Abstract
Human DNA polymerase ν is one of three A family polymerases conserved in vertebrates. Although its biological functions are unknown, pol ν has been implicated in DNA repair and in translesion DNA synthesis (TLS). Pol ν lacks intrinsic exonucleolytic proofreading activity and discriminates poorly against misinsertion of dNTP opposite template thymine or guanine, implying that it should copy DNA with low base substitution fidelity. To test this prediction and to comprehensively examine pol ν DNA synthesis fidelity as a clue to its function, here we describe human pol ν error rates for all 12 single base-base mismatches and for insertion and deletion errors during synthesis to copy the lacZ α-complementation sequence in M13mp2 DNA. Pol ν copies this DNA with average single-base insertion and deletion error rates of 7 × 10−5 and 16 × 10−5, respectively. This accuracy is comparable to that of replicative polymerases in the B family, lower than that of its A family homolog, human pol γ, and much higher than that of Y family TLS polymerases. In contrast, the average single base substitution error rate of human pol ν is 3.5 × 10−3, which is inaccurate compared to the replicative polymerases and comparable to Y family polymerases. Interestingly, the vast majority of errors made by pol ν reflect stable misincorporation of dTMP opposite template G, at average rates that are much higher than for homologous A family members. This pol ν error is especially prevalent in sequence contexts wherein the template G is preceded by a C-G or G-C base pair, where error rates can exceed 10%. Amino acid sequence alignments based on the structures of more accurate A family polymerases suggest substantial differences in the O-helix of pol ν that could contribute to this unique error signature.
Keywords: error-prone polymerase, infidelity, base substitutions
1. INTRODUCTION
Among the 16 DNA polymerases encoded by the human genome [1–4], three are members of the A family, composed of enzymes with homology to Escherichia coli (E. coli) DNA pol I. The most extensively studied of these three enzymes is DNA polymerase γ (pol γ). Consistent with its roles in replicating and repairing the mitochondrial genome, pol γ is a highly accurate enzyme due to the high nucleotide selectivity of the polymerase and the presence of an intrinsic 3′ exonuclease that can proofread mistakes [5]. In fact, high nucleotide selectivity is a property shared by most other A family DNA polymerases, of which the most extensively studied are the prototypical E. coli DNA polymerase I [6–9], bacteriophage T7 DNA polymerase [10–12], Thermus aquaticus (Taq) DNA polymerase [13–16] and Bacillus fragment (BF) DNA polymerase [17,18]. The structural and kinetic bases for the high nucleotide selectivity of these A family enzymes have been studied in detail [7–15,17–25] and reviewed in [8,26–30], providing a wealth of information for comparison to more recently discovered A family members whose biological functions are currently unknown.
Family A members include two other human enzymes of unknown function, pol θ [31–33], and the subject of the present study, pol ν. Human pol ν was first described three years ago [34], as an A family DNA polymerase that is conserved in vertebrates but not in invertebrates, yeast or fungi. On that basis, it was proposed that pol ν could have a role related to organ systems that are especially developed in vertebrates. As one approach to investigate the possible functions of pol ν in vivo, several biochemical properties of pol ν have been described [34,35]. Pol ν has moderate processivity, it has efficient strand displacement activity, and it can perform translesion synthesis past a thymine glycol in an oligonucleotide template [35]. Pol ν lacks intrinsic 3′ exonuclease activity and is therefore unable to proofread its own mistakes. This inability to proofread is not unprecedented among A family polymerases; e.g., Taq DNA polymerase [13] and BF DNA polymerase [17] are also 3′ exonuclease-deficient polymerases. Nonetheless, Taq and BF polymerases have high nucleotide selectivity [13–18]. In contrast, a recent kinetic analysis of single nucleotide incorporation at two template locations has demonstrated that pol ν discriminates very poorly against misinsertion of dTTP and dGTP opposite template G or T in an undamaged DNA oligonucleotide [35]. If the resulting T-G and T-T mismatches, or any others not yet examined, were extended by naturally proofreading-deficient pol ν, then synthesis by pol ν would be error prone for base substitutions, thus representing an exception among usually highly accurate A family polymerases. Here we test this prediction using an assay that monitors a wide variety of DNA synthesis errors in numerous sequence contexts. The results reveal that pol ν is the least accurate A family enzyme studied to date. For base substitutions, the low fidelity primarily pertains to one particular mismatch that is preferentially generated in certain sequence contexts. We discuss the possibility that this unusual error specificity may result from differences in the geometry of the nascent base pair binding pocket of pol ν, perhaps as determined by its O-helix, whose putative amino acid composition varies from those of more accurate A family members.
2. MATERIALS AND METHODS
Enzymes, reagents, strains
Recombinant human DNA pol ν was purified to near homogeneity as described [35]. The exonuclease-deficient, large Klenow fragment of E. coli DNA pol I and T4 polynucleotide kinase were purchased from New England BioLabs. [γ-32P]ATP (4500 Ci/mmol) and unlabeled deoxyribonucleotide-triphosphates were from GE Healthcare Biosciences. All strains and materials for processivity analysis [36] and the M13mp2 fidelity assay were from sources described previously [37].
