Abstract
A large-effect QTL for divergence in sex-comb tooth number between Drosophila simulans and D. mauritiana was previously mapped to 73A–84AB. Here we identify genes that are likely contributors to this divergence. We first improved the mapping resolution in the 73A–84AB region using 12 introgression lines and 62 recombinant nearly isogenic lines. To further narrow the list of candidate genes, we assayed leg-specific expression and identified genes with transcript-level evolution consistent with a potential role in sex-comb divergence. Sex combs are formed on the prothoracic (front) legs, but not on the mesothoracic (middle) legs of Drosophila males. We extracted RNA from the prothoracic and mesothoracic pupal legs of two species to determine which of the genes expressed differently between leg types were also divergent for gene expression. Two good functional candidate genes, Scr and dsx, are located in one of our fine-scale QTL regions. In addition, three previously uncharacterized genes (CG15186, CG2016, and CG2791) emerged as new candidates. These genes are located in regions strongly associated with sex-comb tooth number differences and are expressed differently between leg tissues and between species. Further supporting the potential involvement of these genes in sex-comb divergence, we found a significant difference in sex-comb tooth number between co-isogenic D. melanogaster lines with and without P-element insertions at CG2791.
THERE has been renewed interest in the genetics of speciation, yielding a number of new theoretical and empirical results (e.g., Sun et al. 2004; Kirkpatrick and Barton 2006; Scannell et al. 2006; Sweigart et al. 2006), but only a few genes responsible for morphological differences among species have been identified (e.g., Doebley et al. 1995; Sucena and Stern 2000; Gompel et al. 2005). Complex morphological traits are frequently important in the process of speciation, both in the formation of reproductive isolation and during ecological diversification (e.g., Bradshaw et al. 1998; Albertson et al. 2003). However, progress in the quantitative genetics of species differences has been slowed by the inherent difficulties involved in the requisite mapping studies. Although much effort has gone into QTL studies of species differences in complex traits, factors responsible for interspecific quantitative morphological differences have been mapped mostly with limited resolution (e.g., Macdonald and Goldstein 1999; Zeng et al. 2000; Lexer et al. 2005). Individual genes responsible for species divergence in morphological traits remain largely unknown. Thus, a well-developed model system for studying the quantitative genetics of morphological divergence is highly desirable.
The Drosophila sex comb is an excellent model trait for such a system. The role of the sex comb in mating and the potential contribution of divergence in secondary sexual traits to reproductive isolation has generated much interest in its evolution (Spieth 1952; Coyne 1985; Kopp and True 2002). Evolutionary genetic studies have the highest likelihood of success when they are built on a solid genetic and biological foundation. The formation of the sex comb during leg bristle development is well understood and there are a large number of candidate genes that have mutant alleles that disrupt sex-comb development (True et al. 1997; Nuzhdin and Reiwitch 2000; Kopp and True 2002; Schawaroch 2002; Barmina et al. 2005). It is also especially fortunate that established hybrid mapping populations in the Drosophila simulans/D. mauritiana model system are available as a genetic tool (True et al. 1996b; Tao et al. 2003). This system, in combination with previous mapping data and developmental and genetic studies, forms a strong basis for detailed analyses.
Early observational studies of mating in Drosophila suggested that the sex comb might be involved in grasping the female during mounting or in spreading the wings after intromission and that the role of the sex comb in mating varies among species groups (Sturtevant et al. 1942; Spieth 1952). The primary evidence for the importance of the sex comb in male mating success comes from ablation experiments. Ablation of the sex comb by either mechanical or genetic means severely affects the ability of males to inseminate females (Spieth 1952; Cook 1977; Coyne 1985; C. S. Ng and A. Kopp, personal communication). Removal of the sex comb results in increased rejection of ablated males by females and in difficulty in maintaining a mounted position during copulation, possibly because of problems with grasping and positioning the female.
The phylogenetic patterns of morphological differences among species in sex-comb morphology are also consistent with sex combs playing a role in mating. Sex-comb tooth number is diverse in the closely related species of the D. melanogaster complex (Coyne 1985; True et al. 1997). Among the species studied, mean sex-comb tooth number varies from ∼7 teeth in D. teissieri to 14 teeth in D. mauritiana. D. melanogaster itself is intermediate between the two extremes with 10–11 mean sex-comb teeth/leg. Large differences in sex-comb morphology are also observed among more distantly related groups (Kopp and True 2002; Schawaroch 2002). For example, Kopp and True (2002) examined sex-comb morphology within the Oriental Drosophila and found differences in the degree of morphological modification, rotation, number of teeth, and position on particular leg segments.
Studies of sexual selection within species also indicate an association between the sex comb and mating success. The average number of sex-comb teeth was found to differ significantly between mating and nonmating males in natural populations of D. simulans (Markow et al. 1996). Mating D. simulans males have fewer sex-comb teeth, which may indicate a selective advantage for fewer sex-comb teeth in this species. Alternatively, males with fewer teeth may be disadvantaged in mating and are found disproportionately among mating males because they spend a longer time copulating. Additionally, slight differences in sex-comb tooth number were observed to occur in selection experiments examining the effects of presence or absence of male–male mating competition in D. melanogaster (Promislow et al. 1998). Together these studies suggest that species differences in number, form, and position of sex combs in Drosophila may have arisen as a result of sexual selection.
The genetic and developmental bases of species differences in sex-comb morphology remain unknown. Speculation, as always, must begin with what is known about the trait in D. melanogaster. The D. melanogaster sex comb arises from a transverse bristle row (TBR) on the basitarsus of the prothoracic leg. In males, these bristles are modified by rotation, becoming roughly perpendicular to the regular TBRs, and by morphological changes that thicken and darken the bristles (Tokunaga 1962; Held et al. 2004). Two genes involved directly in the regulation of sex-comb development have been identified for the primary control of leg segmental identity (Sex combs reduced, Scr) and sex (doublesex, dsx) specification (Baker and Ridge 1980; Pattatucci et al. 1991). Mutations in a large number of segmentation and polycomb group genes also cause aberrant sex-comb phenotypes (Tokunaga 1962; Stern and Tokunaga 1967). The details of how these genes interact in the sex-comb determination pathway are unknown, but the size and position of the sex comb are likely the result of joint regulation by the major leg-patterning pathways (Hays et al. 1999; Rauskolb 2001; Galindo et al. 2002).
Which of these genes, if any, are involved in the evolution of species differences in the sex comb? Large chromosomal regions that contribute to intra- and interspecific differences in sex-comb tooth number have been identified in a number of species of the D. melanogaster complex, but a combination of low mapping resolution and clustered distributions of sex-comb-development genes has resulted in numerous genes remaining on the list of potential candidates (True et al. 1997; Macdonald and Goldstein 1999; Nuzhdin and Reiwitch 2000; Coyne et al. 2004; Tatsuta and Takano-Shimizu 2006). The largest difference in sex-comb tooth number between sister species amenable to genetic mapping is between D. simulans and D. mauritiana, which differ by approximately three to five sex-comb teeth (Figure 1). True et al. (1997) mapped this difference in D. simulans and D. mauritiana isogenic lines using a set of 17 molecular markers. By composite interval mapping, they located a single QTL on chromosome III in the region 73A-84AB, which accounted for approximately half of the species difference in sex-comb tooth number. There are many candidate genes in this region, including the favorites dsx and Scr, and several polycomb group genes, such as kohtalo and Su(z)12.
Figure 1.—
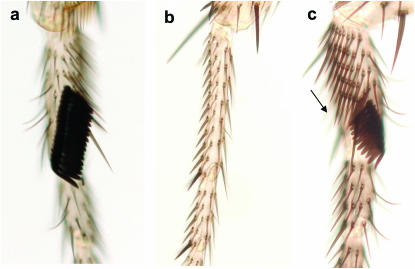
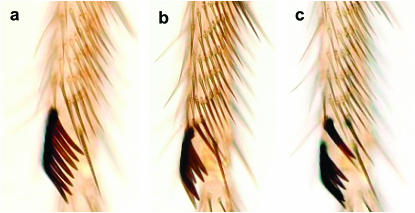
Prothoracic and mesothoracic male legs. (a) D. mauritiana prothoracic leg; the mean sex-comb tooth number in this species is ∼14 teeth. (b) D. mauritiana mesothoracic leg; there are no TBRs and no sex combs. (c) D. simulans prothoracic leg; the mean sex-comb tooth number in this species is ∼10. The arrow points to a TBR.
In this study, we have combined fine-scale mapping of the 73A–84AB third chromosome D. simulans and D. mauritiana sex-comb QTL with species comparisons of gene expression. Because the specific place and probable time of sex-comb determination are known, we were able to move beyond a simple adult whole-body expression approach to an assessment of tissue-specific differences at the time of sex-comb determination. This enabled us to refine the D. melanogaster-based candidate gene list by identifying divergent potential sex-comb-development genes in D. simulans and D. mauritiana. Our approach resulted in identifying new genes that potentially contribute to sex-comb development and are also divergent between species.
MATERIALS AND METHODS
Initial introgression line mapping:
Introgression line stocks:
We used 12 D. mauritiana introgressions marked with P{lacW} in an isogenic D. simulans background to survey the previously identified third chromosome QTL region 73A–84AB. Lines with D. mauritiana introgressions in the region 66D–89D (16.3, 16.9, 26.2, 32.1, 32.2, 32.6, 33.3, 33.5, 33.9, 18.24, 21.12, and 26.14) were obtained from the collection developed by Tao et al. (2003). This set of introgression lines is derived directly by recombination from introgression stocks made by True et al. (1996b) and the introgression lengths were previously estimated by Tao et al. (2003) using a set of allele-specific oligonucleotide markers. Introgression stocks are maintained without recombination by backcrossing heterozygous D. mauritiana introgression males to D. simulans (SimB) chromosome III isogenic females (Tao et al. 2003). The SimB first and second chromosomes are derived from a number of different D. simulans stocks and these chromosomes are potentially genetically heterogeneous. However, the third chromosome is isogenic and derived from the 13w JJ isogenic D. simulans line used in the original True et al. (1997) QTL mapping crosses.
