Abstract
The construction of a dense genetic map for Vitis vinifera and its anchoring to a BAC-based physical map is described: it includes 994 loci mapped onto 19 linkage groups, corresponding to the basic chromosome number of Vitis. Spanning 1245 cM with an average distance of 1.3 cM between adjacent markers, the map was generated from the segregation of 483 single-nucleotide polymorphism (SNP)-based genetic markers, 132 simple sequence repeats (SSRs), and 379 AFLP markers in a mapping population of 94 F1 individuals derived from a V. vinifera cross of the cultivars Syrah and Pinot Noir. Of these markers, 623 were anchored to 367 contigs that are included in a physical map produced from the same clone of Pinot Noir and covering 352 Mbp. On the basis of contigs containing two or more genetically mapped markers, region-dependent estimations of physical and recombinational distances are presented. The markers used in this study include 118 SSRs common to an integrated map derived from five segregating populations of V. vinifera. The positions of these SSR markers in the two maps are conserved across all Vitis linkage groups. The addition of SNP-based markers introduces polymorphisms that are easy to database, are useful for evolutionary studies, and significantly increase the density of the map. The map provides the most comprehensive view of the Vitis genome reported to date and will be relevant for future studies on structural and functional genomics and genetic improvement.
VITIS vinifera L. is the only European species representative of the genus Vitis. The species is widely cultivated and is the most important fruit crop in the world. V. vinifera has a disomic inheritance with 2n = 38 and a relatively small genome (475 Mbp, Lodhi and Reisch 1995). Recent studies carried out in V. vinifera range from genetic diversity analysis (Aradhya et al. 2003) to cultivar and clonal identification (Faria et al. 2004; This et al. 2004), disease resistance gene identification (Donald et al. 2002; Barker et al. 2005; Di Gaspero et al. 2007), and quantitative trait analysis (Doligez et al. 2002; Fischer et al. 2004; Fanizza et al. 2005) in addition to gene expression and metabolic profiling (Terrier et al. 2005; Waters et al. 2005). The development of tools for genome analysis has included the putting together of a large collection of expressed sequence tags (ESTs) (da Silva et al. 2005; Moser et al. 2005), the lengthy undertaking of creating a physical map and whole-genome sequencing, and proteomics (Sarry et al. 2004; Castro et al. 2005; Pereira et al. 2005; Castellarin et al. 2006).
Genetic linkage maps are a prerequisite to studying the inheritance of both qualitative and quantitative traits and to integrating molecular information that is necessary for marker-assisted selection (MAS) and map-based gene cloning techniques (Morgante and Salamini 2003). Thus, a key resource in support of classical genetic and genomics of V. vinifera is the construction of a dense genetic map based on well-characterized, gene-specific molecular markers.
Over the last 15 years genetic linkage maps have been prepared for several plant species (van Os et al. 2006), but high-resolution genetic maps are nevertheless limited to major crop species: the most dense map has been reported for potato, which contains >10,000 markers (van Os et al. 2006). For fruit trees, a total of 840 markers are available in an apple map (Liebhard et al. 2003) and 562 are included in the reference map of Prunus spp. (Dirlewanger et al. 2004). Peach ESTs are currently being anchored to both a genetic and a physical map (Horn et al. 2005), while a growing collection of ESTs from apple and almond has been released to public databases. Several genetic maps are already available for the Vitis genome (Grando et al. 2003; Adam-Blondon et al. 2004; Riaz et al. 2004; Lowe and Walker 2006), primarily based on simple sequence repeats (SSRs) and AFLPs, with the highest marker density (515 markers) present in an integrated linkage map that includes data from five different crosses (Doligez et al. 2006).
SSRs are highly prized as molecular markers due to their codominance and high levels of polymorphism, but a significant effort is required to develop SSR-based maps. AFLP markers are easy to use and reveal large sets of genetic loci, but their transferability between detection platforms (for instance, polyacrylamide gel electrophoresis, gel-based sequencers, and capillary sequencers) can sometimes be difficult and cumbersome (Papa et al. 2005). In addition, the marker nomenclature used does not always allow easy transfer between labs, especially with a very dense fingerprint. Single-nucleotide polymorphism (SNP)-based genetic markers have attracted significant attention when creating dense genetic linkage maps. SNPs are the most abundant class of polymorphisms and they also provide gene-based markers that may prove useful in identifying candidate genes of interest to be associated with quantitative trait loci (Rafalski 2002).
The current study on V. vinifera utilizes a large number of SNP-based genetic markers and maps them in a framework of loci defined by SSR markers in the Syrah × Pinot Noir cross. The markers were derived from V. vinifera collections of ESTs and BAC-end sequences (BESs) available at NCBI (149,691 EST sequences clustered into 15,194 unigenes and 30,832 BESs; http://www.ncbi.nlm.nih.gov/). In addition, SSR and AFLP markers were employed to increase the number of bridges between the genetic and the physical map, considering specific markers used by the international grapevine community.
MATERIALS AND METHODS
Plant material:
The mapping population consisted of 94 progeny plants from a cross between V. vinifera cvs. Syrah and Pinot Noir (clone 115) obtained and grown at the Istituto Agrario di San Michele all'Adige (IASMA).
Genomic DNA was extracted from young leaves following the protocol described by Doyle and Doyle (1990) with slight modification (Grando et al. 2003).
Pinot Noir BAC contigs:
For the clone 115 of Pinot Noir, two BAC libraries were constructed (Adam-Blondon et al. 2005; Keygene laboratory, Wageningen, The Netherlands). A total of 49,536 BAC clones (representing ∼11.4 genome equivalents) were fingerprinted using a fluorescence-based high-throughput method developed by Luo et al. (2003) and assembled into a physical map using FPC 7.2 (http://www.agcol.arizona.edu/software.fpc/) following Nelson et al. (2005) (WebFPC for the grapevine FPC map is available at http://genomics.research.iasma.it).
EST resources and PCR primer design:
A total of 454 EST clusters (Moser et al. 2005) were selected on the basis of homology with transcription factors or coding sequences putatively involved in sugar, flavonoids, and defense-related metabolic pathways. PCR primers were designed using the Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/_www.cgi), adopting the following criteria: (1) expected size of the amplified fragment between 200 and 600 bp, (2) primer size between 18 and 25 bases, (3) primer melting temperature (Tm) between 59° and 61°, and (4) alignment score and global alignment score for self-complementarity and complementarity between primer pairs between 8 and 13. For successfully amplified and mapped ESTs, primer sequences and IASMA cluster names are reported in supplemental Table S1 (http://www.genetics.org/supplemental/).
EST marker nomenclature:
The EST markers were denoted by two letters identifying the tissue-specific cDNA library of origin (BA, berry; FO, leaf; F2, second leaf library; GM, bud; GR, shoot tip; RA, root; IN, inflorescence) followed by four numbers.
BAC-end sequence resources and PCR primer design:
A total of 905 primer pairs defining in silico unique BESs were selected. Primer pairs were designed on BESs using software following the criteria described for the EST sequences. Primer sequences of successfully amplified and mapped BESs are reported in supplemental Table S1 (http://www.genetics.org/supplemental/).
BES marker nomenclature:
BES markers were denoted by the BAC clone number (plate number and position on the plate) followed by F or R to indicate the two end sequences of the BAC clone.
Identification of polymorphic sequences and SNP-based marker development:
Parental genomic DNAs were amplified by PCR using oligonucleotide primers designed from ESTs and BESs as described. Amplification was carried out in a volume of 25 μl, with 20 ng genomic DNA, 1× PCR reaction buffer (QIAGEN, Valencia, CA), 1.5 mm MgCl2, 0.2 mm for each dNTP, 1 unit HotStartTaq DNA polymerase (QIAGEN), and 0.4 μm of each specific primer. PCR reactions were performed using a touchdown PCR protocol (Don et al. 1991) with a 15-min initial denaturation/activation step (hot start). DNA amplification was carried out by denaturing at 94° for 30 sec, annealing for 1 min, and extending at 72° for 1 min. In initial cycles, the annealing temperature was progressively lowered from 62° to 57° by 1° every cycle. Samples were subjected to an additional 30 cycles of amplification after reaching the final annealing temperature of 57° and a final extension step of 7 min at 72°. PCR products were evaluated by gel electrophoresis in 1.5% agarose and visualized by ethidium bromide staining.
