Abstract
The indica rice variety Kasalath carries Pi36, a gene that determines resistance to Chinese isolates of rice blast and that has been located to a 17-kb interval on chromosome 8. The genomic sequence of the reference japonica variety Nipponbare was used for an in silico prediction of the resistance (R) gene content of the interval and hence for the identification of candidate gene(s) for Pi36. Three such sequences, which all had both a nucleotide-binding site and a leucine-rich repeat motif, were present. The three candidate genes were amplified from the genomic DNA of a number of varieties by long-range PCR, and the resulting amplicons were inserted into pCAMBIA1300 and/or pYLTAC27 vectors to determine sequence polymorphisms correlated to the resistance phenotype and to perform transgenic complementation tests. Constructs containing each candidate gene were transformed into the blast-susceptible variety Q1063, which allowed the identification of Pi36-3 as the functional gene, with the other two candidates being probable pseudogenes. The Pi36-encoded protein is composed of 1056 amino acids, with a single substitution event (Asp to Ser) at residue 590 associated with the resistant phenotype. Pi36 is a single-copy gene in rice and is more closely related to the barley powdery mildew resistance genes Mla1 and Mla6 than to the rice blast R genes Pita, Pib, Pi9, and Piz-t. An RT–PCR analysis showed that Pi36 is constitutively expressed in Kasalath.
THE filamentous ascomycete Magnaporthe grisea (Hebert) Barr, which is the causal agent of rice blast, remains the most important pathogen in most rice-producing (Oryza sativa L.) regions (Ou 1985). The use of resistance (R) genes in crop breeding programs has been, and will undoubtedly remain, the major means for its control. However, because of the instability of the pathogen and a high level of variability in pathogenicity between isolates (Ou 1979; Valent and Chumley 1994), host resistance is typically only short-lived in disease-prone environments (Kiyosawa 1981; Mackill and Bonman 1992). The establishment of durable resistance requires the isolation of multiple R genes, as this simplifies the process of R gene stacking into elite cultivars, via either marker-aided breeding or transgenesis.
The rice–rice blast interaction has long served as a model system to study plant–pathogen interactions (Valent 1990). Race-specific resistance closely follows the classical gene-for-gene relationship (Silué et al. 1992; Jia et al. 2000). The isolation and subsequent characterization of R genes will help to unravel the molecular mechanisms underlying the interaction between host and pathogen. Although more than 50 rice R genes have been documented to date (Chen et al. 2005; Liu et al. 2005), only 6 (Pib, Pita, Pi9, Pid2, Pi2, and Pizt) have as yet been isolated (Wang et al. 1999; Bryan et al. 2000; Qu et al. 2006; Chen et al. 2006; Zhou et al. 2006). The sequences of 5 of these (Pib, Pita, Pi9, Pi2, and Pizt) include both nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains, while Pid2 encodes a receptor-like kinase.
Plants use R genes to detect the presence of pathogen, and then to induce a spectrum of defense responses. The interaction between R gene products and pathogen elicitors has been established by a variety of direct and indirect experimental evidence (Jia et al. 2000; Gu et al. 2005; Dodds et al. 2006). The commonest class of R gene encodes proteins containing an NBS–LRR domain (Bent 1996; Hammond-Kosack and Jones 1997; Hulbert et al. 2001). These have been classified into two types on the basis of the presence/absence of an N-terminal TIR domain. Genes in the TIR group are only known among the dicotyledonous species (Meyers et al. 1999; Pan et al. 2000; Bai et al. 2002). The non-TIR group typically includes a coiled-coil (CC) domain at the N terminus. The NBS region is thought to be involved in signal transduction cascades involving phosphorylation/dephosphorylation events with either ATP or GTP (Traut 1994; Dangl and Jones 2001), whereas the CC domain may facilitate homodimerization of the proteins themselves or heterodimerization with other proteins, generating interactions that lead to the repression of signaling (Moffett et al. 2002; Hwang and Williamson 2003). Several studies have identified the LRR domain as the major determinant of recognition specificity for the pathogen avirulence factor(s) (Meyers et al. 1998). LRR-containing sequences are prone to adaptive evolution (Parniske et al. 1997; Mcdowell et al. 1998; Ellis et al. 2000; Sun et al. 2001), and in particular, their insertions and deletions have been shown to be responsible for both R gene loss of function and recognition specificity (Anderson et al. 1997; Wulff et al. 2001). For example, particular loss-of-function alleles of the Arabidopsis thaliana genes RPS2 and RPM1 differ from the effective wild type by only one amino acid residue in the LRR domain (Bent et al. 1994; Grant et al. 1995).
The indica rice variety Kasalath (formerly coded as Q61) confers a stable and high level of partial resistance against Chinese isolates of rice blast. The resistance gene Pi36 has recently been mapped to a location on chromosome 8 (Liu et al. 2005). In this paper, we describe the positional cloning of Pi36 gene based on a prior bioinformatics analysis, long-range PCR (LR–PCR), and an efficient transformation-competent artificial chromosome (TAC) vector-based transformation technique. We believe that this approach should be widely applicable within rice and also other plant species. The cloned Pi36 gene represents an important resource for the development of durable resistance to rice blast, and along with other R genes its sequence should inform the molecular basis of disease resistance in plants.
MATERIALS AND METHODS
Candidate gene prediction:
Candidates for Pi36 were identified in silico from the output of the gene prediction programs FGENESH, RiceGAAS, and Gramene, using as a reference the Nipponbare sequence for the 17-kb genomic region defined by the flanking markers CRG2 and RM5647 (Figure 1A). To verify which, if any, of these are true candidates for Pi36, the sequence from the same genetic interval was derived from the blast-resistant variety Kasalath and two blast-susceptible varieties, Aichi Asahi and Lijiangxintuanheigu (LTH), using a PCR walking approach. The interval was divided into 11 overlapping amplifiable fragments, and sequence comparisons were performed between the alleles from the resistant and susceptible genotype at each candidate gene following a sequence assembly generated by DNAStar software (http://www.DNAStar.com).
