Abstract
Introduction
Translational systems biology approaches can be distinguished from mainstream systems biology in that their goal is to drive novel therapies and streamline clinical trials in critical illness. One systems biology approach, dynamic mathematical modeling (DMM), is increasingly used in dealing with the complexity of the inflammatory response and organ dysfunction. The use of DMM often requires a broadening of research methods and a multidisciplinary team approach that includes bioscientists, mathematicians, engineers, and computer scientists. However, the development of these groups must overcome domain-specific barriers to communication and understanding.
Methods
We present four case studies of successful translational, interdisciplinary systems biology efforts, which differ by organizational level from an individual to an entire research community.
Results
Case 1 is a single investigator involved in DMM of the acute inflammatory response at Cook County Hospital, in which extensive translational progress was made using agent-based models of inflammation and organ damage. Case 2 is a community-level effort from the University of Witten-Herdecke in Cologne, whose efforts have led to the formation of the Society for Complexity in Acute Illness. Case 3 is an institution-based group, the Biosystems Group at the University of California, San Francisco, whose work has included a focus on a common lexicon for DMM. Case 4 is an institution-based, trans-disciplinary research group (the Center for Inflammation and Regenerative Modeling at the University of Pittsburgh, whose modeling work has led to internal education efforts, grant support, and commercialization.
Conclusion
A transdisciplinary approach, which involves team interaction in an iterative fashion to address ambiguity and is supported by educational initiatives, is likely to be necessary for DMM in acute illness. Community-wide organizations such as the Society of Complexity in Acute Illness (SCAI) must strive to facilitate the implementation of DMM in sepsis/trauma research into the research community as a whole.
Keywords: transdisciplinary approaches, mathematical modeling, complexity, education programs, interactive model, glossary, sepsis, inflammation, trauma, computer simulation
INTRODUCTION
“The scale and complexity of today’s biomedical research problems demand that scientists move beyond the confines of their individual disciplines and explore new organizational models for team science. Advances in molecular imaging, for example, require collaborations among diverse groups—radiologists, cell biologists, physicists, and computer programmers…”
--NIH Roadmap for Medical Research: Research Teams for the Future (1)
“The current scientific understanding of both physiology and pathophysiologic processes is of necessity reductionistic (e.g., is knowledge at the gene, gene expression or pathway level) and does not constitute knowledge at the level of the systems biology of the cell, organ, or whole organism, and certainly does not reach a systems understanding of the pathophysiology of particular diseases. Reaching a more systemic and dynamic understanding of human disease will require major additional scientific efforts as well as significant advances in bioinformatics….”
--Food and Drug Administration White Paper: Innovation or Stagnation: Challenge and Opportunity of the Critical Path to New Medical Products (2)
There is a general recognition in the field of critical care that there has been a failure of many therapies for sepsis and related diseases, and that new approaches should be attempted. The two statements quoted above demonstrate recognition by both the primary governmental organization responsible for fostering the science behind the development of new therapeutics in the U.S. (the National Institutes of Health) and the primary U.S. governmental organization responsible for safeguarding the delivery of therapeutics to the public (the Food and Drug Administration) that the complexity* [Footnote: * Terms that are in underlined italics are terms referenced in the Online Modeling Glossary accesible at http://biosystems.ucsf.edu/Researc/dictionary.html] inherent to many biological processes presents significant challenges to the current biomedical research paradigm. The multiple interconnections, parallel pathways, and feedback loops seen in many biological systems transcend cognitive evaluations of cause and effect, contributing to the situation referred to by the two quotes above. From within the critical care community Buchman (3) and Neugebauer (4) recognized in the mid-1990s that the acute inflammatory system (AIR) behaved in this dynamically complex fashion. However, despite the recognition that synthesis deserves the same degree of attention and structure as analysis (4,5), it has proved challenging to identify how this can be accomplished in a formal way. Systems biology approaches, in which extensive data are combined to understand (and ideally predict) the behavior of an entire system, shows promise with regards to unraveling this complexity.
Dynamic mathematical modeling (DMM) is one aspect of systems biology shared with many other disciplines, such as physics, chemistry, and engineering, and is central to classical physiology. More recently, several groups (see below) have attempted to utilize DMM in a translational fashion, with the idea that the current field of systems biology is oriented more towards gaining basic insights than to the generation of novel therapies and the streamlining of clinical trials. While the notion of translational systems biology and DMM may be attractive in concept to clinicians and researchers in the field of critical illness, significant hurdles stand in the way. Most bioscientists find themselves in unfamiliar territory when trying to utilize even basic mathematical modeling tools. Therefore, these efforts need to incorporate mathematicians, engineers, and computer scientists with whom bioscientists are generally unacquainted.
