mechanism maintaining the high polymorphisms (Haldane 1949; Clarke and Kirby 1966; Potts and Wakeland 1990; Hughes and Nei 1992; Parham and Ohta 1996; Apanius et al. 1997; Hedrick and Kim 2000). The three leading forms of pathogen-driven balancing selection acting on the MHC are (1) spatial and temporal changes in selection pressures by pathogens, (2) frequency-dependent selection, and (3) heterozygote advantage through overdominant selection. It has been controversial as to which is most important in contributing to the maintenance of MHC diversity (Hedrick 2002; Borghans et al. 2004; De Boer et al. 2004). The heterozygote advantage (HA) hypothesis postulates that the MHC polymorphism could be maintained by selective advantage of heterozygotes over homozygotes due to enhanced resistance to pathogens (Doherty and Zinkernagel 1975). MHC heterozygous individuals are expected to be superior to both parental homozygotes (i.e., show allele-specific overdominant resistance) especially when infected with multiple species or strains of pathogens, assuming they are able to present a wider array of pathogen-derived peptides to T-cells (Hughes and Nei 1992). The empirical evidence for the HA hypothesis, however, is mixed and ambiguous (Apanius et al. 1997; Penn 2002; Lipsitch et al. 2003).
Some correlative population-level studies in humans (Thursz et al. 1997; Carrington et al. 1999; Jeffery et al. 2000) and fish (Arkush et al. 2002) have found a general resistance advantage for heterozygotes against viral infections. Yet, most of these studies were not able to distinguish whether the advantage was due to MHC heterozygosity or genomewide heterosis (Penn 2002), and as many other population studies have failed to find any support for the HA hypothesis (Hill et al. 1991; Paterson et al. 1998; Langefors et al. 2001; Lohm et al. 2002). Another serious limitation of population-level studies is that comparing the average performance of homozygotes against heterozygotes leaves it unclear whether the observed population-level HA is due to allele-specific overdominance or dominance (heterozygotes are as resistant as the best parental homozygote) (Penn 2002) or is simply a result of a confounding effect of allele-frequency distribution in a host population (Lipsitch et al. 2003).
Experimental infection studies in MHC congenic mice, which differ genetically only at certain MHC loci, provide a common method to control for confounding effects of background loci and allow allele-specific comparisons. Infection studies in congenic mice have shown that MHC heterozygosity often increases pathogen resistance (at least 9 of 16 pathogens tested) and provides HA when resistance is dominant or overdominant (10 of 17 studies reviewed in Penn 2002). However, many of these studies suffer from several serious problems that obscure the generality of the results and leave them inconclusive (Penn 2002). Because previous laboratory studies looked only at single strains of single pathogens and did not measure host fitness, a recent study examined multiple-strain infections and found that heterozygotes had better resistance and survival in seminatural enclosures during avirulent Salmonella infections (Penn et al. 2002). HA was found to be due to resistance being dominant (masking of susceptible allele) rather than overdominant. This indicates that, contrary to common belief, the observed HA in population studies might be due to dominance rather than overdominance. HA due to dominance does not promote diversity, although it could explain the adaptive function of MHC-disassortative mating preferences, another mechanism capable of maintaining MHC diversity (Penn 2002; Penn et al. 2002). It has also been suggested that overdominance could emerge over multiple infections under conditions where resistance is generally dominant and susceptibility profiles of two pathogens are opposite or reciprocal (Mcclelland et al. 2003). However, this would hold only when lifetime fitnesses for all homozygotes and heterozygotes are equal within these two genotypic classes.
Although the current experimental evidence from congenic mice gives some support to the HA through allele-specific dominance, it is relevant to ask what the applicability of these results is to natural outbred populations where individuals bear highly diverse background genes (Apanius et al. 1997; Bernatchez and Landry 2003; Sommer 2005). In congenic inbred mice, MHC effects can be artificially amplified, because their background genes are highly homogenous. It is also known from studies on humans and mice that there are several background genes that either alone or in interaction with MHC genes can influence resistance and the outcome of disease (Vidal et al. 1993; Vukusic et al. 1995; Apanius et al. 1997; Jepson et al. 1997; Mitchison and Roes 2002; Greve et al. 2004) and even overwhelm the MHC effects or change the resistance patterns (Apanius et al. 1997). Therefore, our goal was to determine how MHC and MHC heterozygosity influence resistance on a wild background. This is difficult to test in wild mice because of high MHC diversity, so we crossbred MHC congenic C57BL/10 mice with wild ones (Mus musculus domesticus) to produce mice with characterized MHC haplotypes and half of their background genes coming from wild-derived mice. We infected these mice with single and multiple strains of Salmonella both in the laboratory and in large population enclosures. MHC genes still influenced variation in resistance despite diverse background genes, but resistance was mostly recessive rather than dominant and never overdominant. Heterozygotes did not show any advantage; rather, a significant reproduction disadvantage in MHC heterozygous females was observed. Our results do not support the HA hypothesis and question the applicability of results from MHC congenic mice to wild genetically diverse mouse populations.