Two different approaches were explored for the identification of SNPs: SSCP and resequencing. SSCP electrophoresis (Orita et al. 1989) was carried out both on a nondenaturant gel, as in Salmaso et al. (2004), and on fluorescence-based capillary electrophoresis on the ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA) following the protocol reported by the manufacturer (http://docs.appliedbiosystems.com/search.taf). Amplification reactions that yielded single, well-defined bands were selected for resequencing and SNP discovery. PCR products were sequenced using the ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Sequencing reactions of 10 μl volume were prepared with 10–50 ng of PCR product, 4 μl of ABI Prism BigDye terminator sequencing ready reaction kit (Applied Biosystems), and 5 pmol of the forward primer. Sequencing thermocycling was performed with a 1-min initial denaturation step at 96°, followed by 35 cycles at 96° for 10 sec, 55° for 5 sec, and 60° for 4 min.
DNA sequence electropherograms were aligned with the Pregap4/Gap4 software (Staden Package, Staden et al. 2000) and used to survey parental alleles for polymorphic sites. SNPs were converted to cleaved amplified polymorphic sequences (CAPS) by identifying mutations conferring differential restriction enzyme sites between the two parental alleles. When suitable restriction enzyme sites were missing, oligonucleotide primers with a single-nucleotide mismatch were designed adjacent to the polymorphic position, such that a restriction site was created in the PCR product of one parent, but not in the other (dCAPS) (Michaels and Amasino 1998; Neff and Kalishman 2002). Restriction enzymes were used in CAPS and dCAPS reactions according to the manufacturer's recommendations (Fermentas). CAPS and dCAPS restriction products were visualized on a 2% agarose gel with standard UV transillumination following ethidium bromide staining.
The largest part of the SNP-based marker polymorphisms was assessed by the multiplex minisequencing technique on the ABI Prism 3100 Genetic Analyzer (Applied Biosystems) (Troggio et al. 2007). For each locus under investigation primers flanking the SNP mutations, revealed from resequencing, were designed with the computer program GeneRunner v3.04 (Hastings Software, Hudson, NY) and one matching the following conditions was chosen. Specific parameters were considered as follows: (1) primer size between 18 and 26 bases, (2) primer Tm between 55° and 60°, (3) GC content >40%, and (4) occurrence of hairpin loops and dimers. The minisequencing reaction followed the ABI Prism SNaPshot Multiplex Kit protocol reported at http://docs.appliedbiosystems.com/search.taf.
SSR analysis:
Microsatellite marker primer sequences were obtained from the Vitis Microsatellite Consortium (VMC) coordinated by AgroGene S. A. (Moissy Cramayel, France) and from previously published studies on grape (Thomas and Scott 1993; Bowers et al. 1996, 1999; Sefc et al. 1999; Scott et al. 2000; Merdinoglu et al. 2005).
The selection of suitable markers was based on their presence in genetic linkage maps of V. vinifera (Adam-Blondon et al. 2004; Riaz et al. 2004). The 143 SSR primer pairs selected were first tested on the parents of our maps and on six F1 individuals. Resulting polymorphic markers were run on the entire mapping population of 94 F1 individuals. PCR amplifications were performed in 12.5-μl reactions of 25 ng genomic DNA, 0.5 μm of forward and reverse primer, 0.2 mm of each dNTP (Promega, Madison, WI), 1× Gold PCR buffer (Applied Biosystems), 1.5 or 2 mm MgCl2, and 0.25 unit AmpliTaq Gold DNA Polymerase (Applied Biosystems). Three different fluorescent dyes (6-FAM, HEX, and NED) were used to label the forward primers. An annealing temperature of 50°, 52°, or 56° was used on the basis of previous amplification optimization. Amplification conditions were the same for all primer pairs (7 min at 95° to activate AmpliTaq Gold and 35 cycles of 45 sec denaturation at 94°, 45 sec annealing at 50°, 52°, or 56° and 1 min extension at 72°, followed by 7 min of additional extension at 72°).
PCR products were separated by capillary electrophoresis performed on an ABI Prism 3100 genetic analyzer (Applied Biosystems) using Performance Optimized Polymer 4 (POP 4, Applied Biosystems). Samples were prepared with 9.6 μl of deionized formamide, 0.1 μl of GeneScan 500 ROX size standard (Applied Biosystems), and 0.3 μl of 10× diluted PCR product. The mixture was denaturated (95° for 3 min) and placed 5 min on ice prior to injection. Alleles were separated at 15,000 V for ∼45 min at 60°. The data were analyzed using Genescan 3.7 (Applied Biosystems) for internal standard and fragment size determination. Allelic designations were assigned using Genotyper 3.7 (Applied Biosystems).
SSR marker nomenclature:
SSR markers are identified with the following prefixes: VVS (Thomas and Scott 1993), VVMD (Bowers et al. 1996, 1999), VRZAG (Sefc et al. 1999), SCU (Scott et al. 2000), VMC (Vitis Microsatellite Consortium), and VVI (Merdinoglu et al. 2005).
AFLP analysis:
AFLP analysis was carried out as described by Vos et al. (1995), using EcoRI/MseI enzyme combinations. A total of 16 primer combinations (PCs) used for genetic mapping were selected on the basis of the total number of bands and the level of polymorphism observed in the two parents and in four F1 individuals.
A two-step amplification strategy was followed. In a selective preamplification, both AFLP primers had a single selective nucleotide at the 3′ end. Further selective amplification was carried out using primers having three selective nucleotides at the 3′ end. The EcoRI primer was end labeled using [γ-33P]ATP and T4 polynucleotide kinase. Gel images were electronically scanned and all AFLP markers were scored codominantly using software for AFLP analysis (Keygene). This software displays and analyzes pixel images of phosphorimager scans. For the analysis of pixel images, the software includes tools to navigate through the image and individual band signals and to size and quantify the AFLP bands with great accuracy. Each band of a specific marker is classified with respect to its intensity using a mixture model of normal distributions, as described by Jansen (1993).
AFLP marker nomenclature:
Each polymorphic AFLP fragment was identified by the enzyme combination, the primer selective nucleotides, followed by the estimated molecular size of the DNA fragment in nucleotides. A 10-bp ladder DNA from SequaMark (Research Genetics, Huntsville, AL) was used to estimate the molecular weight of AFLP fragments.
Segregation analysis and map construction:
Markers were tested against the expected segregation ratio using a chi-square goodness-of-fit and the P-value was recorded. Distorted markers were used for linkage analysis unless they affected the order of neighboring markers. Individual maps were constructed for each parental genotype following a double pseudo-testcross strategy (Grattapaglia and Sederoff 1994). Markers of “ab × ab” type were scored in the progeny as “aa” = A, “bb” = H, “ab” = missing data. Marker phase was determined with the aid of an algorithm implemented in “Phasing” (http://math.berkeley.edu/∼dustin/tmap/, Cartwright et al. 2007, accompanying article in this issue). Linkage group assignment and ordering of loci were established on the basis of newly developed software that finds the maximum-likelihood map using an error-compensating model (Cartwright et al. 2007). Linkage groups were determined using the “Grouping” application with a minimum LOD of 8.0 and a maximum distance of 35 cM. An initial order was built using the “Builder” application. A maximum-likelihood order was then obtained using the “Improve” command from either the Builder application or the “MapViewer” application. Markers were interactively removed from the map to see which ones distorted the distances greatly. Recombination rates for all pairwise groups of common markers between the two parents were compared using a χ2-test. Homologous linkage groups between the two parental maps were identified and a consensus map was constructed following the procedure described above. Recombinational values among markers were established by considering the F1 population of the cross Syrah × Pinot Noir as a cross-pollinator population. The Kosambi mapping function (Kosambi 1944) was used to convert the recombination frequencies into map distances (centimorgans). MapChart v2.1 software (Voorrips 2002) was used for the graphical visualization of the linkage groups.
Marker distribution:
To test differences among linkage groups, the relative frequency of marker classes in each group was compared using a χ2-test applied to their frequency in the complete map.
Marker distribution was based on the number of markers in each marker class separately within each 10-cM interval over the total length of each linkage group. Intervals at the end of a linkage group were taken into account only when >7.5 cM. EST sequences and BESs were grouped in two classes representing either coding or noncoding sequences. The distribution was analyzed for each marker class and was compared to the Poisson distribution function P(x) = e−μμx/x!, where the parameter μ is the average number of markers in a 10-cM interval over the entire map and x is the actual marker count in each interval. Observed and expected frequencies were compared with a χ2-test.