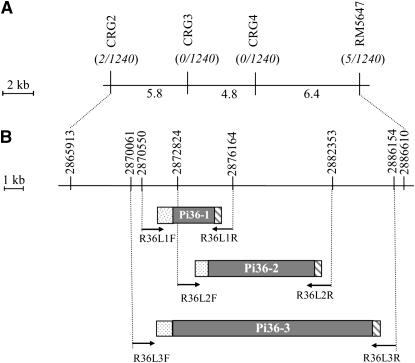
Figure 1.—
In silico map-based cloning of Pi36. (A) Physical and genetic map surrounding the Pi36 locus. The numbers below the map represent distances in kilobases, as estimated from the Nipponbare genome sequence. The numbers in parentheses represent the number of recombinants/gametes in the mapping population previously reported (Liu et al. 2005). (B) Pi36 candidate genes. Pi36-1 and Pi36-2 were predicted by RiceGAAS and Pi36-3 by both FGENESH and Gramene. The shaded box represents the coding region, and the hatched boxes represent predicted 5′ promoter and 3′ poly(A) regions, respectively. The numbers above the map refer to location on the reference Nipponbare genomic sequence. The targets for the LR–PCR primers are indicated.
Candidate gene cloning:
Primer sets were designed to amplify each candidate gene, including their promoter and terminator regions, using the above-mentioned gene annotation. Restriction sites to enable cloning were identified from the Kasalath genomic sequence. Pi36 candidates were amplified from Kasalath genomic DNA by LR–PCR. The 50-μl LR–PCR reactions contained 2.5 units TaKaRa LA Taq (TaKaRa, Dalian, China), 1× GC reaction buffer I, 400 μm dNTP, 100-ng template, and 0.2 μm of each primer. The primer sequences, PCR conditions, and restriction enzymes used are listed in Table 1. Purified LR–PCR products of candidates Pi36-1 and Pi36-2 from three independent reactions were ligated into the BamHI and PstI sites of the vector pCAMBIA1300 to form R36L1CAM and R36L2CAM, respectively. For the longest candidate, Pi36-3, the LR–PCR products were cloned into the AscI site of TAC vector pYLTAC27 to form R36L3TAC. To improve transformation efficiency, the Pi36-3 insert was later recloned into the vector pCAMBIA1300AscI, in which an AscI site was engineered into the multiple cloning sites. This construct was named R36L3CAM. The constructs were validated by restriction analysis and sequenced from both ends using the vector primers CAM1300F and CAM1300R. Details of the vectors, constructs and primers used are listed elsewhere (Table 1; supplemental Table S1 at http://www.genetics.org/supplemental/).
TABLE 1.
Primer sequences used for the amplification of candidate genes for Pi36 by long-range PCR
| Candidate gene | Primer | Sequence (5′–3′)a | Expected size (kb) | Restriction siteb | PCR conditionsc | Vector |
|---|---|---|---|---|---|---|
| Pi36-1 | R36L1F | CGGGATCCTCAACAGACTTGACATGCACATGCTGATT | 5.9 | BamHI | A | pCAMBIA1300 |
| R36L1R | CGGGATCCAAGAATGTAACGTGTGCCTCAGACTCGGTG | |||||
| Pi36-2 | R36L2F | GCAGTCACTGCAGGTCCTACGACATGGAGGATATCATCGACGCCTTC | 9.5 | PstI | B | pCAMBIA1300 |
| R36L2R | GTCAGGTCTGCAGCATGTGGCCAGACTCTGTTGGTGGATTGAAGC | |||||
| Pi36-3 | R36L3F | GCTAGCATGGCGCGCCCTTCGACACGCAAACGTGCACACAGCCACCTATC | 16.1 | AscI | C | pYLTAC27/pCAMBIA1300-AscI |
| R36L3R | GCTTGCTAGGCGCGCCTGCGATGCCACTTCGCTCTTTGCCGATCTGGTTG |
Nucleotides corresponding to a restriction enzyme recognition site are underlined.
Enzyme used for cloning.
All PCRs included an initial denaturation step (94°/2 min), followed by 30 amplification cycles under the conditions indicated. At the end of the cycling procedure, a final incubation of 72°/10 min was given. A, 94°/30 sec, 65°/6.5 min; B, 94°/30 sec, 63.8°/10.5 min; C, 94°/30 sec, 62.8°, 17 min.
Complementation analysis:
Constructs containing each candidate gene were transformed into Agrobacterium tumefaciens strain EHA105 by electroporation (GenePulser Xcell, Bio-Rad, Hercules, CA). Clone stability was tested as per Qu et al. (2003), and stable constructs were transformed into the highly blast-susceptible rice variety Q1063, as described by Hiei et al. (1994). Selfed T1 and T2 progeny were tested for reaction to blast infection with pathogen isolates CHL39 and CHL273, using the spray method described elsewhere (Pan et al. 2003; Liu et al. 2005). For these phenotyping tests, Kasalath and Q1063 represented, respectively, the positive and negative controls. A number of resistant transgenic individuals were randomly selected and subjected both to PCR verification for the presence of the transgene, using the gene-specific primers CRG4F and CRG4R (supplemental Table S1 at http://www.genetics.org/supplemental/), and to Southern hybridization analysis to estimate the transgene copy number. For the latter procedure, ∼3 μg genomic DNA was digested to completion with HindIII; the products were separated by 0.8% agarose gel electrophoresis and were then transferred to a nylon membrane (Hybond-N+, Amersham, Buckinghamshire, UK). A part of the HptII sequence, amplified from the vector pCAMBIA1300 by primers HptF and HptR (see supplemental Table S1), was labeled with α-[32P] by random primer labeling (TaKaRa) for use as a hybridization probe. Southern hybridization was also used to infer the copy number of Pi36-like genes in rice. Genomic DNA of Kasalath and the blast-susceptible variety AS20-1 was digested to completion with EcoRI, KpnI, or BamHI and probed with sequences amplified from the 5′-untranslated region (UTR), 3′ UTR, and a part of the largest intron (L-intron) of the Pi36 gene (see supplemental Table S1).