Processivity Analysis
Processivity was monitored using single-stranded M13mp2 DNA primed with a [γ32P]ATP 5′end labeled DNA oligonucleotide complementary to positions +173 to +192 of the lacZα coding sequence. Reaction mixtures (30 μl) for pol ν contained 20 mM Tris-HCl (pH 8.8), 8 mM magnesium acetate, 2 mM dithiothreitol, 4% glycerol, 80 μg/ml bovine serum albumin and 100 μM each dATP, dCTP, dGTP and dTTP. Reaction mixtures for exonuclease-deficient Klenow fragment (Klenow fragment exo-) pol contained 10 mM Tris (pH 7.5), 7.5 mM dithiothreitol, 10 mM MgCl2 and 25 μM each dATP, dCTP, dGTP and dTTP. Both reaction mixtures contained primed M13mp2 ssDNA substrate in excess (200 fmol) of pol ν (40 fmol) or Klenow fragment exo- pol (160 fmol). Reaction mixtures were incubated at 37°C and 10 μl aliquots were removed at 3, 6 and 9 min for pol ν and at 3 and 5 min for Klenow fragment exo- pol. Samples were mixed in a 1:1 ratio with stop buffer (99% formamide, 5mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue) and DNA products were analyzed by electrophoresis on a 12% denaturing polyacrylamide gel. Products were quantified by phosphorimagery using a Molecular Dynamics Typhoon 9400 and the ImageQuant software. Conditions were such that termination probabilities reflected one cycle of processive synthesis. The termination probability for each template position is defined as the ratio of products of a given length to the sum of that product plus all longer DNA products [38].
M13mp2 Fidelity Assay
DNA synthesis reaction mixtures for pol ν (25 μl) were performed as described above but contained 0.2 nM M13mp2 (407-nucleotide gap) DNA substrate (from nucleotide −216 through +191 of lacZ gene) and 1 mM each dATP, dCTP, dGTP and dTTP. Polymerization reactions were initiated by the addition of pol ν (46 nM), incubated at 37°C for 30 min and terminated by the addition of 20 mM EDTA. Half of the reaction mixture was mixed with 3 μl of SDS buffer (20 mM Tris pH 8.0, 5 mM EDTA, 5% SDS, 0.5 % bromophenol blue and 25% glycerol) and analyzed by agarose gel electrophoresis [37]. Pol ν reactions completely filled the gap (e.g., Fig. 1A). DNA products were also examined for the frequency of lacZ mutants by electroporation and plated onto indicator plates as previously described [37]. Errors were scored as light blue or colorless mutant plaques, while correct synthesis yields plaques that are dark blue. DNA from independent mutants was sequenced in order to identify the errors made during gap-filling synthesis. For sequence changes that yield light blue and colorless plaques, error rates per detectable nucleotide incorporated were calculated as described [37]. Error rates were also calculated by simply dividing the number of observed mutations of a particular type by the total number of nucleotides sequenced in the lacZ clones, a calculation used previously for particularly inaccurate polymerases [39].
Figure 1.
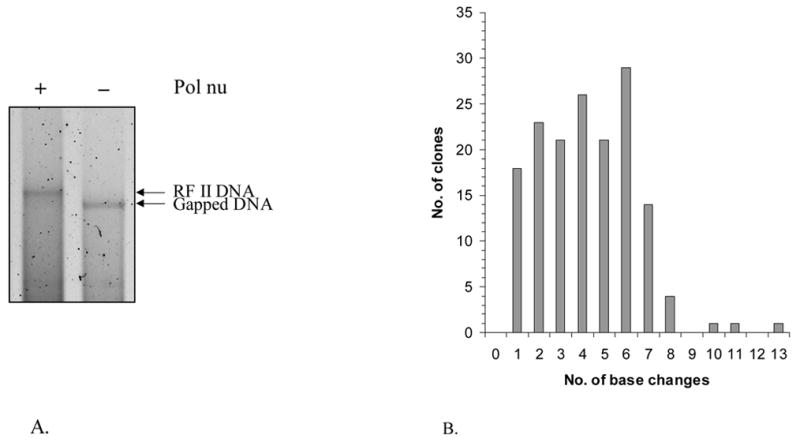
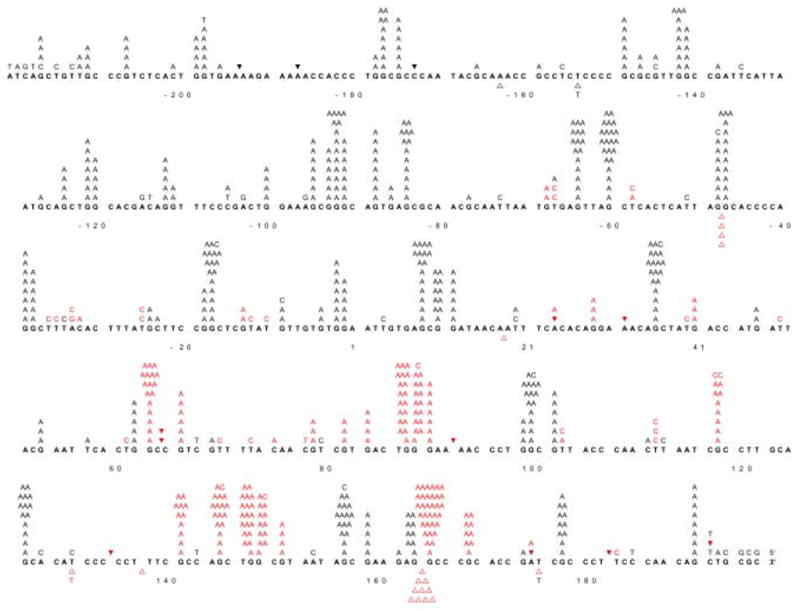
(A) Analysis of gap-filling by agarose gel electrophoresis. Half of the reaction mixture was mixed with 3 μl of SDS dye mix and analyzed by electrophoresis on an 0.8% agarose gel in TAE buffer. Lane A, gap-filling reaction with pol ν and lane B, includes the same reaction in the absence of pol ν. (B) Distribution of base substitutions per clone. 162 independent M13mp2 clones were sequenced and a total of 673 base substitutions were observed. (C) Spectrum of base substitution errors by human pol ν. The 407 template nucleotides within the single-strand gap of the M13mp2 substrate are shown as 5 lines of the template sequence. Base substitutions are indicated by letters above the target sequence. Deletion of a base is depicted by an open triangle whereas addition of a base is represented by a closed inverted triangle above the target sequence. Red characters represent phenotypically detectable changes in the gap region while black characters represent phenotypically undetectable changes found in association with detectable changes. +1 represents the first transcribed nucleotide of the lacZα-complementation region.