Physical and genetic map integration and distance estimation:
To integrate the physical into the genetic map, two complementary strategies were adopted. The first made use of markers derived from BESs for which the determination of their genetic position on the linkage map followed. In the second strategy BAC pools (Barillot et al. 1991) were constructed according to Klein et al. (2000). A total of 24,576 BAC clones (5× genome coverage) were arranged in a stack that was sampled in six distinct ways and a total of 184 BAC pools were obtained. The BAC pools were screened with EST, SSR, and AFLP primers to identify mapped fragments following the amplification conditions described before for the linkage analysis. SSR and EST amplification products from BAC pools, after adding SYBR Gold, were run on a 1.5% agarose gel. BAC clones hosting SSR and EST markers were identified by a Unix-based application with a web interface (Klein et al. 2000). AFLP amplification products from BAC pools were analyzed on acrylamide gels along with amplification products from the two parents and the mapping population as a control (AFLP Quant-Pro, Keygene). BACs containing AFLPs were identified in the same way as the other markers.
Marker order between the genetic linkage map and the physical map was verified for anchored contigs containing three or more genetically mapped markers, considering the two parental maps separately and the consensus map. For the same contigs it was possible to establish the relationship between physical distance and genetic distance.
RESULTS
Generation of marker data set:
SNP-based markers:
Polymorphic SNP loci were identified following PCR amplification and sequencing of alleles from the cultivars Syrah and Pinot Noir. Additional SNP loci were characterized after PCR amplification by SSCP.
Of 454 EST primer pairs tested, 363 (80%) yielded single PCR products and were further considered for SNP discovery, 70 (15%) did not amplify a product, 7 yielded PCR products that were not consistently amplified, and 14 amplified more than one product. A total of 160 PCR products, with the minimum size, were preliminarily assayed as described in materials and methods, by SSCP electrophoresis: 35 showed a clear polymorphic profile and were screened with the 94 F1 progeny, 71 showed a complex profile, and 54 were monomorphic. Of the latter group of SSCP-tested products, 49 monomorphic and 25 with a complex profile were resequenced, along with the remaining 203 PCR products (363 − 160) not assayed by SSCP (277 in total). Twenty-four of the 49 products having a monomorphic SSCP pattern, as well as 13 of 25 with a complex SSCP profile, and 112 untested EST PCR products yielded high-quality sequences that were polymorphic in either parent. Poor-quality sequence data were obtained in 49 cases, while 79 were monomorphic in either parent. Of the 149 polymorphic sequences, 42 were converted in CAPS, 1 in dCAPS, and 106 were analyzed with the multiplex minisequencing technique.
Single PCR products derived from 903 BES primer pairs were sequenced: 316 were readable sequences showing segregating polymorphisms and 174 were monomorphic, while 413 produced low-quality reads, probably due to multiple amplifications. Minisequencing primers flanking the identified SNP mutations were designed for the 316 polymorphic regions in a multiplex design. Of the 413 low-quality sequences, 88 were analyzed by SSCP electrophoresis, but only 8 showed a clear polymorphic profile in either parent.
The marker details for EST and BES regarding parental genotypes at the targeted SNPs, restriction enzymes for CAPS and dCAPS, and SSCP conditions are reported in supplemental Table S1 at http://www.genetics.org/supplemental/.
SSR analysis:
Of the 143 SSR markers tested, 135 segregated in at least one parent in the Syrah × Pinot Noir F1 population. VVS5, VVMD34, VMC8A7, VMC3A9, VMC3F3, and VMC1F10 primer pairs generated monomorphic markers, while the loci VMC4G6 and VMC5E11 were homozygous in both Syrah and Pinot Noir and thus did not segregate in the F1 progeny.
AFLP analysis:
AFLP bands showed either single- or double-dose intensities. The 94 F1 progenies of the Syrah × Pinot Noir population were analyzed with 16 EcoRI/MseI PCs. In total, 391 markers were codominantly scored.
Linkage map:
Segregation distortion and linkage analysis:
A total of 1034 polymorphic markers were scored: 184 from ESTs, 324 from BESs, 135 SSRs, and 391 AFLP markers. Twenty-eight markers (10 from ESTs, 8 from BESs, 2 SSRs, and 8 AFLP markers) with low-quality data were discarded from linkage analysis. A significant deviation (P < 0.05) from the expected Mendelian ratio was observed for 117 (11.6%) of 1006 markers (of these, 69 had P-values between 0.05 and 0.01 and 48 with P < 0.01), including 16 EST markers, 34 BES markers, 27 SSRs, and 40 AFLP markers. Compared to the other marker classes, SSRs showed the highest presence of segregation distortion (20.3%). The number of markers for each type of segregation—1:1 with Syrah or Pinot heterozygous and the other parent homozygous (respectively, types ab × aa and aa × ab in Table 1), 1:2:1 when both parents were heterozygous for the same markers (types ab × ab in Table 1), and 1:1:1:1 when in the cross three or four alleles were segregating (respectively, types ab × ac and ab × cd in Table 1)—and for each marker class is reported in Table 1.
TABLE 1.
Molecular marker segregation types in the Syrah × Pinot F1 population of 94 individuals
| Heterozygous state present in
|
||||||
|---|---|---|---|---|---|---|
| Marker type | Syrah: | Pinot: | Syrah and Pinot
|
|||
| ab × aaa | aa × aba | ab × abb | ab × acc | ab × cdc | Total | |
| EST | 65 | 70 | 36 | 3 | — | 174 |
| BES | 99 | 100 | 117 | — | — | 316 |
| SSR | 22 | 13 | 21 | 54 | 23 | 133 |
| AFLP | 124 | 164 | 95 | — | — | 383 |
| Total | 310 | 347 | 269 | 57 | 23 | 1006 |
EST, expressed sequence tag marker; BES, BAC end-sequence marker; SSR, simple sequence repeat marker.
Marker segregating according to the 1:1 ratio.
Marker segregating according to the 1:2:1 ratio.
Marker segregating according to the 1:1:1:1 ratio.
All maps were constructed at LOD 8.0. Linkage groups were defined according to Adam-Blondon et al. (2004) and Doligez et al. (2006). Linkage analysis for the Syrah map included 659 markers. Fourteen remained unlinked at LOD 6.0. Of the remaining 645 markers, 596 were placed into 19 linkage groups covering 1113 cM. For the Pinot Noir map, linkage analysis included 696 markers. Sixteen remained unlinked at LOD 6.0. Of the remaining 680 markers, 634 were placed into 19 linkage groups covering 1204 cM. There were 49 and 46 markers assigned to linkage groups of the Syrah and the Pinot Noir maps respectively, but not placed on the multipoint map because they were linked at a lower LOD (between 6 and 8), they excessively increased the linkage group end distances, or they affected the order of their neighbor markers. Among them, 12 and 9, respectively, showed distorted segregation ratios. The two parental maps are presented in supplemental Figure S1 at http://www.genetics.org/supplemental/. More details on marker number, linkage group size, and number of gaps for each parental map are reported in supplemental Table S2. Marker order was highly conserved between the two parental maps, with only a few local inversions of closely linked markers (<5 cM) and proximal markers. Of the 99 pairs of linked markers in which parental recombination rates were compared, statistically significant (0.01 < P < 0.05) differences were observed in only 10: VMC8D1-VVIT60, VMC6F1-VMC3B10, VMC2G2-VMC5C5, VVIS58-VMC8D11, ZAG25-VMC8D3, VVIV61-VMC3D12, VMC2B11-VMC6E1, VMC2B11-ZAG112, VMC6E1-ZAG112, and VMCNG1B9-VMC8B5.
Linkage analysis for the consensus map included 1006 markers: 174 from ESTs, 316 from BESs, 133 SSRs, and 383 AFLP markers. Twelve markers remained unlinked at LOD 6.0 (4 from ESTs, 3 from BESs, 1 SSR, and 4 AFLP markers). The remaining 994 markers (170 from ESTs, 313 from BESs, 132 SSRs, and 379 AFLP markers) generated a genetic map comprising 19 linkage groups and covering 1245 cM. Of all mapped markers, 63 were assigned to linkage groups but not placed on the multipoint map because they were linked at a lower LOD (between 6 and 8), they excessively increased the linkage group end distances, or they affected the order of their neighbor markers (Figure 1). Among them 13 showed distorted segregation ratios. Linkage groups ranged in size from 45.8 (LG3) to 92.7 (LG18) cM. The average distance between markers was 1.25 cM and only one gap >20 cM was noted (LG17).
