Abstract
The best-studied cytoskeletal system is the inner surface of the erythrocyte membrane, which provides an erythrocyte with the structural support needed to be stable yet flexible as it passes through the circulation. Current structural models predict that the spectrin–actin-based cytoskeletal network is attached to the plasma membrane through interactions of the protein ankyrin, which binds to both spectrin and the cytoplasmic domain of the transmembrane protein band 3. The crystal structure of the cytoplasmic domain of band 3 predicted that the ankyrin binding site was located on a β-hairpin loop in the cytoplasmic domain. In vitro, deletion of this loop eliminated ankyrin affinity for band 3 without affecting any other protein–band 3 interaction. To evaluate the importance of the ankyrin–band 3 linkage to membrane properties in vivo, we generated mice with the nucleotides encoding the 11-aa β-hairpin loop in the mouse Slc4a1 gene replaced with sequence encoding a diglycine bridge. Mice homozygous for the loop deletion were viable with mildly spherocytic and osmotically fragile erythrocytes. In vitro, homozygous ld/ld erythrocytes were incapable of binding ankyrin, but contrary to all previous predictions, abolishing the ankyrin–band 3 linkage destabilized the erythrocyte membrane to a lesser degree than complete deficiencies of either band 3 or ankyrin. Our data indicate that as yet uncharacterized interactions between other membrane proteins must significantly contribute to linkage of the spectrin–actin-based membrane cytoskeleton to the plasma membrane.
Keywords: cytoskeleton, membrane, transgenic, spherocytosis
The erythrocyte membrane skeleton provides the stability and deformability required by the erythrocyte as it travels through the circulation, and is the paradigm for study of membrane structure and function in many cell types. It is composed of a spectrin–actin-based network of cytoskeletal proteins linked to the inner surface of the erythrocyte plasma membrane. In current models, the critical connection between the spectrin–actin cytoskeleton and the plasma membrane is through the peripheral membrane protein ankyrin, which binds to both spectrin and the cytoplasmic domain of the transmembrane protein band 3 (cdb3) (1).
Ankyrin, originally discovered in erythroid cells, is the prototype of a family of proteins present in many diverse organelles, cell types, and tissues. Ankyrins function in a wide variety of processes, including membrane organization, cell signaling, maintenance of cellular polarity, ion transport, and cell–cell adhesion. Thus, interactions between the cell membrane and the cytoskeleton play a fundamental role in cell membrane structure, function, and other cellular processes in all cells. In the erythrocyte, rupture of the ankyrin–cdb3–spectrin linkage with antibodies against cdb3 leads to loss of membrane stability and spontaneous erythrocyte fragmentation (2).
In addition to interacting with each other, both band 3 and ankyrin interact with other membrane proteins. Ankyrin binds integral membrane proteins of the Rhesus (Rh)-Rh-associated glycoprotein (RhAG) complex (3), Na+/K+-ATPase (4), the Na+/Ca2+ exchanger (5), CD47 (6, 7), and the voltage-dependent Na+ channel (8). Band 3 associates with the integral membrane proteins, glycophorin A and members of the Rh family, as well as cytosolic protein 4.1, protein 4.2, several kinases and phosphatases, glycolytic enzymes, adducin, and hemoglobin (9–11). However, at this point, none of these linkages have been demonstrated to participate in the linkage of the cytoskeleton to the erythrocyte membrane.
In human erythrocytes, perturbations of the ankyrin–cdb3 interaction lead to hereditary spherocytosis (HS) (12–16), a common inherited hemolytic anemia characterized by decreased membrane stability and altered cell morphology. Defects of ankyrin or band 3 cause ≈75% of HS cases (12, 13, 17–19). Erythrocytes from ankyrin-deficient (nb/nb) and band 3-deficient (Slc4a1−/−) mice exhibit a severe loss of mechanical stability, altered morphology, and dramatically reduced survival (20–22). We and others have recently solved the crystal structure of cdb3 (23) and the cdb3 binding domain of ankyrin (24), respectively. The crystal structures predict that an 11-aa β-hairpin loop in cdb3 (residues 188–198 in the mouse and 175–185 in human) binds to the D3D4 domains of ankyrin (24). Deletion of the β-hairpin loop specifically abolishes ankyrin binding to cdb3 in vitro, leading to the conclusion that ankyrin and band 3 interact specifically through this loop (23).
These observations strongly support current models of the erythrocyte membrane skeleton in which the ankyrin–band 3 interaction is the primary linkage between the spectrin–actin cytoskeletal network and the plasma membrane (1, 25, 26). However, whether the ankyrin–band 3 linkage is solely responsible for this critical role in vivo cannot be demonstrated by the binding of protein fragments in vitro or in animals in which one of the proteins is absent.
