Abstract
QTL mapping experiments yield heterogeneous results due to the use of different genotypes, environments, and sampling variation. Compilation of QTL mapping results yields a more complete picture of the genetic control of a trait and reveals patterns in organization of trait variation. A total of 432 QTL mapped in one diploid and 10 tetraploid interspecific cotton populations were aligned using a reference map and depicted in a CMap resource. Early demonstrations that genes from the non-fiber-producing diploid ancestor contribute to tetraploid lint fiber genetics gain further support from multiple populations and environments and advanced-generation studies detecting QTL of small phenotypic effect. Both tetraploid subgenomes contribute QTL at largely non-homeologous locations, suggesting divergent selection acting on many corresponding genes before and/or after polyploid formation. QTL correspondence across studies was only modest, suggesting that additional QTL for the target traits remain to be discovered. Crosses between closely-related genotypes differing by single-gene mutants yield profoundly different QTL landscapes, suggesting that fiber variation involves a complex network of interacting genes. Members of the lint fiber development network appear clustered, with cluster members showing heterogeneous phenotypic effects. Meta-analysis linked to synteny-based and expression-based information provides clues about specific genes and families involved in QTL networks.
MOST naturally occurring genetic variation in populations reflects polymorphic alleles that individually have relatively small effects but collectively result in continuous variation among members of the population. Through genetic mapping, the number and location of loci associated with complex trait variation, i.e., quantitative trait loci or QTL, can be estimated and used to infer the genetic basis of traits that differ between varieties and/or species (Paterson et al. 1988). DNA markers linked to QTL can also be used as diagnostic tools in the selection of desirable genotypes (marker-assisted selection) and as a starting point for cloning of QTL. For these reasons, vast numbers of QTL representing a myriad of traits have been mapped in agronomically important crops, and also in botanical models and animals. A handful of genes underlying QTL have been cloned (e.g., Frary et al. 2000) based largely on fine mapping (Paterson et al. 1990).
A recurring complication in the use of QTL data is that different parental combinations and/or experiments conducted in different environments often result in identification of partly or wholly nonoverlapping sets of QTL. The majority of such differences in the QTL landscape are presumed to be due to environment sensitivity of genes. The use of stringent statistical thresholds to infer QTL while controlling experiment-wise error rates (Lander and Botstein 1989; Churchill and Doerge 1994) implies that only a small fraction of these nonoverlapping QTL can be attributed to false-positive results. Small QTL with opposite phenotypic effects might occasionally be closely linked in coupling in early-generation populations, and separated only in advanced-generation populations after additional recombination.
Comparison of multiple QTL mapping experiments by alignment to a common reference map offers a more complete picture of the genetic control of a trait than can be obtained in any one study. One such trait, the genetic control of variation in growth and development of seed-borne epidermal “lint” fibers, is a natural priority in cotton genome analysis. All 50 Gossypium species have seed-borne epidermal trichomes, often referred to as “fuzz” fibers. “A” genome diploid cottons are distinct from their sister “F” and “B” genomes in that both wild and cultivated forms have longer lint fibers with secondary thickening, which are spinnable; this feature can thus be inferred to have evolved after the divergence of the A, B, and F genomes from a common ancestor ∼5–7 MYA (Wendel 1989). Some view lint as distinct from fuzz fibers based on the secondary thickening, while others view the two as extremes in a continuum. However, all agree that economically important lint fibers are much longer, reaching lengths of ∼20 mm in cultivated A-genome diploids Gossypium herbaceum and G. arboreum. Wild tetraploid cottons have fibers resembling those of wild diploids; however, domestication and scientific improvement of AD tetraploids G. hirsutum and G. barbadense (from different tetraploid clades) have increased fiber length to as much as ≈ mm or more, and also improved other aspects of fiber quality (strength, fineness, and elongation) as well as higher yield, even in environments to which both have been adapted by scientific plant breeding (Jiang et al. 1998).
Accurate measurement of cotton fiber quality requires complex instrumentation and therefore has been a high-priority trait for which to establish diagnostic DNA markers, in view of the fact that cotton provides ∼45% of the raw material for the ∼$500 billion/year worldwide textile industry. Cotton fiber quality is a complex characteristic with many components affecting both the efficacy of processing by textile spinning machinery and the comfort attributes associated with cotton fabrics. To date, >200 QTL related to various aspects of fiber quality have been mapped using different kinds of populations including F2 (Jiang et al. 1998; Paterson et al. 2003; Mei et al. 2004; Lin et al. 2005; Shen et al. 2005; Udall et al. 2006), advanced backcross (Chee et al. 2005a,b; Draye et al. 2005; Lacape et al. 2005), and cytogenetic stocks (Saha et al. 2004).
Meta-analysis of cotton fiber QTL promises to contribute to our understanding of fundamental questions and to expedite crop improvement. Gossypium is an especially good model in which to advance understanding of the consequences of polyploidy in that both derived tetraploids and their ancestral diploids (or at least very close relatives) are known and remain extant, yet the polyploidization event is sufficiently old [∼1–2 million years (Senchina et al. 2003)] for adaptive evolution to have occurred. One particularly exciting opportunity is to shed light on why tetraploid cottons consistently have higher yield and quality than the modern descendants of their diploid progenitors, even in environments to which both have been adapted by scientific plant breeding (Jiang et al. 1998).
Research into the genetic control of cotton fiber development may also benefit from progress in understanding the growth and development of hair-bearing epidermal cells (trichomes) in Arabidopsis. Indeed, Gossypium and Arabidopsis are thought to have shared common ancestry ∼83–86 MYA (Benton 1993), and cotton may be the best crop outside of the Brassicales in which to employ translational genomics from Arabidopsis.
Using a high-density reference genetic map which consists of 3475 loci in total, herein we report the alignment of 432 QTL involving cotton fiber quality (Jiang et al. 1998; Paterson et al. 2003; Chee et al. 2005a,b; Draye et al. 2005), yield (Jiang et al. 1998; Saranga et al. 2001, 2004; Rong et al. 2005b), leaf morphology (Jiang et al. 2000; Waghmare et al. 2005), flower morphology, resistance to bacteria (Wright et al. 1998), trichome distribution and density (Wright et al. 1999), and other traits that were mapped in 11 populations. All QTL were also projected onto a consensus map, which was inferred to resemble the DNA marker arrangement of the hypothetical ancestor of the two subgenomes of tetraploid cotton (Rong et al. 2005a). The consensus map has improved our ability to deduce cotton-Arabidopsis synteny relationships and thus fosters study of correspondence between the cotton QTL and fiber or trichome-related Arabidopsis genes. To encourage further utilization and on-line community access to these data, a CMap resource was developed and can be accessed at our website.