MATERIALS AND METHODS
Experimental mice:
To produce experimental mice with characterized MHC genotypes on a wild, outbred background, we crossbred MHC congenic mice (strains C57BL/SnJ-H2b, B10.D2-H2d, B10.BR-H2k, and B10.Q-H2q) with wild ones. Congenic mice were obtained from the Jackson Laboratories (West Grove, PA), whereas the wild ones originated from the natural populations in the surroundings of Gainesville, Florida. The congenic strains we used in this study carry four different MHC haplotypes (b, d, k, and q), whose alleles have large genetic differences at all class I and II loci (She et al. 1991; Pullen et al. 1992; Edwards et al. 1997). To be able to differentiate wild MHC from known congenic MHC, we first determined alleles at the d17mit63 locus for both the congenic and the wild mice by using microsatellite markers (Saha and Cullen 1986). We used only wild mice possessing nonoverlapping d17mit63 alleles with congenic ones to produce F1's. We genotyped offspring of F1's for their alleles at d17mit63 and chose individuals possessing MHC alleles derived only from congenic types as F2 breeders. We MHC genotyped F2 breeders by using two microsatellite markers, d17mit34 and a tetramer described elsewhere (Saha and Cullen 1986). We used offspring of F3's in infection experiments. To confirm MHC genotypes, we genotyped all experimental mice. Mice were housed in standard colony conditions with the same-sex littermates, three to five per cage.
Seminatural enclosure populations:
To investigate pathogen resistance and fitness under naturalistic conditions, we established four enclosure populations of 30 mice (10 males and 20 females). In each population there were one male and two female mice carrying one of the 10 different MHC genotypes (BB, DD, KK, QQ homozygotes and the respective BD, BK, BQ, DK, DQ, KQ heterozygotes). Age of the mice varied in the beginning of the experiment (171 ± 5 days old), but did not differ among populations or between homozygotes and heterozygotes. We infected mice in two populations and sham-infected them with PBS in the other two. To avoid possible confounding effects of kin-based behaviors, males did not have any relatives and females had no sibs in the same population.
We released mice into large (∼22.2 m2) seminatural enclosures meant to mimic their natural human commensal habitat and social environment. Each of the four enclosures was divided into six equal subsections with 46-cm-high hardware cloth fences. Each subsection contained an additional cylinder of hardware cloth and two nest boxes. Hardware fences and cylinders provide environmental complexity important for natural behavior in mice, and males tend to use these as boundaries for territories. We also set up three refuge boxes (small nest boxes hanging ∼1.5 m from the ground) per each enclosure, which mice could reach only by climbing a wire. Refuge boxes provided shelter for the subordinate individuals from dominant males, and some females used these as nests. Within each subsection, water, food pellets, nesting material, and wood chip bedding were available ad libitum. We conducted regular weekly checks throughout the 21-week experiment to identify the dead adults and to pick up the newborn pups from their nests. We identified the adults with unique ear-puncture marks and genotyped the pups to determine reproductive success of homozygous and heterozygous females (see below).
Pathogens and infections:
We infected mice with different strains of Salmonella enterica serovar Typhimurium. This is an enteric mouse pathogen that becomes systemic by invading the intestinal mucosa and by replicating intracellularly within macrophages and dendritic cells. Host defenses against Salmonella require both the innate and the acquired arms of the immune system (Ravindran and McSorley 2005). Resistance to Salmonella is under polygenic control and both MHC and non-MHC genes are involved (Roy and Malo 2002).
We cultured frozen bacteria in 25 ml of heart–brain infusion at 37° for 12 hr while shaking. We diluted the culture and verified the concentration of bacteria in the inoculum by using quantitative plate counts in duplicates. For each experimental infection, we inoculated 30 μl [108 colony-forming units (CFU)/ml] orally, which is a natural infection route for Salmonella. All mice were food and water restricted 3–4 hr prior to infections. In laboratory experiments, we separated the mice from their littermates and caged them singly 2 days prior to infections.
In the first laboratory experiments, we infected male mice (MHC haplotypes DD, KK, QQ, DK, DQ, and KQ) with a single LT2 strain (Xu and Hsu 1992). In the second experiment, we infected equal numbers of males and females (haplotypes BB, DD, QQ, BD, BQ, and DQ) first with a single LT2 strain. Two weeks after primary infection, we killed some of the mice to assay their pathogen loads and infected the rest of the mice for a second time (experiment 3) with a mixture containing equal numbers of three Salmonella strains: LT2, PMAC45 (Charbit et al. 1997), and M525 (Hormaeche et al. 1981). In the fourth laboratory experiment, we infected equal numbers of males and females (haplotypes BD, BK, BQ, DK, DQ, and KQ) first with a single M525 strain and 2 weeks later with a mixture of strains M525, PMAC51 (Charbit et al. 1997), and χ4665 (Sukupolvi et al. 1997).
In population enclosures we infected mice sequentially in 3-week intervals with seven different Salmonella strains, so that during each subsequent infection we added a novel strain to the inoculum [LT2, PMAC45, M525, PMAC51, χ4665, 628 (Hormaeche et al. 1985), and ATCC 14028 (in an order added to the inoculum)], but kept the volume and the dosage the same. We chose to sequentially increase the number of bacterial strains to imitate natural overlapping epidemics, where novel pathogen strains emerge, as a result of mutation or invasion, while the host population is still going through epidemics from the earlier infections (Penn et al. 2002). The rationale for using sequential infections with multiple strains is that MHC-dependent immune responses have been found to be more pronounced against secondary infections in congenic mice (Penn et al. 2002), and MHC heterozygosity is thought to provide particularly enhanced resistance against infections with multiple pathogens (Hughes and Nei 1992; Apanius et al. 1997; Penn et al. 2002; Mcclelland et al. 2003).