3. RESULTS AND DISCUSSION
Fidelity of human pol ν in the M13mp2 forward mutation assay
We measured the fidelity of human pol ν during synthesis to fill a 407-nucleotide single-stranded template encoding the lacZ α-complementation gene within gapped M13mp2 DNA. In this assay, polymerization errors are detected as light blue and colorless lacZ mutant plaques among dark blue plaques resulting from correct synthesis. Since the assay monitors loss of a gene function that is not essential for M13 plaque formation, it scores a broad range of errors, including all 12 possible single-base substitutions in a variety of sequence contexts, additions and deletions of many different template nucleotides, and more complex errors [37]. The fidelity of DNA synthesis by other DNA polymerases has been determined using this assay, allowing comparisons to pol ν.
The products of two independent gap-filling synthesis reactions by pol ν (e.g. see Fig. 1A) yielded an average lacZ mutant frequency of 18%, (see Table 1 for mutant frequencies for independent experiments). These values are higher than observed for synthesis by other exonuclease-deficient A family polymerases, including Klenow fragment, Taq, bacteriophage T7 and pol γ. To determine the types and locations of errors generated by pol ν, 162 independent lacZ mutants were sequenced. The results (Table 1 and Figure 1B,C) were used to calculate specific error rates (Tables 1–4) and these were compared to the error rates of other polymerases (Table 2). The types of errors observed are discussed here in inverse order of prevalence.
Table I.
Summary of sequence changes generated by Pol ν
| LacZ Mutant Frequency | 0.18a | ||
| Total mutants sequenced | 162 | ||
| Total bases sequenced | 65, 934 | ||
| Total sequence changes | 712 | ||
| Total changes | Detectable changes | Error Rate (×10−4)b | |
|
| |||
| Base substitutions | 673 | 226 | 35 |
| Frameshifts (−1) | 20 | 16 | 1.6 |
| Frameshifts (+1) | 12 | 7 | 0.7 |
| Otherc | 7 | ||
Error rates and mutant frequencies were calculated using data from two independent experiments. Experiment 1: Mutant frequency was 17.6% (317 mutant plaques from a total of 1797). Experiment 2: Mutant frequency of 18.0% (1278 mutant plaques from a total of 7081).
Error rate calculated from only detectable changes (see Materials and Methods).
These errors are further discussed in text.
Table IV.
Sequence Context Dependence for G→A base substitutions by Pol ν
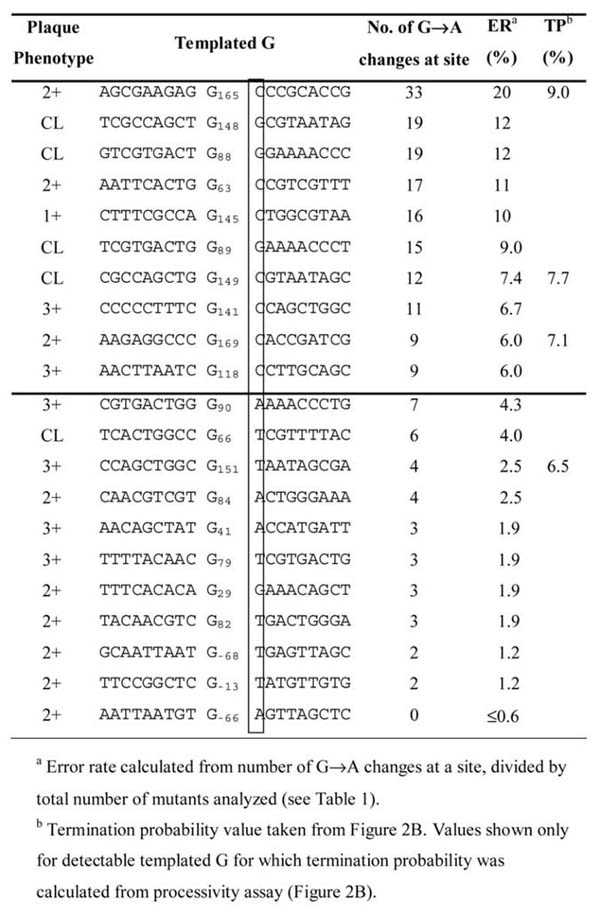
Table II.