To specifically evaluate the importance of the ankyrin–cdb3 interaction in the linkage of the erythrocyte membrane to its cytoskeleton, we generated knockin mice in which the β-hairpin loop in band 3 was deleted and replaced by a flexible diglycine bridge to preserve the structure of cdb3. Erythrocytes from mice homozygous for the loop deletion (ld/ld) were mildly spherocytic and osmotically fragile, and inside-out vesicles from ld/ld erythrocytes were incapable of binding ankyrin. Despite the lack of ankyrin binding, the erythrocyte membrane proteins were assembled in normal amounts, and ld/ld erythrocytes were not as osmotically fragile and survived longer in the circulation than erythrocytes from nb/nb or Slc4a1−/− mice (20–22). We conclude from these data that in addition to the ankyrin–cdb3 linkage, additional proteins are involved in stabilizing the linkage of the spectrin–actin complex to the erythrocyte membrane.
Results
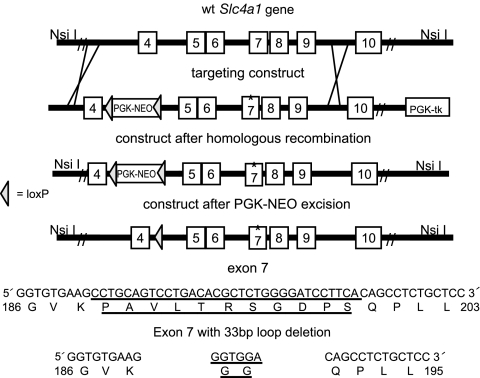
We used homologous recombination in ES cells to introduce a deletion in exon 7 of the murine Slc4a1 (AE1; band 3) gene that replaced the sequence encoding the 11 aa of the β-hairpin loop in cdb3 (Fig. 1; residues 188–198), with sequence encoding two glycine residues. In addition, a phosphoglycerate kinase (PGK)-neo cassette was inserted into intron 4 of the Slc4a1 gene for positive selection of ES cells. Correctly targeted ES cells were injected into blastocysts and fertile male chimeras were bred to female prion-Cre transgenic mice, which express the Cre recombinase in the oocyte (27). F1 progeny from this cross that were negative for Cre, negative for the PGK-neo cassette (the PGK-neo cassette was flanked by loxP sites for later removal with Cre recombinase), and heterozygous for the loop deletion mutation were identified. These animals were mated to generate animals homozygous for the β-hairpin loop deletion.
Fig. 1.
Homologous recombination to delete an 11-amino acid β-hairpin loop in the cytoplasmic domain of band 3. Strategy for targeting the murine Slc4a1 locus (Upper). The amino acid sequence of the β-hairpin loop and the amino acid sequence after the loop deletion (Lower).
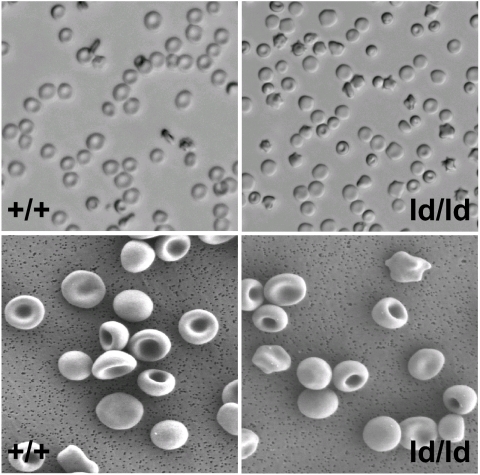
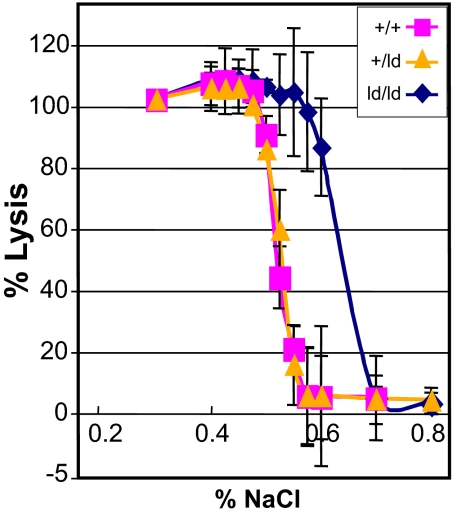
ld/ld mice were born in a normal Mendelian ratio and have a normal life span. Light and SEM comparison of wild-type (+/+) and ld/ld erythrocytes demonstrated that ld/ld cells were smaller than +/+ cells and ld/ld cells had a higher percentage of spherocytes and stomatocytes (Fig. 2). Forty-eight percent of ld/ld cells exhibited altered morphology (nonbiconcave disk shape), whereas <5% the wild-type cells showed altered red cell morphology. Hematologic indices were similar for wild-type, heterozygous +/ld, and homozygous ld/ld mice, with the exception that ld/ld mice had a significantly higher percentage of reticulocytes in the peripheral blood (12.87% versus 2.18% P < 0.001) and Ter119+ erythroid cells in the bone marrow and spleen (1.5- and 1.8-fold, respectively; P < 0.02; Table 1). Consistent with the presence of spherocytic erythrocytes in ld/ld peripheral blood, ld/ld erythrocytes showed a significant increase in osmotic fragility compared with wild-type or heterozygous +/ld erythrocytes (Fig. 3). Spleen weights of the ld/ld mice were significantly greater than wild type, 0.39 ± 0.05 versus 0.18 ± 0.02 (P < 0.01; Table 1). The higher level of reticulocytes, the increased osmotic fragility, the relative increase in erythroid cells in the bone marrow and spleen, and the increased spleen weights of ld/ld mice predict a significant decrease in the estimated lifespan of ld/ld erythrocytes.