MATERIALS AND METHODS
Populations:
QTL and mutant data used in this study were published previously except for those from populations segregating for n2 and im mutants (see below), as described in Table 1.
TABLE 1.
Crosses and QTL of different populations and their file names at the CMap website (http://www.plantgenome.uga.edu/cmap)
| Crossa | Population (no. individuals) | Categoryb | CMap db identifiers | Total QTL | QTL on Atc | QTL on A | QTL on Dtc | Reference |
|---|---|---|---|---|---|---|---|---|
| Pima S-7 (GB) × im (GH) | F2 (124) | EL, FF, FL, FS, FU, YC | im | 17 | 10 | 7 | Herein | |
| FM | 36 | 16 | 20 | A. Desai, P. W. Chee, O. L. May, J. Rong, G. J. Pierce, C. J. Rogers, V. N. Waghmare and A. H. Paterson (unpublished results) | ||||
| Tamcot 2111(GH) × Pima S6 (GB) | BC3F2 (24 families, 33–191 plants/family) | EL, FF, FL, FU | ABHB | 108 | 45 | 63 | Chee et al. (2005a,b); Draye et al. (2005) | |
| CAMD-E (GH) × Sea Island Seaberry (GB) | F2 (271) | EL, FF. FL. FS, YC | HBJF | 14 | 4 | 10 | Jiang et al. (1998) | |
| Pima S-7 (GB) × n2 (GH) | F2 (124) | EL, FF, FL, FS, FU, YC | n2 | 22 | 11 | 11 | Herein | |
| Siv′on (GH) × F-177 (GB) | F2 (406) | EL, FC, FF, FL, FS, FU | HBPS | 79 | 36 | 43 | Paterson et al. (2003) | |
| F3 (208) | DP, YC | 80 | 40 | 40 | Saranga et al. (2004) | |||
| Deltapine 61 (GH) × Sea Island Seaberry (GB) | F2 (180) | LM | HBJL | 21 | 7 | 14 | Jiang et al. (2000) | |
| TMS-22 (GH) × acc. WT936 (GT) | F2 (82) | LM | HT | 11 | 7 | 4 | Waghmare et al. (2005) | |
| Pima S-7 (GB) × Empire B2b6, B2, B3, S295 (GH) | F2 (119–150) | BB | BHW | 7 | 1 | 6 | Wright et al. (1998) | |
| TR | 8 | 5 | 3 | Wright et al. (1999) | ||||
| Pima S-7 (GB) × Fbl (GH) | F2 (140) | YC | Fbl | 1 | 1 | Rong et al. (2005b) | ||
| Pima S-7 (GB) × N1 (GH) | F2 (143) | YC | N1 | 1 | 1 | Rong et al. (2005b) | ||
| Total (tetraploid) | 405 | 184 | 221 | |||||
| Acc. PI529740 (GA) × Acc. PI529670 (GHe) | F2 (167) | FM | A | 27 | 27 | A. Desai, P. W. Chee, O. L. May, J. Rong, G. J. Pierce, C. J. Rogers, V. N. Waghmare and A. H. Paterson (unpublished results) |
All populations are crosses between G. hirsutum and G. barbadense except for A (between G. arboreum acc. PI529740 and G. herbaceum acc. PI529670) and HT (between G. hirsutum acc. TMS-22 and G. tomentosum acc. WT936).
EL, elongation; FF, fineness; FL, length; FS, strength; FU, uniformity; YC, yield component; FM, flower morphology; FC, color; DP, drought response; LM, leaf morphology; BB, bacterial resistance; TR, trichome.
At, Dt: Tetraploid chromosomes derived from A-genome and D-genome diploid progenitors, respectively.
QTL mapping of n2 and im populations:
Two new F2 populations derived from crosses between G. barbadense var. Pima S-7 and G. hirsutum cv. Texas Marker-1 (TM-1) isogenic lines with n2 and im mutants, respectively, were mapped.
A total of 124 F2 plants from each of n2 and im crosses were grown in a field at the Coastal Plain Experiment Station, Tifton, GA during the 2002 season. Five traits reflecting fiber quality parameters were determined by the Cotton Incorporated Textile Services Laboratory (Cotton Incorporated, Cary, NC) using the High-Volume Precision Instrument (HVI), including upper-half mean length (HVuhm), percentage short fiber by weight (HVsfc), uniformity index (HVui), fiber strength (FS), fiber elongation (ELO), and micronaire (Mic). In addition, two fiber yield components, lint percentage and lint index, were measured as reported (Rong et al. 2005b).
DNA markers were selected from a high-density reference map (Rong et al. 2004). Procedures for linkage analysis were reported previously (Rong et al. 2004). Composite interval mapping (Zeng 1994) was performed using Windows QTL Cartographer (Wang et al. 2005) (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). A stringent LR threshold of 13.8 (equivalent to LOD score 3.0) was used to declare significant QTL to keep the experiment-wise likelihood of even one false positive below 5% in the large genome of cotton (Jiang et al. 1998). Effects and percent of phenotypic variance (PV) explained by single QTL (R2) were estimated with Windows QTL Cartographer 2.0 at likelihood peaks.
Integration of QTL from different populations:
For convenience of description, three names were used for different types of genetic maps: individual map, reference map (Rong et al. 2004), and consensus map (Rong et al. 2005a). Individual maps were the original QTL maps summarized in Table 1. To align QTL from different individual maps, the reference map was used, from which subsets of informative markers were drawn to make the individual maps. Finally, some analyses also used a “consensus map” that depicts the marker arrangement along the genome of a hypothetical common ancestor that gave rise to the diploid progenitors of polyploid cotton, inferred as described elsewhere (Rong et al. 2005a). Most of the QTL locations in reference and consensus maps were determined by BioMERCARTOR (Version 2) (Arcade et al. 2004). QTL in regions where the marker order was different between the individual map and reference map were plotted based on the location of the marker nearest to the likelihood peak on the reference map. The individual maps were made over several years, during which changes in the reference map occurred; marker and chromosome nomenclature was all synchronized with that of the current reference map (Rong et al. 2004). Redundant probes, identified as detailed elsewhere (Rong et al. 2005a), were removed. Nomenclature of QTL in the reference map was standardized to the system reported (Chee et al. 2005a,b; Draye et al. 2005), that is, trait name first, then the name of the chromosome on which the QTL was located, followed by a number indicating the loci on this chromosome. If two QTL were reported by Chee et al. (2005a,b) and Draye et al. (2005) on a chromosome, additional QTL for the same trait mapped in the same chromosome of other individual maps will be given a number with two digits to represent their relative locations. For example, because two QTL for fiber length were mapped on linkage group D08 by Chee et al. (2005b) and named FLD08.1 and FLD08.2 respectively, another QTL mapped on the same chromosome was named FLD08.15 representing the relative location of its likelihood peak between FLD08.1 and FLD08.2. The original name for each QTL used in each map, and the name in CMap of each trait as well as their category and source were summarized in supplemental Table 9 (http://www.genetics.org/supplemental/).