Single base error rates for Pol ν and others
| Polymerase Error rates (× 10−6) | ||||
|---|---|---|---|---|
| DNA Polymerase | Single base deletions | Base Substitutions | G:dTMP | T:dGMP |
| hPol νa | 160 | 3500 | 17000 | 1300 |
| hPol γ (Exo-) (+p140 and p55)b | 8 | 41 | 16 | 21 |
| Kf (Exo-)c | 6 | 25 | 5 | 45 |
| hPol αd | 28 | 75 | 13 | 62 |
| yPol δ (Exo-)e | 57 | 130 | 56 | 140 |
| yPol ε (Exo-)f | 56 | 240 | 51 | 75 |
| hPol β* | 140 | 230 | 220 | 100 |
| hPol λ# | 4500 | 900 | 310 | 1400 |
| hPol η§ | 2400 | 35000 | 1200 | 5900 |
| hPol κ‡ | 1800 | 5800 | 4400 | 2100 |
hPol ν: Error rates reported are from this study.
hPol γ (Exo-) (+p140 and p55): single base deletion error rate from Longley et al., 2001. Base substitution error rates (average and G:dTMP and T:dGMP) are unpublished results from Pursell, ZF and Kunkel TA.
Kf (Exo-): single base deletion error rate from Table III, Minnick et al., 1996. Base substitution error rates (average and G:dTMP and T:dGMP) are from Bebenek et al., 1990.
hPol α: Error rates are unpublished results from Kokoska RJ and Kunkel TA.
yPol δ (Exo-): Error rates are taken from Fortune et al., 2005.
yPol ε (Exo-): Error rates are taken from Shcherbakova et al., 2003.
hPol β: Error rates are taken from Bebenek and Kunkel, 2003.
hPol λ: Error rates are taken from Bebenek et al., 2003.
hPol η: Error rates are taken from Matsuda et al., 2001 and 2002.
hPol κ: Error rates are taken from Ohashi et al., 2001.
Deletions and insertions of multiple nucleotides
Among 162 lacZ mutants sequenced, several contained deletions and/or insertions of multiple bases. Four mutants harbored a 317 base deletion that included loss of one of two direct repeats of 5′-CCCGC plus the intervening 312 bases. This deletion has been observed in the error spectra of several other DNA polymerases [40–42]. It can be explained by a model in which the first repeat sequence encountered by pol ν is copied, then the primer frays and relocates to the second repeat sequence to create a duplex primer-template for continued polymerization. This results in a misaligned intermediate that contains a loop of unpaired template strand bases that are eventually deleted. The high frequency with which pol ν generates this particular deletion (4/162 × 18% = 4.4 × 10−3) may be facilitated by an inverted repeat of three G-C base pairs present at the deletion junction that could stabilize the misaligned intermediate through formation of a stem at the base of the single-stranded loop (see Figure 2 in [41,43]). The ability to delete large numbers of nucleotides is consistent with the possibility that pol ν may contribute to larger forms of genome instability than simple point mutations. Other multiple-base errors include a 6-base deletion (+137 through +143), one large, complex error wherein 349 bases (−183 through +166, where +1 is the first transcribed base of the lacZ gene) were replaced with ‘GAC’, and another clone with ‘AC’ inserted at +172.
Figure 2.

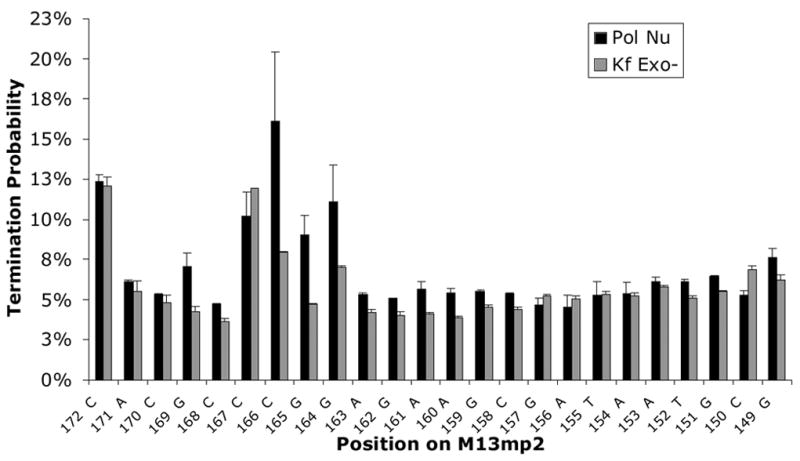
Processive synthesis by pol ν and Klenow fragment exo−. Reactions were performed as described in Materials and Methods. (A) Representative phosphor image of the reaction products of processive DNA synthesis resolved on a 12% denaturing polyacrylamide gel. Lanes 2–4, primer extension products by pol v at 3, 6, and 9-min. Lanes 6–7, primer extension products by Klenow fragment exo− at 3 and 5-min. Lanes 1 and 5 represent negative control reactions. The sequence of the template strand is shown on the right. The numbers on the left indicate the positions along the lacZ gene. (B) Termination probability per nucleotide at each template position, 172 to 149. Numbers along the X-axis indicate nucleotide positions along the lacZ gene. The termination probability is defined as the amount of product of a given length divided by the sum of products of that length plus all greater length products. The values plotted are the average of six values for pol ν (for time points 3, 6, and 9 minutes for pol ν DNA at two different ratios) and an average of four values for Klenow fragment exo− (time points 3 and 5 minutes at one ratio) and the error bars represent the standard deviation.
Error rates for single base insertions and deletions
Among many single base errors made by pol ν, a minority were single base additions and deletions (Figure 1C). These were generated at rates of 7.0 × 10−5 and 16 × 10−5, respectively (Table 1). The single base deletion error rate for pol ν is 3–5 fold higher than those for exonuclease-deficient B family polymerases (Table 2) and ~20-fold higher than for exonuclease-deficient pol γ. On the other hand, the error rate is ~10-fold lower for pol ν compared to Y family pol η and pol κ (Table 2). The number of occurrences of additions and deletions is too few to suggest which of several possible mechanisms (reviewed in [44]) might be responsible for generating these deletion and addition errors, other than to note that a substantial proportion of these errors occur at non-repetitive sequences, and therefore do not result from classical strand slippage [35].