Gene expression analysis:
Two-week-old seedlings of Kasalath and the blast-susceptible variety LTH were inoculated with pathogen isolate CHL39 and maintained in a greenhouse. Leaf samples were collected at 0, 6, 12, 24, 48, and 72 hpi. Total RNA was isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. The assessment of gene expression levels was obtained in a two-step reverse transcription PCR (RT–PCR) process. The initial RT reaction used the SuperScript II reverse transcriptase kit (Invitrogen), following the manufacturer's instructions. For the second PCR reaction, a 0.5–2 μl aliquot of the first reaction was used as template. Each experiment was performed in replicate. To enable discrimination between the various RT–PCR amplicons, the RT–PCR primers (see supplemental Table S1 at http://www.genetics.org/supplemental/ and Figure 4C) were designed from exonic sequence flanking the predicted Pi36 introns, and genomic DNA was included as a negative control. Primers for rice actin (supplemental Table S1) were used as a positive RT–PCR control. Semiquantitative RT–PCR was performed with 23, 26, 29, 32, and 35 cycles.
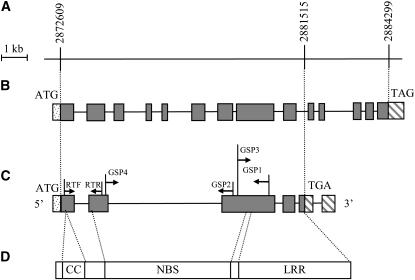
Figure 4.—
The structure of Pi36, as deduced from its genomic and cDNA sequences. (A) The target gene region. The numbers above the map show genomic positions in the Nipponbare genomic sequence. (B) Gene structure as deduced from the genomic DNA sequence. (C) Gene structure as deduced from the cDNA sequence. The shaded box indicates an exon, and the line an intron. The positions of 5′ and 3′ UTR (hatched boxes), the translation start codon (ATG), and the translation stop codon (TGA or TAG) are also shown. The annealing targets of the RACE and RT-PCR primers are indicated. (D) Structure of the Pi36-encoded protein, in which three tandem conserved domains are shown.
Rapid amplification of cDNA ends:
The 5′ and 3′ end sequences of the cDNA were determined by rapid amplification of cDNA ends (RACE) using the GeneRacer kit (Invitrogen), following the manufacturer's instructions. We used total RNA extracted from the leaves of resistant (Kasalath) and susceptible (LTH and AS20-1) plants, harvested 24 hpi. The same RACE primers were used for both the resistant and susceptible templates. The full-length cDNA was divided into three amplifiable fragments. The 5′ RACE was generated by a nested PCR using the primary primer set of GeneRacer 5′ primer and GSP1, and the second set of the GeneRacer 5′ nested primer and GSP2; similarly, the 3′ RACE was generated by the set GSP3 and the GeneRacer 3′ primer, and the intermediate RT–PCR fragment was obtained with the set GSP4 and GSP1 (see supplemental Table S1 at http://www.genetics.org/supplemental/). The relative locations of all the gene-specific primers are shown in Figure 3C (note that the intermediate RT–PCR fragment overlaps both the 5′ and 3′ RACE fragments). The RACE products were ligated into the pGEM-T vector (Promega, Madison, WI), following the manufacturer's instructions, and sequenced.
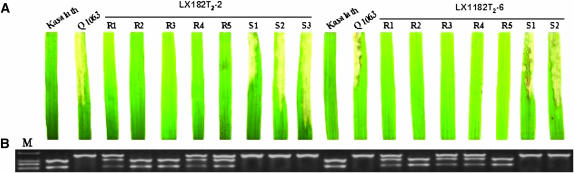
Figure 3.—
Pi36 gene complementation test and molecular analysis of the transgenic lines. (A) Resistance phenotypes of the Pi36 gene donor cv. Kasalath and its receptor cv. Q1063 as well as transgenic plants of two T2 lines segregated into 3R:1S against isolate CHL39. R, resistant; S, susceptible. (B) Cosegregation analysis of the resistance phenotype with the transgenes. The amplified fragment with the primer pair CRG4F and CRG4R was subsequently digested with HaeIII, and the digested product was subjected to 1.5% agarose gel electrophoresis. M represents standard molecular weight marker DL2000.
DNA and protein sequence analysis:
DNA sequence similarity analysis was performed using software BLASTN and BLASTX (Altschul et al. 1997; http://www.ncbi.nlm.nih.gov/BLAST). The promoter and polyadenylation regions were analyzed using TSSP and POLYAH, respectively (http://www.softberry.com/berry.html). Genomic sequence comparisons were performed with pairwise BLAST (http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/bl2.html), and protein sequence similarity analysis was performed with BLASTP (Altschul et al. 1997). Multiple sequence alignments were obtained with ProbCons (Do et al. 2005), and based on these outputs, a phylogenetic tree was generated using the molecular evolutionary genetic analysis (MEGA) program (Nei and Kumar 2000). Bootstrapping was used to provide a confidence estimate for each branch point. The theoretical isoelectric point and protein molecular weight were computed using DNAStar software. The CC structure was predicted by COILS (http://www.ch.embnet.org/software/COILS_form.html).