Fig. 2.
Homozygous ld/ld erythrocytes have altered morphology. (Upper) Comparison of whole erythrocytes under a light microscope from wild-type (+/+) and homozygous loop-deleted (ld/ld) mice. (Lower) Comparison of erythrocytes from +/+ and ld/ld mice by using SEM.
Table 1.
Hematologic indices of wild-type (+/+), heterozygous (+/ld), and loop-deleted (ld/ld) mice and quantitative-RT-PCR analysis of Slc4a1 mRNA levels in +/+, +/ld, and loop-deleted (ld/ld) mutant mouse tissues
| Values | +/+ | +/ld | ld/ld |
|---|---|---|---|
| Hematologic indices | |||
| WBCs, × 103/μl | 7.68 ± 0.23 | 6.85 ± 1.88 | 5.67 ± 1.3 |
| Platelets, × 103/μl | 917.67 ± 27.58 | 781.14 ± 89.3 | 829.33 ± 134.86 |
| RBCs, × 106/μl | 9.28 ± 0.86 | 8.9 ± 0.46 | 8.75 ± 0.45 |
| Hematocrit, % | 51.2 ± 0.97 | 50.87 ± 1.63 | 50.52 ± 1.38 |
| Hb, g/dl | 15.3 ± 0.57 | 13.8 ± 0.49 | 13.32 ± 0.55 |
| Mean cell volume, fl | 55.17 ± 2.19 | 57.15 ± 1.49 | 57.73 ± 1.14 |
| Ter119+ cells in bone marrow, % | 35.42 ± 3.42 | 37.06 ± 2.53 | 54.66 ± 2.90** |
| Ter119+ cells in spleen, % | 12.86 ± 1.67 | 13.01 ± 3.10 | 23.42 ± 4.86** |
| Reticulocytes, % | 2.18 ± 1.22 | 5.47 ± 1.84* | 13.87 ± 4.42*** |
| QPCR | |||
| Bone marrow | 1.0 | 0.92 ± 0.14 | 1.6 ± 0.25**** |
| Spleen | 0.05 ± 0.02 | 0.05 ± 0.03 | 0.081 ± 0.02**** |
| Kidney | 0.002 ± 0.001 | 0.005 ± 0.003 | 0.006 ± 0.003 |
The level of Slc4a1 mRNA in +/+ bone marrow was arbitrarily designated as 1.0. *, P < 0.04; **, P < 0.002; ***, P < 0.001; ****, P < 0.02.
Fig. 3.
RBCs from ld/ld mice are osmotically fragile. Erythrocytes from +/+, heterozygous (+/ld), and ld/ld mice were exposed to increasing amounts of saline (x axis). The percent of cells lysed is shown on the y axis.
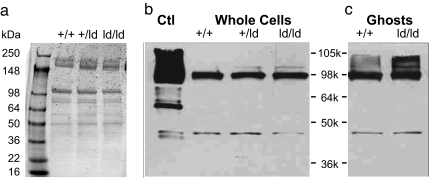
Quantitative RT-PCR analysis of Slc4a1 mRNA showed that the level of Slc4a1 mRNA was greater in ld/ld bone marrow and spleen than the level of Slc4a1 mRNA in wild-type or +/ld bone marrow and spleen (Table 1). The apparent increase in Slc4a1 mRNA levels in ld/ld bone marrow and spleen correlated with the increase in the percentage of Ter119+ erythroid cells in ld/ld bone marrow and spleen. There were no significant differences in the relative abundances of band 3 protein in wild-type, +/ld, and ld/ld erythrocytes. Comparison of wild-type, +/ld, and ld/ld erythrocyte ghost membranes demonstrated normal levels of band 3 and other red cell membrane proteins [spectrin-to-actin ratio: +/+ versus +/ld = 1.00 ± 0.12, +/+ versus ld/ld = 1.11 ± 0.11 (not significant); band 3-to-actin ratio: +/+ versus +/ld = 1.17 + 0.18, +/+ versus ld/ld = 1.16 ± 0.08 (not significant); Fig. 4a]. The levels of other membrane proteins, including glycophorin, RhAg, and CD47, were similar between +/+ and ld/ld erythrocytes (data not shown). These observations were confirmed by Western blot analysis of whole-cell lysates and ghosts (Fig. 4 b and c). These data demonstrate that deletion of the β-hairpin loop deletion does not significantly alter the level of band 3 protein in ld/ld erythrocytes.