Error rates for single base substitutions
A total of 673 single base substitutions were recovered among 162 lacZ mutants sequenced (Table 1, Figure 1B,C). Of these, 226 were substitutions known to individually result in reduced plaque color (those depicted by the red letters in Figure 1C). From this information, the average base substitution error rate per phenotypically detectable nucleotide incorporated by pol ν was calculated to be 3.5 × 10−3. A slightly higher error rate (1 × 10−2) was calculated (see Materials and Methods) for all substitutions regardless of phenotype (i.e., detectable changes plus “silent hitchhikers”). These average error rate values indicate that pol ν is less accurate than other exonuclease-deficient DNA polymerases that have been examined in this same fidelity assay (Table II). For example, pol ν is about as inaccurate as Y family pol η and pol κ (Table II) that are implicated in TLS, consistent with the suggestion that pol ν may participate in bypass of lesions that have abnormal geometry [14]. In fact, it was because those TLS reactions [35] were conducted at pH 8.8 that we performed the current fidelity measurements at that same pH. Note however, that the base substitution error rates of several polymerases have been shown to vary with pH. For example, it has been demonstrated that the fidelity of two A family enzymes, Taq DNA pol [45] and the large Klenow fragment of E. coli pol I [35] decreases by several fold with increasing pH. Thus, when we succeed in obtaining more substantial quantities of purified pol ν, we plan to investigate pol ν fidelity and error specificity at lower pH, which may be more physiologically relevant and may reveal higher fidelity and/or altered specificity.
Mismatch and sequence context dependence of error prone synthesis by pol ν
Despite the fact that 12 different single base-base mismatches are possible, the vast majority of single base substitutions generated by pol ν (196 of 226 occurrences, 87%, Table 3) result from stable misincorporation (i.e., misinsertion followed by mismatch extension) of dTMP opposite template guanine. Such highly biased error specificity has not been seen before with any other polymerase when examined using this broad-spectrum fidelity assay. This error specificity, measured here in a complete polymerization reaction containing all four dNTPs in direct competition, is consistent with a very high relative rate of misinsertion of dTMP opposite one particular template guanine as determined by kinetic analysis of reactions containing single nucleotides [35]. In the present study, the average error rate for the 22 template guanines at which this error can be scored in the M13mp2 forward mutation assay is 1.7 × 10−2 (Table 2). The error rate for the second most common pol ν error, the reciprocal T-dGMP mismatch, is 1.3 × 10−3 (Table 3). Both rates are higher than for other A family members.
Table III.
Base substitution errors generated by Pol ν
| Base | Mutation | Mispair | Number | Pol Nu Error rate (×10−5)a |
|---|---|---|---|---|
| From → To Template ∙dNMP | ||||
| A | A → G | A · dCMP | 1 | 10 |
| A → T | A · dAMP | 0 | ≤8 | |
| A → C | A · dGMP | 1 | 11 | |
|
| ||||
| T | T → C | T · dGMP | 18 | 130 |
| T → A | T · dTMP | 2 | 24 | |
| T → G | T · dCMP | 0 | ≤8 | |
|
| ||||
| G | G → A | G · dTMP | 196 | 1700 |
| G → C | G · dGMP | 5 | 51 | |
| G → T | G · dAMP | 0 | ≤8 | |
|
| ||||
| C | C → T | C · dAMP | 1 | 8 |
| C → G | C · dCMP. | 0 | ≤22 | |
| C → A | C · dTMP. | 2 | 24 | |
Base substitution error rates were calculated using results from two independent experiments. All data are from the forward mutation assay. Error rates for individual base substitutions were calculated as previously described (36).
Among many template guanines in the lacZ template where the G-dTMP error can be scored, the error rate varies by more than 30-fold, from 0.6% at template guanine number −66 to an amazingly high 20% at template guanine +165 (Table 4 and Figure 1C). Very high rates for G to A substitutions were also seen at more than 20 phenotypically silent guanines (black letters above target sequence see Figure 1C). Given these hotspots and the wide variation in rates, we tested whether the error rate for the responsible G-dTMP mismatch might be higher or lower at sites where processive synthesis by pol ν terminates. Pol ν has previously been demonstrated to have moderate processivity similar to that of its A family homolog, the large Klenow fragment of E. coli DNA polymerase I [34]. That observation was confirmed here during processive synthesis to copy the lacZ template (Figure 2A). Moreover, the relative intensities of termination bands generated at some template positions (e.g., those bracketed in Figure 2A) during processive synthesis by pol ν (lanes 2–4) and Klenow fragment Exo- pol (lanes 6–7) differ, indicating that these two A family DNA polymerases interact somewhat differently with undamaged primer-templates during DNA synthesis. When we calculated (see Materials and Methods) the probability of termination of processive synthesis at each template base (Figure 2B), no obvious correlation was observed between error rates and termination probabilities for pol ν (Table 4). Nonetheless, when we considered the identity of the bases flanking the template guanines at which errors were observed, the highest rates were observed when the base pair located 3′ to the template guanine was C-G or G-C (Table 4, top), whereas rates were lower when the base pair located 3′ to the template guanine was T-A or A-T. This implies that sequence context-dependent differences in the rate at which pol ν misincorporates dTMP opposite G partly depends on the identity, and perhaps the stability, of the preceding, i.e., primer terminal base pair. We also note that there are some exceptions to this trend (e.g., guanine at −64, Figure 1C) that could indicate more distant and/or complex influences. For example, in 9 of 10 sequences in which the G to A change is prevalent, there is a pyrimidine two nucleotides 5′ to the template G, whereas there is a purine in 7 of 11 sequences in which the G is A change is less prevalent. A similar trend is seen four nucleotides 5′ of the template G. At this position, 9 of 10 nucleotides are purines when the G to A change is prevalent, while 7 of 10 nucleotides are pyrimidines at this location when the G to A change is less prevalent.