CMap display of genetic maps:
CMap v0.16 was downloaded from the Generic Components for Model Organism Database project (GMOD; http://www.gmod.org). Genetic mapping data were prepared in spreadsheets and imported into CMap.
RESULTS
A CMap visualization tool for meta-analysis:
To streamline comparisons of QTL and other genomic data among chromosomes, subgenomes, populations, and syntenic locations in Arabidopsis, we developed a cotton CMap resource (publicly available at http://www.plantgenome.uga.edu/cmap). Using BioMERCARTOR (Arcade et al. 2004), a total of 432 QTL from 11 individual maps (Table 1) were aligned to the reference map (Rong et al. 2004) and to the consensus map representing inferred gene (marker) arrangements along the chromosomes of a common ancestor of the A and D genomes (Rong et al. 2005a). Among them, 224 QTL from five populations were related to fiber quality, 64 from four populations were related to yield components, 63 from two populations (one tetraploid and one diploid A genome) were related to flower morphology, 33 from one population were related to drought response, 32 from two populations were related to leaf morphology, 8 from four populations were related to trichome density, 7 from four populations were related to bacterial blight resistance, and 1 from one population was related to earliness. Available from the CMap resource are individual QTL maps, the reference map, and the consensus map, plus relationships of cotton chromosomes to inferred gene orders for a hypothetical common ancestor of cotton and Arabidopsis (Rong et al. 2005a) based on correspondence inferred using CSII (Levine 2002) and FISH (Calabrese et al. 2003).
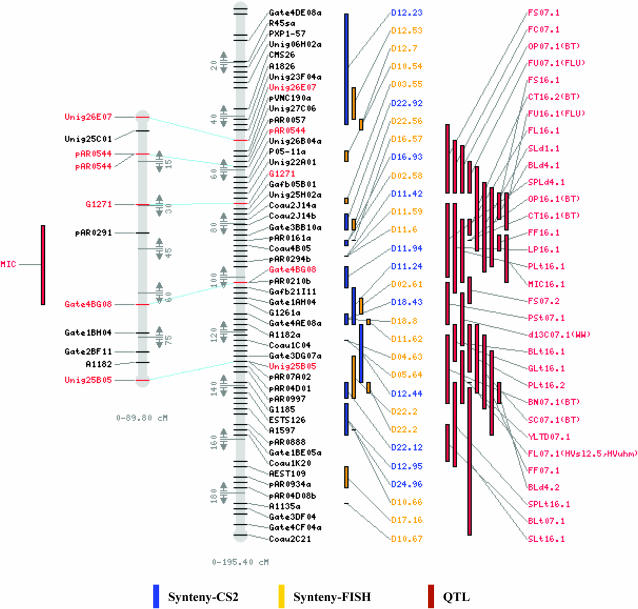
In an example of the visualizations possible with this resource (Figure 1), a QTL on one chromosome (Chr. 16) of an individual population (n2) is aligned to the corresponding “chromosome” of the consensus map, also indicating corresponding regions of Arabidopsis and showing all QTL known from all populations for the region shown. Thus, the CMap resource expedites the comparison of QTL between different homeologous chromosomes as well as between cotton QTL and Arabidopsis orthologs in the syntenic regions.
Figure 1.—
A CMap display of the comparison between an individual map and a consensus map. The left map is chromosome 16 from the n2 population including one QTL (MIC) shown on the left side of the map. The right map is consensus chromosomes C4, reconstructed from the At and Dt maps as described (Rong et al. 2005a). To the immediate right of this map, blocks of inferred synteny with Arabidopsis are shown. D (or Ds) number indicates correspondence to the accordingly numbered Arabidopsis α-duplicated segment (Bowers et al. 2003), and the number after the dot indicates the span of this particular segment of correspondence in the α-duplicated segment (which can be visualized at our CMAP website; also see Rong et al. 2005a for further details). To the right of blocks of inferred synteny, QTL likelihood intervals are plotted, labeled as described in the text. Where appropriate, parenthetical information indicates experimental treatments (for example, ww, well-watered, and bt, both treatments: Paterson et al. 2003), or alternate name of the trait if different in prior publications are presented in parenthesis after each QTL name. Detailed explanation of these descriptions can be found in the references cited.
Cotton fiber QTL are enriched in the D subgenome:
More QTL were detected in the tetraploid D (hereafter Dt) than in the tetraploid A (hereafter At) subgenome, specifically 221 vs. 184 (Table 1). Of special interest were QTL related to lint fiber quality characteristics, in that the At progenitor produces spinnable fiber but the Dt progenitor does not. We focused this analysis on fiber quality characteristics because of their relatively high heritability (May and Wofford 2000) and economic importance. QTL for fiber yield components from a small number of studies are also in the CMap database, but in view of the complexity and low heritability of this trait we considered that more extensive testing would be needed before a meaningful meta-analysis could be performed on these.
Based on three well-studied crosses for which both parents are elite cultivars or breeding lines (Table 2), the Dt subgenome contained 112 lint fiber-related QTL vs. 84 in the At (Tables 2 and 3), a marginally significant difference (P = 0.0455). The 196 genes and QTL mapped in these elite crosses were further classified into the following trait categories: elongation (EL), fiber color (FC), fiber fineness (FF), fiber length (FL), fiber strength (FS), fiber uniformity (FU), micronaire (MIC), and short fiber content (SF) (Table 3). All traits except FC and FL have more QTL on Dt than on At, but the major contributors to the subgenome bias are FS and MIC, for which the Dt subgenome contains significantly more QTL than does the At subgenome (P = 0.041, P = 0.005, respectively; Table 3). FF is controlled by the largest number of loci (57), followed by FL and EL (34, 33). SF and FC are controlled by the fewest loci, 8 and 11 respectively.
TABLE 2.
Chi-square test of fiber-related QTL distributed on At and Dt subgenome in three non-mutant populations
| Populationa | QTL | Atb | Dtb | Goodness of fit to 1:1 | Reference |
|---|---|---|---|---|---|
| CAMD-E (GH) × Sea Island Seaberry (GB) | 9 | 3 | 6 | 0.317 | Jiang et al. (1998)c |
| Siv′on (GH) × F-177 (GB) | 79 | 36 | 43 | 0.431 | Paterson et al. (2003) |
| Tamcot 2111 (GH) × Pima S6 (GB) | 108 | 45 | 63 | 0.083 | Chee et al. (2005a,b); Draye et al. (2005) |
| Total | 196 | 84 | 112 | 0.045 |
GH, G. hirsutum; GB, G. barbadense
At, Dt: Tetraploid chromosomes derived from A-genome and D-genome diploid progenitors, respectively.