Fig. 4.
ld/ld erythrocytes are not deficient in band 3. (a) SDS/PAGE of erythrocyte ghost membranes from +/+, +/ld, and ld/ld mouse blood. (b and c) Western blot of whole RBCs and ghosts from +/+, +/ld, and ld/ld mutant mice. Ctl, human erythrocyte ghosts. Double antibody (anti-band 3 and anti-actin) staining was used.
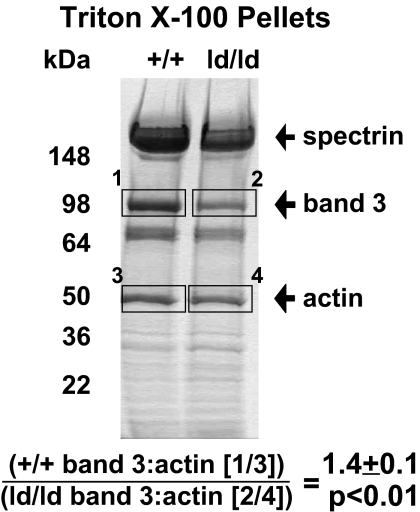
To measure attachment of loop-deleted band 3 (ld-B3) to the spectrin–actin–erythrocyte membrane cytoskeleton, wild-type and ld/ld whole RBCs and erythrocyte ghosts were extracted with 2% Triton X-100. Analysis of the insoluble spectrin–actin skeletons showed a 1.4-fold reduction (P < 0.01) in the retention of band 3 in the membrane skeletons of ld/ld erythrocytes compared with wild-type cells, whereas the ratio of spectrin to actin in wild-type and ld/ld erythrocytes was unaffected (0.98 not significant; Fig. 5). These data are consistent with reduced attachment of the ld-B3 to the spectrin–actin skeleton.
Fig. 5.
Band 3 retention in membrane skeletons is reduced in ld/ld erythrocytes. SDS/PAGE of whole RBCs extracted with 2% Triton X-100. Bands were quantified with densitometry scanning. One milliliter of packed RBCs was washed three times with PBS and solubilized with 1 ml of 2% Triton X-100 containing 20 μg/ml pepstatin A, 40 μg/ml PMSF, 20 μg/ml leupeptin, 0.5 mM DTT, and 1 mM EDTA, pH = 6.5. The experiment was repeated three times and the average ratio of band 3 to actin from these experiments is shown.
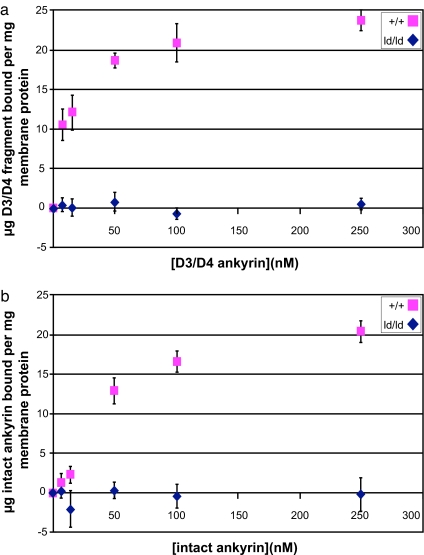
Bennett and colleagues (28) demonstrated that the 45.6-kDa D3/D4 subdomains of ankyrin contain the primary binding site for band 3. To directly measure ankyrin binding to membrane associated band 3, KI-stripped inside-out vesicles (KI-IOVs) were prepared from wild-type and homozygous ld/ld erythrocytes and confirmed to be devoid of endogenous ankyrin and other skeleton proteins by SDS/PAGE (data not shown). Ankyrin binding was measured by using an 125I-labeled fragment containing the ankyrin D3/D4 subdomains (29). Wild-type KI-IOVs bound the D3/D4 subunit in a concentration-dependent fashion, reaching saturation at ≈100 nM (Kd = 14.22 nM). In contrast, KI-IOVs from ld/ld erythrocytes bound only background levels of the labeled D3/D4 ankyrin fragment, indicating that amino acid residues 188–198 of cdb3 are responsible for virtually all of the band 3-D3/D4-ankyrin interaction (Fig. 6a).
Fig. 6.
KI-IOVs from ld/ld mice do not bind ankyrin. KI-IOVs (30 μg/ml) were incubated for 3 h on ice with increasing amounts of D3/D4 125I-ankyrin (a) or intact 125I-ankyrin (b). Membranes were pelleted, washed, and assayed in γ counter for radioactive ankyrin (n = 5). The resulting graph is a plot with subtracted heat-denatured controls. Mutant mice KI-IOVs exhibit negligible binding to either the D3/D4 ankyrin fragment of full-length ankyrin.
To test whether ankyrin domains other than D3/D4 ankyrin fragment were capable of interacting with ld-B3, the experiments were repeated with 125I-labeled full-length ankyrin (2.1) purified from human erythrocytes. Similar to the results with the D3/D4 ankyrin subdomains, no full-length ankyrin bound to homozygous ld/ld KI-IOVs (Fig. 6b). KI-IOVs from +/ld erythrocytes bound ankyrin with the same affinity as wild-type band 3 (data not shown). We conclude that the β-hairpin loop formed by residues 188–198 of cdb3 is essential for ankyrin binding in murine RBCs.