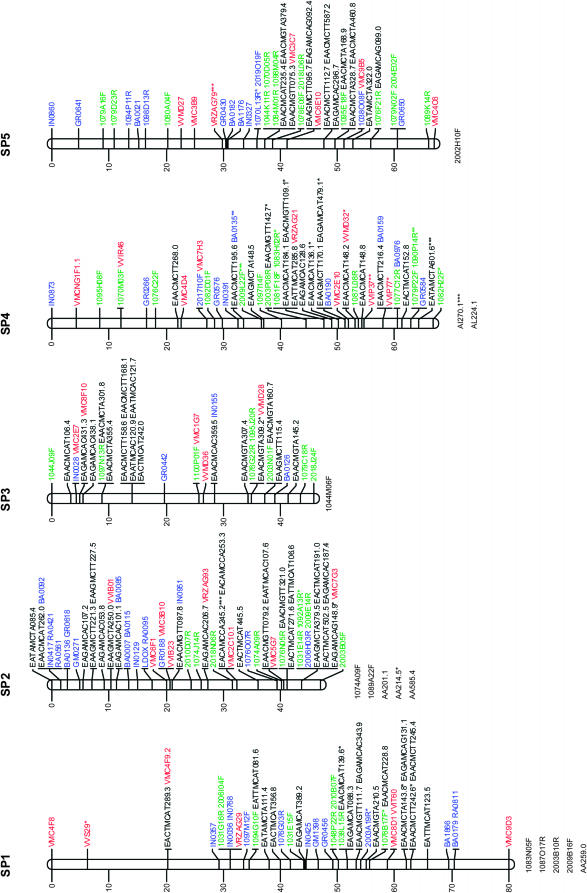
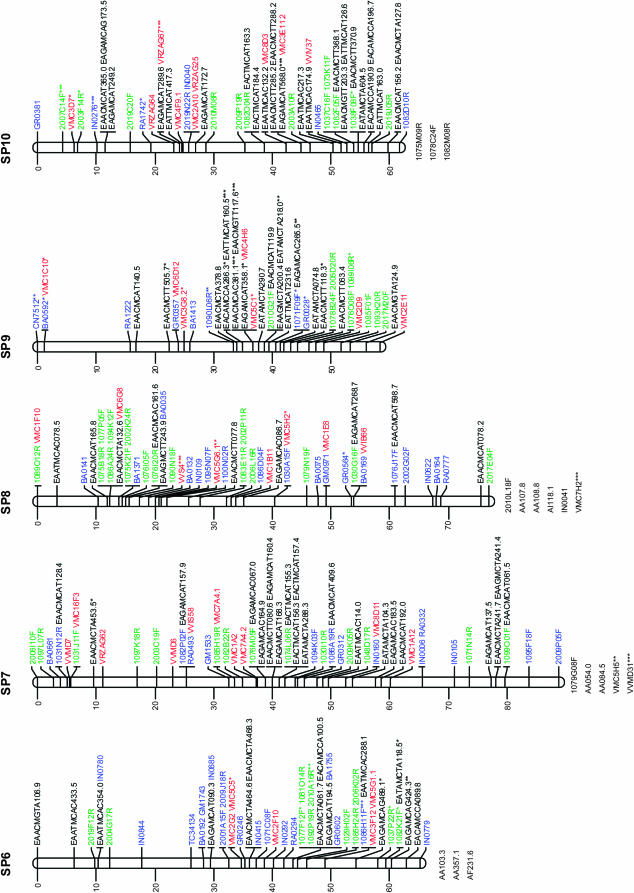
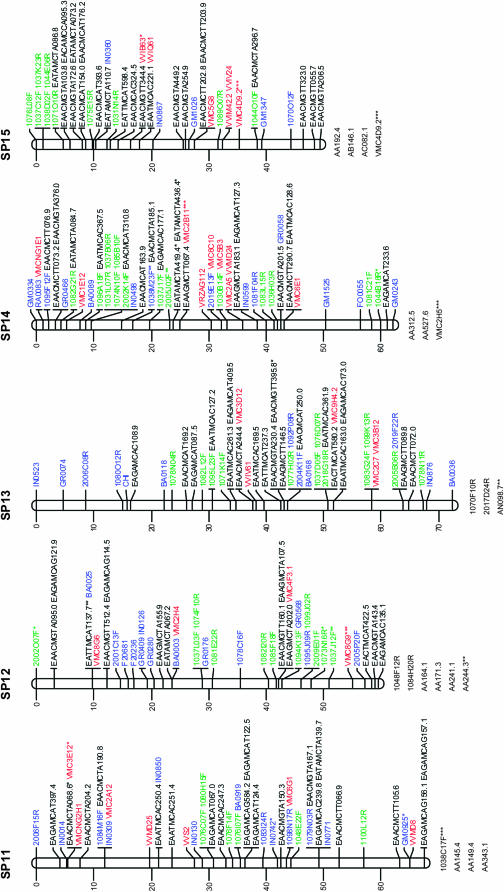
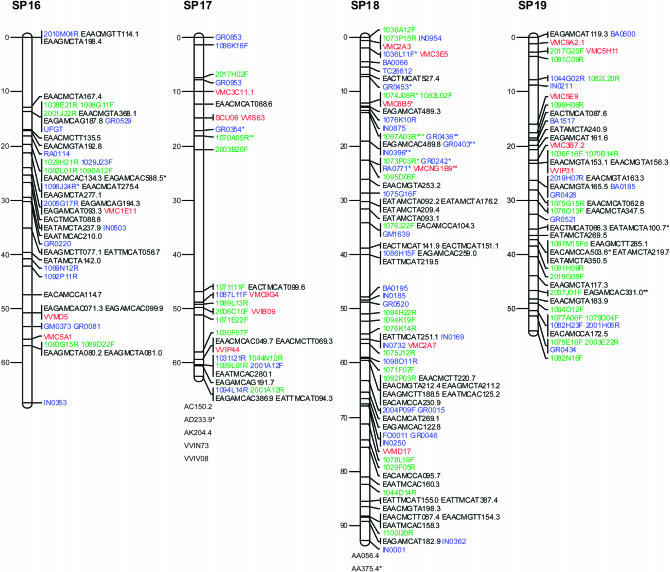
Figure 1.—
Consensus linkage map of Vitis vinifera from the Syrah × Pinot Noir cross. Linkage groups are numbered according to the V. vinifera map of Adam-Blondon et al. (2004). Markers from EST and from coding BES regions are shown in blue, noncoding BESs in green, SSRs in red, and AFLP markers in black. Loci with a distorted segregation ratio are marked by asterisks (*P < 0.05; **P < 0.01; ***P < 0.001). Distances of markers from the top are indicated on the left in centimorgans. Linked markers that excessively increased the linkage group end distances or that affected the order of neighbors in the group were not included in the map and are listed below each linkage group.
Table 2 reports the distribution of marker types in linkage groups together with the map length in centimorgans for each linkage group and between adjacent markers. In addition, Table 2 includes the number of BAC contigs anchored to each linkage group, as well as the megabases of physical coverage of each linkage group provided by the assigned BAC contigs.
TABLE 2.
Distribution of marker types by linkage group, map length by linkage group, distance between adjacent markers, number of contigs anchored to the genetic map, and physical coverage per linkage group
| Map length (cM)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Linkage group | No. of markers
|
Average distance between adjacent markers | No. of contigs anchoreda | Coverage of anchored contigs (Mbp) | |||||
| Total | EST | BES | SSR | AFLP | Total | ||||
| 1 | 48 | 9 | 15 | 7 | 17 | 80.0 | 1.67 | 20 | 17.68 |
| 2 | 62 | 15 | 13 | 8 | 26 | 47.0 | 0.76 | 15 | 16.17 |
| 3 | 35 | 5 | 9 | 5 | 16 | 45.8 | 1.31 | 13 | 16.09 |
| 4 | 51 | 9 | 15 | 9 | 18 | 66.8 | 1.31 | 18 | 16.95 |
| 5 | 49 | 8 | 21 | 7 | 13 | 67.0 | 1.37 | 21 | 21.84 |
| 6 | 51 | 13 | 16 | 5 | 17 | 65.2 | 1.28 | 19 | 15.05 |
| 7 | 64 | 8 | 21 | 12 | 23 | 88.8 | 1.39 | 25 | 25.12 |
| 8 | 60 | 13 | 23 | 9 | 15 | 76.9 | 1.28 | 23 | 21.78 |
| 9 | 42 | 7 | 10 | 7 | 18 | 58.3 | 1.39 | 14 | 11.68 |
| 10 | 55 | 5 | 17 | 9 | 24 | 61.7 | 1.12 | 24 | 17.43 |
| 11 | 49 | 8 | 12 | 7 | 22 | 67.5 | 1.38 | 17 | 17.20 |
| 12 | 47 | 9 | 17 | 4 | 17 | 59.6 | 1.27 | 16 | 22.25 |
| 13 | 51 | 7 | 19 | 5 | 20 | 72.4 | 1.42 | 27 | 22.42 |
| 14 | 58 | 10 | 18 | 10 | 20 | 62.1 | 1.07 | 20 | 25.01 |
| 15 | 45 | 4 | 11 | 6 | 24 | 49.4 | 1.10 | 13 | 8.42 |
| 16 | 49 | 8 | 14 | 3 | 24 | 67.3 | 1.37 | 19 | 12.72 |
| 17 | 38 | 3 | 16 | 8 | 11 | 62.4 | 1.64 | 15 | 17.69 |
| 18 | 85 | 22 | 24 | 6 | 33 | 92.7 | 1.09 | 27 | 27.94 |
| 19 | 55 | 7 | 22 | 5 | 21 | 54.1 | 0.98 | 21 | 18.29 |
| Total | 994 | 170 | 313 | 132 | 379 | 1245 | 1.25 | 367 | 351.73 |
EST, expressed sequence tag marker; BES, BAC end-sequence marker; SSR, simple sequence repeat marker. Linkage groups are numbered according to the Vitis vinifera map of Adam-Blondon et al. (2004). Distances are in Kosambi centimorgans.