Four QTL for yield components and one for earliness not included, as discussed in text.
TABLE 3.
Chi-square test of the QTL distribution between tetraploid At and Dt subgenomes for different fiber traits
| Traita | QTL | Atb | Dtb | Goodness of fit to 1:1 |
|---|---|---|---|---|
| EL | 33 | 15 | 18 | 0.6015 |
| FC | 11 | 7 | 4 | 0.3657 |
| FF | 57 | 25 | 32 | 0.3538 |
| FL | 34 | 17 | 17 | 1.0000 |
| FS | 24 | 7 | 17 | 0.0412 |
| FU | 21 | 10 | 11 | 0.8273 |
| MIC | 8 | 0 | 8 | 0.0047 |
| SF | 8 | 3 | 5 | 0.4795 |
| Total | 196 | 84 | 112 | 0.0455 |
EL, elongation; FC, color; FF, fineness; FL, length; FS, strength; FU, uniformity; MIC, micronaire; SF, short fiber content.
At, Dt: Tetraploid chromosomes derived from A-genome and D-genome diploid progenitors, respectively.
Cotton fiber QTL are clustered:
QTL were non-randomly distributed across chromosomes and chromosomal regions (supplemental Table 1 at http://www.genetics.org/supplemental/). Chromosome length (from the genetic map, in centimorgans) had a significant relationship with the number of total QTL and fiber QTL in the Dt subgenome (r = 0.62, P < 0.01; and r = 0.51, P < 0.05, respectively), but not in the At (r = −0.11, r = 0.15). As a result, although the Dt genome as a whole carries more QTL for fiber quality than does the At genome, the distributions of QTL between different pairs of homeologs varied widely. Seven Dt chromosomes or linkage groups (Chr. 14, 17, 20, 22, 23; LG D03 and D08) had more fiber-related QTL than their homeologous partners, with the largest difference between linkage group D08 (21 QTL) and Chr. 4/5 (8 QTL). Five At chromosomes or linkage groups (Chr. 1, 6, 7; LG A01 and A03) have more fiber QTL than their homeologous counterparts, with the largest difference between Chr. 06 (the shortest At chromosome, 9 QTL), and Chr. 25 (4 QTL).
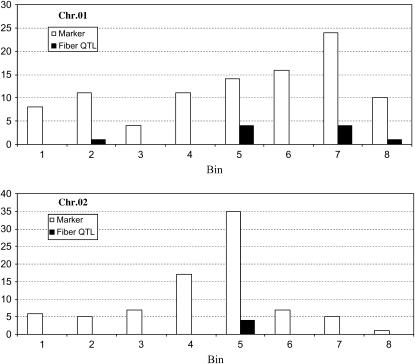
When each chromosome was divided into bins of 20 cM and the center of the QTL likelihood interval used as the QTL location, fiber-related QTL were found to be clustered in some chromosomal regions (Figure 2; supplemental Table 2 at http://www.genetics.org/supplemental/). Based on the total of 235 bins across the cotton genome and 196 fiber QTL, the Poisson probability distribution function indicated that the presence of three or more QTL in a bin was significantly (P < 0.05) higher than the random expectation. A total of 87 (44.5%) QTL were located in 24 such bins (supplemental Table 2). The three bins with the largest numbers of fiber QTL (5 or 6) were on Chr. 17 and linkage group D08 (2 bins). In no cases were statistically significant QTL clusters found in corresponding regions of homeologous chromosomes. These clusters are further elucidated below as examples of how integration of positional, comparative, expression, and functional data might narrow the lists of candidate genes that may represent the QTL.
Figure 2.—
Number and distribution of genetic markers and fiber-related QTL in 20-cM bins along chromosomes 1 and 2 of a reference genetic map (Rong et al. 2004). Open bars represent marker number and solid bars are 2-LOD likelihood intervals for fiber QTL.
Homology and homeology:
Integration of QTL from different populations into a common map facilitates exploration of their allelic and homeologous relationships, albeit at a level of resolution limited by comparative marker densities, variation in recombination rates in different crosses, variation in gene densities across the genome, and other factors. QTL for the same trait from different crosses, that had a likelihood peak within the same 20-cM bin, were considered potential alleles (albeit requiring further study to confirm). Considering the different fiber-related traits individually, with only a maximum of 57 QTL per trait (FF), the occurrence of even 2 QTL in the same bin was statistically unlikely to occur by chance (at P < 0.05). A total of four such clusters were found, each comprising 2 QTL for FF (three cases) and EL (one case) (supplemental Table 3 at http://www.genetics.org/supplemental/). The 8 QTL compose 4.1% of the total. All of these clustered QTL were located in fiber QTL-rich bins identified above (bin 5 of Chr. 02, bin 2 of Chr. 12, bin 6 of Chr. 15 and bin 8 of D08), although these bins collectively compose only 3.6% of the genome. The two EL QTL in bin 8 of D08 were detected in the same population (ELD08.3 and ELD08.4). However, the members of the three pairs of FF QTL, respectively, were detected in different experiments.
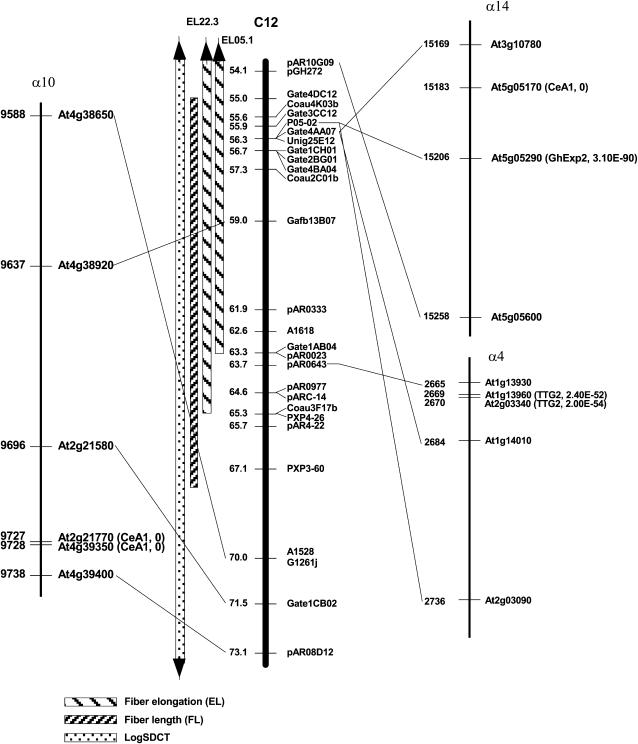
Likewise, QTL for the same trait were considered to be potentially homeologous if their likelihood peaks fell within pairs of bins inferred to be homeologous based on nonrandom alignments of duplicated DNA markers (Rong et al. 2004). Based on these criteria, a total of 41 (20.9%) QTL from six fiber traits were found to be potentially homeologous (supplemental Table 4 at http://www.genetics.org/supplemental/). Fiber fineness was unusual in that 24 (42.1%) of 57 QTL were potentially homeologous. Figure 3 displays a middle region of consensus chromosome C12 with two potentially homeologous loci for EL that were originally mapped on Chr. 05 by Paterson (EL05.1) (Paterson et al. 2003) and Chr. 22 by Chee (EL22.3) (Chee et al. 2005a). This is an example of homeologous loci for a fiber related trait detected in different experiments.