Discussion
The specific linkages involved in the formation of erythrocyte membrane skeleton have served as prototypes for the assembly of related cytoskeletons in other cell types. Unlike the studies with knockout mice, in which a gene is modified to create a nonfunctional or null allele, our approach introduces a minimal modification into the band 3 protein. The mouse developed here allowed us to specifically examine the role of the β-hairpin loop in cdb3 in the ankyrin–band 3 linkage. Our findings establish that additional linkages between ankyrin and the red cell membrane exist, which has implications for models of both the erythrocyte membrane skeleton and other cytoskeletons involving ankyrin linkages.
Although deletion of the β-hairpin loop comprising residues 188–198 of cdb3 abolished ankyrin binding in ld/ld mice, heterozygous +/ld erythrocytes had the same ankyrin binding and osmotic fragility profiles as wild-type erythrocytes. Because ankyrin binds tetrameric band 3 (30, 31), in heterozygous erythrocytes, only 6.25% of band 3 tetramers would be expected to be composed of four ld-B3 proteins. Our ankyrin binding and osmotic fragility assays are not sensitive enough to detect a difference of this magnitude.
KI-IOVs retain integral membrane proteins, including other possible ankyrin anchors, such as the Rh-RhAG complex, the Na+/Ca2+ exchanger, CD47, and the Na+/K+-ATPase. Because we did not observe any binding of either intact ankyrin or its D3/D4 fragment to ld/ld KI-IOVs, we conclude that the association of ankyrin with these other possible anchors is too weak to exist in the absence of a functional band 3 and/or other peripheral membrane proteins.
In the ld/ld mouse, only the major band 3–ankyrin interaction is lost. Despite a total absence of ankyrin binding, erythrocytes from ld/ld mice are more stable than nb/nb, Slc4a1−/−, or wild-type erythrocytes resealed with competing fragments of ankyrin or cdb3. Our observations contradict several hypotheses that the ankyrin–band 3 bridge alone is critical for red cell morphology, resulting in unstable spherocytic cells whenever the band 3–ankyrin linkage is compromised.
We hypothesize that the increased instability of nb/nb and Slc4a1−/− erythrocytes, as well as erythrocytes resealed with competing fragments of ankyrin or cdb3, is caused by the elimination of multiple stabilizing interactions. For example, deficiency of band 3 causes concomitant loss of Rh complex proteins and CD47 (neither of which bind ankyrin), as well as glycophorin A and protein 4.2 from the membrane (21, 22). These additional protein deficiencies clearly contribute to membrane instability. Similarly, our data predict that protein deficiencies beyond ankyrin occur in nb/nb erythrocytes (20).
Further evidence for additional linkages contributing to membrane stability comes from analysis of adducin and protein 4.2-deficient knockout mice, as well as rare patients with protein 4.2 deficiency. In all cases, a mild hemolytic anemia with an intact red cell membrane skeleton, similar to our ld/ld mice, is observed (32–35). It is possible that ankyrin binds to cdb3 at a second, low-affinity site (36). A recent finding by W. A. Anong and P.S.L. (unpublished data) demonstrates that band 3 interacts directly with adducin in IOVs. Together, these data demonstrate that although the ankyrin–cdb3 linkage is important for red cell membrane stability, the roles of other cytoskeleton-to-membrane linkages and/or additional proteins that contribute to the ankyrin–cdb3 bridge (e.g., CD47, protein 4.2) must be much greater than previously proposed. Identification of these protein interactions has important implications for our understanding of cytoskeleton biology in both erythroid and nonerythroid cells, as well as in the pathogenesis of membrane-linked inherited hemolytic anemia.
Methods
Materials.
Na125I was obtained from PerkinElmer (Waltham, MA). Leupeptin and pepstatin were purchased from Sigma–Aldrich (St. Louis, MO). Triton X-100 was bought from Pierce Chemical (Rockford, IL). The TRIzol reagent and SuperScript III Platinum Two-Step quantitative RT-PCR kit with Syber Green were from Invitrogen (Carlsbad, CA). The anti-rabbit secondary antibody was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). All other chemicals were obtained from Sigma–Aldrich.
Homologous Recombination Strategy in ES Cells.