Crosses and scoring:
For each of the 12 introgression lines, we crossed males and females heterozygous for the introgression and scored mean sex-comb tooth number for 10 progeny males from each line and each genotype. Third chromosome genotypes (in the SimB background) were +/+, +/P{lacW}, and P{lacW}/P{lacW}, where “+” is the wild-type D. simulans background and P{lacW} indicates 1 of 12 P-element-marked D. mauritiana introgressed regions. The probability of recombination occurring within an introgression in a single hybrid cross is very small (True et al. 1996a), and in subsequent analyses we assume complete linkage between the visible marker and the D. mauritiana introgressed region.
Statistical analysis:
Each of the 15 informative markers spanning the set of introgression lines was tested for a significant effect [F-test, type III sum of squares (SS)] on variation in sex-comb tooth number in single-marker ANOVA models using the procedure GLM (SAS Institute, Cary NC). We then corrected for multiple tests using the Benjamini and Hochberg (BH; 1995), step-down procedure to control the false discovery rate (FDR) with a cutoff of FDR < 0.05. A number of simulation and theoretical studies have shown that this simple procedure can be used in single-marker analysis to correct for multiple testing in QTL analysis. It is especially advantageous in fine-scale mapping experiments, where markers are likely to be positively associated due to linkage disequilibrium. This is because the BH procedure is robust for correlation of tests and increases the power to detect multiple QTL relative to other correction methods, while remaining reasonably conservative as marker density increases (Weller et al. 1998; Sabatti et al. 2003; Simonsen and McIntyre 2004; Benjamini and Yekutieli 2005). To account for possible epistatic effects on sex-comb tooth number, we also tested every possible pair of markers in full two-factor ANOVA models using the same methods as for single-marker analysis.
Fine-scale mapping:
Stock construction:
To further increase resolution in regions identified as significantly affecting variation in sex comb, we used a single P-element-marked D. mauritiana line, P{lacW}, at 76C (3Z4 76C, True et al. 1996b). By crossing this D. mauritiana stock to a D. simulans stock with appropriate visible mutations, we created a set of lines with recombination breakpoints between P{lacW}76C and a right-flanking visible marker, ebony (93C7). The positions of the two markers were chosen to flank the QTL regions identified in the initial introgression line mapping.
The P{lacW}76C marker and e+ D. mauritiana allele were backcrossed into an isogenic w; st, e D. simulans background for 20 generations (in the w; P{lacW}76C background, st is not visible), resulting in an introgression containing the D. mauritiana 76C–93C region plus ∼5 cM to either side of the selected markers (Fisher 1949; Naveira and Barbadilla 1992). From the cross w; P{lacW}76C, e+/P{lacW}76C+, e females to w; st, e males, we collected recombinant w; P{lacW}76C, e and w; P{lacW}76C+, e+ males. The resulting nearly isogenic recombinant introgressions were maintained by backcrossing heterozygous recombinant introgression males to w; st, e.
Crosses and scoring:
Sixty-two of these lines were scored for sex-comb tooth number. For each line, we crossed males and females that were heterozygous for the D. mauritiana introgression. Male progeny were then divided into D. simulans and D. mauritiana introgression classes by the appropriate visible marker, P{lacW}76C or ebony. The expression of mini-white in different P{lacW} inserts varies depending on position and this causes eye color to range from a very light peach to a dark red, almost indistinguishable from wild type. Most of the P inserts are semidominant, making it possible to distinguish the heterozygous and homozygous classes. We found, however, that P{lacW}76C was difficult to score as one copy vs. two copies, such that all introgression males with D. mauritiana introgressed regions to the left of the centromere had varying shades of bright orange eyes, regardless of heterozygosity or homozygosity at P{lacW}76C. The other visible marker, e+, is dominant, so introgression males with D. mauritiana regions to the right of the centromere all have wild-type body color. Because we were unable to distinguish the heterozygous and homozygous visible marker classes, we scored 30 males from each line with one or the other D. mauritiana visible marker, homozygous or heterozygous for the D. mauritiana introgression (w; P{lacW}76C, e/P{lacW}76C+, e or w; P{lacW}76C, e/P{lacW}76C, e and P{lacW}76C+, e+/P{lacW}76C+, e or P{lacW}76C+, e+/P{lacW}76C+, e+), and 15 homozygous D. simulans males (w; P{lacW}76C+, e/P{lacW}76C+,e). We then analyzed a contrast between D. simulans males and their D. mauritiana introgression siblings.
Molecular markers:
To map the position of recombination breakpoints, we developed five pairs of PCR-based species-specific SNP markers in the region 76C–84A (Table 1). For each species and for each SNP, we designed a primer pair to amplify ∼100–200 bp in one species, but not in the other. In the first step, we identified and aligned by BLAST available D. simulans sequences for 14 genes from the Drosophila Population Genomics Project D. simulans syntenic assembly (http://www.dpgp.org/syntenic_assembly) of the seven sequenced D. simulans lines (Altschul et al. 1990). Gene regions with assembled sequence from at least three of the D. simulans lines were used to identify matching preliminary D. mauritiana genome trace sequences (∼100–150 bp) available from the NCBI trace archive (454 platform, Margulies et al. 2005; sequences available at http://www.ncbi.nlm.nih.gov/Traces). Primer pairs were designed from aligned sequences such that one primer was complementary to at least one 3′ species-specific SNP (a few primers contained one or more internal sequence SNPs) and the other primer was always designed from conserved sequence. To increase the probability of a match to the isogenic D. simulans background used in the mapping experiment, we used only D. simulans D. mauritiana SNPs that were fixed within the sequenced D. simulans lines. We were unable to determine if the SNP differences were fixed (or at high frequency) in D. mauritiana because only one sequence was available. We tested each pair of primers by PCR with template DNA from the two parental lines (w; st, e and 3Z4 76C) and chose five pairs for five genes from a total of 22 primer pairs designed, which reliably produced species-specific amplification in the parental lines.
TABLE 1.
Species-specific primer pairs used in fine-scale mapping
| Species | Gene | Forward primer | Reverse primer | Annealing temperature |
|---|---|---|---|---|
| D. mauritiana | rdgC | agtggcagcaggtattttgcgat | tgggcttgcactcgtgcgat | 63° |
| D. simulans | rdgC | gtggcagcaggtattttgcggg | tgggcttgcactcgtgcgat | 63° |
| D. mauritiana | Ten-m | gccagcctaacacttgcttctt | gccaagaaacggcggtttgg | 65° |
| D. simulans | Ten-m | gctgggaacgggaaagaacgca | gccaagaaacggcggtttgg | 66.8° |
| D. mauritiana | CG11367 | ctgacggggtatatcagctttctg | ccaagtggctcctgttaaacttc | 63° |
| D. simulans | CG11367 | ctgacggggtatatcagctttctg | ctccaagtggctcctgttaaacttg | 63° |
| D. mauritiana | Dip2 | ccacgaaaacgcgctgacca | ctttggatgcacttggccgtttg | 66.8° |
| D. simulans | Dip2 | ccacgaaaacgcgctgacca | ctttggatgcacttggccgtttt | 66.8° |
| D. mauritiana | Scr | gccagggctcgtttgggtag | ttatggggcgaacaaagcgcc | 66.8° |
| D. simulans | Scr | gccagggctcgtttgggtac | ttatggggcgaacaaagcgcc | 66.8° |
To determine the position of the internal breakpoint for each recombinant nearly isogenic line (NIL), three males carrying the D. mauritiana visible marker (P{lacW}76C or e+) were selected and pooled for DNA extractions. Flies were frozen at −80° prior to DNA extraction and genomic DNA was isolated from homogenized flies by a modified alkaline lysis protocol (Ashburner 1989). Each SNP marker was then scored three times using standard PCR and gel electrophoresis, with the two parental species lines as additional positive and negative controls.
Statistical analysis:
The procedure for statistical analysis of the fine-scale mapping data was similar to that of the initial mapping study (Proc GLM; F-test, type III SS; SAS Institute). However, because the number of lines largely exceeded the number of markers, we were able to fit a multiple marker ANOVA model, including the main effects for all five SNP-based markers and the two visible markers. Note that males were binned by dominant visible markers into D. mauritiana or D. simulans classes, collapsing the heterozygous and homozygous D. mauritiana marker males into a single D. mauritiana class. The main effect of each marker was estimated as its linear effect, the difference between the least square means of the dominant visible marker class (heterozygous and homozygous D. mauritiana) and the recessive visible marker class (homozygous D. simulans). On average, we expect a two-to-one ratio of D. mauritiana introgression heterozygotes to homozygotes among the males scored, causing mean sex-comb tooth number of the D. mauritiana class males to be weighted toward the heterozygous class. While dominance effects and deviations from Hardy–Weinberg cannot produce false positives for significant marker trait association, the estimated effects of these markers are only approximate.
Combining D. mauritiana genotypes into a single class could cause the assumption of homogeneity of variance to be violated. Homogeneity of variance tests for single-marker models were implemented using SAS proc GLM (data not shown; Levene's test, Levene 1960; Proc GLM, SAS Institute). As a subset of tests were significant, we implemented a simple permutation test randomizing mean sex comb over marker state (Sokal and Rohlf 2001). For 10,000 permuted data sets, the multiple-marker model was fit and the F-statistic was calculated.
To test for epistatic interactions, while accounting for possible linkage effects, we fit models for each possible pairwise interaction between our markers separately. Each model included the interaction term for a single pair and the main effects of all seven markers. In all cases, the BH procedure was used with cutoff FDR < 0.05 to correct for multiple testing. Although we have no evidence for epistatic effects on viability in this region, it is possible that associated deviations in marker class frequency could produce false positives for epistasis.
Gene expression for male prothoracic and mesothoracic legs in D. simulans and D. mauritiana:
Dissection and RNA extraction:
The tarsal segments and distal portion of the tibia of 16-hr-old pupal male prothoracic and mesothoracic legs were collected from males of isogenic D. simulans and isogenic D. mauritiana lines (D. simulans w; st, e and D. mauritiana iso 207, >20 generations of full sib mating). Dissections were performed on ice in Trizol and stored at −80° until RNA extraction, as described in Barmina et al. (2005). Samples for each leg of each species, consisting of 31–34 individual males, were pooled to form three independent biological replicates of first and second leg samples for the initial RNA extraction.