Pathogen load assays:
To assess resistance, we examined bacterial loads, mortality, change in body weight, and spleen weight. In laboratory infection experiments the mice were euthanized 7–15 days after the primary infection with a single strain [experiments (exp.) 1 and 2] or 14 days after the secondary infection with multiple strains (exp. 3 and 4). The mice that survived in enclosure populations until the end of the 21-week-long experiment were euthanized to assay their pathogen loads. We dissected and homogenized spleens in 1 ml of PBS under sterile conditions, cultured 50 μl of each homogenate on selective agar plates, and incubated the plates overnight (37°). We determined bacterial loads by counting the number of CFU per milliliter of spleen homogenates on the plates (the mean of two replicate plates per mouse).
Female reproductive success:
We estimated the reproductive success of MHC homozygous and heterozygous females in the enclosure populations by using molecular markers. To determine female reproductive success we genotyped the offspring and used length variation in the control region of the mitochondrial genome (5′-TTGGTTTCACGGAGGAGGATGGT and 5′-CACCACCAGCACCCAAAGCT). For each population, we used female founders such that homozygotes and heterozygotes possessed nonoverlapping alleles for mitochondrial markers. We reversed the markers characterizing the heterozygous and homozygous females in half of the populations to avoid any possible confounding of the effects of heterozygosity with effects caused by marker alleles or closely linked genes. We did not determine the parentage for the pups, but the markers enabled us to determine whether the mother was MHC homozygous or heterozygous.
Statistical analyses:
We always tested the data for assumptions of normality and equality of variances before conducting parametric tests. The Salmonella loads were log10-transformed to meet the requirements for parametric tests. We always controlled statistically the effects of age, sex, preinfection weight, and duration of infection on response variables by including them into AN(CO)OVA models either as factors or as covariates. We determined the haplotype-specific resistance profiles by comparing the mean bacterial loads of each heterozygous genotype separately against the means of the corresponding homozygotes (i.e., BD vs. BB and BD vs. DD) with independent sample t-tests. Resistance was classified to be dominant, if the mean of heterozygotes was statistically significantly lower than in less resistant homozygotes, but did not significantly differ from more resistant homozygotes, and recessive, if the mean of heterozygotes was significantly higher than in more resistant homozygotes, but did not significantly differ from less resistant homozygotes. This method can potentially lead to biased results, if the sample sizes for homozygous genotypes differ. Because statistical power increases with increasing sample size, it is more likely to detect a statistically significant difference between heterozygotes and homozygous genotype with a larger sample size than between heterozygotes and homozygous genotype with a smaller sample size. To control for this possible caveat we made sample sizes for homozygous genotypes equal by randomly excluding individuals from homozygous groups with a bigger sample size and reran the pairwise comparisons. This did not change the classification of resistance profiles or our interpretation of the results. In enclosure populations, we used nonparametric Kaplan–Meier tests to analyze adult survivorship and tested for between-group differences in prevalence and pup production by using logit loglinear models or chi-square tests. We conducted all statistical tests using SPSS for Windows statistical software version 12.01 and present the results as mean ± SEM, with two-tailed probability values.
RESULTS
Laboratory experiments:
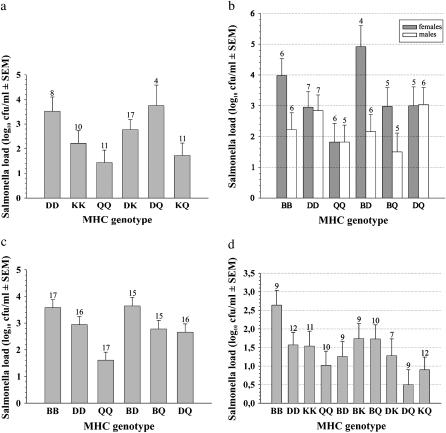
In the first laboratory experiment with males, there was a statistically significant MHC effect on resistance against single LT2 strain infection (pathogen load F = 5.58, d.f. = 2, P = 0.01; Figure 1a). This was due to QQ males having significantly better clearance compared to the DDs (Tukey's pairwise test: P = 0.007), whereas the QQs did not differ significantly from the KKs (P = 0.12). Neither Salmonella load (t-test: d.f. = 59, P = 0.57) nor spleen size (d.f. = 60, P = 0.88) differed between homozygotes and heterozygotes. However, there was a tendency for homozygotes to gain more weight than heterozygotes [mean increase in weight (grams), 1.43 ± 0.32 and 0.64 ± 0.30, respectively; heterozygosity, F = 3.38, P = 0.07; covariate preinfection body weight, F = 5.2, P = 0.03].