Controlling error prone synthesis by pol ν
One important implication of low fidelity synthesis by human pol ν is that its activity in cells should be tightly regulated. This could occur at several levels, including regulating mRNA expression in a tissue specific manner [46–48]. Based on the suggestion that pol ν may participate in TLS and exciting precedents with inaccurate Y family pols involved in TLS [49], it will be interesting to search for protein partners (e.g., polymerase clamps) and for post-translational modification of pol ν. These could perhaps involve residues in the amino terminal half of the pol ν open reading frame, whose functions are yet to be explored. It is also possible that pol ν fidelity could be modulated in cells, either by accessory proteins that might enhance fidelity or via error correction by extrinsic proofreading [50,51] or by DNA mismatch repair [35]. The ability of pol ν to generate G-dTMP mismatches at a high rate also leads one to wonder if specialized forms of mismatch repair, e.g., initiated by thymine DNA glycosylase, might process pol ν errors in cells.
Implications for possible functions of pol ν
Its moderate processivity and strand displacement activity [35,52] suggest that pol ν operates in transactions requiring more extensive DNA synthesis than those repair pathways (e.g., short patch BER, NHEJ) performed by X family polymerases. In considering what these transactions might be, it is remarkable that pol ν shares several properties in common with Y family pol κ. Both enzymes are moderately processive (Figure 2, [35,53]), both can bypass thymine glycol lesions [52], and both are error prone, especially for G-dTMP and T-dGMP errors (Table 2 and [54]). It is worth noting that, in addition to the idea that both pols may participate in TLS, pol κ has recently been implicated [55] in gap-filling DNA synthesis during nucleotide excision repair (NER). This may include pol κ involvement in gap-filling during specialized NER to repair inter-strand crosslinks in DNA [56,57], a process thought to require lesion bypass [58,59]. Notably, pol ν shares some sequence homology to Drosophila melanogaster Mus308, defects in which lead to sensitivity to killing by DNA crosslinking agents.
The unique error signature of pol ν could simply reflect use of a substrate in vivo that in some way mimics the structure of a G-dTMP mismatch. It is also possible that pol ν contributes to error prone DNA synthesis that is selectively advantageous to vertebrates. One currently unexplored possibility is that pol ν might participate in somatic hypermutation of immunoglobulin genes. Interestingly, the pol ν homolog DNA polymerase theta (pol θ) has already been proposed to introduce substitutions at G-C base pairs in immunoglobulin genes [60–62], and the TLS enzyme pol η has been implicated in generating substitutions at A-T base pairs during SHM [22,23,63].
The putative nascent base pair binding pocket of pol ν
Possible explanations for the unusual error specificity of pol ν can be considered in light of substantial structural and functional information on other, more accurate A family polymerases. The crystal structures of T7 DNA pol [17,64], BF DNA pol [15,65] and Taq DNA pol [18,66] all reveal correct dNTPs that are tightly accommodated in a binding pocket at the active site, such that incorrect base pairs are expected to sterically clash. These and other structures involving mismatched bases [26–28], along with substantial biochemical evidence (reviewed in [27]) indicate that these A family members selectively incorporate correct dNTPs based partly on geometric complementarity in the nascent base pair binding pocket. We therefore asked which of the amino acid residues that contribute to the binding pocket in high fidelity A family members are conserved in low fidelity pol ν. The binding pocket of Taq pol I that is assembled by correct dNTP-induced conformational changes (Figure 3A) is comprised of a number of amino acids that are highly conserved in bacteriophage T7 DNA pol, Klenow fragment pol I, BF DNA pol I and human pol γ (Figure 3B). Numerous studies (reviewed in [67]), have shown that fidelity is reduced by non-conservative amino acid replacements for certain residues of this binding pocket (shown in blue in Figure 3B). As one example, a single Glu710Ala substitution in KF pol strongly reduces nucleotide selectivity [68,69]. Perhaps cogent to the unusual error specificity of pol ν, replacing KF Glu710 with alanine had an unanticipated and highly unusual error specificity, in that fidelity was reduced only for a subset of the 12 mismatches, especially those involving misinsertion of dTTP and dCTP. Kinetic analysis suggested that the Glu710 side chain in KF pol has an important role in excluding wobble base pairs in the binding pocket. Glu710 in KF pol is conserved as Glu628 in pol ν, and therefore does not itself account for the unique error signature of pol ν, but the Glu710Ala results nevertheless serve as proof-of-principle that even a single amino acid difference could potentially account for low fidelity involving a subset of mismatches, as reported here for G-dTTP and T-dGTP mispairs generated at high rates by pol ν. Studies with Taq also highlight the importance of the O-helix and how perturbations of this helix affect the fidelity of the enzyme. On these bases, we note that among many amino acids that contribute to the binding pocket of the accurate A family polymerases, several that are located in the O-helix that contributes to binding pocket geometry of accurate A family polymerases are not conserved in pol ν (Figure 3B). Probing these differences may be useful for future attempts to understand the remarkable error specificity, and perhaps the in vivo functions, of pol ν.
Figure 3.