Figure 3.—
Genetic map and Arabidopsis syntenic regions of a segment of cotton consensus chromosome C12 (from 54.1 to 73.1 cM) showing the locations of fiber-related QTL, Arabidopsis orthologs of cotton fiber-related cDNAs, and Arabidopsis genes functioning in trichome development. Vertical bars on the left side of C12 represent the locations of QTL for fiber elongation (Paterson et al. 2003), fiber length (Chee et al. 2005b), and mass of seed cotton (log transformed) (Jiang et al. 1998). Solid lines link putative orthologs between cotton and Arabidopsis which were used to infer correspondence in this region. The numbers on the left side of the Arabidopsis segments are consecutive numbers assigned to genes in each α-duplicated segment, provided to permit one to know (by subtraction) the number of intervening genes between two anchor points. Values in parentheses to the right of Arabidopsis gene designations indicate gene identifier and e-value for match to a candidate gene identified based on expression pattern (as detailed in supplemental Table 7 at http://www.genetics.org/supplemental/) or a role in Arabidopsis trichome development (supplemental Table 8).
Impact of mutants in fiber development on the QTL landscape:
We postulated that the QTL landscape might be altered by nonlinear interactions among unlinked genes in the fiber-development program. Most QTL mapping, however, has only modest power to resolve such epistasis. As a test case we explored how different discrete single-gene mutations alter the spectrum of QTL explaining variation in a trait. Specifically, we compared the QTL maps resulting from crosses of one common parent, Pima S7, to two closely-related genetic stocks containing different discrete mutations implicated in lint fiber development.
n2 is a recessive mutant expressing completely or partially naked seed, previously assigned to Chr. 26 using aneuploid stocks (Percy and Kohel 1999), but recently suggested based on DNA markers to map to its homeolog, Chr. 12 (Rong et al. 2005b). A total of 242 probes detecting 370 loci mapped to 41 linkage groups covering 3116.2 cM of the n2 population, at an average spacing of 8.42 cM.
im is a recessive mutant causing immature fiber, characterized by reduced secondary cell wall development and fiber weight reduction of 40% or more (Kohel and McMichael 1990), assigned to Chr. 03 based on aneuploid stocks (Kohel et al. 2002). A total of 233 probes detecting 363 loci mapped to 37 linkage groups in the im population, at an average spacing of 9.6 cM. Both n2 and im maps have similar marker orders and recombination distances, and are available in the CMap database.
Nine traits, including two lint yield components, seed weight, and six fiber quality components, were measured (see materials and methods). While all traits varied over a considerable range, no significant difference between the n2 and im mapping populations was found for any measured trait (supplemental Table 5 at http://www.genetics.org/supplemental/).
A total of 22 QTL were detected on 14 chromosomes in the n2 population (supplemental Table 6a at http://www.genetics.org/supplemental/). More than half of the QTL were on only 4 chromosomes, with 4 on Chr. 12, 3 on its homeolog (Chr. 26), 3 on Chr. 17 and 2 on linkage group D08. Ten other chromosomes each contained single QTL (supplemental Table 6a). Two QTL for lint index each explain a remarkably high percentage of phenotypic variation (66.5%), with other QTL ranging from 10.4% to 25.3% of PV explained.
In the im population, only 17 QTL were detected for the same traits measured in n2. One fiber strength QTL was detected in both populations (FS02.1 and FS02.2) in bin 5 of Chr. 02. The other 16 QTL were all located on different chromosomes than their n2 counterparts (supplemental Table 6b at http://www.genetics.org/supplemental/). For example, no QTL were detected in the im population on Chr. 12, 17, and 26, which each had 3–4 QTL in n2. Chr. 02 and A02, which show 1 or 0 QTL in the n2 population, had 4 and 3 QTL in the im population, respectively. One large-effect QTL (explaining 46.7% of phenotypic variation) was detected on linkage group D02 for FUD02.1(HVui) in the im population, but no corresponding QTL was found in n2. Likewise, the strong QTL for lint index in the n2 population showed no corresponding effects in the im population, although a relatively weak QTL was detected for this trait on Chr. 14 of im. While the two study populations were only of sufficient size to detect the largest-effect segregating QTL, the use of stringent significance thresholds supports the validity of the QTL that were claimed, and even these relatively large-effect QTL did not correspond in the two populations. Since these two populations involved very similar genetic backgrounds tested in the same environment, the non-overlapping QTL detected suggest that the differing qualitative mutations cause different pathways and/or networks to be limiting factors of fiber quality.
Although only one correspondence (FS02.1 and FS02.2) was found in fiber QTL between the n2 and im populations, considerable correspondence was found between the n2 or im fiber QTL and those detected in the three non-mutant populations introduced above. Seven QTL found in the mutant populations map to bins containing QTL for the same fiber traits from other populations, increasing the number of nonrandom QTL clusters from 4 (based only on the three non-mutant populations) to 12 (including the QTL from the mutant populations) (supplemental Table 3 at http://www.genetics.org/supplemental/). Clusters had 2 QTL each, for EL (three clusters), FF (three), FL (two), FS (three), and SF (one) and composed 10.9% (24 of 224) of all fiber QTL. This correspondence to QTL from other larger populations provides further support for the validity of at least a subset of the inferred QTL.
The QTL for different fiber characters located on the same chromosome were generally clustered both in n2 and im. For example, 3 QTL on Chr. 26 of n2 were all in bin 5. Four QTL on Chr. 02 of im were centered in bins 4 and 5 and 2 more in bin 2 of A02. Many of the QTL, scattered on different bins of Chr. 12 in n2, or different chromosomes, were clustered with QTL detected in the elite populations. In total, inclusion of mutant population data increased QTL clusters to 27 bins of 105 QTL vs. 24 bins of 87 QTL based on the elite populations alone, with 6 bins containing 5–7 fiber QTL on Chr. 1, 2, 16, 17, and D08 (2 bins). As found in the three non-mutant populations, no statistically significant QTL clusters occurred in corresponding regions of the homeologous chromosomes.