Figure 1.—
Salmonella resistance of congenic/wild mice in laboratory infection experiments. (a) Exp. 1: MHC genotype-specific bacterial loads (mean of log10 CFU/ml + SEM) in males challenged with a primary infection with single LT2 strain. (b) Exp. 2: Bacterial loads separately for females and males infected with a primary, single LT2 strain. (c) Exp. 3: Bacterial loads in females and males challenged with a primary single LT2 and with a secondary multiple-strain infection with LT2, PMAC45, and M525. (d) Exp. 4: Bacterial loads in females and males challenged with a primary single M525 and with a secondary multiple-strain infection with M525, PMAC51, and χ4665. Sample sizes are indicated above the error bars.
In the second laboratory experiment, when using both male and female mice and different genotypic combinations than in the first experiment, there was a marginally significant MHC effect on ability to clear a single LT2 strain infection (F = 3.06, d.f. = 36, P = 0.06; Figure 1b). This was because the males bearing the B haplotype showed significantly lower loads compared to females (sex, F = 7.75, n = 68, P = 0.007; presence of B, F = 1.0, P = 0.32; sex × presence of B interaction, F = 8.36, P = 0.005). When testing separately for each sex, the MHC effect was significant in females (F = 5.47, n = 19, P = 0.02), but not in males (F = 0.79, n = 18, P = 0.47). MHC effect in females was due to the QQs having lower loads compared to the BBs (P = 0.01), whereas the other pairwise comparisons were nonsignificant. On average, neither bacterial loads (t-test; d.f. = 31, P = 0.24), nor spleen weight (P = 0.23), nor weight gain (P = 0.19) differed significantly between heterozygous and homozygous females.
In the third laboratory experiment MHC genotype had a significant effect on resistance against secondary multiple-strain infection with LT2/PMAC45/M525 (F = 13.59, n = 50, P < 0.001; Figure 1c). The mice did not show a similar sex × B haplotype interaction effect as in a previous experiment (sex, F = 0.31, n = 96, P = 0.58; presence of B, F = 12.7, P = 0.001; sex × presence of B interaction, F = 0.12, P = 0.73). The QQs had statistically significantly lower Salmonella loads compared to the DDs (P = 0.004) and the BBs (P < 0.001), whereas loads in the BBs and the DDs did not differ (P = 0.24). On average, heterozygotes did not differ from homozygotes in bacterial loads (P = 0.27), change in body mass (P = 0.97), or spleen weight (P = 0.58).
In the fourth laboratory experiment, there was also a significant MHC effect on resistance to secondary multiple-strain infection with M525/PMAC51/χ4665 (F = 3.32, P = 0.03; Figure 1d). The QQs had statistically significantly lower loads compared to the BBs (P = 0.02), whereas the other pairwise comparisons were nonsignificant. There was a statistically nonsignificant tendency for heterozygotes to have lower mean Salmonella loads compared to homozygotes (log10 of CFU/ml, 1.22 ± 0.17 and 1.66 ± 0.19, respectively; d.f. = 96, P = 0.09), whereas there were no statistically significant differences in change of body mass (P = 0.19) or spleen weight (P = 0.29).
When we conducted haplotype-specific comparisons (i.e., compared the mean loads in heterozygotes separately against the respective parental homozygotes) to examine the resistance patterns in all four laboratory experiments, half were nonsignificant (7/15), and among the 8 significant comparisons the resistance was mostly recessive (6/8) rather than dominant (2/8), but never overdominant (0/8) (Table 1).
TABLE 1.
The summary of the results of laboratory infection experiments
| Resistance profiles of genotypic combinations
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Experiment | N | MHC | HA | BD | BK | BQ | DK | DQ | KQ |
| 1. 1°, single LT2 (♂) | 61 | * | NS | — | — | — | NS | R | NS |
| 2. 1°, single LT2 (♂ and ♀) | 68 | * (♀) | NS | R | — | R | — | R | — |
| 3. 1° LT2 + 2° mix (♂ and ♀) (LT2, PMAC45/M525) | 96 | *** | NS | NS | — | R | — | R | — |
| 4. 1° M525 + 2° mix (♂ and ♀) (M525/PMAC51/χ4665) | 98 | * | NS | D | NS | NS | NS | D | NS |
“MHC” denotes MHC effect on resistance and “HA” denotes heterozygote advantage (the average of all heterozygous genotypes vs. the average of all homozygous ones). Resistance profiles of genotypic combinations were tested by comparing the mean Salmonella loads of each heterozygous genotype separately against the means of the corresponding homozygotes (i.e., BD vs. BB and BD vs. DD) with independent samples t-tests. *P < 0.05; ***P < 0.001; NS, nonsignificant. Resistance: D, dominant; R, recessive; —, not tested.
Population enclosures:
Salmonella-infected mice showed significantly lower survival rates over 21 weeks in enclosure populations compared to sham-treated control mice (57 and 80%, respectively; n = 120, Kaplan–Maier P = 0.007). Females showed higher survival rates compared to males in control populations (P = 0.004), but not in infected ones (P = 0.78). Survival rates for homozygous and heterozygous mice did not differ in infected (P = 0.49; Figure 2) or in sham-treated control populations (P = 0.46; Figure 2).
Figure 2.—
Survival rates (percentage of individuals alive) for homozygous and heterozygous congenic/wild mice in enclosures for Salmonella-infected and control populations.