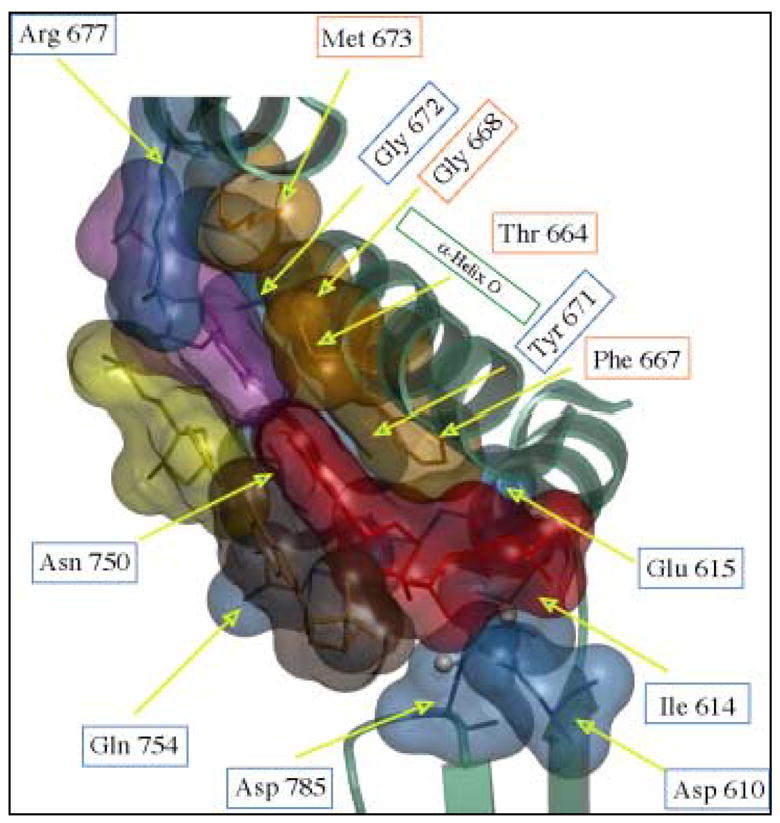

(A) The nascent base pair binding pocket at the active site of Taq DNA pol I. Amino acids of Taq DNA pol I that interact with the incoming nucleotide (red) and the templating base (magenta) that are conserved in pol ν are colored in blue. Amino acids that are not conserved are represented in orange. The figure was created based on the structure of Taq DNA pol I in a ternary complex with DNA and incoming ddGTP (Protein Data Bank accession number 1QSS). (B) Conserved and non-conserved amino acids in Family A DNA polymerases including: Human pol ν, Taq DNA pol I, E. coli DNA pol I (Kf-), Bacteriophage T7 DNA pol, Bacillus stearothermophilus DNA pol I (BF) and human pol γ. Amino acids of Taq DNA pol I that are conserved in pol ν are white in a blue background. Amino acids that are not conserved are white in an orange background. Conserved amino acids with respect to pol ν are indicated in bold.
Acknowledgments
We thank Drs. Katarzyna Bebenek, William C. Copeland and Stephanie Nick McElhinny for reading and critically commenting on manuscript. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, by grant CA101980 from the NIH to RDW, and by the University of Pittsburgh Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 2.Shcherbakova PV, Bebenek K, Kunkel TA. Functions of eukaryotic DNA polymerases. Sci Aging Knowledge Environ. 2003;2003:RE3. doi: 10.1126/sageke.2003.8.re3. [DOI] [PubMed] [Google Scholar]
- 3.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 4.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. ASM Press; 2006. [Google Scholar]
- 5.Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. J Biol Chem. 2001;276:38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- 6.Bebenek K, Joyce CM, Fitzgerald MP, Kunkel TA. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J Biol Chem. 1990;265:13878–13887. [PubMed] [Google Scholar]
- 7.Carroll SS, Cowart M, Benkovic SJ. A mutant of DNA polymerase I (Klenow fragment) with reduced fidelity. Biochemistry. 1991;30:804–813. doi: 10.1021/bi00217a034. [DOI] [PubMed] [Google Scholar]
- 8.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 9.Kim TW, Brieba LG, Ellenberger T, Kool ET. Functional evidence for a small and rigid active site in a high fidelity DNA polymerase: probing T7 DNA polymerase with variably sized base pairs. J Biol Chem. 2006;281:2289–2295. doi: 10.1074/jbc.M510744200. [DOI] [PubMed] [Google Scholar]
- 10.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 11.Wong I, Patel SS, Johnson KA. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 12.Kim TW, Delaney JC, Essigmann JM, Kool ET. Probing the active site tightness of DNA polymerase in subangstrom increments. Proc Natl Acad Sci U S A. 2005;102:15803–15808. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tindall KR, Kunkel TA. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- 14.Eckert KA, Kunkel TA. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1990;18:3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Waksman G. Crystal structures of a ddATP-, ddTTP-, ddCTP, and ddGTP- trapped ternary complex of Klentaq1: insights into nucleotide incorporation and selectivity. Protein Sci. 2001;10:1225–1233. doi: 10.1110/ps.250101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel PH, Suzuki M, Adman E, Shinkai A, Loeb LA. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. J Mol Biol. 2001;308:823–837. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- 17.Kiefer JR, Mao C, Hansen CJ, Basehore SL, Hogrefe HH, Braman JC, Beese LS. Crystal structure of a thermostable Bacillus DNA polymerase I large fragment at 2.1 A resolution. Structure. 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 18.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 19.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 20.Beese LS, Friedman JM, Steitz TA. Crystal structures of the Klenow fragment of DNA polymerase I complexed with deoxynucleoside triphosphate and pyrophosphate. Biochemistry. 1993;32:14095–14101. doi: 10.1021/bi00214a004. [DOI] [PubMed] [Google Scholar]
- 21.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 22.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 23.Doublie S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure. 1999;7:R31–35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. Embo J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow) Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 26.Echols H, Goodman MF. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 27.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 28.Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 29.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 30.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 31.Sharief FS, Vojta PJ, Ropp PA, Copeland WC. Cloning and chromosomal mapping of the human DNA polymerase theta (POLQ), the eighth human DNA polymerase. Genomics. 1999;59:90–96. doi: 10.1006/geno.1999.5843. [DOI] [PubMed] [Google Scholar]