Relationships between QTL and fiber-related cDNAs/genes:
A consensus map that depicts the inferred marker arrangement along the genome of the common ancestor that gave rise to the diploid progenitors of tetraploid cotton (Rong et al. 2005a) sets the stage for exploring relationships between cotton QTL and genes from taxa such as Arabidopsis that diverged from cotton prior to polyploid formation. All QTL from the reference map were plotted to corresponding locations on the consensus map. In parallel, we determined the locations in the Arabidopsis genome of a total of 203 candidate genes representing up to the four best matches (at E < 10−10) for each of 78 cDNAs/genes known from prior published work to be preferentially expressed during cotton lint fiber growth and development (supplemental Table 7 at http://www.genetics.org/supplemental/, which includes their protein family affiliation, GenBank accession number, expression pattern if known, and citations of published descriptions) and 28 Arabidopsis genes known to be critical for trichome and/or fiber development (supplemental Table 8). The Arabidopsis orthologs were plotted to their locations in an inferred ancestral gene order that mitigates the effects of a whole-genome duplication in Arabidopsis since its divergence from cotton (i.e., α-duplicated segments). Established syntenic relationships between cotton and Arabidopsis (Rong et al. 2005a) were used to determine correspondence between the QTL and candidate genes. QTL locations were plotted based on the midpoint of the QTL likelihood interval.
We found evidence of a general association between concentrations of candidate genes and cotton fiber-related QTL. Based on synteny inferred using CrimeStat II (Rong et al. 2005a), a significant correlation (r = 0.260, N = 167, P = 0.0003) was found between QTL number in cotton and candidate gene number in Arabidopsis. When we considered only the smaller number (largely a subset) of regions of synteny inferred using FISH (Rong et al. 2005a), the correlation was weaker (r = 0.072, N = 189) and fell short of significance (P = 0.164). In both CSII and FISH-based models, the correlations between numbers of fiber QTL and all (not just candidate) Arabidopsis genes in the syntenic regions detected by CrimestatII or FISH were not significant, suggesting that the relationship is specific to the population of candidate genes. Figure 3 presents an example in which four fiber-related QTL on a region of consensus chromosome C12 ranging from 54.1 to 73.1 cM showed synteny with Arabidopsis α 4, 10, and 14 duplicated segments. At least six Arabidopsis sequences in this region matched two fiber-related cDNAs (CesA1 and GhExp1) and one Arabidopsis trichome gene (TTG2). Numbers and names of all Arabidopsis orthologs in the syntenic regions can be found in CMap.
DISCUSSION
The subgenome from the non-fiber-producing ancestor plays a large role in the genetic control of fiber growth and development in polyploid cotton:
Tetraploid cottons have higher lint fiber yield and quality than the modern descendants of their diploid progenitors, even in environments to which both have been adapted by plant breeding (Jiang et al. 1998). While the genetic potential for lint fiber development appears to have been transmitted to tetraploid cottons by their A genome progenitor, early demonstrations that Dt-subgenome loci have been recruited to contribute further to lint fiber genetics in tetraploids (Jiang et al. 1998) gain further support from the more extensive data now available from larger numbers of QTL in additional independent populations that include a higher degree of replication across environments (Paterson et al. 2003), and advanced-generation studies able to detect QTL of smaller phenotypic effect (Chee et al. 2005a,b; Draye et al. 2005; Lacape et al. 2005). This finding implicates the Dt-genome (from the non-fiber-producing ancestor) in evolution of the transgressive fiber quality and yield of polyploid cottons relative to their diploid progenitors.
A wealth of research has revealed that gene copies from genome duplications (polyploidizations) experience different fates during their evolution including gene loss, subfunctionalization, and neofunctionalization (Lynch and Force 2000; Rastogi and Liberles 2005). This raises the possibility that differential evolution of homeologous fiber-related genes duplicated by polyploid formation (Cronn et al. 1999) is partly responsible for modern cotton fiber quality. Randomly chosen genes show similar evolutionary rates in the two genomes before and after polyploid formation. However, lack of knowledge of the gene network involved in lint fiber morphogenesis has precluded study of its specific constituents; perhaps the Dt alleles of key lint fiber-related genes have evolved more quickly. In other words, selection may have played a different role in genetic changes of homeologous genes in At and Dt genomes, respectively, after reunion of these two genomes ∼1–2 MYA.
Meta-analysis reveals a complex QTL landscape:
Although the sets of QTL found for the same fiber-related trait in different experiments showed nonrandom correspondence, we were surprised that correspondence was only observed for ∼10% of the total QTL set. Comparison in the same environment of two new mapping populations which shared one identical parent, Pima S-7, and had closely related alternative parents that differed in carrying specific fiber mutants (n2 and im, respectively) showed virtually no common QTL. However, the validity of many QTL in each of these populations is supported by their correspondence to QTL found in other populations. This suggests that lint fiber development may involve a complex gene network in which perturbations at one point have widespread consequences. The discrete mutants studied may impose particularly large perturbations; if small differences in QTL alleles among populations also cause the same phenomenon, then the generally low level of correspondence among QTL found in different populations might be explained.
These findings have several implications. In terms of basic genetics, the heterogeneity in QTL revealed by the different experiments reiterates the need for study of a broad sampling of germplasm, in a wide range of environments, to gain a representative picture of the true genetic complexity of a trait in a particular taxon. From an applied standpoint, the data highlight the need to validate QTL specifically in each genetic background in which they are to be deployed (for example, in mainstream breeding).
Members of the lint fiber development network appear clustered, but cluster members show heterogeneous phenotypic effects:
Several lines of evidence suggest nonrandom distribution of genes implicated in lint fiber development. First, 105 (46.9%) lint fiber-related QTL were concentrated in 27 bins composing 12.1% of the genome. Further, the corresponding regions of the Arabidopsis genome also contain nonrandom concentrations of genes that are implicated in fiber/trichome development based on either expression patterns or functional information.
Most of these clusters show heterogeneous phenotypic consequences. In only a small subset of cases (12) did more than one QTL affect the same phenotype. Moreover, in many cases the genes were detected in different experimental populations, precluding pleiotropy as a general explanation (although it may contribute in some cases).
A tantalizing hypothesis is that concentrations of cotton lint fiber QTL may represent groups of coordinately regulated genes and/or groups of small gene families that have undergone proximal duplication followed by sub- or neofunctionalization. Progress in physical mapping of several members of the Gossypium genus may permit this hypothesis to be tested in the near future.