Fifty-nine percent (20/34) of the experimentally infected mice that survived were still Salmonella infected at the end of the 21-week enclosure experiment. The proportion of Salmonella-infected mice did not significantly differ between homozygotes and heterozygotes (9/14 and 11/20, respectively; χ2 = 0.29, d.f. = 1, P = 0.59), and there were no significant differences in mean bacterial loads between homozygotes and heterozygotes (d.f. = 32, P = 0.61).
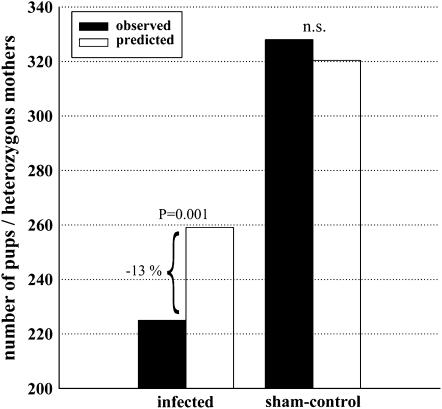
A total of 977 pups were produced in enclosure populations during the experiment, and infected mice produced 19% fewer offspring compared to sham controls (χ2 = 13.1, d.f. = 1, P < 0.001). In Salmonella-infected populations heterozygous females showed a statistically significant 13% deficiency in reproductive success relative to that expected (χ2 = 11.3, d.f. = 1, P = 0.001; Figure 3). In sham-treated populations, the relative proportion of pups produced by heterozygous females did not significantly differ from that expected (χ2 = 0.04, d.f. = 1, P = 0.85; Figure 3).
Figure 3.—
Reproductive success of MHC heterozygous females in enclosures separately in Salmonella-infected and control populations. The solid bars indicate the observed and the open ones the predicted number of pups produced by heterozygous females. The predicted number of pups is based on the relative proportion of heterozygous to homozygous females (0.6 and 0.4, respectively) within the populations.
DISCUSSION
Our results from laboratory infection tests in MHC homozygous congenic/wild mice showed similar Salmonella resistance patterns to those found previously in inbred C57BL/10 congenic mice (QQ > KK ≥ DD > BB; e.g., Penn et al. 2002). These provide the first experimental evidence that interacting wild-type background genes do not in general override the MHC effects. However, unexpectedly the presence of diverse interacting background genes drastically changed the resistance profiles for homozygous and heterozygous haplotypes. Contrary to previous studies in congenic mice using Salmonella (Penn et al. 2002; McClelland et al. 2003) and most other pathogens (Penn 2002; McClelland et al. 2003), haplotype-specific resistance profiles in congenic/wild mice were mostly recessive (i.e., heterozygotes were as susceptible as the worst corresponding homozygote), rather than dominant or overdominant. These results emphasize the importance of interacting background genes in determining the expressed resistance profiles and suggest that results obtained from the MHC congenic mice with low background gene diversity may not hold in more genetically diverse natural populations.
Interestingly, the resistance pattern also differed depending on the Salmonella strain and/or genotype combinations. When using a virulent strain (LT2) in single primary infections and in a secondary multiple-strain infection, the resistance was recessive, whereas in a secondary infection with a mixture of avirulent strains it was dominant (Table 1). This is consistent with an earlier finding, in which MHC heterozygous congenic mice showed, at first, a slight survival advantage when infected with avirulent strains, but the advantage disappeared during subsequent infections with more virulent strains (Penn et al. 2002). Although the exact mechanism is still not known, it could be related to virulence-dependent differences in ability of Salmonella to interfere with host T-cell responses. Salmonella, like many other pathogens, has evolved sophisticated mechanisms to manipulate host T-cell responses by interfering with antigen processing and presentation and by inducing apoptosis in antigen-presenting cells (Ravindran and Mcsorley 2005). To manipulate host T-cell responses bacteria must first infect host cells. Virulent strains with fast growth rates are likely to spread and invade host macrophages and dendritic cells much faster and in higher concentrations compared to avirulent ones and may consequently be more efficient in interfering with host T-cell responses. If the ability of Salmonella to manipulate host T-cell effector function was dependent solely on the virulence, one could assume that both homozygous and heterozygous resistance allele combinations would have been equally affected. However, resistance was altered only in heterozygous genotypes, whereas the homozygotes showed similar resistance patterns regardless of the virulence of the strain. Therefore, it is plausible that some kind of threshold-dependent dominance can explain why heterozygotes were not able to respond as effectively as the corresponding homozygote.