Samples were thawed and homogenized in 1 μl linear polyacrylamide and 1 ml Trizol. After addition of 200 μl chloroform, the samples were shaken for 20 sec and left to incubate for 5 min. They were then centrifuged at 14,000 rpm for 20 min at 4° and the upper phase was collected. Samples were transferred to a new tube and 500 μl isopropanol was mixed in. They were then incubated overnight at −20°. The next day RNA was precipitated by centrifugation for 20 min at 4°, supernatant was removed, and the pellet was washed in 1 ml of 75% EtOH. After drying, the RNA was resuspended in 20 μl of DEPC water and stored at −20°. The concentration was then determined by fluorometry (Quant-iT RiboGreen kit, Invitrogen, San Diego) and quality control and size measurements were taken using the Bioanalyzer from Agilent Technologies (RNA 6000 Nano LabChip kit). Yield from the initial extraction ranged from 1288 to 2279 ng. To obtain the requisite concentration of RNA needed for hybridizations, 100 ng from the initial RNA extraction was amplified and labeled as described in the eukaryotic target preparation two-cycle in vitro transcription Affymetrix protocol (for the protocol, see Eukaryotic Sampling and Array Processing documentation at http://www.affymetrix.com/). Fifteen micrograms of labeled fragmented cRNA was then hybridized to the Drosophila Genome 2.0 Affymetrix GeneChip array. Hybridization and quantification was done at the University of California, Davis, Affymetrix core facility, using GCOS software to analyze the array images. A total of 12 chips, 3 chips/leg/species, were analyzed.
Quality control:
Chips were examined using the RACE web tool (http://race.unil.ch) to implement quality control methods from the Bioconductor affy and affyPLM packages (Gautier et al. 2004; Psarros et al. 2005). The Pearson correlation coefficients of the normalized intensity scores across chips were generally high. For MAS 5.0 GC robust multi-array average (GCRMA) normalization, they ranged from 0.976 to 0.993 (0.989–0.998) within species and from 0.938 to 0.964 (0.979–0.989) between species. Relative log expression and the distribution of intensity scores after normalization were consistent across chips and no outliers were identified by these measures. However, one chip (replicate one, leg two) had higher prenormalization intensity scores than the other chips and, consequently, higher standard error across probe sets as indicated by a normalized unscaled standard error (NUSE) >1.05. Because other indicators of chip quality were good and including or excluding the data from this chip in preliminary analyses had no significant effect on the results, we did not exclude this chip in the final analysis.
Statistical analysis:
To allow for direct comparison between these analyses and those of Barmina et al. (2005), we used the same normalization procedure, MAS 5.0 combined with scaling by the slide median and log transformation, and significance cutoffs of FDR < 0.2 or P < 0.001. FDR was implemented as described in Benjamini and Hochberg (1995) for cutoffs of FDR < 0.2, < 0.1, and < 0.05. Additionally, we used the GCRMA procedure, which includes a sequence-specific background correction, quantile normalization, and median polish to normalize and summarize expression scores (Wu and Irizarry 2004). GCRMA has been shown to be among the best methods for normalizing data when analyzing gene expression differences (Irizarry et al. 2006). It is not possible to know what the best normalization procedure is for every gene in every array data set. It is probable that there is a range of best procedures for different genes, samples, and chip designs with similar expression score distributions and probe set properties. By including both the Affymetrix standard method and GCRMA, which performs well in overall measurements of both accuracy and precision, we hope to provide a robust analysis. All data normalization and presence/absence calls were done using the bioconductor R affy package (http://www.bioconductor.org). We report genes found to be significantly different in gene expression in leg and species comparisons for both normalization procedures.
Features annotated as transposons and control features were eliminated from the analysis (261 of 18,952 probe sets). We also masked any features called as absent by the MAS 5.0 procedure on all 12 chips (7345). The combined leg and species data set was analyzed by ANOVA, with model yijk = μ + Lij+ Sik +(LS)ijk + εijk, where y is the normalized expression score for the ith gene, μ is the mean intensity score, L is the effect of leg (j = leg 1 or leg 2), S is the effect of species (k = D. simulans or D. mauritiana), LS is the interaction of leg type and species, and ε is the error variance (procedure GLM, SAS Institute). We also analyzed the data separately within each species for leg differences, yijs = μ + Lijs+ εijs (jth leg for species s), and within each leg tissue type for species differences, yijl = μ + Sijl + εijl (jth species for leg l). Note that while this gives us some idea of how differences are distributed within classes, each individual species or leg-type analysis has reduced sample size in comparison to the combined analysis and therefore reduced power. For each effect and each gene, the significance was determined by an F-test with type III SS (procedure GLM, SAS Institute).
Signal differences between species can sometimes arise as a result of reduced hybridization due to sequence divergence. However, average sequence divergence between D. simulans and D. mauritiana is low, ∼0.5%, and the two species are roughly equidistant from D. melanogaster (Hey and Kliman 1993). Although signal loss due to sequence divergence may occur when D. simulans or D. mauritiana cRNA is hybridized to the D. melanogaster GeneChip, in the majority of cases such signal loss will be shared by both species. However, in any specific case, it is possible that sequence divergence and expression divergence may be confounded.
To account for possible effects of sequence divergence in tests for species differences in gene expression for our identified candidate genes, we conducted a probe-level analysis of the data. As for the gene-level analysis, we used the bioconductor R affy package (http://www.bioconductor.org) to normalize the data. Normalized probe-level signal was generated by simply carrying out the standard GCRMA and MAS 5.0 normalization procedures, but eliminating the gene-level summarization steps. The normalized probe-level scores were then scaled to the slide median and GCRMA normalized scores were log transformed. As Mas 5.0 probe-level scores (PM − MM/PM + MM) can be negative, they were not log transformed in the probe-level analysis. We then masked outlier probes by hand, eliminating probe pairs with obvious loss of signal in one species but not in the other. The individual probe signal data were analyzed fitting the model yijkl = μ + Lij+ Sik + Pil + (LS)ijk + (LP)ijl + (SP)ikl + εijkl, where the additional term P is the effect of probe (l = 1 … 14), LP is the interaction of leg type and probe pair, and SP is the interaction of species and probe pair (procedure GLM, SAS Institute). Additional, nonmasked, species-specific signal attenuation and/or cross-hybridization effects on subsets of probe pairs in a gene's probe set are expected to contribute to the species by probe variance rather than the main species effect.
P-element insertions at CG2791:
We examined five different D. melanogaster insertions at CG2791 for effects on sex-comb morphology (P{SUPor-P}KG01932, PBac{RB}CG2791e04491, PBac{5HPw+}A146, P{RS5}5-SZ-3716, and PBac{WH}CG2791f01853; Bellen et al. 2004; Ryder et al. 2004; Thibault et al. 2004). We then scored sex-comb tooth number for two of the insertion lines, P{SUPor-P}KG01932 and P{RS5}5-SZ-3716, which had weakly penetrant qualitative effects on sex-comb morphology (see Figure 5 for locations). Sex-comb tooth number was scored for 20 homozygous insertion males of each type. For P{SUPor-P}KG01932, mean sex-comb tooth number and qualitative differences were compared to both the isogenic genetic background stock, y1; ry506 (Bloomington stock no. 4405), and the heterozygous y1 w67c23; P{SUPor-P}KG01932 ry506/TM3, Sb males (20 males each). For P{RS5}5-SZ-3716, mean sex-comb tooth number was compared to the isogenic genetic background stock, w1118 iso (DrosDel stock DSK 001). We also examined males and females for additional effects of the insertions on sexually dimorphic traits (abdominal pigmentation, loss of the vaginal teeth, and rotation or other abnormalities of the genitalia).
Figure 5.—
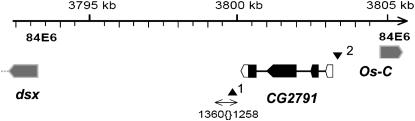
Overview of the CG2791 gene region. Transcript structure is shown only for CG2791 (solid boxes indicate coding sequence). 1360{}1258 is a natural transposable element located 34 bp downstream of CG2791. Triangles mark the locations of the two P-element insertions that we examined: 1, P{SUPor-P}KG01932 located 100 bp from CG2791 in 1360{}1258; 2, P{RS5}5-SZ-3716 located 7 bp 5′ of exon 1.
Enhancer trap expression pattern:
Twenty-four-hour-old D. melanogaster PBac{GAL4D,EYFP}CG2791PL00285/UAS-lacZ male pupae were halved and ventral sides were fixed in 100 mm NaH2PO4, pH 7.0, 150 mm NaCl, 1 mm MgCl2, 0.1% Triton 100×, and 1% glutaraldehyde for 30 min (PBac{GAL4D,EYFP}CG2791PL00285; Bellen et al. 2004). Ventral sides were washed three times for 10 min each in the same solution, minus glutaraldehyde. Legs were dissected in washing solution and incubated in 10 mm NaH2PO4, pH 7.0, 150 mm NaCl, 0.1% Triton 100×, 3.3 mm K3[Fe(CN)6], 3.3 mm K4[Fe(CN)6], 0.1% X-gal. After color development, legs were rinsed well in PBS and mounted in 80% glycerol.
RESULTS
Initial introgression line mapping:
We first analyzed sex-comb tooth number in a set of 12 introgression lines from the Tao et al. (2003) collection to confirm the position of the True et al. (1997) chromosome III QTL. Of 15 informative markers used in the analysis, 8 markers corresponding to 70D–84AB were significant at FDR < 0.05 in single-marker ANOVA (Table 2; Figure 2). Although somewhat larger, this corresponds approximately to the 73A–84AB region identified by True et al. (1997). The slight difference in position is not unexpected due to differences in cross design and marker density between the two mapping studies.
TABLE 2.