The targeting construct was generated in a modified version of the pPNT vector (37), which contains a PGK-Neo gene flanked by loxP sites and a PGK-hsv-tk gene. The 5′ arm was a 3,227-bp BamHI/XhoI fragment (coordinates chromosome 11: 102,472,363 to 102,469,136) of the murine Slc4a1 (AE1, band 3) gene that extends into exon 4. The 3′ arm was a 5,232-bp XhoI/SphI fragment of the murine Slc4a1 gene (coordinates chromosome 11: 102,469,136 to 102,464,304). The 3′ arm was modified to delete the 33-bp sequence encoding the β-hairpin loop (coordinates chromosome 11: 102,467,497 to 102,467,529) while adding a 6-bp sequence encoding a diglycine bridge in exon 7. The pPNT vector was linearized with NotI and transfected into TC1 ES cells as described I refs. 38 and 39. A total of 67 G418/gancyclovir-resistant ES cell clones were collected and analyzed for homologous recombination events by Southern blot analysis. Eight clones (11.9%) were correctly targeted. PCR analysis with primers spanning the deleted region (Slc4a1 forward: 5′-CAATGGGAAACTCAATCCAAATAG-3′; reverse: 5′-TCCATCCAGCCCTTCCTTC-3′) demonstrated that four of eight clones had the deletion, whereas in the other clones, the recombination event was between the loop-deletion mutation and the neo gene in exon 4.
All animal procedures were approved by the National Human Genome Research Institute (NHGRI) Animal Care and Use Committee (ASP# G-04-2). Approximately 12 cells from two different loop deletion clones were injected into C57BL/6J blastocysts to generate chimeric mice. Male chimeras were bred to 129 female mice carrying the prion-Cre transgene (27) for germ line removal of the neomycin resistance gene cassette in the targeting construct. PCR analysis of DNA from tail biopsies identified offspring that were heterozygous for both the loop deletion and the prion-Cre transgene (Cre forward: 5′-CCGGGCTGCCACGACCA-3′; reverse: 5′-GGCGCCGCAACACCATTTTT-3′) that were crossed to generate an F2 generation. PCR analysis of F2 mice identified animals that contained the deletion but were negative for both the prion-Cre transgene and the neo cassette (neo forward: 5′-GGACCTTGCACAGATAGCGT-3′; reverse: 5′-CTGTTCCTGACCTTGATGTG-3′). These animals were bred to generate homozygous loop deletion and control mice for analysis.
Blood Analysis.
Blood was collected from the retroorbital sinus. Osmotic fragility was determined as described in ref. 40. Reticulocyte counts were determined manually by staining of peripheral blood smears with crystal violet.
RNA Analysis.
RNA was extracted from the bone marrow, spleen, and kidney by using TRIzol Reagent according to the manufacturer's instructions. The relative levels of Slc4a1 mRNA (Slc4a1 RNA forward: 5′-TCGAGGATCAGATTCGGCCT-3′; reverse: 5′-GAGGGTCCTTCTGAGCCCC-3′) and β2-microglobulin (β-2 forward: 5′-TGCTATCCAGAAAACCCCTC-3′; reverse: 5′-GTCATGCTTAACTCTGCAGG-3′) were compared by using the SuperScript III Platinum Two-Step qRT-PCR kit with Syber Green on an ABI Prism 7700 Sequence Detector (Applied Biosystems, Foster City, CA) machine.
Preparation of KI-IOVs.
KI-IOVs were prepared from fresh blood as described in ref. 41. Briefly, RBCs were washed with PBS (137 mM NaCl/2.7 mM KCl/8.1 mM K2HPO4/1.5 mM KH2PO4, pH 7.4) and ghosts were prepared by hypotonic lysis in 5 mM sodium phosphate/1 mM EDTA, pH = 8.0, in the presence of 30 μg/ml PMSF. The lysate was centrifuged at 25,000 × g for 20 min and the supernatant was removed. The membrane pellets were resuspended and washed three additional times in lysis buffer. The white ghosts were then resuspended in a 100-fold volume excess of buffer containing 0.5 mM EDTA, 0.5 mM DTT, 40 μg/ml PMSF, pH = 8.0, at 4°C, warmed to 37°C over 30 min, and pelleted at 25,000 × g for 30 min to remove spectrin and actin. The remaining peripheral proteins were removed by incubation for 30 min at 37°C in a 100-fold excess of 1 M KI, 25 mM sodium phosphate, 1 mM EDTA, 0.5 mM DTT, and 40 μg/ml PMSF, pH = 7.5. The resulting KI-IOVs were pelleted for 30 min at 25,000 × g then washed twice with 5 mM sodium phosphate/1 mM EDTA, pH = 8.0/0.5 mM DTT. Membrane concentrations were determined by using the Bradford assay.
Detergent Extraction Assay and Densitometry Scan.
RBCs were isolated from blood from wild-type and mutant mice and washed with PBS three times. One milliliter of packed RBCs was then solubilized by the addition of 1 ml of 2% Triton X-100, which was supplemented with 20 μg/ml pepstatin A, 40 μg/ml PMSF, 20 μg/ml leupeptin, 0.5 mM DTT, and 1 mM EDTA. To achieve separation between solubilized membrane components and insoluble membrane skeletons, samples were layered onto a 35% sucrose cushion and spun at 85,000 × g for 90 min. Pellets were dissolved in SDS and separated by SDS/PAGE, and bands were quantified by scanning densitometry.
Binding of 125I-ankyrin to RBC Membranes.