The poor performance of heterozygotes could be due to gene dose effects (Dorf et al. 1979; Moore et al. 1980) or differences in T-cell repertoire selection during early ontogeny (Vukusic et al. 1995). The latter explanation is of particular relevance for our congenic/wild mice because the heterozygous individuals were heterozygous at all MHC loci with highly disparate haplotypes. It has been suggested that there is an optimal number of MHC molecules, so that having a higher number could have diminishing returns on the diversity of the T-cell repertoire generated during thymic selection, because of a need to eliminate T-cells that react to self-peptide-MHC molecule combinations (Lawlor et al. 1990). This idea was proposed to explain the lack of MHC duplication, although it was later suggested that there might also be optimal levels of MHC heterozygosity (Nowak et al. 1992; De Boer and Perelson 1993; but see Borghans et al. 2003) and, if so, that mating preferences might function to optimize the heterozygosity of offspring (Penn and Potts 1998). Recent work on fish found evidence for an optimal number of MHC molecules (Wegner et al. 2003), although it is still unclear whether this is due to heterozygosity or number of loci. Similar but less convincing results have also been found in birds (Bonneaud et al. 2004). Our MHC heterozygous congenic/wild mice had maximal heterozygosity at all class I and II loci with high allelic disparity and therefore may have expressed excess levels of heterozygosity resulting in a reduced T-cell repertoire and suboptimal resistance. Trade-offs in increasing heterozygosity are expected to be more important in mice with a wild background than in lab strains, as there would be higher diversity of self-antigens presented during the development of the T-cell repertoire. Our experimental design does not allow differentiating the relative impact of background heterozygosity from that of MHC heterozygosity, but earlier findings indicate that both have an additive effect on T-cell repertoire selection and effector function (Vukusic et al. 1995).
We also found a sex-dependent effect on resistance against primary single-strain infection in exp. 2; the males bearing the susceptibility haplotype B, either as a homozygote or in any heterozygous combination, showed significantly better resistance than females carrying the same haplotype (Figure 1b). However, this sex × B haplotype interaction vanished when the mice were given a secondary infection with a mixture of multiple strains (exp. 3). Although difficult to interpret, this result shows that determination of resistance is a complex phenomenon involving haplotype, sex, and infection type (primary vs. secondary and single vs. multiple strains) specific effects. These complex interactions often make the outcome of infectious disease highly unpredictable.
Since resistance in congenic/wild mice was mostly recessive (Table 1), this explains why there was no general resistance advantage for heterozygotes. In the colony, heterozygotes had a statistically nonsignificant advantage in resistance against secondary infection with multiple strains (exp. 4), whereas in exp. 1, heterozygous males showed a statistically nonsignificant disadvantage in weight gain during a primary infection with a single strain. Furthermore, in an enclosure experiment survival rates of heterozygous and homozygous individuals did not differ, but infected heterozygous females showed a statistically significant disadvantage in reproductive success compared to homozygotes (Figure 3). Although the surviving heterozygous females did not show higher bacterial loads compared to homozygous ones, it is likely that differences in ability to cope with infection were involved in causing the observed heterozygote disadvantage in reproduction for two reasons. First, infection reduced breeding success as seen in lower pup production in infected vs. sham-treated control populations and second, reproduction disadvantage for heterozygous females was found in infected but not in control populations.
In conclusion, our results do not support the HA hypothesis or the idea that an adaptive function of MHC-based mating preferences would be to produce MHC heterozygous offspring. Moreover, our finding for altered resistance type in congenic/wild mice with diverse background loci implies that congenic models may be overly simplified to capture the complex and unpredictable mode of genetic determination of resistance in natural populations. In the future, there is clearly a need for studies investigating determinants of pathogen resistance and modes of selection in wild genetically diverse host populations. Our conclusions largely hinge on results from haplotype q, and there might be something peculiar about these particular haplotypes. Therefore, to make generalizations, it is still necessary to test wild genotypes. These studies should also use other pathogens, or preferably a mixture of different strains or species, to determine the generality of our results with Salmonella. Our conclusions are based on data from a single (although mixed strains) pathogen species. The addition of multiple pathogens has been shown to yield HA even though no HA was found during pure infections of each single pathogen (McClelland et al. 2003). Furthermore, it will be of particular interest to determine the relative influence of MHC heterozygosity and background diversity on resistance and to investigate whether other vertebrates show similar immunogenetic optimality found in fish (Wegner et al. 2003). These studies should also be designed to allow allele-specific comparisons to define whether the possible population-level HA is due to resistance being dominant or overdominant or just a result of a confounding effect of allele frequency distribution in a given population (Lipsitch et al. 2003). Overrepresentation of MHC heterozygotes among individuals carrying lower pathogen loads has often been interpreted as evidence for overdominant selection in population-level studies. However, on the basis of our results, it is evident that neither general HA nor dominance of resistance can be assumed.
Acknowledgments
All experiments complied with federal regulations and the University of Utah's Institutional Animal Care and Use Committee guidelines. This research was supported by National Science Foundation grants IBN-9904609 and IBN-0344907, National Institutes of Health grant GM39578, The Academy of Finland, and the Austrian Academy of Sciences.