RESULTS
Pi36 candidate genes:
The Pi36 locus has been mapped within a 17-kb interval (Figure 1A). To identify candidates for the gene, the Nipponbare version of the 17-kb interval was scanned by gene prediction software, revealing the three NBS–LRR-type sequences Pi36-1, Pi36-2, and Pi36-3. Both Pi36-1 and Pi36-2 were identified by RiceGAAS (Figure 1B), while Pi36-3 was identified by both Gramene and FGENESH. The Pi36-3 sequence includes both Pi36-1 and Pi36-2. LR–PCR products, each representing one of the alleles of the three candidate sequences, were successfully generated; these had the anticipated lengths of 5.9, 9.5, and 16.1 kb. To exclude PCR artifact as a source of sequence variation, three independent LR–PCR products were cloned for each of the candidate genes from each of the templates, and these were independently sequenced. The allelic versions of each of the three candidate genes were then compared. The Kasalath alleles differ from those in the susceptible varieties by a number of base substitutions and small indels (data not shown). The level of nucleotide sequence similarity between the alleles was 97% (between Kasalath and either Nipponbare or Aichi Asahi) and 98% (Kasalath vs. LTH). Gene prediction identified the same set of three candidate genes in each of the three alternative versions of the Nipponbare 17-kb segment.
Complementation analysis of the candidate genes:
The constructs containing each candidate gene were individually transformed into the highly susceptible variety Q1063. A total of 259 and 39 independent T0 transformants, respectively, were generated using R36L1CAM and R36L2CAM. For the analysis of Pi36-3, 24 (R36L3TAC) and 63 (R36L3CAM) independent T0 individuals were obtained. The pattern of segregation for rice blast resistance was observed among the T1 progeny of each primary transgenic. All T1 individuals derived from a T0 parent carrying R36L1CAM or R36L2CAM were highly susceptible to blast isolates CHL39 and CHL273 (which are both avirulent on Kasalath and virulent on Q1063). Ten of 24 tested T1 families derived from a T0 parent carrying R36L3TAC segregated resistant vs. susceptible in a ratio between 1:3.5 and 2.8:1, while the segregation ratio shown by 33 T1 families derived from a T0 parent carrying R36L3CAM varied from 1:3 to 4:1 (data not shown, supplemental Figure S1 at http://www.genetics.org/supplemental/).
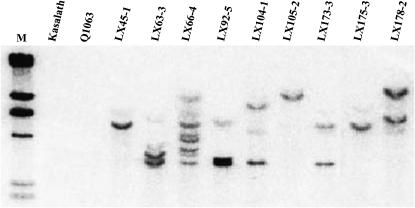
To confirm the presence and stable integration of the transgene Pi36-3, molecular assay was first conducted by Southern blot analysis. The results showed that all the resistant transgenic plants harbored the transgene of interest, most of which contained between one and three copies of the transgene Pi36-3, although few transgenic plants contained multiple copies (Figure 2). To further verify steady inheritance of the transgene Pi36-3, two T2 lines, LX182 T2-2 and LX182 T2-6, whose progeny segregated 3:1 for resistance, were chosen for a cosegregation analysis between blast resistance and the presence of the marker CRG4, which lies within Pi36 (Figure 1A). Since resistance cosegregated perfectly with the presence of CRG4 (Figure 3), Pi36-3 must represent a functional gene.
Figure 2.—
Southern blot analysis of blast resistant transgenic plants. Genomic DNA was isolated from resistant transformed and susceptible non-transformed plants. A fragment of hptII was used as a probe. Lane 1, molecular weight marker λHindIII; lane 2, Kasalath (resistant); lane 3, Q1063 (susceptible); lanes 4–12, transgenic T1 progeny.
Structure of Pi36:
Six RACE products from both the 5′ and 3′ end of the genomic sequence of Kasalath were sequenced to identify the size and structure of Pi36. The 5′ and 3′ RACE products as well as an intermediate RT–PCR fragment overlapped one another, thereby providing complete coverage of the transcribed region (Figure 4C). The size and structure of Pi36 were determined by comparing the full-length cDNA sequence with the genomic DNA sequence. Pi36 contains a 3171-bp coding region, interrupted by four introns (433, 5069, 124, and 259 bp) and flanked by a 65-bp 5′ UTR and a 725-bp 3′ UTR. A 123-bp intron is present within the 3′ UTR (Figure 4C). The size and structure of Pi36 are rather different from those predicted by annotation of the genomic sequence (Figure 4, B and C), which consists of 13 introns and 14 exons, with the translation stop codon at position 2,884,299. In the cDNA-derived structure, the stop codon is shifted 5′ by 2784 bp. Both start codons, however, lie at position 2,872,609 (Figure 4, A–C).