The markers with segregation distortion (shown in Figure 1 with *, **, or ***, respectively, when P < 0.05, P < 0.01, or P < 0.001) were randomly distributed throughout the genome, except for LG9 and LG18 where some clusters of markers were observed.
Marker order was generally consistent between homologs from the parental and the consensus maps, with only a few local inversions of closely linked markers and terminal inversions present on linkage groups 1, 13, and 17. On the basis of 118 SSR markers distributed across all linkage groups, the marker order of the map presented is always consistent with the one observed in the integrated map based on five segregating populations of V. vinifera (Doligez et al. 2006).
Marker distribution:
For each type of marker (from ESTs and BESs, SSRs, and AFLP markers), the χ2-test applied to the number of polymorphic loci mapping to each linkage group was not significant (P < 0.05). This result indicated a homogeneous distribution of marker types among linkage groups.
Under the assumption of random marker distribution, the number of 10-cM intervals containing from 0 to 10 or more markers should also follow a Poisson distribution. The ratio between the variance and the mean (coefficient of dispersion) of the distribution of the number of markers per 10-cM interval provides a rapid method to test if the observed frequencies are distributed following a Poisson distribution. The coefficient of dispersion was very close to 1 when coding loci and SSR markers were considered, suggesting a random distribution for these types of markers. When considering AFLP markers and noncoding DNA loci alone or AFLP markers and noncoding loci together, the coefficients of dispersion were 2.3, 2.0, and 3.2, respectively, indicating that these markers tended to cluster in the map.
The observed and expected number of markers/10-cM interval (when considering AFLP markers and noncoding loci together) for all linkage groups deviate significantly from a Poisson distribution (χ2 = 149, d.f. = 9, P < 0.0001). The deviation was greatest in those intervals containing 1, 2, 9, or >10 markers. Similar results were obtained when considering the parental maps separately (supplemental Table S2 at http://www.genetics.org/supplemental/).
Physical and genetic distance estimation:
A total of 623 markers (123 from ESTs, 293 from BESs, 93 SSRs, and 114 AFLP markers) were used to anchor to the genetic linkage map 367 BAC contigs covering 352 Mbp of a V. vinifera physical DNA map (Table 2; http://genomics.research.iasma.it; supplemental Figures S2 and S3 at http://www.genetics.org/supplemental/).
The marker position on BAC contigs was used to verify the accuracy of marker order estimated with meiotic methods (supplemental Figures S2a and S2b at http://www.genetics.org/supplemental/). This was possible for 167 markers assigned to 44 anchored BAC contigs with >3 markers per contig. The linear order of the genetically mapped markers based on FPC analysis of the contig agreed closely with their order in the consensus map and the two parental maps of V. vinifera presented here. Only 4 markers showed a small inversion when comparing their physical distances in the contig with their genetic map position.
Estimates of the grape genome size are 1245 cM and 475 Mbp, meaning that, on average, 1 Mbp should correspond to 2.6 cM. The comparison of the genetic and physical distances separating neighboring markers also present in anchored contigs made it possible to compare the recombination frequency at different genome positions. Three examples had markedly higher recombination frequencies of 41, 19, and 8 cM/Mbp. At the other extreme, there were two pairs of markers separated by 440 and 600 kbp, respectively, which had no observable recombinations between them (Table 3).
TABLE 3.
Physical and genetic comparative distances
| Genetic distance: | Physical distance
|
||||
|---|---|---|---|---|---|
| Linked markers | LG | cMa | Contigb | Kbpc | cM/Mbp |
| 2006F15R-EAGAMCAT397.4 | 11 | 4.1 | Ctg 93 | 100 | 41 |
| RA0811-VMC9D3 | 1 | 7.6 | Ctg 336 | 400 | 19 |
| VVIT60-EAACMCTT242.6 | 1 | 3.2 | Ctg 4744 | 400 | 8 |
| ZAG112-VMC6C10 | 14 | 1.2 | Ctg 1054 | 300 | 4 |
| EAACMCTT080.6-EAGAMCAT160.4 | 7 | 0.0 | Ctg 550 | 440 | 0 |
| 2010A16R-1077F12F | 6 | 0.0 | Ctg 185 | 600 | 0 |
Distance in centimorgans in the linkage map (Pinot Noir map) between two distal markers presented in an anchored contig.
Anchored contig listed at http://genomics.research.iasma.it.
Physical distance between the two markers of the contig.
DISCUSSION
This work represents the most comprehensive linkage analysis in grapevine to date. The map spans 1245 cM divided into 19 linkage groups, corresponding to the number of grapevine chromosomes, with an average distance of 1.3 cM between adjacent markers. SNP-based markers were developed from both EST sequences and genomic BESs with a similar efficiency rate (38.3% of polymorphic markers from 454 selected EST sequences vs. 35% from 903 selected BESs). The low efficiency in developing SNP-based markers can be explained by multiple factors including the amplification rate, sequencing failure, and the presence of monomorphic regions.
The amplification efficiency was higher when primers were designed from genomic sequences compared with EST sequences (100% vs. 80%). Zhu et al. (2003) reported a similar amplification rate when amplifying soybean genomic DNA with primers designed from EST sequences. These authors attributed PCR failures to sequence variations at primer annealing sites. Additional explanations may include the presence of large introns within PCR amplicons and the positioning of primers at the intron–exon junctions.
On the other hand, resequencing of amplified products gives rise to a higher percentage of failed sequences from BES (46%) compared with ESTs (17%). The poor quality of BES data from PCR amplification resulting in simple fragments of DNA is likely to be a result of heterogeneous DNA templates due to duplicated genes or multigene families, but could also be the result of heterozygous allelic variations generating sequencing artifacts. This is supported by the higher percentage of failed sequences from BESs, which are mostly noncoding and therefore have a high probability of being polymorphic (Gaafar et al. 2005). Primers designed on predicted protein-coding BESs showed, in fact, a lower percentage of failed sequences aligned to the frequency of ESTs (23.5%).
In grape, polymorphic DNA loci are relatively frequent. Salmaso et al. (2004) found 1 SNP every 116 bp in the coding regions of 25 genes, using EST-derived primers in the analysis of seven V. vinifera cultivars. C. Segala (personal communication) found a value of 5.88 SNPs/1000 bases within noncoding regions of 183 BESs, when comparing the cultivars Syrah and Pinot Noir. The high percentage of monomorphic regions (28% for EST, 19% for BES) is quite unexpected when compared with what is reported in the literature on grape and can be explained in part by preferential PCR amplification of one allele due to mismatches between the PCR primer and the second allelic template (Walsh et al. 1992). On the other hand, coding sequences have a higher probability of being monomorphic due to a direct effect of selection in favor of sequence conservation.
AFLP technology was very useful in the generation of a high number of molecular markers and, therefore, for the integration of the physical and genetic maps. An average of 24.4 AFLP markers per primer combination were detected between the two parents of our map. This is comparable to what is reported by Grando et al. (2003) for an interspecific cross of grape, but higher than the average of 8.0 markers per primer combination reported by Doucleff et al. (2004) for a cross of two interspecific hybrids of grape. The high detection rate of AFLP markers reported here is possibly due to the accurate selection of primer combinations facilitated by the previous work carried out in our group (Grando et al. 2003).