References
- Apanius, V., D. Penn, P. R. Slev, L. R. Ruff and W. K. Potts, 1997. The nature of selection on the major histocompatibility complex. Crit. Rev. Immunol. 17: 179–224. [DOI] [PubMed] [Google Scholar]
- Arkush, K. D., A. R. Giese, H. L. Mendonca, A. M. McBride, G. D. Marty et al., 2002. Resistance to three pathogens in the endangered winter-run chinook salmon (Oncorhynchus tshawytscha): effects of inbreeding and major histocompatibility complex genotypes. Can. J. Fish. Aquat. Sci. 59: 966–975. [Google Scholar]
- Bernatchez, L., and C. Landry, 2003. MHC studies in nonmodel vertebrates: What have we learned about natural selection in 15 years? J. Evol. Biol. 16: 363–377. [DOI] [PubMed] [Google Scholar]
- Bonneaud, C., J. Mazuc, O. Chastel, H. Westerdahl and G. Sorci, 2004. Terminal investment induced by immune challenge and fitness traits associated with major histocompatibility complex in the house sparrow. Evol. Int. J. Org. Evol. 58: 2823–2830. [DOI] [PubMed] [Google Scholar]
- Borghans, J. A., A. J. Noest and R. J. De Boer, 2003. Thymic selection does not limit the individual MHC diversity. Eur. J. Immunol. 33: 3353–3358. [DOI] [PubMed] [Google Scholar]
- Borghans, J. A., J. B. Beltman and R. J. De Boer, 2004. MHC polymorphism under host-pathogen coevolution. Immunogenetics 55: 732–739. [DOI] [PubMed] [Google Scholar]
- Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov et al., 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283: 1748–1752. [DOI] [PubMed] [Google Scholar]
- Charbit, A., S. M. Newton, P. E. Klebba, J. M. Clement, C. Fayolle et al., 1997. Expression and immune response to foreign epitopes in bacteria. Perspectives for live vaccine development. Behring Inst. Mitt. 98: 135–142. [PubMed] [Google Scholar]
- Clarke, B., and D. R. Kirby, 1966. Maintenance of histocompatibility polymorphisms. Nature 211: 999–1000. [DOI] [PubMed] [Google Scholar]
- De Boer, R. J., and A. S. Perelson, 1993. How diverse should the immune system be? Proc. Biol. Sci. 252: 171–175. [DOI] [PubMed] [Google Scholar]
- De Boer, R. J., J. A. Borghans, M. van Boven, C. Kesmir and F. J. Weissing, 2004. Heterozygote advantage fails to explain the high degree of polymorphism of the MHC. Immunogenetics 55: 725–731. [DOI] [PubMed] [Google Scholar]
- Doherty, P. C., and R. M. Zinkernagel, 1975. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature 256: 50–52. [DOI] [PubMed] [Google Scholar]
- Dorf, M. E., J. H. Stimpfling and B. Benacerraf, 1979. Gene dose effects in Ir gene-controlled systems. J. Immunol. 123: 269–271. [PubMed] [Google Scholar]
- Edwards, S. V., K. Chesnut, Y. Satta and E. K. Wakeland, 1997. Ancestral polymorphism of Mhc class II genes in mice: implications for balancing selection and the mammalian molecular clock. Genetics 146: 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve, B., J. Reddy, H. P. Waldner, R. A. Sobel and V. K. Kuchroo, 2004. Dissimilar background genes control susceptibility to autoimmune disease in the context of different MHC haplotypes: NOD.H-2(s) congenic mice are relatively resistant to both experimental autoimmune encephalomyelitis and type I diabetes. Eur. J. Immunol. 34: 1828–1838. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1949. Disease and evolution. Ric. Sci. 19: 68–75. [Google Scholar]
- Hedrick, P. H., and T. J. Kim, 2000. Genetics of complex polymorphisms: parasites and maintenance of the histocompatibility complex variation, pp. 204–234 in Evolutionary Genetics: From Molecules to Morphology, edited by R. S. Singh and C. B. Krimbas. Cambridge University Press, Cambridge.
- Hedrick, P. W., 2002. Pathogen resistance and genetic variation at MHC loci. Evol. Int. J. Org. Evol. 56: 1902–1908. [DOI] [PubMed] [Google Scholar]
- Hill, A. V. S., C. E. M. Allsopp, D. Kwiatkowski, N. M. Anstey, P. Tumasi et al., 1991. Common West-African HLA antigens are associated with protection from severe malaria. Nature 352: 595–600. [DOI] [PubMed] [Google Scholar]
- Hormaeche, C. E., M. C. Fahrenkrog, R. A. Pettifor and J. Brock, 1981. Acquired immunity to Salmonella typhimurium and delayed (footpad) hypersensitivity in BALB/c mice. Immunology 43: 547–554. [PMC free article] [PubMed] [Google Scholar]
- Hormaeche, C. E., K. A. Harrington and H. S. Joysey, 1985. Natural resistance to salmonellae in mice: control by genes within the major histocompatibility complex. J. Infect. Dis. 152: 1050–1056. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., and M. Nei, 1992. Models of host-parasite interaction and MHC polymorphism. Genetics 132: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway, C. A., P. Travers, M. Walport and M. Sclomchik, 2005. Immunobiology: The Immune System in Health and Disease. Garland Science, New York.
- Jeffery, K. J., A. A. Siddiqui, M. Bunce, A. L. Lloyd, A. M. Vine et al., 2000. The influence of HLA class I alleles and heterozygosity on the outcome of human T cell lymphotropic virus type I infection. J. Immunol. 165: 7278–7284. [DOI] [PubMed] [Google Scholar]
- Jepson, A., W. Banya, F. Sisay-Joof, M. Hassan-King, C. Nunes et al., 1997. Quantification of the relative contribution of major histocompatibility complex (MHC) and non-MHC genes to human immune responses to foreign antigens. Infect. Immun. 65: 872–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, J., 1986. Natural History of the Histocompatibility Complex. Wiley, New York.