The challenge, then, turns to the faciliation and development of collaborative transdisciplinary teams. However, the compartmentalization of the scientific community has led to disparity with respect to the language and vocabulary that is used in each community, a fact that was dramatically evident in one of the first attempts to initiate a transdisciplinary dialog between biomedical researchers and applied mathematicians in the setting of acute illness initiated through the University of Cologne/Witten-Herdecke University at the 1st International Conference on Complexity in Acute Illness (6). The primary conclusion from that Conference was that establishing a common basis of communication was of the utmost importance and that a recurrent, iterative process was needed and would be necessary to reconcile the different paradigms and expectations from within each community. Despite the growing recognition of the need for transdisciplinary teams, and of some of the barriers to their development, there is a paucity of guidance regarding the actual process of their creation (7). Therefore, we present four case reports of the experiences of interdisciplinary groups of various sizes and interests, all of which are involved in DMM related to acute inflammation. The report for each group is divided into their Background, Methodology, Accomplishments, and Limitations and Lessons Learned..
Case 1: The sole interdisciplinary scientist
Background
The first case is that of Gary An, who was a trauma surgeon in a clinical setting at Cook County Hospital in Chicago at the time of the initiation of his modeling program in 1999. Accordingly, his approach to DMM was prompted by clinical observation of the inadequacies in the treatment of sepsis, and the gulf between the known cellular/molecular pathophysiology and effective therapeutic options. Personal investigation led to contact with the concepts of Complexity and Complex Systems Analysis (CAS) and the development in an interest in DMM. Due to his clinical involvement and lack of formal training in mathematics or computer science, An sought out methodologies and tools that were amenable to a novice in both those fields. As a result, he focuses his research on utilizing Agent-based Modeling (ABM) in the fashion described below.
Methodology
Agent-based Modeling is a modeling technique that consists of viewing a system as a series of components that can be classified based on their rules of behavior. (For a more extensive discussion see references 8, 9). This modeling technique has an advantage for modelers not extensively trained in mathematics by allowing agent rule systems to be expressed as conditional statements (“if-then”) rather than equations. This property of ABM also makes it well suited to translating the results of basic science research into an in-silico framework. An uses an ABM software tool, Netlogo (10), which is semantic and similar to natural language. Like many modelers, An functions without the support of a basic science laboratory; accordingly, he relies upon published literature for the basis of his models. Additionally, he is constrained by the properties of Netlogo software which, while it facilitates model construction, is limited relative to the large computing facilities available to academic institutions or industry. His modeling focus is heavily influenced by the use of ABM insomuch it is hierarchical and class-oriented, primarily treating cells as the core agent level.
Accomplishments
An’s initial models focused on the interaction between endothelial cells and circulating inflammatory cells (8, 9, 11–13). These models followed the premise that disordered systemic inflammation was propagated by interactions at this level and looked at this interaction as a global proxy for total system behavior. Prompted by the unsuccessful anti-cytokine sepsis trials in the 1990s, he simulated and prospectively reproduced a series of those clinical trials using a model based on the state of knowledge at the time of their design (9). The primary conclusion of this paper was the potential use of DMM/ABM as a “proof of concept” test early in the drug discovery/clinical trial design process. The endothelial ABM, however, could not recreate patterns of organ-specific behaviors. Accordingly, he developed an ABM of an in vitro gut epithelial model (14), which led to combining the systemic communication endothelial model with more organ-specific epithelial barrier function models for both the gut (15) and the lung (16), and linked multi-organ failure in an abstracted way (17). In parallel, he introduced a syntactical grammar to bind together the results from different researchers into a community-wide, unified whole (18). An also developed a framework for modeling intracellular signaling and synthetic pathways (19), in order to develop more finely grained rule systems for cellular agents in trans-hierarchical modular models, and also to provide a platform for discovering and evaluating intracellular targets for therapeutic intervention (20).
Limitations and Lessons Learned
The limitations affecting An are the paucity of time, resources, and expertise to perform all the “real-world” tasks necessary to sufficiently determine the validity and verity of a model, and thus move the model from the conceptual, qualitative stage to a more applicable, quantitative one. Recognizing these limitations, An has since moved to an academic environment at Northwestern University. This new environment is expected to increase access to laboratory reference data sets, the ability to interact with bioscientists, and entry into the academic network will all be facilitated by the move to Northwestern, as well as providing the institutional infrastructure and track record that would be more attractive to funding agencies.