Sequence analysis of the Pi36-encoded protein:
A comparison of the deduced amino acid sequence of the Pi36 alleles from two susceptible japonica varieties [LTH (pi36j1) and Nipponbare (pi36j2)] and the resistant Kasalath identified nine substitutions and two deletions among the alleles. In addition, six substitutions distinguish the alleles Pi36 and pi36i from the blast-susceptible indica variety AS20-1. A global analysis suggests that just one substitution at residue 590 defines the functional Pi36 gene. The deduced 1056-amino acid sequence of the Pi36-encoded protein has a molecular mass of 120 kDa and a calculated isoelectric point of 6.61, and contains six conserved motifs typical of NBS proteins (Figure 5). The GMGGLGKTT sequence (beginning at residue 206) conforms to the kinase 1a (P loop) consensus, while IVIDDIWD (beginning at residue 286) and GSKILVTTRK (beginning at residue 310) correspond, respectively, to the kinase 2 and kinase 3a consensus motifs (Traut 1994; Grant et al. 1995). In addition, GVPLAIITIAS (beginning at residue 372) and LKNCLLYL (beginning at residue 427) correspond, respectively, to the conserved R gene NBS domains 2 and 3 consensus motifs (Traut 1994; Grant et al. 1995). The final conserved NBS motif VHD (beginning at residue 501) corresponds to the conserved MHD (methionine–histidine–aspartate) motif. The C-terminal region of the protein includes 17 imperfect LRR repeats (residues 578–1056), composed of ∼15% leucine. The repeats, which are based on an LxxLxxLxxLxL consensus, vary in length between 22 and 44 amino acids. LRRs 14, 15, 16, and 17 show little or no similarity to the LRR consensus. The primary structure of the LRR-containing domain is illustrated in Figure 5. Finally, a COIL analysis (Lupas et al. 1991; http://www.ch.embnet.org/software/COIL_form.html) showed that a CC region is probably present (P > 0.95) between amino acids 24 and 52. The CC region contains three perfect hxxhxxh and one hxxhxxx motif (where h represents one of L, I, M, V, or F, and x is any residue). Overall the evidence suggests strongly that Pi36 belongs to the CC–NBS–LRR family of R genes.
Figure 5.—
Deduced amino acid sequence of the Pi36 encoded protein. The seven conserved motifs forming the NBS region are underlined. Residue 590, the single amino acid substitution distinguishing the blast-resistant from the blast-susceptible form of the protein, is double underlined. The C-terminal LRR region is shown separately from the rest of the sequence.
Phylogenetic analysis of Pi36:
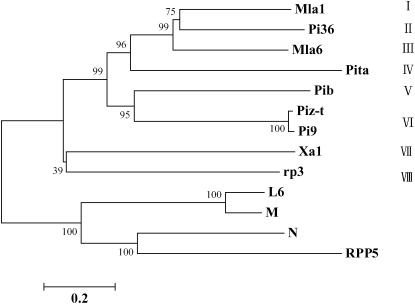
Southern hybridization analysis was employed to estimate the copy number of Pi36-related genes in rice. Only a single hybridizing fragment was present in both resistant and susceptible genotypes, indicating that Pi36 is a single-copy gene (supplemental Figure S2 at http://www.genetics.org/supplemental/). Interestingly, only a single copy is present in both reference sequences of varieties Nipponbare and 93-11 by BLAST analysis, suggesting that Pi36 is a single-copy gene in the rice genome. The evolutionary relationship between Pi36 and twelve R genes related was assessed by a phylogenetic amino acid-based sequence analysis using ProbCons and MEGA (Figure 6). The degree of homology shared by these genes varies considerably, and two heterogeneous groups can be recognized, reflecting an early divergence in the evolution of the R gene family. Cereal R genes are largely clustered into one of these groups, whereas those derived from the dicotyledonous species fall into the other group. The cereal R genes were further classified into eight distinct subgroups: Mla1 (I), Pi36 (II), Mla6 (III), Pita (IV), Pib (V), Piz-t/Pi9 (VI), Xa1 (VII), and rp3 (VIII) (Figure 6). Pi36 appears to be more closely related to the barley powdery mildew R genes Mla1 and Mla6 than to the rice blast R genes Pita, Pib, Pi9, and Piz-t. Pi36, Mla1, and Mla6 are well conserved in their NBS region, but diverge significantly in the LRR region (data not shown).
Figure 6.—
Phylogenetic analysis of Pi36 with other 10 R genes. Multiple amino acid alignments were conducted using ProbCons and a neighbor-joining phylogenetic tree was generated using MEGA. Numbers on branches indicate the percentage of 1000 bootstrap replicates which support the adjacent node. The unit branch length is 0.2 nucleotide substitutions per site, as indicated by the bar.
Expression pattern of Pi36:
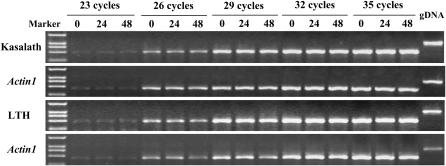
Only a faint band of the expected size was detected when 23 cycles were used, most likely due to the low expression level of the Pi36 gene. However, a stronger band was observed when 29, 32, and 35 cycles were applied (Figure 7). The results revealed that no detectable differences in expression could be observed either in a time course postinoculation with blast pathogen, or between resistant and susceptible hosts. Thus, the expression of Pi36 is likely to be constitutive, and is not induced by blast infection.
Figure 7.—
Expression patterns of Pi36 assayed by semiquantitative RT–PCR. Two-week-old resistant Kasalath and susceptible LTH seedlings were inoculated with blast (isolate CHL39). The expression of Pi36 was assayed at various time points postinoculation. Genomic DNA (gDNA) serves as a control to distinguish PCR products from cDNA and gDNA. The rice Actin1 gene acted as a positive control.