Significant markers for initial introgression line mapping
| Marker | Pa | Pb |
|---|---|---|
| 70D (fz) | 0.001c | 0.77 |
| 76B (Ash1) | 0.0001c | 0.78 |
| 77B (rdgC) | 5.70 × 10−5c | — |
| 77E (pka-R1) | 0.0008c | 0.90 |
| 78C (Edg78E) | 0.0043c | 0.83 |
| 82C (5-Ht2) | 0.0014c | 0.87 |
| 83B (Rga) | 0.0001c | 0.0006c |
| 84B (Antp) | 0.0139c | 0.003c |
Single-marker ANOVA.
Including marker 77B as a cofactor.
FDR < 0.05.
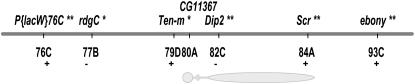
Figure 2.—
Mapping results for the initial 12 introgression lines. Parallel lines indicate the left breakpoint of the 84E–93F inversion. A single asterisk indicates markers that were significant at FDR < 0.05. Double asterisks indicate markers that were significant at FDR < 0.05 in two marker models with marker 77B. Shaded boxes mark the two QTL regions associated with marker 77B and 83B–84B (bounded by markers that were not significant in tests with marker 77B as a cofactor—70D–77E and 82C–93E). Paired tick marks show the location of the left 84E–93F inversion breakpoint. (a) Interactions between marker 68E and markers 77E–82C have recessive negative epistatic effects (see Figure 3A). (b) Interactions between markers 68E–78C and markers 93D–89D have positive synergistic epistatic effects (see Figure 3B). (c) Interactions between 68E–70D and 77E–78C with one or more of markers of 83B, 84B, or 93E also have a slight positive synergistic effect (see Figure 3C).
When markers are analyzed individually, their correlations may cause effects associated with one marker to be attributed to other markers (Zeng 1994). To account for the effect of the region linked to 77B (rdgC), the most significant marker, we retested each of the seven other significant markers with the 77B marker included as a cofactor (Table 2). Two markers, 83B (Rga) and 84B (Antp), retained significance (FDR < 0.05; 83B, P = 0.0006; 84B, P = 0.0032). We conclude that there are at least two separate QTL in this region, one on either side of the chromosome III centromere, associated with 77B and with 83B–84B (∼70D–77E and 82C–93E).
Epistatic effects are commonly observed in species hybrids and could potentially contribute to the observed phenotypic variation in sex-comb tooth number among introgression lines. To test for epistasis, we analyzed 56 of 105 possible pairwise interaction terms between markers (remaining pairs were impossible to test due to few or no observations in one of the four possible classes). A total of 31 marker pairs had significant interaction terms at FDR < 0.05 (supplemental Table 1 at http://www.genetics.org/supplemental/). However, a small number of genes might result in many significant tests due to linkage disequilibrium among markers. Examining the pattern of significant interactions (see Figures 2 and 3 and supplemental Table 1), we conclude that a minimum of two genetic interactions could explain nearly all of the results.
Figure 3.—
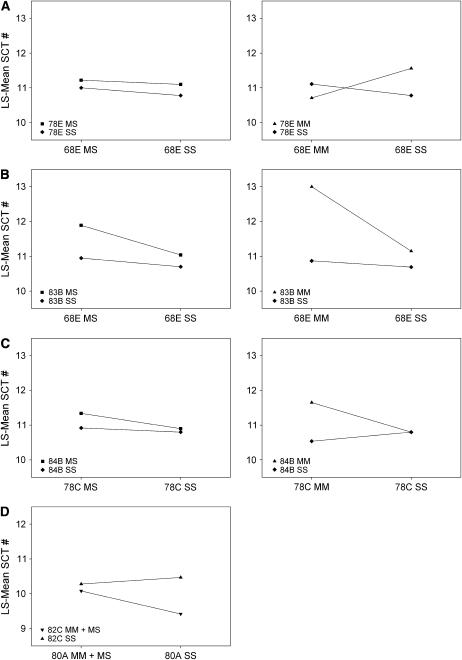
(A–D) Significant interaction terms between markers shown as reaction norms. Plots show least-square means of sex-comb tooth number for D. simulans and D. mauritiana marker genotypes. (A–C) Examples of epistasis in the introgression lines. Graphs on the left show heterozygous D. mauritiana, D. simulans, and homozygous D. simulans genotypes for one marker in combination with those for the second marker. Graphs on the right show homozygous D. mauritiana and D. simulans genotypes. (A) Interactions between markers 68E and 77E–82C have similar plots. (B) Interactions between markers 68E–78C and 93D–89D have similar plots. (C) Example of interactions between 68E–70D and 77E–78C with one or more of 83B, 84B, or 93E. (D) The 80A/82C interaction detected in the recombinant nearly isogenic lines. The plot shows homozygous D. simulans marker classes and the combined D. mauritiana class.
Fine-scale introgression line mapping:
The resolution of the initial mapping study was not sufficient to narrow the list of possible candidates, due to the small number of recombination breakpoints in the lines available. To increase the mapping resolution in the region containing the two most significant markers from the initial mapping experiment, 77B and 84B, we created 62 additional lines with recombination breakpoints between P{lacW} at 76C and ebony at 93C. The larger data set and small number of markers allowed us to analyze the five internal SNP markers and two flanking visible markers as cofactors in a multiple-factor ANOVA model. All left-side visible marker lines (P{lacW}76C) share the D. mauritiana introgressed region to the left of the marker, but vary in length of the introgressed region to the right of the marker. Conversely, all lines with the visible marker to the right of the centromere (ebony) share the D. mauritiana region to the right of the marker and vary in the length of the left-flanking region. We account for the effects of possible QTL associated with the left- and right-flanking shared regions by including the two visible markers in the model. The two flanking visible markers were significant at FDR < 0.05, with ebony having the most significant association of all the markers in the model (Table 3). Four of the internal SNP markers, 77B (rdgC), 79D (Ten-m), 82C (Dip2), and 84A (Scr), are also significantly associated with variation in sex-comb tooth number (Table 3; Figure 4).
TABLE 3.
Significant markers and effects for recombinant NIL lines
| Marker | Type III SS | F-value | P | Effectc | SEc | t-valuec |
|---|---|---|---|---|---|---|
| 93C (ebony)a | 79.67 | 157.54 | 4.23 × 10−35* | 0.54 | 0.043 | 12.55 |
| 84A (Scr)b | 39.29 | 77.70 | 2.18 × 10−18* | 0.58 | 0.065 | 8.81 |
| P{lacW}76Ca | 25.52 | 50.47 | 1.57 × 10−12* | 0.41 | 0.058 | 7.10 |
| 82C (Dip2)b | 12.25 | 24.22 | 9.15 × 10−7* | −0.43 | 0.087 | −4.92 |
| 77B (rdgC)b | 7.25 | 14.34 | 0.0002* | −0.24 | 0.064 | −3.79 |
| 79D (Ten-m)b | 2.2 | 4.36 | 0.04 | 0.14 | 0.065 | 2.09 |
P < 0.0001 in permutation test (F-statistic, 10,000 permutations).
Visible markers associated with flanking regions.
Molecular markers.
Linear effect, as the difference between the D. simulans and D. mauritiana class least-square means.
Figure 4.—
Results of recombinant nearly isogenic line mapping. A single asterisk indicates markers that were significant at FDR < 0.05. Double asterisks indicate markers significant at P ≪ 0.0001. Gray ovals mark significant interactions. Although the 80A marker did not have a significant main effect on sex-comb tooth number, there were two significant interactions associated with this marker—80A/82C and 80A/84A.
The effects of the significant markers, as the difference between the D. mauritiana visible mutant class and the homozygous D. simulans class, are also listed in Table 3. The D. mauritiana regions linked to the two visible markers and to two of the internal markers, at 84A and 79D, increase sex-comb tooth number, with SCR having the largest effect of the three markers. Two markers at 77B and 82C had significant negative effects on sex-comb tooth number.
The left-flanking region of the 76C–93C introgression is associated with a positive effect on sex-comb tooth number. This is probably due to the same factor or factors underlying the 70D–77E QTL detected in the initial mapping. We cannot refine the exact position of this QTL because the visible marker segregates with the entire left-flanking introgressed region in the fine-scale mapping lines. The first definable 3L QTL is located between 76C and 79D and is associated with a negative effect on sex-comb tooth number (∼3.22 Mbp; bounded by opposite-effect markers and/or by nonsignificant markers; Whittaker et al. 1996). In the same way, we can show that a negative-effect factor or factors are located in the 80A–84A region (associated with the 82C marker, ∼2.92 Mbp) and a slight positive effect is located between 77B and 80A (associated with the 77D marker, ∼2.01 Mbp). The overall pattern of marker effects indicates that at least one sex-comb divergence gene or group of genes is also located close to Scr (84A), as the left-flanking marker, 82C, is associated with a negative effect and the right-flanking marker, 93C, is associated with a positive effect of similar magnitude. This can be explained either by a single gene or by a group of genes located between 84A and 93C or by two regions contributing to sex-comb tooth divergence, one located near 84A and the other in the right-flanking D. mauritiana region that segregates with 93C. We are unable to formally distinguish between the two possibilities with the current data. Note that 93C is much closer to 84A in these species than it is in D. melanogaster, because of the third chromosome 84E–93F inversion (see Figure 2). Consequently, while the physical size of the region between these two markers in D. melanogaster is quite large, ∼14.41 Mbp, in D. simulans and D. mauritiana, it is much smaller (∼1.7 Mbp).
We tested all possible pairwise interaction terms to determine the extent of epistasis. As the number of lines that we generated is many more than the number of markers, we were able to fit interaction terms in addition to main effects. In each model, we included the interaction term for the marker pair tested, its main effects, and the main effects of all additional markers. Inclusion of these main effects should eliminate false positives for significant marker interactions that can occur due to linkage effects in simple two-marker models (Zeng 1994). With this model, only two interactions were significant at FDR < 0.05, one between the 80A marker (CG11367) and 82C (Dip2) and the other between 80A and 84A (Scr) (P = 8.11 × 10−11 and P = 5.60 × 10−4, respectively; Figure 4). The simplest explanation is that there is a strong interaction between a gene near the chromosome III centromere and one located between the other two markers.