Case 2: The University of Witten-Herdecke/Cologne Experience
Background
The group at the University of Witten-Herdecke in Cologne, Germany (formerly at the University of Cologne) operates at the other end of the academic and organizational spectrum, yet was driven by the frustration of Dr. Edmund Neugebauer with the status quo of sepsis research (21, 22). The emphasis of this group has been on attempting to educate and shift the views of an entire community, with the idea that this could only be accomplished with leadership from the upper levels of the community’s hierarchy (23, 24). In the critical care community, the role was fulfilled by Timothy Buchman from Washington University.
Methodology
Starting from the 1990s, Neugebauer and Lefering sought collaboration within his own institution with the Departments of Traumatology (Tjardes, Maegele) and Applied Mathematics in Cologne (Seydel), Witten-Herdecke (an der Heyden) and Munich (Müller), and attempted to increase awareness for the need for synthesis within the shock research community. An interdisciplinary working group with regular meetings (IAG Math-Med) was established with regular meetings.
Accomplishments
The research at the University of Witten-Herdecke has taken several trajectories: efforts to standardize evaluation of the quality of data (meta-analysis and decision tree approaches on experimental data) (22), the establishment, refinement, and utilization of national injury/trauma scoring systems (25–27), as well as projects on modeling the auto regulation of blood-brain barrier, inflammation and coagulation using DMM. Additionally, this group spent considerable energy on mentoring and raising awareness for the need to involve complex systems in critical illness research. Ultimately, and with collaboration with Dr. An the University of Pittsburgh group (see below), the Society for Complexity in Acute Illness (SCAI; www.scai-med.org) and its annual meeting (the International Conference on Complexity in Acute Illness [ICCAI; www.iccai.org]) were formed.
Limitations and Lessons Learned
SCAI serves as a center for discussion and collaboration for the critical care modeling community, and also as a forum for outreach to integrate modeling into general medical research. To be effective, however, SCAI requires institutional trans-disciplinary research centers to produce the quality data needed to sway opinion and provide the basis for accepting DMM as a valid biomedical research tool.
Case 3: The Biosystems Group at the University of California, San Francisco
Background
The Biosystems Group was formed in 1999 by Anthony Hunt as a primary site for computational systems biology research at UCSF. The primary rationale for its creation was to facilitate the development of new therapeutic options in the treatment of acute illness by lowering the boundaries to the application of DMM and simulation towards this goal.
Methodology
The group’s research focuses on new methods for building in silico analogues of in vitro and in vivo systems. Having a primary focus on education from the outset, effort was also directed at establishing a means by which student researchers from different domains could become aware of particular approaches of systems biology, and to access tools and expertise to assist in their areas of research interest. As such, this group has emphasized communication among established academic groups to enable evolution and growth of new ideas and concepts. An intranet framework with an internal “wiki” (http://en.wikipedia.org/wiki/Wiki) process, similar to the CIRM SharePoint® tool (see below), helps foster communication and distribution of knowledge within the group. On the research side, the group relies in part on modeling and simulation frameworks developed for use in other domains (Mason http://cs.gmu.edu/~eclab/projects/mason/, Swarm http://swarm.org/wiki/Main_Page, and RePAST http://repast.sourceforge.net/).
Accomplishments
The group quickly came to the same realization described by Neugebauer above (6): the establishment of a common lexicon was necessary because ambiguities in terminology strained trans-domain communication. This realization provided the initial impetus for the development of an online Dictionary of Mathematical Modeling Terms, whose goal has been to provide a means by which ambiguity could be either resolved (in circumstances of simple lack of knowledge) or cataloged based on context. The dictionary/glossary is being utilized in this manuscript for terms that are underlined and italicized, and can be accessed at http://biosystems.ucsf.edu/Researc/dictionary.html. The Glossary is constantly evolving, using iterative negotiation to resolve tensions as they emerge from differences in language use.
As mentioned above their models are designed to be working analogues of referent biological systems. Components of their in silico liver (28) and in silico hepatocytes (29, 30) map logically to identifiable biological components. Their epithelial morphogenesis models (31, 32) are beginning to be used by wet-lab colleagues to bring clarity to the principles that govern how this important cell type functions under stress, both in vitro and in vivo.