As reported for sorghum (Klein et al. 2000), AFLP technology and a BAC DNA pooling strategy provide an inexpensive and efficient way to build high-quality genetic and physical genome maps. The AFLP markers in this study offered a low-cost anchoring approach to align the maternal and paternal maps.
Segregation distortion:
Segregation distortion has often been observed in fruit and forest trees (Liebhard et al. 2003; Ma et al. 2004; Yin et al. 2004). The proportion of SSR markers with distorted segregation observed in this study (20.3%) was higher than that reported by Doligez et al. (2002), Adam-Blondon et al. (2004), and Riaz et al. (2004) (9.9%). On the other hand, it is similar to the values reported by Grando et al. (2003) for V. riparia (22.4%). The proportion of AFLP markers with distorted segregation (10.4%) is very similar to that of Grando et al. (2003) (8.5% for V. vinifera cv. Moscato and 16.4% for V. riparia) and Doucleff et al. (2004) (9%). In our case, during the process of map construction only 11 of the 117 markers deviating significantly from the expected Mendelian ratio were discarded because they affected the order of their neighboring markers.
Clustering of markers with distorted segregation was observed for LG9 and LG18. Similar cases have been repeatedly reported in mapping studies in crops and fruit trees (Vuylsteke et al. 1999; Cervera et al. 2001; Yin et al. 2004), suggesting the action of a causal mechanism (e.g., linkage to genes affecting seed or seedling viability). However, no evidence of marker segregation distortion was found in the regions of the other maps of V. vinifera corresponding to those we report here for LG9 and LG18 (Doligez et al. 2006).
Distribution of marker types in the linkage map:
Clustering of AFLP and noncoding BES-derived markers was observed across all linkage groups, with more than one large cluster (>10 markers) per linkage group when considering AFLP and noncoding BES markers together. The distribution of such clusters deviated significantly from Poisson expectations. Clustering of markers, particularly of AFLP markers, which mostly represent noncoding sequences (Young et al. 1999), has been observed in several species, including grapevine (Tanksley et al. 1992; Vuylsteke et al. 1999; Doucleff et al. 2004). It is known that DNA markers are clustered in the centromeric region due to the suppression of recombination in the heterochromatic regions surrounding centromeres (Young et al. 1999; Vuylsteke et al. 1999; van Os et al. 2006).
The preferential finding of AFLP loci in recombinationally suppressed regions implies that aspects of the AFLP technique (for instance, the restriction enzymes used) may favor the finding of these markers in a specific region of the genome, like centromeric regions. The high frequency of polymorphism in noncoding sequences, and the effect of the restriction enzyme MseI that cuts more frequently in regions with a high A + T content, may explain this result. Noncoding sequences have a higher A + T content than coding sequences (Young et al. 1999): the selective +3/+3 nucleotide composition of marker clusters—for instance, LG7 and LG18—is consistent with this model (Alonso-Blanco et al. 1998). Moreover, the use of HindIII-based BAC libraries may partially explain the clustering of markers designed from BES regions. Due to their nucleotide composition, HindIII cleavage sites are, in fact, nonrandomly distributed in the genome and more frequent in AT-rich regions. In addition, clustered markers may not necessarily be physically close even if they appear that to be so on a recombination-based map because of the lack of recombination in the region (Keim et al. 1997). In fact, for V. vinifera we identified (with the limits of the size of our segregating population) regions with suppressed recombination spanning physical sizes up to 600 kbp.
Our results (see Table 3 and the CMap tool available at http://genomics.research.iasma.it) suggest that regions of both increased and reduced recombination exist within the grape genome, although they are not conclusive because of significant uncertainties in both the genetic and the physical distances. Variations in the correspondence between physical and genetic distance along chromosomes are well known and have already been well documented in several species (Tanksley et al. 1992; Chen et al 2002; King et al. 2002). When available, such variations add important information to map-based cloning projects. A high-resolution genetic map, a prerequisite for chromosome walking, is much easier to generate in the presence of high-recombination values. In regions of suppressed recombination, a larger progeny size is needed to recover the number of crossovers necessary for constructing detailed genetic maps (Tanksley et al. 1992).
In contrast to AFLP and noncoding BES-derived markers, SSR and other markers based on coding sequences appeared to be randomly distributed throughout the map. Random distribution of SSR loci has also been reported by other authors for grape and other fruit-tree maps (Testolin et al. 2001; Doligez et al. 2006).
Comparison with the parental maps and other grapevine maps:
The map presented here was generated using a two-step strategy. Parental linkage groups were first constructed using different types of segregating markers and then homologous linkage groups were merged. The accessibility of common markers and especially codominant ones allowed not only for the identification of homologous linkage groups but also for the integration of both parental maps. The integration of the two parental linkage maps into a single reference map was possible due to the colinearity of fully informative loci (few local inversions of closely linked markers and proximal markers) and the homogeneous recombination rates between homologous pairs of loci of both individual linkage maps. This result was expected since the two cultivars from the pedigree reconstruction reported in Vouillamoz and Grando (2006) are 3° relatives and it also justified the use of Syrah markers to anchor Pinot Noir BAC contigs. Local inversions of closely linked markers were probably due to statistical inaccuracy linked to the limited number of individuals studied, as reported in Di Gaspero et al. (2007), rather than the consequences of true chromosomal inversion. Differences at the terminal regions and between closely linked markers cannot be resolved since usually a number of almost equivalent marker orders exists (Yan et al. 2005). However, due to the availability of a physical map we are able to resolve some of these inversions (for instance, marker SCU06 in linkage group 17, supplemental Figures S1 and S2b at http://www.genetics.org/supplemental/). Only a few cases of heterogeneous recombination rates between Syrah and Pinot Noir were observed. Few statistically significant differences in recombination rates have also been found in other published grapevine maps (Adam-Blondon et al. 2004; Lowe and Walker 2006).
A total of 118 SSR markers are present in our map and in the integrated V. vinifera linkage map of Doligez et al. (2006). This allows accurate cross-referencing between the different maps and offers the future opportunity of exploiting a much larger number of markers for a given genomic region in both basic and applied projects. SSR marker loci/positions were well conserved in all linkage groups between the map presented here and the integrated map of Doligez et al. (2006). Due to the positioning of common markers with other maps produced for V. vinifera where QTL intervals have been identified, it is now possible to transfer this information to ours as well as to other genetic crosses.
The linkage map sizes produced in other mapping experiments are larger than the one reported here, despite a lower number of markers mapped in the former cases. The reported values are: 1676 cM (502 markers) for the composite map of two pseudo-testcrosses, Cabernet Sauvignon × Bianca and Cabernet Sauvignon × Vitis breeding line 20/3 (Di Gaspero et al. 2007); 1647 cM (515 markers) for the integrated map of five different populations of of V. vinifera (Doligez et al. 2006); 1406 cM (220 markers) for the Syrah × Grenache map (Adam-Blondon et al. 2004); and 1728 cM (153 markers) for the Riesling × Cabernet Sauvignon map (Riaz et al. 2004). Differences in the size of the linkage maps may derive both from the use of different marker types and from genotyping errors. They are most likely, however, to be the result of the different mapping programs used for map construction.
To build the map presented here, a recently developed mapping program (Cartwright et al. 2007, this issue) was used that considers genotyping errors and reduces the inflationary effect caused by increasing the number of markers. Errors inflate the number of recombinations and considerably expand map intervals (Harald et al. 2000). In this respect, our map is more reliable in terms of marker order and in marker distance estimation as demonstrated in Cartwright et al. (2007). The accuracy of marker order estimated by meiotic methods was also verified using the physical distance information for the genetically mapped markers contained in the anchored BAC contigs with three or more mapped markers. Moreover, the use of different marker types enhances the chances of sampling different parts of the genome (Yin et al. 2004).
Previous low-density genetic linkage maps of V. vinifera were based primarily on SSR and AFLP markers (Grando et al. 2003; Adam-Blondon et al. 2004; Riaz et al. 2004; Doligez et al. 2006). The addition of SNP-based markers introduces polymorphisms that are easy to database and useful for evolutionary studies and that significantly increase the density of the map. The result is an improved resource for fine mapping quantitative trait loci, identifying candidate genes, and map-based gene isolation.
Acknowledgments
The authors thank Jessica Zambanini, Marco Facci, and Diego Micheletti for their technical support; Paolo Fontana and Alessandro Cestaro for web site design and maintenance; and Francesco Salamini for helpful discussions and critical reading of the manuscript. This work was supported by the “Grapevine Physical Mapping” and “A.M.I.CA. Vitis'” projects funded by the Autonomous Province of Trento. AFLP is a registered trademark and the AFLP technology is covered by patents and patent applications of Keygene N.V.