Gene expression in prothoracic and mesothoracic legs in D. simulans and D. mauritiana:
There is inherent uncertainty in using genetic data from D. melanogaster alone to determine a function-based list of candidate genes. Given the large number of genes involved, development of the sex comb is likely to be quite complex and some differences in sex-comb development may have evolved between D. melanogaster and its sister species. Accordingly, we augmented the initial list of D. melanogaster sex-comb genes by determining which genes with a potential role in sex-comb development in the sister species were also divergent for gene expression between them. To develop a species-specific list of sex-comb development genes, we followed the methodology of Barmina et al. (2005), who used a tissue- and time-specific comparison of transcript level to discover potential sex-comb development genes in D. melanogaster.
The rotation of the nascent sex-comb bristle cells occurs early in the development of the sex comb, prior to formation of the sex-comb teeth, beginning somewhere between 16 and 17 hr postpupariation (Held et al. 2004). Following the reasoning of Barmina et al. (2005), the expression pattern in the pupal leg necessary to specify which bristle cells will eventually differentiate into sex-comb teeth should be present at the time of rotation of the precursor cells. The set of genes expressed at different levels between the prothoracic and the mesothoracic leg will include those involved in specification of the sex comb because no transverse bristle rows develop on the mesothoracic leg, while the most distal row of bristle precursor cells on the basitarsi of the prothoracic leg develop into the sex comb. To determine which genes with a potential role in sex-comb development were also expressed differently between species, we compared D. simulans and D. mauritiana gene expression levels in the distal portion of prothoracic and mesothoracic legs.
Our expression data are from 12 chips in total, consisting of three biological replicates for each of the two leg tissues, prothoracic and mesothoracic, in the two species. Of 14,419 annotated genes (FlyBase and/or Ensembl, D. melanogaster gene databases) represented as one or more transcripts on the Affymetrix Drosophila 2.0 GeneChip, 9967 were included in the analysis of leg-tissue expression of D. simulans and D. mauritiana (determined as present in 1 or more of 12 chips by MAS 5.0). While ∼69% of fly genes were included in our analysis, the actual proportion of genes expressed at functional levels in the leg is probably less. Our criteria for inclusion was intentionally liberal, so as to not miss potential presence/absence differences in gene expression. Some of the genes included in our analyses may be false positives for leg gene expression. One more conservative approximation for leg-expressed genes would be the number of genes determined as present on at least half of the chips for a single species. For D. simulans, the species with the greatest number of detectable transcripts, using this cutoff reduces the number of “present” genes to 6922 or ∼48% of annotated genes. We have included all of the presence and absence calls in supplemental Table 2 at http://www.genetics.org/supplemental/.
Here we report annotated genes found to be significantly different in gene expression in leg and species comparisons using the cutoffs of FDR < 0.2 or P < 0.001 for either or both the MAS 5.0 and GCRMA normalization procedures. For the full leg-type and species ANOVA model, using the MAS 5.0 (GCRMA given in parentheses) normalization procedure, there were 13 (10) genes with a significant effect of leg type on expression level, 3050 (2793) genes with a significant effect of species, and 3 (2) genes significant for an interaction between leg and species (Tables 4–6; Complete species lists are reported in supplemental Tables 3 and 4 at http://www.genetics.org/supplemental/).
TABLE 4.
Genes expressed differently between prothoracic and mesothoracic legs in D. simulans and D. mauritiana
| Gene | Band | Biological process | Molecular function | FDRab | Pa | Pb |
|---|---|---|---|---|---|---|
| CG4962 | 72E1 | Unknown | Unknown | 0.05ab | <0.0001a | <0.0001b |
| Scrcd | 84A5 | A/P axis specification | Transcription factor | 0.05ab | <0.0001a | <0.0001b |
| Nordcd | 60C3 | Olfactory learning | Fibronectin type III | 0.2a, 0.05b | <0.0001a | <0.0001b |
| CG2791cd | 84E6 | Unknown | Unknown | 0.05b | 0.0001a | <0.0001b |
| CG14191 | 18A3 | Unknown | Unknown | 0.05a | <0.0001a | 0.0003b |
| CG13857cde | 94A11 | Unknown | Unknown | — | 0.0002a | 0.0002b |
| CG4766 | 5D1 | Unknown | MAB-21 related | — | 0.0006a | 0.005b |
| phr | 43E18 | DNA repair | Nucleic acid binding | 0.2b | — | <0.0001b |
| ara | 69C8 | Morphogenesis | Transcription factor | — | 0.0065a | 0.0002b |
| kal-1 | 95E1 | Cell adhesion | Fibronectin type III | — | 0.0044a | 0.0008b |
| CG2016 | 82E4 | Unknown | Takeout family | — | 0.0003a | 0.0018b |
| CG15186 | 83E8 | Unknown | Unknown | — | — | 0.0002b |
| CG16772 | 38A8 | Unknown | Unknown | — | 0.0002a | — |
| CG6640 | 67D8 | Transport | Transporter activity | — | 0.0004a | — |
| CG14795 | 2B1 | Unknown | Unknown | — | 0.0008a | — |
| UbcD2 | 32A5 | Ubiquitin cycle | Ubiquitin conjugation | — | 0.0008a | — |
| Dll | 60E2 | Morphogenesis | Transcription factor | — | 0.0008a | — |
FDR < 0.2 or P < 0.001 for full-species and leg-type ANOVA.
MAS 5.0 normalization procedure.
GCRMA normalization procedure.
Male first to second legs (Barmina et al. 2005).
Female first legs to second legs (Barmina et al. 2005).
Male first legs to female first legs (Barmina et al. 2005).
TABLE 6.
Genes with significant leg type by species interaction terms
| Gene | Band | Biological process | Molecular function | FDRab | Pa | Pb |
|---|---|---|---|---|---|---|
| phr | 43E18 | DNA repair | Nucleic acid binding | 0.05b | — | <0.0001b |
| CG6230 | 32D4 | Calcium ion homeostasis | ATPase | 0.05a | 0.0002a | — |
| CG3402 | 61C8 | Unknown | Protein binding | 0.05b | — | 0.0003b |
| CG6640 | 67D8 | Transport | Transporter activity | 0.05a | 0.0006a | — |
| CG14096 | 76B3 | Unknown | Unknown | 0.05a | 0.0009a | — |
FDR < 0.2 or P < 0.001 in the full-species and leg-type ANOVA.
MAS 5.0 normalization procedure.
GCRMA normalization procedure.
TABLE 5.
Genes differentially expressed between species and between leg types
| Gene | Band | Biological process | Molecular function | FDRab | Pa | Pg |
|---|---|---|---|---|---|---|
| CG4962 | 72E1 | Unknown | Unknown | 0.05ab | <0.0001a | <0.0001b |
| CG14191 | 18A3 | Unknown | Unknown | 0.05ab | <0.0001a | 0.0007b |
| CG6640 | 67D8 | Transport | Transporter activity | 0.05a, 0.1b | <0.0001a | 0.0143b |
| ara | 69C8 | Morphogenesis | Transcription factor | 0.05a, 0.1b | 0.0026a | 0.0068b |
| UbcD2 | 32A5 | Ubiquitin cycle | Ubiquitin conjugation | 0.05a, 0.1b | 0.0003a | 0.0123b |
| kal-1 | 95E1 | Cell adhesion | Fibronectin type III | 0.05ab | 0.0014a | 0.0014b |
| CG4766 | 5D1 | Unknown | MAB-21 related | 0.1a, 0.2b | 0.0068a | 0.0420b |
| phr | 43E18 | DNA repair | Nucleic acid binding | 0.05b | — | 0.0003b |
| CG15186 | 83E8 | Unknown | Unknown | 0.05b | — | <0.0001b |
| CG16772 | 38A8 | Unknown | Unknown | 0.05a | 0.0002a | — |
| CG2791cd | 84E6 | Unknown | Unknown | 0.1b | — | 0.006b |
| CG2016 | 82E4 | Unknown | Takeout family | 0.2a | 0.0282a | — |
| CG13857cde | 94A11 | Unknown | Unknown | 0.2a | 0.03a | — |
FDR < 0.2 or P < 0.001 for species and leg in the full-species and leg-type ANOVA. P-values are for species differences; see Table 4 for leg-type values.
MAS 5.0 normalization procedure.
GCRMA normalization procedure.
Male first to second legs (Barmina et al. 2005).
Female first legs to second legs (Barmina et al. 2005).
Male first legs to female first legs (Barmina et al. 2005).
To understand how many genes are differentially expressed between species in the prothoracic leg relative to the mesothoracic leg, we also examined genes that were expressed differently between species in the first or second leg alone and those that were expressed differently between leg tissues in D. simulans or D. mauritiana alone. For analysis of species differences in gene expression in prothoracic or in mesothoracic legs, 1698 (655) prothoracic leg expressed genes were expressed differently between the two species, but only 444 (243) mesothoracic leg genes were significantly divergent for gene expression (supplemental Tables 5 and 6 at http://www.genetics.org/supplemental/). The difference in the proportion of divergent genes was significant, for both normalization procedures, at P < 0.0001 (chi-square, Yates' correction; Sokal and Rohlf 2001). Comparisons of prothoracic vs. mesothoracic leg gene expression within D. simulans or D. mauritiana, using the MAS 5.0 (GCRMA) normalization procedure, revealed 4 genes (6), for D. simulans and 5 genes (5) for D. mauritiana, differentially expressed between legs (supplemental Tables 7 and 8). None of these genes met the FDR < 0.2 cutoff and the gene list mostly overlaps with genes found in the analysis of overall leg expression differences.
As has been found in most comparisons of different normalization procedures, the MAS 5.0 and GCRMA procedures produced somewhat different gene lists (Verhaak et al. 2006). In general, about half of the genes found using one normalization procedure were also significant using the other. Of the genes found to differ between leg types using the MAS 5.0 normalization procedure (12 genes) or GCRMA normalization procedure (10 genes), 6 were significant for both (Table 4). For genes significantly different between species, there were 1804 genes in common between the two normalization procedures (supplemental Tables 3 and 4 at http://www.genetics.org/supplemental/). However, the handful of genes found to have a significant interaction between leg and species effects on gene expression were all specific to the normalization procedure used (Table 6).