Intact ankyrin and D3D4 ankyrin fragment were iodinated with 1 mCi of Na125I (as described in ref. 41) to a specific activity of between 22,000 and 40,000 cpm/μg. KI-IOVs (30 μg/ml final concentration) prepared from wild-type and mutant RBCs were incubated for 3 h on ice with increasing amounts of 125I-ankyrin (7–250 nM final concentration) in a buffer consisting of 5% sucrose, 50 mM sodium phosphate, 50 mM boric acid, 30 mM NaCl, 1 mM EDTA, 0.2 mM DTT adjusted to pH 6.5. Two washes with the same buffer were performed to remove unbound 125I-ankyrin. The washed KI-IOVs were centrifuged at maximum speed in an Eppendorf 5415D table top centrifuge, and the pellets were assayed for 125I-ankyrin incorporation into the membranes by using a γ counter. To determine the background, before the addition of 125I-ankyrin, wild-type and mutant KI-IOVs were heat denatured (65°C, 5 min). These values were subtracted from the respective binding curves.
Acknowledgments
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK62039 (to P.G.G.), National Institute of General Medical Sciences Grant GM24417-28 (to P.S.L.), and National Human Genome Research Institute intramural funds (to D.M.B.).
Abbreviations
- cdb3
cytoplasmic domain of band 3
- ld
loop deletion
- PGK
phosphoglycerate kinase
- KI-IOVs
KI-stripped inside-out vesicles.
Footnotes
The authors declare no conflict of interest.
References
- 1.Lux SE. Nature. 1979;281:426–429. doi: 10.1038/281426a0. [DOI] [PubMed] [Google Scholar]
- 2.Van Dort HM, Knowles DW, Chasis JA, Lee G, Mohandas N, Low PS. J Biol Chem. 2001;276:46968–46974. doi: 10.1074/jbc.M107855200. [DOI] [PubMed] [Google Scholar]
- 3.Nicolas V, Le Van Kim C, Gane P, Birkenmeier C, Cartron JP, Colin Y, Mouro-Chanteloup I. J Biol Chem. 2003;278:25526–25533. doi: 10.1074/jbc.M302816200. [DOI] [PubMed] [Google Scholar]
- 4.Nelson WJ, Veshnock PJ. Nature. 1987;328:533–536. doi: 10.1038/328533a0. [DOI] [PubMed] [Google Scholar]
- 5.Li ZP, Burke EP, Frank JS, Bennett V, Philipson KD. J Biol Chem. 1993;268:11489–11491. [PubMed] [Google Scholar]
- 6.Bruce LJ, Ghosh S, King MJ, Layton DM, Mawby WJ, Stewart GW, Oldenborg PA, Delaunay J, Tanner MJ. Blood. 2002;100:1878–1885. doi: 10.1182/blood-2002-03-0706. [DOI] [PubMed] [Google Scholar]
- 7.Dahl KN, Parthasarathy R, Westhoff CM, Layton DM, Discher DE. Blood. 2004;103:1131–1136. doi: 10.1182/blood-2003-04-1331. [DOI] [PubMed] [Google Scholar]
- 8.Malhotra JD, Kazen-Gillespie K, Hortsch M, Isom LL. J Biol Chem. 2000;275:11383–11388. doi: 10.1074/jbc.275.15.11383. [DOI] [PubMed] [Google Scholar]
- 9.Bruce LJ, Beckmann R, Ribeiro ML, Peters LL, Chasis JA, Delaunay J, Mohandas N, Anstee DJ, Tanner MJ. Blood. 2003;101:4180–4188. doi: 10.1182/blood-2002-09-2824. [DOI] [PubMed] [Google Scholar]
- 10.Low PS. Biochim Biophys Acta. 1986;864:145–167. doi: 10.1016/0304-4157(86)90009-2. [DOI] [PubMed] [Google Scholar]
- 11.Low PS, Rathinavelu P, Harrison ML. J Biol Chem. 1993;268:14627–14631. [PubMed] [Google Scholar]
- 12.Lux SE, Tse WT, Menninger JC, John KM, Harris P, Shalev O, Chilcote RR, Marchesi SL, Watkins PC, Bennett V, et al. Nature. 1990;345:736–739. doi: 10.1038/345736a0. [DOI] [PubMed] [Google Scholar]
- 13.Savvides P, Shalev O, John KM, Lux SE. Blood. 1993;82:2953–2960. [PubMed] [Google Scholar]
- 14.Iolascon A, Miraglia del Giudice E, Camaschella C, Pinto L, Nobili B, Perrotta S, Cutillo S. Br J Haematol. 1991;78:551–554. doi: 10.1111/j.1365-2141.1991.tb04487.x. [DOI] [PubMed] [Google Scholar]
- 15.Hanspal M, Yoon SH, Yu H, Hanspal JS, Lambert S, Palek J, Prchal JT. Blood. 1991;77:165–173. [PubMed] [Google Scholar]
- 16.Pekrun A, Eber SW, Kuhlmey A, Schroter W. Ann Hematol. 1993;67:89–93. doi: 10.1007/BF01788132. [DOI] [PubMed] [Google Scholar]