References
- Adam-Blondon, A.-F., C. Roux, D. Claux, G. Butterlin, D. Merdinoglu et al., 2004. Mapping 245 SSR markers on the Vitis vinifera genome: a tool for grape genetics. Theor. Appl. Genet. 109: 1017–1027. [DOI] [PubMed] [Google Scholar]
- Adam-Blondon, A.-F., A. Bernole, G. Faes, D. Lamoureux, S. Pateyron et al., 2005. Construction and characterization of BAC libraries from major grapevine cultivars. Theor. Appl. Genet. 110: 1363–1371. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco, C., A. J. M. Peeters, M. Koornneef, C. Lister, C. Dean et al., 1998. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 14: 259–271. [DOI] [PubMed] [Google Scholar]
- Aradhya, M. K., G. S. Dangl, B. H. Prins, J. M. Boursiquot, M. A. Walker et al., 2003. Genetic structure and differentiation in cultivated grape, Vitis vinifera L. Genet. Res. 81: 179–192. [DOI] [PubMed] [Google Scholar]
- Barillot, E., B. Lacroix and D. Cohen, 1991. Theoretical analysis of library screening using a N-dimensional pooling strategy. Nucleic Acids Res. 19: 6241–6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, C. L., T. Donald, J. Pauquet, M. B. Ratnaparkhe, A. Bouquet et al., 2005. Genetic and physical mapping of the grapevine powdery mildew resistance gene, Run1, using a bacterial artificial chromosome library. Theor. Appl. Genet. 111: 370–377. [DOI] [PubMed] [Google Scholar]
- Bowers, J. E., G. S. Dangl, R. Vignani and C. P. Meredith, 1996. Isolation and characterization of the new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 45: 1142–1149. [DOI] [PubMed] [Google Scholar]
- Bowers, J. E., G. S. Dangl and C. P. Meredith, 1999. Development and characterization of additional microsatellite DNA markers for grape. Am. J. Enol. Vit. 50: 243–246. [Google Scholar]
- Cartwright, D. A., M. Troggio, R. Velasco and A. Gutin, 2007. Genetic mapping in the presence of genotyping errors. Genetics 176: 2521–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin, S. D., G. Di Gaspero, R. Marconi, A. Nonis, E. Peterlunger et al., 2006. Colour variation in red grapevines (Vitis vinifera L.): genomic organisation, expression of flavonoid 3′-hydroxylase, flavonoid 3′,5′-hydroxylase genes and related metabolite profiling of red cyanidin-/blue delphinidin-based anthocyanins in berry skin. BMC Genomics 7: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, A. J., C. Carapito, N. Zorn, C. Magne, E. Leize et al., 2005. Proteomic analysis of grapevine (Vitis vinifera L.) tissues subjected to herbicide stress. J. Exp. Bot. 56: 2783–2795. [DOI] [PubMed] [Google Scholar]
- Cervera, M. T., V. Storme, B. Ivens, J. Gusmão, B. H. Liu et al., 2001. Dense genetic linkage maps of three Populus species (Populus deltoides, P. nigra and P. trichocarpa) based on AFLP and microsatellite markers. Genetics 158: 787–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., G. Presting, W. B. Barbazuk, J. L. Goicoechea, B. Blackmon et al., 2002. An integrated physical and genetic map of the rice genome. Plant Cell 14: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva, F. G., A. Iandolino, F. Al-Kayal, M. C. Bohlmann, M. A. Cushman et al., 2005. Characterizing the grape transcriptome. Analysis of expressed sequence tags from multiple Vitis species and development of a compendium of gene expression during berry development. Plant Physiol. 139: 574–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gaspero, G., G. Cipriani, A.-F. Adam-Blondon and R. Testolin, 2007. Linkage maps of grapevine displaying the chromosomal location of 420 microsatellite markers and 82 markers for R-gene candidates. Theor. Appl. Genet. 114: 1249–1263. [DOI] [PubMed] [Google Scholar]
- Dirlewanger, E., E. Graziano, T. Joobeur, F. Garriga-Calderé, P. Cosson et al., 2004. Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc. Natl. Acad. Sci. USA 101: 9891–9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doligez, A., A. Bouquet, Y. Danglot, F. Lahogue, S. Riaz et al., 2002. Genetic mapping of grapevine (Vitis vinifera L.) applied to the detection of QTLs for seedlessness and berry weight. Theor. Appl. Genet. 105: 780–795. [DOI] [PubMed] [Google Scholar]
- Doligez, A., A.-F. Adam-Blondon, G. Cipriani, G. Di Gaspero, V. Laucou et al., 2006. An integrated SSR map of grapevine based on five different populations. Theor. Appl. Genet. 113: 369–382. [DOI] [PubMed] [Google Scholar]
- Don, R. H., P. T. Cox, B. J. Wainwrigth, K. Baker and J. S. Mattick, 1991. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19: 4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald, T. M., F. Pellerone, A.-F. Adam-Blondon, A. Bouquet, M. R. Thomas et al., 2002. Identification of resistance gene analogs linked to a powdery mildew resistance locus in grapevine. Theor. Appl. Genet. 104: 610–618. [DOI] [PubMed] [Google Scholar]
- Doucleff, M., Y. Jin, F. Gao, S. Riaz, A. F. Krivanek et al., 2004. A genetic linkage map of grape, utilizing Vitis rupestris and Vitis arizonica. Theor. Appl. Genet. 109: 1178–1187. [DOI] [PubMed] [Google Scholar]
- Doyle, J. J., and J. L. Doyle, 1990. Isolation of plant DNA from fresh tissue. Focus Biotech. 12: 13–15. [Google Scholar]
- Fanizza, G., F. Lamaj, L. Costantini, R. Chaabane and M. S. Grando, 2005. QTL analysis for fruit yield components in table grapes (Vitis vinifera). Theor. Appl. Genet. 111: 658–664. [DOI] [PubMed] [Google Scholar]
- Faria, M., A. M. Beja-Pereira, A. Martins, M. A. Ferreira and M. E. S. Nunes, 2004. Grapevine clones discriminated using stilbene synthase-chalcone synthase markers. J. Sci. Food Agric. 84: 1186–1192. [Google Scholar]
- Fischer, B., I. Salakhutdinov, M. Akkurt, R. Eibach, K. J. Edwards et al., 2004. Quantitative trait locus analysis of fungal disease resistance factor on a molecular map of grapevine. Theor. Appl. Genet. 108: 501–515. [DOI] [PubMed] [Google Scholar]
- Gaafar, R. M., U. Hohmann and C. Jung, 2005. Bacterial artificial chromosome-derived molecular markers for early bolting in sugar beet. Theor. Appl. Genet. 110: 1027–1037. [DOI] [PubMed] [Google Scholar]
- Grando, M. S., D. Bellin, K. J. Edwards, C. Pozzi, M. Stefanini et al., 2003. Molecular linkage maps of Vitis vinifera L. and Vitis riparia Mchx. Theor. Appl. Genet. 106: 1213–1224. [DOI] [PubMed] [Google Scholar]
- Grattapaglia, D., and R. Sederoff, 1994. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudotestcross: mapping strategy and RAPD markers. Genetics 4: 1121–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harald, H., H. Goring and J. D. Terwilliger, 2000. Linkage analysis in the presence of errors II: marker-locus genotyping errors modeled with hypercomplex recombination fractions. Am. J. Hum. Genet. 66: 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, R., A.-C. Lecouls, A. Callahan, A. Dandekar, L. Garay et al., 2005. Candidate gene database and transcript map for peach, a model species for fruit trees. Theor. Appl. Genet. 110: 1419–1428. [DOI] [PubMed] [Google Scholar]
- Jansen, R. C., 1993. Maximum likelihood in a generalized linear finite mixture model by using the EM algorithm. Biometrics 49: 227–231. [Google Scholar]
- Keim, P., J. M. Schupp, S. E. Travis, K. Clayton, T. Zhu et al., 1997. A high-density soybean genetic map based on AFLP markers. Crop Sci. 37: 537–543. [Google Scholar]
- King, J., I. P. Armstead, I. S. Donnison, H. M. Thomas, R. N. Jones et al., 2002. Physical and genetic mapping in the grasses Lolium perenne and Festuca pratensis. Genetics 161: 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, P. E., R. R. Klein, S. W. Cartinhour, P. E. Ulanch, J. Dong et al., 2000. A high-throughput AFLP-based method for constructing integrated genetic and physical maps: progress toward a sorghum genome map. Genet. Res. 10: 789–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi, D. D., 1944. The estimation of map distance from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Liebhard, R., B. Koller, L. Gianfranceschi and C. Gessler, 2003. Creating a saturated reference map for the apple (Malus x domestica Borkh.) genome. Theor. Appl. Genet. 106: 1497–1508. [DOI] [PubMed] [Google Scholar]