DISCUSSION
An efficient cloning strategy for Pi36:
Positional cloning is the accepted means to isolate genes where only phenotype and genomic location are known. The latter requires the prior generation of a high-resolution genetic map in the region surrounding the target. Generally, the map needs to be complemented with large insert (YAC or BAC) libraries. Where these requirements are met, the target can be narrowed down to a single insert (or a contig) based on the presence of closely linked hybridizing markers (Song et al. 1995; Yoshimura et al. 1998; Wang et al. 1999; Bryan et al. 2000; Sun et al. 2004; Qu et al. 2006; Chen et al. 2006). However, in rice, because its almost complete genome sequence is publicly available, much of this process can be performed in silico. With respect to Pi36, the necessary high-resolution genetic map had already been assembled (Liu et al. 2005). Thus the identification of candidates for Pi36 could be reduced to a bioinformatics-based search of the relevant physical genomic segment. Validation of the candidates was then achieved using a transgenic complementation test. The process was accelerated by exploiting the capability of LR-PCR to amplify sequences too large for conventional PCR (Feuillet et al. 2003; Horvath et al. 2003; Song et al. 2003). In the event, the three candidate genes were all recovered by LR–PCR, avoiding the need to subclone from a large insert library.
Insert size in the pCAMBIA1300 vector is limited, making it difficult to clone sequences as large as 10 kb. However, the pLYTAC27 TAC vector tolerates a much larger insert size (Liu et al. 2002) and was successfully used to clone Pi36-3 (>16 kb). We were then able to transfer the target fragment into a modified form of pCAMBIA1300 for the complementation study. This general approach has proven to be an effective means of cloning large genes.
Pi36 belongs to the CC–NBS–LRR family of R genes:
Some 580 NBS-encoding genes have been identified in the rice genome. Of these, ∼490 belong to the CC–NBS–LRR family and 101 are thought to be pseudogenes (Bai et al. 2002; Monosi et al. 2004). Of the three candidates for Pi36, one is a functional copy, while the other two are probably pseudogenes. Interestingly, the structure of Pi36 deduced from the genomic sequence was rather different from that deduced from the cDNA sequence. There are 13 introns in the former, but only 5 in the latter (Figure 4). This resulted in removing stop codons from genomic positions 2,884,229 to 2,881,515. That is, the stop codon in the latter one was shifted from the right flanking marker RM5647 for 2784 bp (Figure 1). This is an additional evidence to suggest that the latter is the actual structure of Pi36, because of the occurrence of recombination in this region within a stretch of 6.4 kb (Liu et al. 2005). Our results can be considered in light of the hypothesis that automatic annotations commonly inserted introns to remove stop codons or frameshift mutations (Monosi et al. 2004). Introns interrupting the NBS domain of R genes are more common in cereals than in dicotyledonous species (Bai et al. 2002). In rice, some six putative R genes (Bai et al. 2002) and three characterized rice blast-resistance genes (Pi-ta, Bryan et al. 2000; Pib, Wang et al. 1999; Pi9, Qu et al. 2006) carry intron(s) in their NBS domain. One intron is present in the NBS region of Pi36 (of length 5069 bp, and beginning at amino acid residue 284; Figure 4C), similar in size to that present in Pi9. Pi36 therefore has a unique structure with respect to intron position and size when compared with other rice genes. Conserved splicing sites (gt and ag) were present at the intron/exon junctions of introns 1, 2, 3, and 5, but at intron 4, the splicing site was ag and ct (Figure 4C). Introns have been commonly detected in the 5′ UTR region of R genes (Vos et al. 1998; Wang et al. 1999; Van Der Vossen et al. 2005) but seldom in the 3′ UTR. Two 3′ UTR introns are present in Pi9 (Qu et al. 2006), and one in Pi36. Whether these features of Pi36 have any biological significance has yet to be determined.
The LRR regions of rice NBS–LRR genes vary considerably in size and sequence, reflecting substantial divergence in the R genes (Bai et al. 2002). The LRRs occupy almost the entire C-terminal region of the CC–NBS–LRR proteins (Meyers et al. 2003), but the repeats are mostly imperfect, with only few conforming to any consensus sequence (Bai et al. 2002). In Pib, Pita, and Pi9, the regions are leucine-rich but have no clearly distinguishable repetitive structure. A similar pattern pertains to the Pi36 sequence, which encodes a 136-residue non-LRR region at its C-terminus. Further research is needed to establish whether the sequences at the Pi36 C-terminus play any role in the determination of specificity with respect to particular pathogen isolates.
Evolutionary relationships between Pi36 and other NBS–LRR R genes:
R genes are involved in the disease resistance response in a wide variety of plant species. They share a common structure and therefore probably act via a common mechanism. In evolutionary terms, it is widely assumed that the R genes have a common origin (Caicedo et al. 1999). The functional and evolutionary analysis of R genes is the focus of much current research. Pi36 is a single-copy R gene, and hence could represent a useful model for such functional and evolutionary studies. At the protein level, the Pi36 product most closely resembles the barley Mla1 and Mla6 proteins, and is less closely related to the rice bast Pib and Pita proteins. Multiple alignment of the amino acid sequences of Pi36 with 10 other R genes has demonstrated that nonconservative residue substitution was most frequent in the LRR domain and least in the NBS domain, supporting the widely held view that the LRR regions are subject to diversifying selection, and that they are responsible for specificity (McDowell et al. 1998; Meyers et al. 1998; Sun et al. 2001).
A single amino acid mutation is responsible for the resistance phenotype:
Pathogen–plant coevolution operates by simultaneous selection for avirulence genes in the pathogen and resistance genes in the host (Stahl and Bishop 2000). The direct interaction between an NBS–LRR protein and a pathogen avirulence gene product was first shown for the rice Pita/Avr-Pita system (Jia et al. 2000). The deduced Pita protein of a susceptible host differs from that of a resistant one by a single substitution of serine for alanine (Bryan et al. 2000). Similarly, we have established that a single amino acid difference distinguishes the resistant and susceptible alleles of the Pi36 product. In this case, the replacement of asparagine by serine determines blast resistance. A possible mechanistic explanation of the large biological effect of this small sequence difference could relate to the finding that when Nectria haematococca mycelia invade host tissue, high levels of free asparagine and homoserine become readily accessible to the fungus, and this induces the expression of pelD, a known virulence factor (Rogers et al. 2000). Thus serine and asparagines residues may be important for determining the resistance or resistance-related response. A more likely reason is that sequence variation at the active site affects molecular interactions and therefore changes function (Hanzawa et al. 2005). We are presently attempting the isolation of Avr-Pi36 to enable the dissection of the interactions between the host and the pathogen.