Limitations and Lessons Learned
Because the BioSystems Group lies on the path from Case 1 (An) to CIRM in Case 4, it has experienced the challenges of both cases. The structure of academe facilitates the flow of resources toward the most vigorous of the established domains. Insuring the intellectual viability of an interdisciplinary group and its projects requires more creativity, time, and energy than would be required of a similar research enterprise within an established domain. The research is transdisciplinary; the methods are interdisciplinary. Consequently, group members—not the colleagues from different, established domains—have learned that they need to take responsibility for establishing and maintaining effective communication between project participants, regarding any aspect of modeling and simulation. Deliberately designing simulations to be interesting and reasonably understandable at the model observer level across all of the represented domains has helped lower barriers to productive communication.
Case 4: The Center for Inflammation and Regenerative Modeling (CIRM), University of Pittsburgh
Background
The multi-disciplinary team for DMM of acute inflammation had its genesis at the University of Pittsburgh in 2000, and was formalized into the Center for Inflammation and Regenerative Modeling (CIRM; www.mirm.pitt.edu) in late 2004. While it was spurred by a group of individuals, the eventual formation and structure of the CIRM required cross-departmental collaboration and support among the Departments of Surgery, Critical Care Medicine, and Mathematics. Important energy towards the establishment of the CIRM was derived from the interdisciplinary McGowan Institute for Regenerative Medicine (www.mirm.pitt.edu). Each of the principals involved in the eventual formation of the CIRM had a fair amount of exposure to another field: a medical doctor with a physics background (Gilles Clermont), a mathematician with a neuroscience background (Carson Chow), and an inflammation biologist with a background in mathematics and computer science (Yoram Vodovotz).
Methods
The CIRM uses both equation-based and agent-based models in its simulations, which are geared towards rapid translation in the form of simulated clinical trials and quantitative predictions. The group has also explored basic insights into the biology of inflammation and organ damage. One mechanism employed at the CIRM in order to increase the efficiency of the team-based process is depicted in Fig. 1. In order to keep the interdisciplinary project on task, CIRM investigators use the Microsoft SharePoint® application and its messaging, tasking, and teleconferencing capabilities. This approach is used widely in industry.
Figure 1. Interdisciplinary mathematical modeling process at the CIRM.
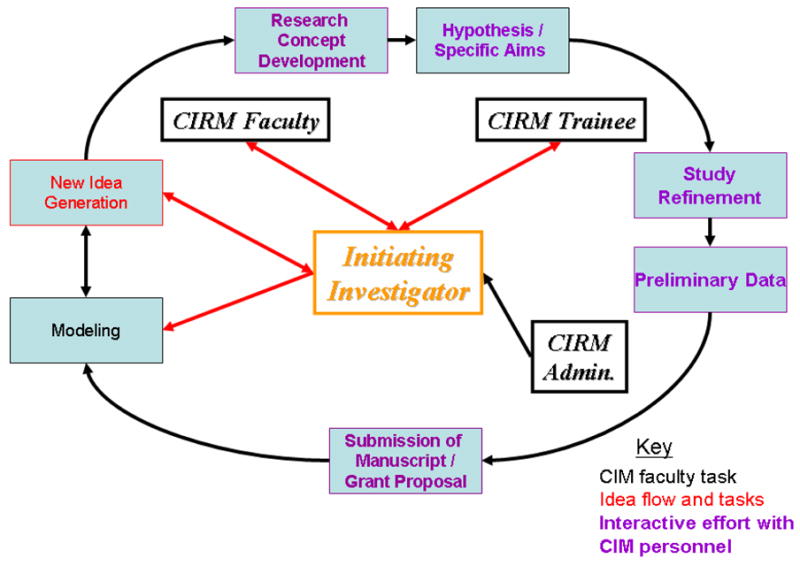
An iterative process of modeling and idea generation is initially established. Modeling is carried out based on the initial concepts of the initiating investigator and later through the interaction of the interdisciplinary group (red arrows indicate idea flow). Purple indicates interactive effort with CIRM personnel.
Accomplishments
The CIRM has concentrated its efforts on translational modeling of inflammation and organ damage in the settings of sepsis and trauma (33–36), using large mathematical models to make quantitative predictions and to simulate clinical trials. In conjunction, however, this group has also utilized smaller models to address basic questions in acute inflammation (37–39) as well as developing methods for calibration of mathematical models to data (40). The group at Pittsburgh has established a graduate level course on the application of mathematical modeling to inflammation (“A Systems Approach to Inflammation” (http://www.pitt.edu/~cler/mscmp3780/mscmp3780.htm), with the intent to develop multi-disciplinarily trained researchers for the future. Importantly, this work has been funded by various funding agencies, including the NIH and Commonwealth of Pennsylvania, and has resulted in the commercial translation in the formation of a company (Immunetrics, Inc.) that has begun to bring DMM to the arena of pharmaceutical companies developing sepsis therapies (41) (Note: GA, YV, and GC are consultants to Immunetrics, Inc.).