- 17.Jarolim P, Murray JL, Rubin HL, Taylor WM, Prchal JT, Ballas SK, Snyder LM, Chrobak L, Melrose WD, Brabec V, Palek J. Blood. 1996;88:4366–4374. [PubMed] [Google Scholar]
- 18.Coetzer TL, Lawler J, Liu SC, Prchal JT, Gualtieri RJ, Brain MC, Dacie JV, Palek J. N Engl J Med. 1988;318:230–234. doi: 10.1056/NEJM198801283180407. [DOI] [PubMed] [Google Scholar]
- 19.Costa FF, Agre P, Watkins PC, Winkelmann JC, Tang TK, John KM, Lux SE, Forget BG. N Engl J Med. 1990;323:1046–1050. doi: 10.1056/NEJM199010113231507. [DOI] [PubMed] [Google Scholar]
- 20.Yi SJ, Liu SC, Derick LH, Murray J, Barker JE, Cho MR, Palek J, Golan DE. Biochemistry. 1997;36:9596–9604. doi: 10.1021/bi9704966. [DOI] [PubMed] [Google Scholar]
- 21.Southgate CD, Chishti AH, Mitchell B, Yi SJ, Palek J. Nat Genet. 1996;14:227–230. doi: 10.1038/ng1096-227. [DOI] [PubMed] [Google Scholar]
- 22.Peters LL, Shivdasani RA, Liu SC, Hanspal M, John KM, Gonzalez JM, Brugnara C, Gwynn B, Mohandas N, Alper SL, et al. Cell. 1996;86:917–927. doi: 10.1016/s0092-8674(00)80167-1. [DOI] [PubMed] [Google Scholar]
- 23.Chang SH, Low PS. J Biol Chem. 2003;278:6879–6884. doi: 10.1074/jbc.M211137200. [DOI] [PubMed] [Google Scholar]
- 24.Michaely P, Tomchick DR, Machius M, Anderson RG. EMBO J. 2002;21:6387–6396. doi: 10.1093/emboj/cdf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agre P, Smith BL, Saboori AM, Asimos A. Soc Gen Physiol Ser. 1988;43:91–100. [PubMed] [Google Scholar]
- 26.Cho MR, Eber SW, Liu SC, Lux SE, Golan DE. Biochemistry. 1998;37:17828–17835. doi: 10.1021/bi981825c. [DOI] [PubMed] [Google Scholar]
- 27.Scheel JR, Garrett LJ, Allen DM, Carter TA, Randolph-Moore L, Gambello MJ, Gage FH, Wynshaw-Boris A, Barlow C. Nucleic Acids Res. 2003;31:e57. doi: 10.1093/nar/gng057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaely P, Bennett V. J Biol Chem. 1995;270:22050–22057. doi: 10.1074/jbc.270.37.22050. [DOI] [PubMed] [Google Scholar]
- 29.Van Dort HM, Moriyama R, Low PS. J Biol Chem. 1998;273:14819–14826. doi: 10.1074/jbc.273.24.14819. [DOI] [PubMed] [Google Scholar]
- 30.Thevenin BJ, Low PS. J Biol Chem. 1990;265:16166–16172. [PubMed] [Google Scholar]
- 31.Casey JR, Reithmeier RA. J Biol Chem. 1991;266:15726–15737. [PubMed] [Google Scholar]
- 32.Gilligan DM, Lozovatsky L, Gwynn B, Brugnara C, Mohandas N, Peters LL. Proc Natl Acad Sci USA. 1999;96:10717–10722. doi: 10.1073/pnas.96.19.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters LL, Jindel HK, Gwynn B, Korsgren C, John KM, Lux SE, Mohandas N, Cohen CM, Cho MR, Golan DE, Brugnara C. J Clin Invest. 1999;103:1527–1537. doi: 10.1172/JCI5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouhassira EE, Schwartz RS, Yawata Y, Ata K, Kanzaki A, Qiu JJ, Nagel RL, Rybicki AC. Blood. 1992;79:1846–1854. [PubMed] [Google Scholar]
- 35.Rybicki AC, Heath R, Wolf JL, Lubin B, Schwartz RS. J Clin Invest. 1998;81:893–901. doi: 10.1172/JCI113400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willardson BM, Thevenin BJ, Harrison ML, Kuster WM, Benson MD, Low PS. J Biol Chem. 1989;264:15893–15899. [PubMed] [Google Scholar]
- 37.Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 38.Nemeth MJ, Cline AP, Anderson SM, Garrett-Beal LJ, Bodine DM. Blood. 2005;105:627–634. doi: 10.1182/blood-2004-07-2551. [DOI] [PubMed] [Google Scholar]
- 39.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2003. [Google Scholar]
- 40.Dacie JV, Lewis SM, Luzzatto L. Practical Haematology. New York: Churchill Livingstone; 1995. pp. 234–238. [Google Scholar]
- 41.Bennett V. Methods Enzymol. 1983;96:313–324. doi: 10.1016/s0076-6879(83)96029-9. [DOI] [PubMed] [Google Scholar]