Which of the genes that are regulated differently in the prothoracic leg (as compared to the mesothoracic leg) are good candidates for species differences in sex-comb tooth number? We found 13 genes, of the 17 significant for differences in leg segment expression, which were also expressed at significantly different levels across species. Several of these were genes of known function (ara, UbcD2, kal-1, and phr) with diverse functions from morphogenesis to DNA repair (Table 5). However, most were relatively uncharacterized CGs with little functional information available. From this list of genes, we are specifically interested in those located in the three main sex-comb QTL that we previously identified by fine mapping (76C–79D, 80A–84A, and 84A–93C). Three of these genes emerged as expression-based candidates, with positions located close to QTL identified in our mapping studies. These were CG15186 (83E8-F1), CG2791 (84E6), and CG2016 (82E). The genes Scr and dsx, which are common favorite candidate genes for sex-comb evolution, were not found to be divergent for gene expression between the species studied.
In addition to analyzing the normalized gene-level summarization scores, we also analyzed the probe-level data for our three candidate genes to account for possible sequence-divergence-based signal loss effects. The probe-level analysis results are reported in Table 7. For GCRMA normalization, all of the genes had significant tests for species differences, but for MAS 5.0 normalization, only CG2791 had a significant effect of species on expression level in the probe-level analysis. Differences between the results of the probe-level analysis and those of the gene-level summarized expression score analysis could be due to sequence divergence in the probe target regions, changes in cross-hybridizing regions, or differences in splicing between species. Another possibility is that there are differences in power between the different analyses and normalization procedures.
TABLE 7.
Probe-level analysis of newly identified candidate genes
| Leg
|
Species
|
||||
|---|---|---|---|---|---|
| Gene | Band | Pa | Pb | Pa | Pb |
| CG2791 | 84E6 | 0.044a | <0.0001b | <0.0001a | <0.0001b |
| CG2016 | 82E4 | — | <0.0001b | — | <0.0001b |
| CG15186 | 83E8 | — | — | — | 0.0002b |
MAS 5.0 probe-level normalization procedure.
GCRMA probe-level normalization procedure.
P-element insertions at CG2791:
In addition to being the only gene for which P-element insertions were already available, CG2791 is particularly interesting among the three genes identified in our experiments because of its proximity to dsx and because this gene is also expressed differently between the first and second legs in D. melanogaster (Barmina et al. 2005).
We found two D. melanogaster insertions at CG2791, P{SUPor-P}KG01932, and P{RS5}5-SZ-3716, which had both quantitative and qualitative effects on sex-comb tooth number (Figures 5 and 6; P{SUPor-P}KG01932, Bellen et al. 2004; P{RS5}5-SZ-3716, Ryder et al. 2004). The three other P insert lines that we examined (PBac{RB}CG2791e04491 and PBac{WH}CG2791f01853, Thibault et al. 2004; PBac{5HPw+}A146, Bellen et al. 2004) do not have obvious effects on the sex comb, but it is possible that they may have subtle effects that are not easily seen. Males from the KG01932 insertion line have a significant reduction in sex-comb tooth number. Homozygous insertion males have a mean sex-comb tooth number of 7.75, while males with only one copy of the insertion have 9.05 mean sex-comb teeth (two-tailed t-test; P < 0.0001). Homozygous males from the genetic background line (no insertion) have 11.28 mean sex-comb teeth (significantly different from males with either one or two copies of the KG01932 insertion, two-tailed t-test; P < 0.0001). Homozygous KG01932 males also sometimes had other defects, such as thin or abnormally pigmented sex-comb teeth at the proximal end of the sex comb (Figure 6). Approximately one-quarter of the sex combs that we examined also had small gaps between sex-comb teeth that were never observed in heterozygous or background males. Males from the P{RS5}5-SZ-3716 insertion line have a slight, but significant, reduction in sex-comb tooth number. Males homozygous for the insertion have a mean of 9.43 sex-comb teeth, while the isogenic w1118 males from the DrosDel genetic background stock have a mean of 10.55 (two-tailed t-test; P = 0.0002). This insertion also had a weakly penetrant qualitative effect on the sex comb. In ∼20% of the legs that we examined, the teeth of the sex comb were tightly packed relative to wild type and the proximal teeth of the comb were often placed at a lesser angle then the other teeth, causing a slight curve at the top (data not shown).
Figure 6.—
Effect of P{SUPor-P} insertion KG01932 on D. melanogaster sex combs. (a–c) The sex comb of males homozygous for the KG01932 insertion. Mean sex-comb tooth number is 7.75 teeth. Approximately a quarter of the sex combs that we scored had more severe sex-comb phenotypes, such as gaps and partially developed teeth.
One caveat to the results for the P{SUPor-P} insertion is that, while the two chromatin insulators in the P{SUPor-P} construct make it a very effective tool for mutagenesis, they may also act to alter gene expression at a distance (Roseman et al. 1995). Doublesex is located ∼7 kb downstream of CG2791 in D. melanogaster (Figure 5), so it is technically possible that the phenotype that we observed could be caused by a subtle effect on dsx expression rather than by disruption of CG2791 expression. However, dsx mutations are usually heavily pleiotropic, causing many intersexual phenotypes, and we did not observe any obvious intersex phenotypes when examining male and female leg bristles and other dimorphic traits (Nothiger et al. 1987).
Enhancer trap expression pattern:
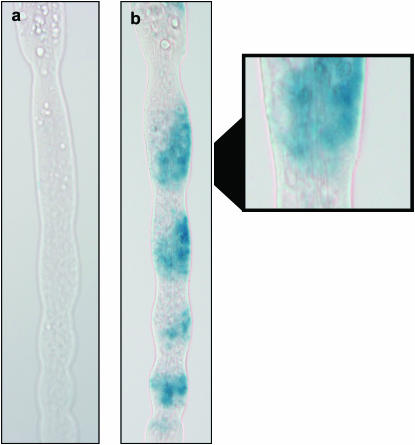
The expression pattern of CG2791 in D. melanogaster pupal legs was determined by β-gal staining of a lacZ reporter under control of the PBac{GAL4D, EYFP} CG2791PL00285 GAL4 enhancer trap insertion in CG2791. We found no visible staining at 24 hr postpupariation in the mesothoracic leg, but expression was detected in multiple segments on the ventral anterior surface of the prothoracic leg, including the region overlapping the developing sex comb (Figure 7).
Figure 7.—
Expression of lacZ under control of a GAL4 enhancer trap insertion in CG2791. Pupal legs of 24-hr-old PBac{GAL4D, EYFP} CG2791PL00285/UAS-LacZ D. melanogaster males were β-gal stained and slide mounted. (a) No staining was observed in the mesothoracic 24-hr pupal leg. (b) Staining on the ventral anterior surface of the prothoracic leg; the magnified region shows the location of developing sex-comb teeth on the basitarsus.
DISCUSSION
Introgression line mapping of the 73A–84AB region:
We have carried out a series of experiments to resolve the genetics of interspecific differences in a sexually dimorphic character, sex-comb tooth number. We started our analyses in pursuit of the gene or genes responsible for the appearance of a single large-effect QTL at 73A–84AB (True et al. 1997). A common limitation of QTL mapping is that a single peak might be caused by superposition of multiple linked QTL of smaller effect or even of opposite sign. We found that such a linked complex of several QTL, in which a combination of main and epistatic terms jointly contribute to between-species differences in sex-comb tooth number, is responsible for the large chromosome III QTL found in lower resolution mapping (Figure 4). We believe that our study is the first to firmly establish a set of closely linked composite effects underlying species divergence in morphology.
The effort to map sex-comb tooth number differences between D. simulans and D. mauritiana began with Coyne's (1985) chromosome-level mapping study. A significant effect on sex-comb tooth number was found for every chromosome arm with the largest increase associated with D. mauritiana 3R (0.73 sex-comb teeth, Coyne 1985). QTL mapping with multiple markers per chromosome arm also found a strong effect associated with the third chromosome and refined this to the region spanning the centromere between the 73A (tra) and 84AB (Antp) markers (True et al. 1997).
We found that both the 84A (Scr) and 93C (ebony) markers were associated with large positive effects on sex-comb tooth number in our nearly isogenic lines. There were also two factors associated with transgressive effects on sex-comb tooth number located between 76C and 93C in the regions 76C–79D and 80A–84A (Figure 4). Several compelling hypotheses might explain opposite-sign QTL for divergence in sex-comb tooth number. It is possible that the negative effects that we have observed are not additive and that instead of QTL with opposing sign, these effects in fact are epistatic effects conditional on the D. mauritiana or D. simulans genetic backgrounds. Abundant, background-dependent epistasis is the rule rather than the exception for other male fertility traits (Orr and Irving 2001). While we cannot say conclusively that these QTL are the result of interspecific epistasis, we did detect one occurrence of diminishing epistasis between the 80A marker and the 82C marker. The combination of the two D. mauritiana regions appears to reduce the expected negative effect of the 82C QTL. This is consistent with the hypothesis that the negative effect associated with the 82C marker is, at least partially, epistatic in nature.
Another explanation for transgressive QTL is that they may arise as a result of within-species evolutionary processes, including stabilizing selection and drift, or from heterogeneity in selection processes when a trait is subject to multiple reversing rounds of directional selection (Long et al. 1995; Bradshaw et al. 1998; Macdonald and Goldstein 1999; Zeng et al. 2000). Although sexual selection has probably played a role in generating the diversity of sex-comb morphology observed among Drosophila species as a whole, the majority of selection on sex-comb tooth number throughout the history of these species may have been stabilizing or sex-comb tooth number per se in these species might be relatively unimportant to mating. Additionally, many D. melanogaster genes have pleiotropic effects on the sex comb (for example, Saget et al. 1998). Some species differences in sex-comb tooth number could be a by-product of selection on other traits regulated by these genes. Indeed, the region on which we have focused also has significant effects on interspecific difference in anal plate bristle number, clasper bristle number, and possibly shape of the posterior lobe (Liu et al. 1996; True et al. 1997; Zeng et al. 2000).