Limitations and Lessons Learned
While in many ways the CIRM approach represents an ideal means of establishing a transdisciplinary research group, it does have some limitations as a model for others to follow. The individual commitment of each of the founders was considerable and the latitude allowed them by their respective departments laudable. As mentioned above, each had at least some experience in the others’ field of expertise. It may be argued that the CIRM experience is the exception that proves the rule that it is difficult to find people willing to invest a large degree of effort into re-educating themselves in a foreign scientific field. Finally, the porosity of internal inter-departmental barriers that was necessary for the development of the group is, sadly, atypical for most academic institutions.
DISCUSSION
The experiences described above take place in vastly different environments and at different scales of organization, but common themes can be identified with respect to the challenges and the approaches to dealing with them. Furthermore, it is useful to identify the limitations of the various experiences, and examine how these can be resolved by returning to the common thematic elements. The following is a list of fostering characteristics at each organizational level:
The Individual: All the researchers involved in this paper have training, either formal or informal, in disciplines outside their primary area of expertise. This gives them a broad base of experience that allows then to decide if they are convinced of the utility of DMM for their own research.
The Institution: In all the institutions there was the recognition, either explicit or implicit recognition of a need for a paradigm shift. However, since institutions are intrinsically conservative, a catalyst was needed to initiate this reaction/process. In each case this came from the type of individual noted above. Furthermore, the institutions needed to have a pre-existing flexibility to act across inter-departmental barriers, either to foster trans-departmental projects, or allow the development of more freestanding research centers/groups that could operated outside those barriers.
The Community: The larger the organization, the greater the resistance to change. However, there is recognition that some “new” way of doing things must come to light. There must be individuals positioned high enough within the organizational hierarchy (such as Drs. Buchman and Neugebauer) to be able to nurture and shepherd the basis of a paradigm shift, and there must be institutions available to produce the quality data to back up the claims. Grassroots support for this effort must also exist: leaders cannot by themselves create such a paradigm shift.
Each organizational level, however, has limitations, and these are briefly summarized below:
The Individual: As mentioned before, the solo practitioner cannot move beyond the conceptual or prototyping stage due to limitations of time, resources and expertise.
The Institution: Inter-departmental barriers and competition result in hierarchies of priority with respect to funding and resource allocation. It may be difficult to convince the power structure to allocate scarce resources to novel, and heretofore, untested, ideas.
The Community: The resistance of an organization to change is exponentially related it its size. The “proof of concept” that is needed to sway such a community requires the efforts and resources of multiple institutional projects.
Therefore, Figure 2 demonstrates the nested, hierarchical and recursive nature of these relationships. Individuals must be the catalyst for these endeavors. Institutions provide the generation of data and establishment of proof. Community level organizations like SCAI provide cohesion and direction for the introduction and advancement of these concepts as we move towards a paradigm shift.
Figure 2. Structural Relationship between Multiple Organizational Levels in the Mathematical Modeling Community.
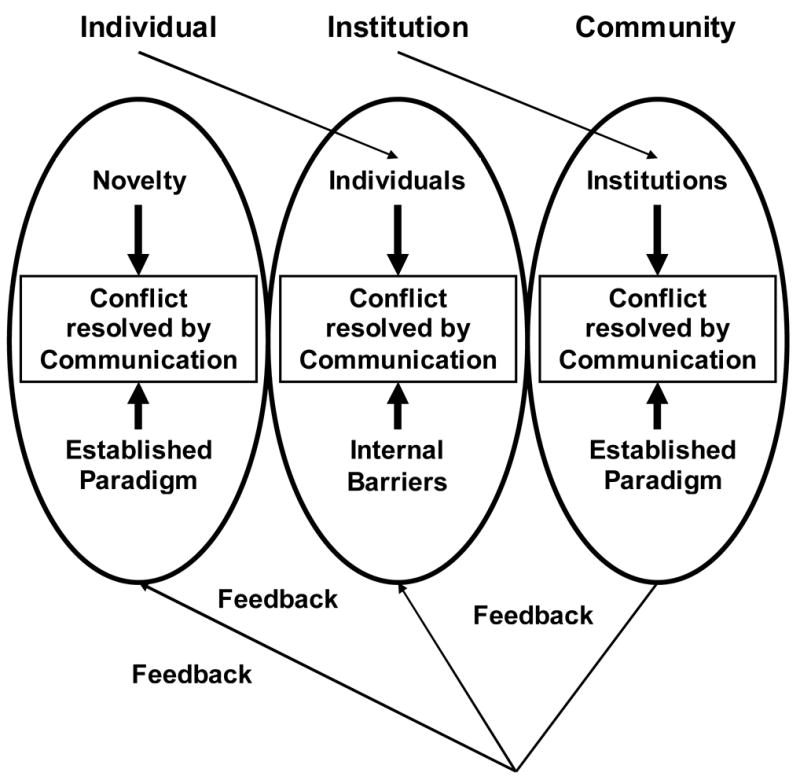
Each organizational level within the modeling community can be seen as having a similar internal structure. At each level the challenge of integrating the new research paradigm is addressed using a communication process based on iterative negotiation. These organizational levels are nested, insomuch the output at each level is the input for the next, higher level organization. The output of the entire community, as the concepts related to modeling gain acceptance, then provides positive feedback at the lower levels by increasing individual interest and supporting institutional commitment.