Acknowledgments
We are grateful to Y Liu for the kind provision of the pYLTAC27 vector and to Robert Koebner for critical reading of the manuscript. This research has been supported by grants from the National 973 project (2006CB1002006), the National 863 projects (2002AA2Z1002; 2006AA10A103; 2006AA100101), the Innovation Research Team Project from the Ministry of Education of China (IRT0448), the Guangdong Provincial Natural Science Foundation (039254), and the Special Project for the Distinguished University Professor from the Department of Education of Guangdong Province.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. DQ900896.
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSLBLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P. A., G. J. Lawrence, B. C. Morrish, M. A. Ayliffe, E. J. Finnegan et al., 1997. Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9: 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, J., L. A. Pennill, J. Ning, S. W. Lee, J. Ramalingam et al., 2002. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 12: 1871–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A. F., 1996. Plant disease resistance genes, function meets structure. Plant Cell 8: 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A. F., B. N. Kunkel, D. Dahlbeck, K. L. Brown, R. Schmidt et al., 1994. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bryan, G. T., K. S. Wu, L. Farrall, Y. L. Jia, H. P. Hershey et al., 2000. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12: 2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, A. L., B. A. Schaal and B. N. Kunkel, 1999. Diversity and molecular evolution of the RPS2 resistance gene in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., L. Wang, Z. Q. Que, R. Q. Pan and Q. H. Pan, 2005. Genetic and physical mapping of Pi37(t), a new gene conferring resistance to rice blast in the famous cultivar St. No. 1. Theor. Appl. Genet. 111: 1563–1570. [DOI] [PubMed] [Google Scholar]
- Chen, X. W., J. J. Shang, D. X. Chen, C. L. Lei, Y. Zou et al., 2006. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46: 794–804. [DOI] [PubMed] [Google Scholar]
- Dangl, J. L., and J. D. G. Jones, 2001. Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Do, C. B., M. S. Mahabhashyam, M. Brudno and S. Batzoglou, 2005. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 15: 330–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P. N., G. J. Lawrence, A. M. Catanzariti, T. Teh, C. I. Wang et al., 2006. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103: 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J. G., P. Dodds and T. Pryor, 2000. The generation of plant disease resistance gene specificities. Trends Plant Sci. 5: 373–379. [DOI] [PubMed] [Google Scholar]
- Feuillet, C., S. Travella, N. Stein, L. Albar, A. Nublat et al., 2003. Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. USA 100: 15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M. R., L. Godiard, E. Straube, T. Ashfield, J. Lewald et al., 1995. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846. [DOI] [PubMed] [Google Scholar]
- Gu, K., B. Yang, D. Tian, L. Wu, D. Wang et al., 2005. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435: 1122–1125. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K. E., and J. D. G. Jones, 1997. Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 575–607. [DOI] [PubMed] [Google Scholar]
- Hanzawa, Y., T. Money and D. Bradley, 2005. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 102: 7748–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y., S. Ohta, T. Komari and T. Kumashiro, 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Horvath, H., N. Rostoks, R. Brueggeman, B. Steffensonm, D. Von Wettstein et al., 2003. Genetically engineered stem rust resistance in barley using the Rpg1 gene. Proc. Natl. Acad. Sci. USA 100: 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert, S. H., C. A. Webb, S. M. Smith and Q. Sun, 2001. Resistance gene complexes, Evolution and utilization. Annu. Rev. Phytopathol. 39: 285–312. [DOI] [PubMed] [Google Scholar]
- Hwang, C., and V. M. Williamson, 2003. Leucine-rich repeat-mediated intramolecular interactions in nematode recognition and cell death signaling by the tomato resistance protein Mi. Plant J. 34: 585–593. [DOI] [PubMed] [Google Scholar]
- Jia, Y. L., S. A. Mcadams, G. T. Bryan, H. P. Hershey and B. Valent, 2000. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19: 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa, S., 1981. Gene analysis for blast resistance. Oryza 18: 196–203. [Google Scholar]
- Liu, X. Q., L. Wang, S. Chen, F. Lin and Q. H. Pan, 2005. Genetic and physical mapping of Pi36(t), a novel rice blast resistance gene located on rice chromosome 8. Mol. Genet. Genomics 274: 394–401. [DOI] [PubMed] [Google Scholar]
- Liu, Y. G., H. Liu, L. Chen, W. Qiu, Q. Zhang et al., 2002. Development of new transformation-competent artificial chromosome vectors and rice genomic libraries for efficient gene cloning. Gene 282: 247–255. [DOI] [PubMed] [Google Scholar]
- Lupas, A., M. Vandyke and J. Stock, 1991. Predicting coiled coils from protein sequences. Science 252: 1162–1164. [DOI] [PubMed] [Google Scholar]
- Mackill, D. J., and J. M. Bonman, 1992. Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82: 746–749. [Google Scholar]
- Mcdowell, J. M., M. Dhandaydham, T. A. Long, M. G. Aarts, S. Goff et al., 1998. Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10: 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B. C., K. A. Shen, P. Rohani, B. S. Gaut and R. W. Michelmore, 1998. Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 11: 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B. C., A. W. Dickerman, R. W. Michelmore, R. M. Pecherer, S. Sivaramakrishnan et al., 1999. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20: 317–332. [DOI] [PubMed] [Google Scholar]
- Meyers, B. C., A. Kozik, A. Griego, H. Kuang and R. W. Michelmore, 2003. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett, M., G. Farnham, J. Peart, and D. C. Baulcombe, 2002. Interaction between domains of a plant NBS-LRR protein in disease resistance related cell death. EMBO J. 21: 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monosi, B., R. J. Wisser, L. Pennill and S. H. Hulbert, 2004. Full-genome analysis of resistance gene homologues in rice. Theor. Appl. Genet. 109: 1434–1447. [DOI] [PubMed] [Google Scholar]
- Nei, M., and S. Kumar, 2000. Molecular Evolution and Phylogenetics. Oxford University Press, Oxford.