- Lodhi, M. A., and B. I. Reisch, 1995. Nuclear DNA content of Vitis species, cultivars and other genera of the Vitaceae. Theor. Appl. Genet. 90: 11–16. [DOI] [PubMed] [Google Scholar]
- Lowe, K. M., and M. A. Walker, 2006. Genetic linkage map of the interspecific grape rootstock cross Ramsey (Vitis champinii) × Riparia Gloire (Vitis riparia). Theor. Appl. Genet. 112: 1582–1592. [DOI] [PubMed] [Google Scholar]
- Luo, M. C., C. Thomas, F. M. You, J. Hsiao, S. Ouyang et al., 2003. High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics 82: 378–389. [DOI] [PubMed] [Google Scholar]
- Ma, H., P. H. Moore, Z. Liu, M. S. Kim, Q. Yu et al., 2004. High-density linkage mapping revealed suppression of recombination at the sex determination locus in papaya. Genetics 166: 419–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdinoglu, D., G. Butterlin, L. Bevilacqua, V. Chiquet, A.-F. Adam-Blondon et al., 2005. Development and characterization of a large set of microsatellite markers in grapevine (Vitis vinifera L.) suitable for multiplex PCR. Mol. Breed. 15: 349–366. [Google Scholar]
- Michaels, S. D., and R. M. Amasino, 1998. A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. Plant J. 14: 381–385. [DOI] [PubMed] [Google Scholar]
- Morgante, M., and F. Salamini, 2003. From plant genomics to breeding practice. Curr. Opin. Biotechnol. 14: 214–219. [DOI] [PubMed] [Google Scholar]
- Moser, C., C. Segala, P. Fontana, I. Salakhudtinov, P. Gatto et al., 2005. Comparative analysis of expressed sequence tags from different organs of Vitis vinifera L. Funct. Integr. Genomics 5: 208–217. [DOI] [PubMed] [Google Scholar]
- Neff, M. M., and E. T. M. Kalishman, 2002. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 18: 613–615. [DOI] [PubMed] [Google Scholar]
- Nelson, W. M., A. K. Bharti, E. Butler, F. Wei, G. Fuks et al., 2005. Whole-genome validation of high-information-content fingerprinting. Plant Physiol. 139: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita, M., H. Iwahana, H. Kanazana, K Hayashi and T. Sekiya, 1989. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc. Natl. Acad. Sci. USA 86: 2766–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa, R., M. Troggio, P. Ajmone-Marsan and F. Nonnis Marzano, 2005. An improved protocol for the production of AFLPTM markers in complex genomes by means of capillary electrophoresis. J. Anim. Breed. Genet. 122: 62–68. [DOI] [PubMed] [Google Scholar]
- Pereira, G. E., J.-P. Gaudillere, C. Van Leeuwen, G. Hilbert, O. Lavialle et al., 2005. 1H NMR and chemometrics to characterize mature grape berries in four wine-growing areas in Bordeaux, France. J. Agric. Food Chem. 53: 6382–6389. [DOI] [PubMed] [Google Scholar]
- Rafalski, A., 2002. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 5: 94–100. [DOI] [PubMed] [Google Scholar]
- Riaz, S., G. S. Dangl, K. J. Edwards and C. J. Meredith, 2004. A microsatellite marker based framework linkage map of Vitis vinifera L. Theor. Appl. Genet. 108: 864–872. [DOI] [PubMed] [Google Scholar]
- Salmaso, M., G. Faes, C. Segala, M. Stefanini, I. Salakhutdinov et al., 2004. Genome diversity and gene haplotypes in the grapevine (Vitis vinifera L.), as revealed by single nucleotide polymorphisms. Mol. Breed. 14: 385–395. [Google Scholar]
- Sarry, J.-M., N. Sommerer, F.-X. Sauvage, A. Bergoin, M. Rossignol et al., 2004. Grape berry biochemistry revisited upon proteomic analysis of the mesocarp. Proteomics 4: 201–215. [DOI] [PubMed] [Google Scholar]
- Scott, K. D., P. Eggler, G. Seaton, M. Rossetto, E. M. Ablett et al., 2000. Analysis of SSR derived from grape ESTs. Theor. Appl. Genet. 100: 723–726. [Google Scholar]
- Sefc, K. M., F. Regner, E. Turetschek, J. Glössl and H. Steinkellner, 1999. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 42: 367–373. [PubMed] [Google Scholar]
- Staden, R., K. F. Beal and J. K. Bonfield, 2000. The Staden package, 1998. Methods Mol. Biol. 132: 115–130. [DOI] [PubMed] [Google Scholar]
- Tanksley, S. D., M. W. Ganal, J. P. Prince, M. C. de Vicente, M. W. Bonierbale et al., 1992. High density molecular linkage maps of tomato and potato genomes. Genetics 132: 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier, N., D. Glissant, J. Grimplet, F. Barrieu, P. Abbal et al., 2005. Isogene specific oligo arrays reveal multifaceted changes in gene expression during grape berry (Vitis vinifera L.) development. Planta 222: 832–847. [DOI] [PubMed] [Google Scholar]
- Testolin, R., W. G. Huang, O. Lain, R. Messina, A. Secchione et al., 2001. A kiwifruit (Actinidia spp.) linkage map based on microsatellites and integrated with AFLP markers. Theor. Appl. Genet. 103: 30–36. [Google Scholar]
- This, P., A. Jung, P. Boccacci, J. Borrego, R. Botta et al., 2004. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor. Appl. Genet. 109: 1448–1458. [DOI] [PubMed] [Google Scholar]
- Thomas, M. R., and N. S. Scott, 1993. Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequence-tagged sites (STSs). Theor. Appl. Genet. 86: 985–991. [DOI] [PubMed] [Google Scholar]
- Troggio, M., G. Malacarne, S. Vezzulli, G. Faes, M. Salmaso et al., 2007. Comparison of different methods for SNP detection in grapevine. Vitis (in press).
- van Os, H. V., S. Andrzejewski, E. Bakker, I. Barrena, G. Bryan et al., 2006. Construction of a 10,000 marker ultra-dense genetic recombination map of potato: providing a framework for accelerated gene isolation and a genomewide physical map. Genetics 173: 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips, R. E., 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee et al., 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vouillamoz, J. F., and M. S. Grando, 2006. Genealogy of wine grape cultivars: ‘Pinot’ is related to Syrah. Heredity 97: 102–110. [DOI] [PubMed] [Google Scholar]
- Vuylsteke, M., R. Mank, R. Antonise, E. Bastiaans, M. L. Senior et al., 1999. Two high-density AFLP® linkage maps of Zea mays L.: analysis of distribution of AFLP markers. Theor. Appl. Genet. 99: 921–935. [Google Scholar]
- Walsh, P. S., H. A. Erlich and R. Higuchi, 1992. Preferential PCR amplification of alleles: mechanisms and solutions. Genome Res. 1: 241–250. [DOI] [PubMed] [Google Scholar]
- Waters, D. L., T. A. Holton, E. M. Ablett, L. S. Lee and R. J. Henry, 2005. cDNA microarray analysis of developing grape (Vitis vinifera cv. Shiraz) berry skin. Funct. Integr. Genomics 5: 40–58. [DOI] [PubMed] [Google Scholar]
- Yan, Z., C. Denneboom, A. Hattendorf, O. Dolstra, T. Debener et al., 2005. Construction of an integrated map of rose with AFLP, SSR, PK, RGA, RFLP, SCAR and morphological markers. Theor. Appl. Genet. 110: 766–777. [DOI] [PubMed] [Google Scholar]
- Yin, T. M., S. P. DiFazio, L. E. Gunter, D. Riemenschneider and G. A. Tuskan, 2004. Large-scale heterospecific segregation distortion in Populus revealed by a dense genetic map. Theor. Appl. Genet. 109: 451–463. [DOI] [PubMed] [Google Scholar]
- Young, W. P., J. M. Schupp and P. Keim, 1999. DNA methylation and AFLP marker distribution in the soybean genome. Theor. Appl. Genet. 99: 785–792. [Google Scholar]
- Zhu, Y. L., Q. J. Song, D. L. Hyten, C. P. Van Tassel, L. K. Matukumalli et al., 2003. Single-nucleotide polymorphisms in soybean. Genetics 168: 123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]