Meta-analysis linked to synteny-based and expression-based information provides clues about specific genes and families involved in QTL networks:
Integration of QTL from individual maps using a common reference map will be helpful to infer possible positional relationships to QTL locations on other maps that share some DNA markers with our reference map (Lacape et al. 2003, 2005; Shen et al. 2005; Han et al. 2006). For example, a QTL governing fiber elongation (ELO) (Lacape et al. 2005) near marker G1058 in Chr. 22 corresponds closely to a QTL for the same trait (EL22.1) on the reference map, suggesting that these two QTL may be the same. Similarly, if new QTL were plotted in maps that share common markers with this high-density map, it can quickly be determined if they might correspond to previously identified QTL.
The integration of QTL, discrete mutants, and regions syntenic with Arabidopsis in a common map can facilitate the exploitation of Arabidopsis sequence information in further study and cloning of fiber genes. Burgeoning information about differential expression patterns of cotton genes, together with extensive knowledge of the molecular control of epidermal trichome development in Arabidopsis, provide valuable clues about possible cotton genes in the fiber development network. The locations of cotton fiber QTL, and information about synteny between cotton and Arabidopsis, make it possible to begin to search for intersections among these various data types that point to specific genes as candidates worthy of further functional testing to determine if they are directly responsible for genetic variation in cotton fiber development.
For example, the collective data in clusters of QTL suggest numerous candidate genes and possible approaches for advancing progress toward identification of the causal gene(s), as follows:
The cluster with five fiber QTL in Chr. 17 was in a region ranging from 2.4 to 11.4 cM on bin 1. These QTL each are based on different measurements of fiber quality, and reported in two different studies [FS17.1 and FC17.1 (Paterson et al. 2003) and FL17.1, FU17.1, and MIC17.1 (Chee et al. 2005a,b; Draye et al. 2005)]. The region harboring these QTL showed synteny (identified by both CrimestatII and FISH) with Arabidopsis duplications α03 and α21. Gene At4g18780 in α21 is CelA1 (E = 0) and two Arabidopsis genes (At1g12560 and At1g62980) in α03 are members of the expansin gene family (best matching AtEXP7, E < 1E-154 and AtEXP18, E < 6E-156) involved in Arabidopsis trichome development.
In linkage group D08, 11 of 22 fiber QTL were clustered in bins 3 and 8. QTL in bin 3 are ELD08.2, FLD08.1(HVsl2.5,HVuhm,Lw), FUD08.1(HVui), SFD08.1(HVsfc, SFCn), FSD08.2(STR), and FFD08.2, and distributed from 40.3 to 53.1 cM, a marker-sparse segment. Except for FSD08.2 (STR) reported by (Jiang et al. 1998), all others were detected in advanced backcross populations (Chee et al. 2005a,b; Draye et al. 2005). No QTL were found in the homeologous region on Chr. 05. These QTL were located in a region (D05.153) showing synteny with a segment of Arabidopsis duplication α5 where four candidate genes were found including paralogs (At1g80350 and At1g80360, E = 8E-118 and 1.4E-56, respectively) of fra2, and homologs (At1g15690 and At1g79840, E = 4.8E-97 and 0) of proton-translocating pyrophosphatase (Ppase) and a homeobox protein (GhHox1). Another 5 fiber-related QTL on D08 were located in a region of bin 8 from 140.3 to 157.4 cM. All five QTL were found in advanced backcross populations (Chee et al. 2005a,b; Draye et al. 2005), including three fiber length loci, FLD08.2 (HVsl2.5, HVuhm, Lw), FUD08.2 (HVui), and SFD08.2 (HVsfc), and two fiber elongation loci, ELD08.3 and ELD08.4. The region showed synteny with part of α07 (D07.160) and one of the Arabidopsis genes, At4g09820, is the paralog of EGL3 (E < 1.5E-38).
Acknowledgments
We thank numerous colleagues for valuable assistance and suggestions, and the United States–Israel Binational Agricultural Research and Development Fund (US-2506-94R to A.H.P. and Y.S.), the BOYSCAST program of the Department of Science and Technology, India (to V.N.W.), United States Department of Agriculture National Research Initiative (02-01412 to A.H.P.), the National Science Foundation Plant Genome Research Program (DBI-9872630, DBI-0211700 to A.H.P. and J.F.W.), the Fonds National belge pour la Recherche Scientifique (research associate grant to X.D.), the Texas and Georgia Agricultural Experiment Stations, Texas Higher Education Coordinating Board, Cotton Incorporated, and United States Department of Agriculture–Initiative for Future Agriculture and Food Systems (00-52100-9685 to A.H.P., P.W.C., J.R.G., O.L.M., C.W.S.) for financial support.
References
- Arcade, A., A. Labourdette, M. Falque, B. Mangin, F. Chardon et al., 2004. BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20: 2324–2326. [DOI] [PubMed] [Google Scholar]
- Benton, M. J., 1993. The Fossil Record, Ed. 2. Chapman & Hall, New York.
- Bowers, J. E., B. A. Chapman, J. Rong and A. H. Paterson, 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438. [DOI] [PubMed] [Google Scholar]
- Calabrese, P. P., S. Chakravarty and T. J. Vision, 2003. Fast identification and statistical evaluation of segmental homologies in comparative maps. Bioinformatics 19(Suppl. 1): i74–i80. [DOI] [PubMed] [Google Scholar]
- Chee, P., X. Draye, C. Jiang, L. Decanini, T. Delmonte et al., 2005. a Molecular dissection of interspecific variation between Gossypium hirsutum and G. barbadense (cotton) by a backcross-self approach: I. Fiber elongation. Theor. Appl. Genet. 111: 757–763. [DOI] [PubMed] [Google Scholar]
- Chee, P., X. Draye, C.-X. Jiang, L. Decanini, T. Delmonte et al., 2005. b Molecular dissection of phenotypic variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: III. Fiber length. Theor. Appl. Genet. 111: 772–781. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative triat mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn, R. C., R. L. Small and J. F. Wendel, 1999. Duplicated genes evolve independently after polyploid formation in cotton. Proc. Natl. Acad. Sci. USA 96: 14406–14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draye, X., P. Chee, C. Jiang, L. Decanini, T. Delmonte et al., 2005. Molecular dissection of interspecific variation between Gossypium hirsutum and G. barbadense (cotton) by a backcross-self approach: II. Fiber fineness. Theor. Appl. Genet. 111: 764–771. [DOI] [PubMed] [Google Scholar]
- Frary, A., T. C. Nesbitt, A. Frary, S. Grandillo, E. v. d. Knaap et al., 2000. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88. [DOI] [PubMed] [Google Scholar]
- Han, Z., C. Wang, X. Song, W. Guo, J. Gou et al., 2006. Characteristics, development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor. Appl. Genet. 112: 430–439. [DOI] [PubMed] [Google Scholar]
- Jiang, C., R. J. Wright, S. S. Woo, T. A. DelMonte and A. H. Paterson, 2000. QTL analysis of leaf morphology in tetraploid Gossypium (cotton). Theor. Appl. Genet. 100: 409–418. [Google Scholar]
- Jiang, C. X., R. J. Wright, K. El-Zik and A. H. Paterson, 1998. Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc. Natl. Acad. Sci. USA 95: 4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohel, R. J., and S. C. McMichael, 1990. Immature fiber mutant of upland cotton. Crop Sci. 30: 419–421. [Google Scholar]
- Kohel, R. J., D. M. Stelly and J. Yu, 2002. Tests of six cotton (Gossypium hirsutum L.) mutants for association with aneuploids. J. Hered. 93: 130–132. [DOI] [PubMed] [Google Scholar]
- Lacape, J.-M., T. B. Nguyen, S. Thibivilliers, B. Bojinov, B. Courtois et al., 2003. A combined RFLP-SSR-AFLP map of tetraploid cotton based on a Gossypium hirsutum × Gossypium barbadense backcross population. Genome 46: 612–626. [DOI] [PubMed] [Google Scholar]
- Lacape, J.-M., T.-B. Nguyen, B. Courtois, J.-L. Belot, M. Giband et al., 2005. QTL analysis of cotton fiber quality using multiple Gossypium hirsutum x Gossypium barbadense backcross generations. Crop Sci. 45: 123–140. [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, N., 2002. CrimeStat: A spatial statistics program for the analysis of crime incident locations (v 2.0). Ned Levine & Associates, Houston, TX, and the National Institute of Justice, Washington, DC.