CONCLUSIONS AND FUTURE DIRECTIONS
As the quotes at the beginning of this article point out, biomedical research has reached a crossroads, and it has become increasingly clear that the road that needs to be taken leads towards greater cooperation, increased collaboration and widened horizons. The complexity of the challenges facing the critical care community requires the intellectual resources of the community as a whole. We hope that the case studies described in this manuscript will provide guidance in the development of transdisciplinary research groups engaged in translational systems biology; from the level of stimulating individual exploration to the establishment of institutional centers and collaborations to utilizing the resources that have been developed for the entire research community in the form of SCAI. These are exciting times that hold the promise for systems-level efforts to break through longstanding barriers to therapies in the field of acute illness. We at SCAI will do our part to support individual intellectual curiosity to embark on these efforts, as well as to encourage institutional and community-wide support and nurturing.
Acknowledgments
This work was supported in part by the National Institutes of Health grants R01-GM-67240-02 (GC, YV), P50-GM-53789-08 (GC, YV), R01-HL-76157-02 (GC, YV), R01-HL-080926 (GC, YV), 2R13-GM072437-02 (GC) as well as grants from the Pittsburgh Lifesciences Greenhouse (YV), the Commonwealth of Pennsylvania (YV) and the CDH Research Foundation (CAH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.NIH Roadmap for Medical Research: Research Teams. [Accessed June 15, 2006]; Available at: http://nihroadmap.nih.gov/researchteams/
- 2.U.S. Food and Drug Administration report. Innovation or Stagnation: Challenge and Opportunity in the Critical Path to New Medical Products. [Accessed June 15, 2006]; Available at: http://www.fda.gov/oc/initiatives/criticalpath/whitepaper.html.
- 3.Buchman TG. Physiologic Stability and Physiologic State. J Trauma. 1996;41:599–605. doi: 10.1097/00005373-199610000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Neugebauer EA, Willy C, Sauerland S. Complexity and non-linearity in shock research: reductionism or synthesis? Shock. 2001;16(4):252–8. doi: 10.1097/00024382-200116040-00003. [DOI] [PubMed] [Google Scholar]
- 5.Ropella GEP, Hunt CA, Sheikh-Bahaei S. Methodological Considerations of Heuristic Modeling of Biological Systems. Proceedings of The 9th World Multi-Conference on Systemics, Cybernetics and Informatics; July 10–13, 2005; Orlando, Florida. Available at: http://biosystems.ucsf.edu/Researc/ [Google Scholar]
- 6.Neugebauer EA, Tjardes T. Multidisciplinary Working Group on Complexity. New approaches to shock and trauma research: learning from multidisciplinary exchange. J Trauma. 2004;56(5):1156–65. doi: 10.1097/01.ta.0000119207.14267.63. [DOI] [PubMed] [Google Scholar]
- 7.Committee on Facilitating Interdisciplinary Research and Engineering and Public Policy Committee on Science. Washington, DC: The National Academies Press; 2005. Facilitating Interdisciplinary Research. http://www.nationalacademies.org/cosepup. [Google Scholar]
- 8.An G. Agent-based computer simulation and SIRS: Building a bridge between basic science and clinical trials. Shock. 2001;16(4):266–273. doi: 10.1097/00024382-200116040-00006. [DOI] [PubMed] [Google Scholar]
- 9.An G. In-silico experiments of existing and hypothetical cytokine-directed clinical trials using agent based modeling. Crit Care Med. 2004;32(10):2050–2060. doi: 10.1097/01.ccm.0000139707.13729.7d. [DOI] [PubMed] [Google Scholar]
- 10.Northwestern University Center for Connected Learning: Netlogo Homepage. [Accessed June 10, 2006]; Available at: http://ccl.northwestern.edu/netlogo/
- 11.An G, Lee I. Complexity, Emergence and Pathophysiology: Using Agent Based Computer Simulation to characterize the Non-Adaptive Inflammatory Response. InterJournal Complex Systems. http://www.interjournal.org. Manuscript # [344]. May, 2000.