Our data do not clearly favor any of these possible explanations. In whole-chromosome QTL mapping experiments, it is possible to use Orr's (1998) sign test to determine, at least, whether the distribution of effects is consistent with primarily directional selection on the trait. Unfortunately, Orr's test assumes that there is no significant contribution of epistatic effects to the overall difference between traits. Given the presence of both epistasis and multiple linked QTL, one would need both high-resolution genomewide mapping results and a statistical test robust for the presence of epistasis to determine if there has been significant directional selection on sex-comb tooth number between these species.
Gene expression in prothoracic and mesothoracic legs in D. simulans and D. mauritiana:
Genes involved in specification of the sex comb will be up- or downregulated in the first leg relative to the second ∼16 hr into pupariation. We examined species differences in transcript level at this time point in the first and second legs of D. simulans and D. mauritiana and found 4 genes (of a total of 10 genes) also found by Barmina et al. (2005). These genes were expressed differently between the first and second legs in D. melanogaster and were also significantly different between male legs in D. simulans and D. mauritiana (Scr, Nord, CG2791, and CG13857; Table 1). Of these 4 genes, Scr is known to specify front-leg identity and CG13857 was identified in the D. melanogaster experiment as a strong potential downstream target of Scr and dsx (Barmina et al. 2005). Doublesex itself was not detected as varying significantly between male first and second legs within D. melanogaster and we also did not detect any leg-type differences in dsx expression in D. simulans and D. mauritiana (Barmina et al. 2005). Several of the D. melanogaster sex-comb development candidate genes were also divergent for leg gene expression between D. simulans and D. mauritiana (ana, CG2791, and CG13857; Table 2).
Our experiment was designed to pick divergent sex-comb determination genes, but other genes involved in interesting first- and second-leg differences may also have been picked up in our screen. Prothoracic legs develop several rows of TBRs, which do not form on mesothoracic legs (Held 2002). A second difference between first and second legs is the development of the male-specific chemosensory bristles. The role of first-leg chemosensory bristles in pheromone detection has generated a strong interest in genes associated with this trait (Bray and Amrein 2003; Robertson et al. 2003). There are no known differences between these species in TBRs or chemosensory bristles. Nevertheless, it is possible that some of the species or leg expression differences that we have detected are associated with these other traits (see supplemental Tables 3 and 4 at http://www.genetics.org/supplemental/ for complete gene lists).
Between-species comparisons of gene expression in entire adult flies have repeatedly shown that genes with male-biased expression diverge faster than other genes (Meiklejohn et al. 2003; Nuzhdin et al. 2004; Zhang et al. 2004). Since most available species comparisons in gene expression have used whole-fly RNA extractions, we do not know how these differences are proportioned among the different tissues. Although many of the observed differences are probably associated with the testes and accessory glands (Arbeitman et al. 2004; Jagadeeshan and Singh 2005; Wagstaff and Begun 2005). We found that genes that were expressed in the sexually dimorphic prothoracic legs were significantly more likely to be divergent for gene expression than genes expressed in the mesothoracic legs, which are not sexually dimorphic (even though roughly equal numbers of genes were detected as present in both leg tissues).
Combined array and mapping results:
Taking a candidate gene approach to identifying the quantitative trait genes underlying the relatively large QTL regions mapped in most experiments can narrow the field from thousands of genes to only a few possibilities. However, two difficulties with this approach are apparent. There are frequently too few or too many candidate genes in a region of interest and there are enough genes of unknown function that taking a strictly function-based approach may result in failing to examine the correct gene or genes. The risk of overlooking the right gene or of unnecessarily including a functional candidate is increased when working with species other than D. melanogaster. The functions of most D. melanogaster named genes are probably conserved, because these are most often genes in which mutations have large, pleiotropic effects on development. However, it is possible that functional divergence has occurred even in the closely related species with which we are working (True and Haag 2001).
We elected to combine a traditional candidate gene approach with a development-based analysis of gene expression to narrow the list of candidate genes and to include genes of unknown function. While our methodology allowed us to quickly identify good candidates, a drawback is that such a specific technique could miss important genes that fall outside its scope. We may well have missed sex-comb tooth number genes with causal coding variation or expression differences manifest at other times, because we limited our study to gene expression differences occurring at a specific time point. However, many of the morphological differences that have been mapped to the gene level have turned out to be caused by differences in cis regulatory regions (Sucena and Stern 2000; Gompel et al. 2005; Clark et al. 2006; see, for review, Wray 2007). Therefore we believe that, in this case, focusing on expression differences is an acceptable limitation on our search for sex-comb tooth number divergence genes.
Of the three genes located in sex-comb tooth number QTL that were also identified in our expression screen, one is located very close to the Scr marker (CG2791, 84E6). The other two genes, CG15186 (83E8–F1) and CG2016 (82E), are located in a QTL associated with both negative and epistatic effects (80A–84A5). There were some differences in the results of the gene-level analysis and the probe-level tests. However, only CG15186 was not significant for both leg and species effects in the probe-level analysis (Table 7).
Very little is known about the genes that we have identified, but what we do know is intriguing. The function of CG2016 itself has not been studied, but it is a known member of the takeout gene family (Dauwalder et al. 2002). Fruitless and dsx appear to regulate takeout, and mutations in this gene severely affect male mating behavior. For CG2791, data and images from the BDGP in situ hybridization project show that this gene is expressed during several stages of embryonic development, beginning with maternal expression (Tomancak et al. 2002). We also confirmed, by a Gal4 enhancer trap insertion in CG2791, that in D. melanogaster this gene is expressed in the prothoracic pupal leg, but not in the mesothoracic leg (confirming the array results), and that the pattern of expression overlaps the area of the developing sex comb. Two P-element insertions proximal to the 3′- and 5′-end of this gene, respectively, also turned out to have an effect on sex-comb development in D. melanogaster. The two insertions had weakly penetrant qualitative effects on sex-comb morphology (KG01932, Figure 6, b and c; P{RS5}5-SZ-3716; see results for description). These insertions into CG2791 not only affect sex-comb morphology, but also specifically result in a reduced sex-comb tooth number phenotype. We found that males homozygous for the KG01932 insertion had approximately three fewer sex-comb teeth than males of the same genetic background who lacked the insertion (Figure 6a) and males homozygous for the P{RS5}5-SZ-3716 insertion had one fewer sex-comb tooth than genetic background males. Thus, at the least, we can functionally demonstrate that variation in this previously unknown gene could underlie a portion of the difference in sex-comb tooth number. On the basis of the associated mutant phenotypes, we propose to name this gene snaggle tooth (snt).
Both major functional candidate genes and one of our expression-based candidates are located in the 84A–93C interval associated with an increase in sex-comb tooth number (Scr, dsx, and snt). The relatively small size and coordinate position of multiple candidate genes make this region a promising location for future study. While we found no evidence for differences between these species in the expression of Scr and dsx, they could still be associated with sex-comb tooth number differences through variation in coding sequence, regulation of translation, or expression differences occurring earlier than the time point measured. Certainly, we cannot rule out a role for these genes in divergence of sex-comb tooth number. However, the fact that all three genes that we identified are basically of unknown function raises the possibility that evolution of sex-comb tooth number may primarily be a story of the evolution of “modifier genes” and not that of “major genes.”
As a secondary sexual character, evolutionary changes in sex-comb development may directly or indirectly play a role in the formation of reproductive isolation. Identifying and functionally characterizing genes that partially account for species differences in sex comb will allow us to examine, at the gene level, how evolutionary processes shape within-species variation in the network of genes underlying sex-comb development and how divergence in these genes produces the complex genetic architecture that we observe for species differences.
Acknowledgments
We thank Yun Tao for providing fly stocks used in initial mapping and John True and Cathy Laurie for their inspiring work on the genetics of species differences in morphology. We also thank Artyom Kopp for advice and assistance with slides and photography and the Kopp lab, as a whole, for allowing us to use their equipment and expertise. This work was funded by National Institutes of Health grant RO161773.
References
- Albertson, R. C., J. T. Streelman and T. D. Kocher, 2003. Directional selection has shaped the oral jaws of Lake Malawi cichlid fishes. Proc. Natl. Acad. Sci. USA 100: 5252–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S., W. Gish, W. Miller, E. Myers and D. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Arbeitman, M. N., A. A. Fleming, M. L. Siegal, B. H. Null and B. S. Baker, 2004. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development 131: 2007–2021. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., 1989. Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Baker, B. S., and K. A. Ridge, 1980. Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics 94: 383–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barmina, O., M. Gonzalo, L. M. McIntyre and A. Kopp, 2005. Sex- and segment-specific modulation of gene expression profiles in Drosophila. Dev. Biol. 288: 528–544. [DOI] [PubMed] [Google Scholar]
- Bellen, H., R. Levis, G. Liao, Y. He, J. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57: 289–300. [Google Scholar]
- Benjamini, Y., and D. Yekutieli, 2005. Quantitative trait loci analysis using the false discovery rate. Genetics 171: 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw, H. D., K. G. Otto, B. E. Frewen, J. K. McKay and D. W. Schemske, 1998. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus). Genetics 149: 367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, S., and H. Amrein, 2003. A putative Drosophila pheromone receptor expressed in male-specific taste neurons is required for efficient courtship. Neuron 39: 1019–1029. [DOI] [PubMed] [Google Scholar]
- Clark, R. M., T. N. Wagler, P. Quijada and J. Doebley, 2006. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 38: 594–597. [DOI] [PubMed] [Google Scholar]
- Cook, R., 1977. Behavioral role of the sexcombs in Drosophila melanogaster and Drosophila simulans. Behav. Genet. 7: 349–357. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., 1985. Genetic studies of three sibling species of Drosophila with relationship to theories of speciation. Genet. Res. 46: 169–192. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., S. Elwyn, S. Y. Kim and A. Llopart, 2004. Genetic studies of two sister species in the Drosophila melanogaster subgroup, D. yakuba and D. santomea. Genet. Res. 84: 11–26. [DOI] [PubMed] [Google Scholar]
- Dauwalder, B., S. Tsujimoto, J. Moss and W. Mattox, 2002. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 16: 2879–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., A. Stec and C. Gustus, 1995. teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141: 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. A., 1949. The Theory of Inbreeding. Hafner, New York.