- Lin, Z., D. He, X. Zhang, Y. Nie, X. Guo et al., 2005. Linkage map construction and mapping QTL for cotton fibre quality using SRAP, SSR and RAPD. Plant Breed. 124: 180–187. [Google Scholar]
- Lynch, M., and A. Force, 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, O. L., and T. J. Wofford, 2000. Breeding transformed cotton expressing enhanced fiber strength. J. New Seeds 2: 1–13. [Google Scholar]
- Mei, M., N. H. Syed, W. Gao, P. M. Thaxton, C. W. Smith et al., 2004. Genetic mapping and QTL analysis of fiber-related traits in cotton (Gossypium). Theor. Appl. Genet. 108: 280–291. [DOI] [PubMed] [Google Scholar]
- Paterson, A. H., E. S. Lander, J. D. Hewitt, S. Peterson, S. E. Lincoln et al., 1988. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 335: 721–726. [DOI] [PubMed] [Google Scholar]
- Paterson, A. H., J. W. DeVerna, B. Lanini and S. D. Tanksley, 1990. Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in an interspecies cross of tomato. Genetics 124: 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A. H., Y. Saranga, M. Menz, C. X. Jiang and R. J. Wright, 2003. QTL analysis of genotype x environment interactions affecting cotton fiber quality. Theor. Appl. Genet. 106: 384–396. [DOI] [PubMed] [Google Scholar]
- Percy, R. G., and R. J. Kohel, 1999. Qualitative Genetics, pp. 319–360 in Cotton: Origin, History, Technology, and Production, edited by C. W. Smith and J. T. Cothren. John Wiley & Sons, Hoboken, NJ.
- Rastogi, S., and D. Liberles, 2005. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol. Bio. 5: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, J., C. Abbey, J. E. Bowers, C. L. Brubaker, C. Chang et al., 2004. A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166: 389–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, J., J. E. Bowers, S. R. Schulze, V. N. Waghmare, C. J. Rogers et al., 2005. a Comparative genomics of Gossypium and Arabidopsis: Unraveling the consequences of both ancient and recent polyploidy. Genome Res. 15: 1198–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, J., G. Pierce, V. Waghmare, C. Rogers, A. Desai et al., 2005. b Genetic mapping and comparative analysis of seven mutants related to seed fiber development in cotton. Theor. Appl. Genet. 111: 1137–1146. [DOI] [PubMed] [Google Scholar]
- Saha, S., J. Wu, J. N. Jenkins, J. J. C. McCarty, O. A. Gutierrez et al., 2004. Effect of chromosome substitutions from Gossypium barbadense L. 3–79 into G. hirsutum L. TM-1 on agronomic and fiber traits. J. Cotton Sci. 8: 162–169. [Google Scholar]
- Saranga, Y., M. Menz, C. X. Jiang, R. J. Wright, D. Yakir et al., 2001. Genomic dissection of genotype x environment interactions conferring adaptation of cotton to arid conditions. Genome Res. 11: 1988–1995. [DOI] [PubMed] [Google Scholar]
- Saranga, Y., M. Menz, C. Jiang, R. Wright, D. Yakir et al., 2004. Genetic and physiological dissection of adaptations associated with cotton productivity under arid conditions. Plant, Cell and Environ. 27: 263–277. [Google Scholar]
- Senchina, D. S., I. Alvarez, R. C. Cronn, B. Liu, J. Rong et al., 2003. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 20: 633–643. [DOI] [PubMed] [Google Scholar]
- Shen, X., W. Guo, X. Zhu, Y. Yuan, J. Z. Yu et al., 2005. Molecular mapping of QTLs for fiber qualities in three diverse lines in upland cotton using SSR markers. Mol. Breed. 15: 169–181. [Google Scholar]
- Udall, J. A., J. M. Swanson, K. Haller, R. A. Rapp, M. E. Sparks et al., 2006. A global assembly of cotton ESTs. Genome Res. 16: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghmare, V. N., J. Rong, C. J. Rogers, G. J. Pierce, J. F. Wendel et al., 2005. Genetic mapping of a cross between Gossypium hirsutum (cotton) and the Hawaiian endemic, Gossypium tomentosum. Theor. Appl. Genet. 111: 665–676. [DOI] [PubMed] [Google Scholar]
- Wang, S., C. J. Basten and Z. B. Zeng, 2005. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC.
- Wendel, J. F., 1989. New world tetraploid cottons contain old-world cytoplasm. Proc. Natl. Acad. Sci. USA 86: 4132–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, R., P. Thaxton, K. El-Zik and A. H. Paterson, 1998. D-subgenome bias of Xcm resistance genes in tetraploid Gossypium (Cotton) suggests that polyploid formation has created novel avenues for evolution. Genetics 149: 1987–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, R., P. Thaxton, A. H. Paterson and K. El-Zik, 1999. Molecular mapping of genes affecting pubescence of cotton. J. Heredity 90: 215–219. [Google Scholar]
- Zeng, Z. B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]