- Ou, S. H., 1979. Breeding rice for resistance to blast, a critical view, pp. 79–137 in Proceedings of the Rice Blast Workshop. International Rice Research Institute, Mantla, Philippines.
- Ou, S. H., 1985. Blast, pp. 109–201 in Rice Diseases, Ed. 2. Commonwealth Mycological Institute, Kew Surrey, UK.
- Pan, Q., Y. S. Liu, O. Budai-Hadrian, M. Sela, L. Carmel-Goren et al., 2000. Comparative genetics of nucleotide binding site-leucine rich repeat resistance gene homologues in the genomes of two dicotyledons, tomato and Arabidopsis. Genetics 155: 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q. H., Z. Hu, T. Tanisaka and L. Wang, 2003. Fine mapping of the blast resistance gene Pi15, linked to Pii, on rice chromosome 9. Acta Bot. Sin. 45: 871–877. [Google Scholar]
- Parniske, M., K. E. Hammond-Kosack, C. Golstein, C. M. Thomas, D. A. Jones et al., 1997. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91: 821–832. [DOI] [PubMed] [Google Scholar]
- Qu, S., G. Coaker, D. Francis, B. Zhou and G. L. Wang, 2003. Development of a new transformation-competent artificial chromosome (TAC) vector and construction of tomato and rice TAC libraries. Mol. Breed. 12: 297–308. [Google Scholar]
- Qu, S., G. Liu, B. Zhou, M. Bellizzi, L. Zeng et al., 2006. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172: 1901–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, L. M., Y. K. Kim, W. Guo, L. González-Candelas, D. Li et al., 2000. Requirement for either a host- or pectin-induced pectate lyase for infection of Pisum sativum by Nectria hematococca. Proc. Natl. Acad. Sci. USA 97: 9813–9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silué, D., J. L. Notteghem and D. Tharreau, 1992. Evidence for a gene-for-gene relationship in the Oryza sativa-Magnaporthe grisea pathosystem. Phytopathology 82: 577–580. [Google Scholar]
- Song, J. Q., J. M. Bradeen, S. K. Naess, J. A. Raasch, S. M. Wielgus et al., 2003. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl. Acad. Sci. USA 100: 9128–9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W. Y., G. L. Wang, L. L. Chen, H. S. Kim, L. Y. Pi et al., 1995. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806. [DOI] [PubMed] [Google Scholar]
- Stahl, E. A., and J. G. Bishop, 2000. Plant-pathogen arms races at the molecular level. Curr. Opin. Plant Biol. 3: 299–304. [DOI] [PubMed] [Google Scholar]
- Sun, Q., N. C. Collins, M. Ayliffe, S. M. Smith, J. Drake et al., 2001. Recombination between paralogues at the rp1 rust resistance locus in maize. Genetics 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. L., Y. L. Cao, Z. F. Yang, C. G. Xu, X. H. Li et al., 2004. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37: 517–527. [DOI] [PubMed] [Google Scholar]
- Traut, T. W., 1994. The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide binding sites. Eur. J. Biochem. 229: 9–19. [DOI] [PubMed] [Google Scholar]
- Valent, B., 1990. Rice blast as a model system for plant pathology. Phytopathology 80: 33–36. [Google Scholar]
- Valent, B., and F. G. Chumley, 1994. Avirulence genes and mechanism of genetic instability in the rice blast fungus, pp. 111–134 in Rice Blast Disease, edited by R. S. Zeigler, S. A. Leong and P. S. Teng. CABI, Wallingford, UK.
- Van Der Vossen, E. A., J. Gros, A. Sikkema, M. Muskens, D. Wouters et al., 2005. The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 44: 208–222. [DOI] [PubMed] [Google Scholar]
- Vos, P., G. Simons, T. Jesse, J. Wijbrandi, L. Heinen et al., 1998. The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotechnol. 16: 1365–1369. [DOI] [PubMed] [Google Scholar]
- Wang, Z. X., M. Yano, U. Yamanouchi, M. Iwamoto, L. Monna et al., 1999. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 19: 55–64. [DOI] [PubMed] [Google Scholar]
- Wulff, B. B. H., C. M. Thomas, M. Smoker, M. Grant and J. D. G. Jones, 2001. Domain swapping and gene shuffling identify sequences required for induction of an Avr-dependent hypersensitive response by the tomato Cf-4 and Cf-9 proteins. Plant Cell 13: 255–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, S., U. Yamanouchi, Y. Katayose, S. Toki, Z. X. Wang et al., 1998. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. USA 95: 1663–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B., S. Qu, G. Liu, M. Dolan, H. Sakai et al., 2006. The eight amino-acid differences within three leucine-rice repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant-Microbe Interact. 11: 1216–1228. [DOI] [PubMed] [Google Scholar]