- Galindo, M., S. Bishop, S. Greig and J. Couso, 2002. Leg patterning driven by proximal-distal interactions and EGFR signaling. Science 297: 256–259. [DOI] [PubMed] [Google Scholar]
- Gautier, L., L. Cope, B. Bolstad and R. Irizarry, 2004. affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315. [DOI] [PubMed] [Google Scholar]
- Gompel, N., B. Prud'homme, P. J. Wittkopp, V. A. Kassner and S. B. Carroll, 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487. [DOI] [PubMed] [Google Scholar]
- Hays, R., K. T. Buchanan, C. Neff and T. V. Orenic, 1999. Patterning of Drosophila leg sensory organs through combinatorial signaling by hedgehog, decapentaplegic and wingless. Development 126: 2891–2899. [DOI] [PubMed] [Google Scholar]
- Held, L. I., Jr., 2002. Imaginal Discs: The Genetic and Cellular Logic of Pattern Formation. Cambridge University Press, Cambridge, UK/London/New York.
- Held, L. I., M. J. Grimson and Z. Du, 2004. Proving an old prediction: the sex comb rotates at 16 to 24 hours after pupariation. Dros. Inf. Serv. 87: 76–78. [Google Scholar]
- Hey, J., and R. M. Kliman, 1993. Population genetics and phylogenetics of DNA sequence variation at multiple loci within the Drosophila melanogaster species complex. Mol. Biol. Evol. 10: 804–822. [DOI] [PubMed] [Google Scholar]
- Irizarry, R., Z. Wu and H. Jaffee, 2006. Comparison of Affymetrix GeneChip expression measures. Bioinformatics 22: 789–794. [DOI] [PubMed] [Google Scholar]
- Jagadeeshan, S., and R. Singh, 2005. Rapidly evolving genes of Drosophila: differing levels of selective pressure in testis, ovary, and head tissues between sibling species. Mol. Biol. Evol. 22: 1793–1801. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick, M., and N. Barton, 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp, A., and J. True, 2002. Evolution of male sexual characters in the Oriental Drosophila melanogaster species group. Evol. Dev. 4: 278–291. [DOI] [PubMed] [Google Scholar]
- Levene, H., 1960. Robust tests for the equality of variance, in Contributions to Probability and Statistics, edited by I. Olkin. Stanford University Press, Stanford, CA.
- Lexer, C., D. M. Rosenthal, O. Raymond, L. A. Donovan and L. H. Rieseberg, 2005. Genetics of species differences in the wild annual sunflowers, Helianthus annuus and H. petiolaris. Genetics 169: 2225–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., J. Mercer, L. Stam, G. Gibson, Z. Zeng et al., 1996. Genetic analysis of a morphological shape difference in the male genitalia of Drosophila simulans and D. mauritiana. Genetics 142: 1129–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A. D., S. L. Mullaney, L. A. Reid, J. D. Fry, C. H. Langley et al., 1995. High resolution mapping of genetic factors affecting abdominal bristle number in Drosophila melanogaster. Genetics 139: 1273–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald, S., and D. Goldstein, 1999. A quantitative genetic analysis of male sexual traits distinguishing the sibling species Drosophila simulans and D. sechellia. Genetics 153: 1683–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader et al., 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow, T., D. Bustoz and S. Pitnick, 1996. Sexual selection and a secondary sexual character in two Drosophila species. Anim. Behav. 52: 759–766. [Google Scholar]
- Meiklejohn, C., J. Parsch, J. Ranz and D. Hartl, 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 100: 9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveira, H., and A. Barbadilla, 1992. The theoretical distribution of lengths of intact chromosome segments around a locus held heterozygous with backcrossing in a diploid species. Genetics 130: 205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothiger, R., M. Leuthold, N. Andersen, P. Gerschwiller, A. Gruter et al., 1987. Genetic and developmental analysis of the sex-determining gene double sex (dsx) of Drosophila melanogaster. Genet. Res. 50: 113–123. [Google Scholar]
- Nuzhdin, S., and S. Reiwitch, 2000. Are the same genes responsible for intra- and interspecific variability for sex comb tooth number in Drosophila? Heredity 84: 97–102. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S. V., M. L. Wayne, K. L. Harmon and L. M. McIntyre, 2004. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol. Biol. Evol. 21: 1308–1317. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1998. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52: 935–949. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., and S. Irving, 2001. Complex epistasis and the genetic basis of hybrid sterility in the Drosophila pseudoobscura Bogota-USA hybridization. Genetics 158: 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattatucci, A. M., D. C. Otteson and T. C. Kaufman, 1991. A functional and structural analysis of the Sex combs reduced locus of Drosophila melanogaster. Genetics 129: 423–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promislow, D., E. Smith and L. Pearse, 1998. Adult fitness consequences of sexual selection in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 95: 10687–10692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psarros, M., S. Heber, M. Sick, G. Thoppae, K. Harshman et al., 2005. RACE: Remote Analysis Computation for gene Expression data. Nucleic Acids Res. 33: W638–W643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb, C., 2001. The establishment of segmentation in the Drosophila leg. Development 128: 4511–4521. [DOI] [PubMed] [Google Scholar]
- Robertson, H. M., C. G. Warr and J. R. Carlson, 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100: 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman, R., E. Johnson, C. Rodesch, M. Bjerke, R. Nagoshi et al., 1995. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics 141: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder, E., F. Blows, M. Ashburner, R. Bautista-Llacer, D. Coulson et al., 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatti, C., S. Service and N. Freimer, 2003. False discovery rate in linkage and association genome screens for complex disorders. Genetics 164: 829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saget, O., F. Forquignon, P. Santamaria and N. Randsholt, 1998. Needs and targets for the multi sex combs gene product in Drosophila melanogaster. Genetics 149: 1823–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell, D., K. Byrne, J. Gordon, S. Wong and K. Wolfe, 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440: 341–345. [DOI] [PubMed] [Google Scholar]
- Schawaroch, V., 2002. Phylogeny of a paradigm lineage: the Drosophila melanogaster species group (Diptera: Drosophilidae). Biol. J. Linn. Soc. 76: 21–37. [Google Scholar]
- Simonsen, K. L., and L. M. McIntyre, 2004. Using alpha wisely: improving power to detect multiple QTL. Stat. Appl. Genet. Mol. Biol. 3: 1–23. [DOI] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 2001. Biometry. W. H. Freeman, San Francisco.
- Spieth, H. T., 1952. Mating behavior within the genus Drosophila (Diptera). Bull. Am. Mus. Nat. Hist. 99: 395–474. [Google Scholar]
- Stern, C., and C. Tokunaga, 1967. Nonautonomy in differentiation of pattern-determining genes in Drosophila, I. The sex comb of eyeless-dominant. Proc. Natl. Acad. Sci. USA 57: 658–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H., W. P. Spencer and J. T. Patterson, 1942. The classification of the genus Drosophila, with descriptions of nine new species. Univ. of Texas Publ. 4213: 5–51. [Google Scholar]
- Sucena, E., and D. L. Stern, 2000. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc. Natl. Acad. Sci. USA 97: 4530–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S., C. T. Ting and C. I. Wu, 2004. The normal function of a speciation gene, Odysseus, and its hybrid sterility effect. Science 305: 81–83. [DOI] [PubMed] [Google Scholar]
- Sweigart, A., L. Fishman and J. Willis, 2006. A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics 172: 2465–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., S. Chen, D. Hartl and C. Laurie, 2003. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics 164: 1383–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta, H., and T. Takano-Shimizu, 2006. Genetic architecture of variation in sex-comb tooth number in Drosophila simulans. Genet. Res. 87: 93–107. [DOI] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. [DOI] [PubMed] [Google Scholar]
- Tokunaga, C., 1962. Cell lineage and differentiation on the male foreleg of Drosophila melanogaster. Dev. Biol. 4: 489–516. [DOI] [PubMed] [Google Scholar]
- Tomancak, P., A. Beaton, R. Weiszmann, E. Kwan, S. Shu et al., 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3: RESEARCH0088.1–RESEARCH0088.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True, J. R., and E. S. Haag, 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 3: 109–119. [DOI] [PubMed] [Google Scholar]
- True, J., J. Mercer and C. Laurie, 1996. a Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142: 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True, J., B. Weir and C. Laurie, 1996. b Genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics 142: 819–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True, J., J. Liu, L. Stam, Z.-B. Zeng and C. Laurie, 1997. Quantitative genetic analysis of divergence in male secondary sexual traits between Drosophila simulans and Drosophila mauritiana. Evolution 51: 816–832. [DOI] [PubMed] [Google Scholar]
- Verhaak, R. G., F. J. Staal, P. J. Valk, B. Lowenberg, M. J. Reinders et al., 2006. The effect of oligonucleotide microarray data pre-processing on the analysis of patient-cohort studies. BMC Bioinformatics 7: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff, B., and D. Begun, 2005. Molecular population genetics of accessory gland protein genes and testis-expressed genes in Drosophila mojavensis and D. arizonae. Genetics 171: 1083–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller, J. I., J. Z. Song, D. W. Heyen, H. A. Lewin and M. Ron, 1998. A new approach to the problem of multiple comparisons in the genetic dissection of complex traits. Genetics 150: 1699–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker, J. C., R. Thompson and P. M. Visscher, 1996. On the mapping of QTL by regression of phenotype on marker-type. Heredity 77: 23–32. [Google Scholar]
- Wray, G.A., 2007. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8: 206–216. [DOI] [PubMed] [Google Scholar]
- Wu, Z., and R. Irizarry, 2004. Preprocessing of oligonucleotide array data. Nat. Biotech. 22: 656–658. [DOI] [PubMed] [Google Scholar]
- Zeng, Z. B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z.-B., J. Liu, L. Stam, C.-H. Kao, J. Mercer et al., 2000. Genetic architecture of a morphological shape difference between two Drosophila species. Genetics 154: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., T. Hambuch and J. Parsch, 2004. Molecular evolution of sex-biased genes in Drosophila. Mol. Biol. Evol. 21: 2130–2139. [DOI] [PubMed] [Google Scholar]