- 32.Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. Embo J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- 35.Takata K-i, Shimizu T, Iwai S, Wood RD. Human DNA Polymerase N (POLN) Is a Low Fidelity Enzyme Capable of Error-free Bypass of 5S-Thymine Glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 36.Abbotts J, Bebenek K, Kunkel TA, Wilson SH. Mechanism of HIV-1 reverse transcriptase. Termination of processive synthesis on a natural DNA template is influenced by the sequence of the template-primer stem. J Biol Chem. 1993;268:10312–10323. [PubMed] [Google Scholar]
- 37.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 38.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J Biol Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 39.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J Mol Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 40.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 41.Fortune JM, Stith CM, Kissling GE, Burgers PM, Kunkel TA. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase {delta} Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kunkel TA, Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]
- 43.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 44.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 45.Eckert KA, Kunkel TA. Fidelity of DNA synthesis catalyzed by human DNA polymerase alpha and HIV-1 reverse transcriptase: effect of reaction pH. Nucleic Acids Res. 1993;21:5212–5220. doi: 10.1093/nar/21.22.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 47.Shcherbakova PV, Fijalkowska IJ. Translesion synthesis DNA polymerases and control of genome stability. Front Biosci. 2006;11:2496–2517. doi: 10.2741/1985. [DOI] [PubMed] [Google Scholar]
- 48.Lehmann AR. Clubbing together on clamps: The key to translesion synthesis. DNA Repair (Amst) 2006;5:404–407. doi: 10.1016/j.dnarep.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 49.Zhong X, Garg P, Stith CM, McElhinny SA, Kissling GE, Burgers PM, Kunkel TA. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 51.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 52.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J Biol Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 53.Fischhaber PL, Gerlach VL, Feaver WJ, Hatahet Z, Wallace SS, Friedberg EC. Human DNA polymerase kappa bypasses and extends beyond thymine glycols during translesion synthesis in vitro, preferentially incorporating correct nucleotides. J Biol Chem. 2002;277:37604–37611. doi: 10.1074/jbc.M206027200. [DOI] [PubMed] [Google Scholar]
- 54.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 55.Kunkel TA, Houten BV. Survival choices. Nat Cell Biol. 2006;8:547–549. doi: 10.1038/ncb0606-547. [DOI] [PubMed] [Google Scholar]
- 56.McHugh PJ, Sarkar S. DNA interstrand cross-link repair in the cell cycle: a critical role for polymerase zeta in G1 phase. Cell Cycle. 2006;5:1044–1047. doi: 10.4161/cc.5.10.2763. [DOI] [PubMed] [Google Scholar]
- 57.Sarkar S, Davies AA, Ulrich HD, McHugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. Embo J. 2006;25:1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, J OW. DNA polymerase theta contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc Natl Acad Sci U S A. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger AJ, III, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase theta plays a dominant role in immunoglobulin gene somatic hypermutation. Embo J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pavlov YI, Rogozin IB, Galkin AP, Aksenova AY, Hanaoka F, Rada C, Kunkel TA. Correlation of somatic hypermutation specificity and A-T base pair substitution errors by DNA polymerase eta during copying of a mouse immunoglobulin kappa light chain transgene. Proc Natl Acad Sci U S A. 2002;99:9954–9959. doi: 10.1073/pnas.152126799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 62.Mayorov VI, Rogozin IB, Adkison LR, Gearhart PJ. DNA polymerase eta contributes to strand bias of mutations of A versus T in immunoglobulin genes. J Immunol. 2005;174:7781–7786. doi: 10.4049/jimmunol.174.12.7781. [DOI] [PubMed] [Google Scholar]
- 63.Doublie S, Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr Opin Struct Biol. 1998;8:704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- 64.Kiefer JR, Mao C, Braman JC, Beese LS. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 65.Li Y, Kong Y, Korolev S, Waksman G. Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates. Protein Sci. 1998;7:1116–1123. doi: 10.1002/pro.5560070505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. Embo J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Minnick DT, Liu L, Grindley ND, Kunkel TA, Joyce CM. Discrimination against purine-pyrimidine mispairs in the polymerase active site of DNA polymerase I: a structural explanation. Proc Natl Acad Sci U S A. 2002;99:1194–1199. doi: 10.1073/pnas.032457899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki M, Avicola AK, Hood L, Loeb LA. Low fidelity mutants in the O-helix of Thermus aquaticus DNA polymerase I. J Biol Chem. 1997;272:11228–11235. doi: 10.1074/jbc.272.17.11228. [DOI] [PubMed] [Google Scholar]
- 69.Suzuki M, Yoshida S, Adman ET, Blank A, Loeb LA. Thermus aquaticus DNA polymerase I mutants with altered fidelity. Interacting mutations in the O-helix. J Biol Chem. 2000;275:32728–32735. doi: 10.1074/jbc.M000097200. [DOI] [PubMed] [Google Scholar]