- Langefors, A., J. Lohm, M. Grahn, O. Andersen and T. von Schantz, 2001. Association between major histocompatibility complex class IIB alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proc. Biol. Sci. 268: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor, D. A., J. Zemmour, P. D. Ennis and P. Parham, 1990. Evolution of class-I MHC genes and proteins: from natural selection to thymic selection. Annu. Rev. Immunol. 8: 23–63. [DOI] [PubMed] [Google Scholar]
- Lipsitch, M., C. T. Bergstrom and R. Antia, 2003. Effect of human leukocyte antigen heterozygosity on infectious disease outcome: the need for allele-specific measures. BMC Med. Genet. 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohm, J., M. Grahn, A. Langefors, O. Andersen, A. Storset et al., 2002. Experimental evidence for major histocompatibility complex-allele-specific resistance to a bacterial infection. Proc. Biol. Sci. 269: 2029–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland, E. E., D. J. Penn and W. K. Potts, 2003. Major histocompatibility complex heterozygote superiority during coinfection. Infect. Immun. 71: 2079–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, N. A., and J. Roes, 2002. Patterned variation in murine MHC promoters. Proc. Natl. Acad. Sci. USA 99: 10561–10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M. J., D. E. Singer and R. M. Williams, 1980. Linkage of severity of experimental allergic encephalomyelitis to the rat major histocompatibility locus. J. Immunol. 124: 1815–1820. [PubMed] [Google Scholar]
- Nowak, M. A., K. Tarczy-Hornoch and J. M. Austyn, 1992. The optimal number of major histocompatibility complex molecules in an individual. Proc. Natl. Acad. Sci. USA 89: 10896–10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham, P., and T. Ohta, 1996. Population biology of antigen presentation by MHC class I molecules. Science 272: 67–74. [DOI] [PubMed] [Google Scholar]
- Paterson, S., K. Wilson and J. M. Pemberton, 1998. Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a large unmanaged ungulate population (Ovis aries L.). Proc. Natl. Acad. Sci. USA 95: 3714–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn, D. J., 2002. The scent of genetic compatibility: sexual selection and the major histocompatibility complex. Ethology 108: 1–21. [Google Scholar]
- Penn, D., and W. Potts, 1998. How do major histocompatibility complex genes influence odor and mating preferences? Adv. Immunol. 69: 411–436. [DOI] [PubMed] [Google Scholar]
- Penn, D. J., K. Damjanovich and W. K. Potts, 2002. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl. Acad. Sci. USA 99: 11260–11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts, W. K., and E. K. Wakeland, 1990. The maintenance of MHC polymorphism. Immunol. Today 11: 39–40. [DOI] [PubMed] [Google Scholar]
- Pullen, J. K., R. M. Horton, Z. L. Cai and L. R. Pease, 1992. Structural diversity of the classical H-2 genes: K, D, and L. J. Immunol. 148: 953–967. [PubMed] [Google Scholar]
- Ravindran, R., and S. J. McSorley, 2005. Tracking the dynamics of T-cell activation in response to Salmonella infection. Immunology 114: 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, M. F., and D. Malo, 2002. Genetic regulation of host responses to Salmonella infection in mice. Genes Immun. 3: 381–393. [DOI] [PubMed] [Google Scholar]
- Saha, B. K., and S. E. Cullen, 1986. Molecular mapping of murine I region recombinants: crossing over in the E beta gene. J. Immunol. 136: 1112–1116. [PubMed] [Google Scholar]
- She, J. X., S. A. Boehme, T. W. Wang, F. Bonhomme and E. K. Wakeland, 1991. Amplification of major histocompatibility complex class II gene diversity by intraexonic recombination. Proc. Natl. Acad. Sci. USA 88: 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, S., 2005. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front. Zool. 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi, S., A. Edelstein, M. Rhen, S. J. Normark and J. D. Pfeifer, 1997. Development of a murine model of chronic Salmonella infection. Infect. Immun. 65: 838–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thursz, M. R., H. C. Thomas, B. M. Greenwood and A. V. Hill, 1997. Heterozygote advantage for HLA class-II type in hepatitis B virus infection. Nat. Genet. 17: 11–12. [DOI] [PubMed] [Google Scholar]
- Vidal, S. M., D. Malo, K. Vogan, E. Skamene and P. Gros, 1993. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell 73: 469–485. [DOI] [PubMed] [Google Scholar]
- Vukusic, B., L. Poplonski, L. Phillips, J. Pawling, T. Delovitch et al., 1995. Both MHC and background gene heterozygosity alter T cell receptor repertoire selection in an antigen-specific response. Mol. Immunol. 32: 1355–1367. [DOI] [PubMed] [Google Scholar]
- Wegner, K. M., M. Kalbe, J. Kurtz, T. B. Reusch and M. Milinski, 2003. Parasite selection for immunogenetic optimality. Science 301: 1343. [DOI] [PubMed] [Google Scholar]
- Xu, H. R., and H. S. Hsu, 1992. Dissemination and proliferation of Salmonella typhimurium in genetically resistant and susceptible mice. J. Med. Microbiol. 36: 377–381. [DOI] [PubMed] [Google Scholar]