- 12.An G, Lee I. Agent-Based Computer simulation (ABCS) and the inflammatory response: Building a tool to study Systemic Inflammatory Response Syndrome (SIRS) Simulation and Gaming. 2001;32(3):344–361. [Google Scholar]
- 13.An G. Synthetic microanalysis, agent based computer simulation and Systemic Inflammatory Response Syndrome: Capturing the complexity of the inflammatory response. Crit Care Med. 2000;28(12S):A64. (supp)(abstr) [Google Scholar]
- 14.An G, Delude R. Agent based model of cell culture epithelial barrier function: Using computer simulation in conjunction with a basic science model. Shock. 2004;21S2(s13) (supp)(abstr) [Google Scholar]
- 15.An G, Feinman R, Xu D-Z, Deitch E. In-silico unification of different basic science models of gut epithelial barrier function using agent based modeling. Crit Care Med. 2004;32(12s):A95. (supp)(abstr) [Google Scholar]
- 16.An G. Agent based models of pulmonary epithelial barrier function. Advance Program from the 102nd Annual International Conference of the American Thoracic Society. 2006 (supp)(abstr) [Google Scholar]
- 17.An G. Computer simulations of multiple organ failure secondary to shock and sepsis with a multi-tissue, endothelial level agent based model. Shock. 2004;21S2(s66) (supp)(abstr) [Google Scholar]
- 18.An G. Concepts for developing a collaborative in-silico model of the acute inflammatory response using agent based modeling. J Crit Care. 2006;21(1):105–110. doi: 10.1016/j.jcrc.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 19.An G. A Synthetic Framework for Investigating Multiple Intracellular Signaling Pathways using Agent Based Modeling. Shock. 2006;25(S1):83. (supp)(abstr) [Google Scholar]
- 20.An G. Integrative Modeling of Inflammation and Organ Function Using Agent Based Modeling. Shock. 2006;26(S1):2. (supp)(abstr) [Google Scholar]
- 21.Tjardes T, Neugebauer EA. Sepsis research in the next millennium: concentrate on the software rather than the hardware. Shock. 2002;17 (1):1–8. doi: 10.1097/00024382-200201000-00001. [DOI] [PubMed] [Google Scholar]
- 22.Neugebauer EA, Holaday JW, editors. Handbook of Mediators in Septic Shock. Boca Raton, FL: CRC Press; 1993. [Google Scholar]
- 23.Neugebauer EAM, Willy C, Sauerland S. Complexity and non-linearity in shock research: reductionism or synthesis. Shock. 2001;16:252–8. doi: 10.1097/00024382-200116040-00003. [DOI] [PubMed] [Google Scholar]
- 24.Willy C, Neugebauer EAM, Gerngroß H. The concept of nonlinearity in complex systems. Eur J Trauma. 2003;29:11–22. [Google Scholar]
- 25.Bouillon B, Lefering R, Vorweg M, Tiling T, Neugebauer E, Troidl H. Trauma score systems: Cologne Validation Study. J Trauma. 1997;42(4):652–8. doi: 10.1097/00005373-199704000-00012. [DOI] [PubMed] [Google Scholar]
- 26.APd DGU, Neugebauer EAM, et al. Das Traumaregister der deutschen Gesellschaft für Unfallchirurgie als Grundlage des interklinischen Qualitätsmanagements in der Schwerverletztenversorgung - Eine Multicenterstudie der DGU. Unfallchirurg. 2000;103:30–37. doi: 10.1007/s001130050005. [DOI] [PubMed] [Google Scholar]
- 27.Rixen D, Raum M, Bouillon B, Lefering R, Neugebauer EAM, APot DGU. Base Deficit Development and its Prognostic Significance in Posttrauma Critical Illness: An Analysis by the Trauma Registry of the Deutsche Gesellschaft für Unfallchirurgie. Shock. 2001;15:83–89. doi: 10.1097/00024382-200115020-00001. [DOI] [PubMed] [Google Scholar]
- 28.Hunt CA, Ropella GEP, Yan L, Hung DY, Roberts MS. Physiologically Based Synthetic Models of Hepatic Disposition. Journal of Pharmacokinetics and Pharmacodynamics. 2006 doi: 10.1007/s10928–006-9031-3. [DOI] [PubMed] [Google Scholar]
- 29.Sheikh-Bahaei S, Ropella GEP, Hunt CA. In Silico Hepatocyte: Agent-based Modeling of the Biliary Excretion of Drugs In Vitro. In: Yilmaz L, editor. Proc 2006 Agent-Direc Simul Symp, Huntsville, AL. SCS Press; San Diego, CA: Apr 3–9, 2006A. pp. 157–163. (ADS’06) [Google Scholar]
- 30.Sheikh-Bahaei S, Hunt CA. Prediction of In Vitro Hepatic Biliary Excretion Using Stochastic Agent-Based Modeling and Fuzzy Clustering. In: Perrone LF, et al., editors. Proc 2006 Winter Simul Conf. 2006B. in press. [Google Scholar]
- 31.Grant MR, Hunt CA. An In Silico Analogue of In Vitro Systems Used to Study Epithelial Cell Morphogenesis. In: Priami C, editor. CMSB 2006, LNBI. Vol. 4210. 2006A. pp. 285–97. [Google Scholar]
- 32.Grant MR, Mostov KE, Tlsty TD, Hunt CA. Simulating Properties of In Vitro Epithelial Cell Morphogenesis. PLoS Computational Biology. 2006B;2(9):e129. doi: 10.1371/journal. pcbi.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clermont G, Bartels J, Kumar R, Constantine G, Vodovotz Y, Chow C. In silico design of clinical trials: a method coming of age. Crit Care Med. 2004;32:2061–2070. doi: 10.1097/01.ccm.0000142394.28791.c3. [DOI] [PubMed] [Google Scholar]
- 34.Chow CC, Clermont G, Kumar R, Lagoa C, Tawadrous Z, Gallo D, Betten B, Bartels J, Constantine G, Fink MP, Billiar TR, Vodovotz Y. The acute inflammatory response in diverse shock states. Shock. 2005;24:74–84. doi: 10.1097/01.shk.0000168526.97716.f3. [DOI] [PubMed] [Google Scholar]
- 35.Lagoa CE, Bartels J, Baratt A, Tseng G, Clermont G, Fink MP, Billiar TR, Vodovotz Y. The role of initial trauma in the host’s response to injury and hemorrhage: Insights from a comparison of mathematical simulations and hepatic transcriptomic analysis. Shock. 2006 doi: 10.1097/01.shk.0000232272.03602.0a. In Press. [DOI] [PubMed] [Google Scholar]
- 36.Prince JM, Levy RM, Bartels J, Baratt A, Kane JM, III, Lagoa C, Rubin J, Day J, Wei J, Fink MP, Goyert SM, Clermont G, Billiar TR, Vodovotz Y. In silico and in vivo approach to elucidate the inflammatory complexity of CD14-deficient mice. Mol Med. 2006;12:88–96. doi: 10.2119/2006-00012.Prince. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar R, Clermont G, Vodovotz Y, Chow CC. The dynamics of acute inflammation. J Theoretical Biol. 2004;230:145–155. doi: 10.1016/j.jtbi.2004.04.044. [DOI] [PubMed] [Google Scholar]
- 38.Reynolds A, Rubin J, Clermont G, Day J, Vodovotz Y, Ermentrout GB. A reduced mathematical model of the acute inflammatory response: I. Derivation of model and analysis of anti-inflammation. J Theor Biol. 2006;242:220–236. doi: 10.1016/j.jtbi.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Day J, Rubin J, Vodovotz Y, Chow CC, Reynolds A, Clermont G. A reduced mathematical model of the acute inflammatory response: II. Capturing scenarios of repeated endotoxin administration. J Theor Biol. 2006;242:237–256. doi: 10.1016/j.jtbi.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 40.Constantine G, Bartels J, Buliga M, Clermont G, Vodovotz Y. An optimization algorithm based on optimal linear codes. J Pure Appl Math. 2006 In Press. [Google Scholar]
- 41.Chang S, Baratt A, Clermont G, Planquois J-M, Yan SB, Williams M, Macias W. Mathematical model predicting outcomes of sepsis patients treated with Xigris(R): ENHANCE trial. Shock. 2006;25:70–71. (supp)(abstr) [Google Scholar]


