Why is Drosophila a good model of cardiac physiology?
The fly heart has been an excellent model of cardiovascular development for over a decade. Since the discovery of the homeobox transcription factor tinman (Azpiazu and Frasch 1993, Bodmer 1993, Bodmer et al. 1990) and the recognition that it is conserved in vertebrates [reviewed in (Bodmer 1995, Harvey 1996)], more and more evidence has corroborated the idea that much of the regulatory genetic network controlling the specification and differentiation of the heart is conserved from flies to mammals [reviewed in (Bodmer and Frasch 1999, Bodmer et al. 2005, Cripps and Olson 2002, Zaffran and Frasch 2002) ], laying the ground work for molecular models of congenital heart disease in humans [reviewed in (Chien and Olson 2002, Prall et al. 2002, Seidman and Seidman 2002, Olson 2004, Srivastava 2006)]. Given the remarkable conservation of molecular and embryological mechanisms underlying cardiogenesis in the animal kingdom, it seems plausible that the genetic control of heart function may also be conserved. Clearly, many proteins that carry out cardiac function, such as ion channels and contractile proteins, are highly conserved (reviewed in Bodmer et al. 2005): contributors to excitation-contraction coupling, such as the ryanodine receptor, SERCA, myosin, troponin, and ion channels likely to be involved in pacemaking, such as Ih/HCN (Monier et al. 2005), are all present in fly cardiomyocytes. Also, plasma membrane invag inations forming T tubules have been observed in the fly’s heart, much like in vertebrates, and the mononucleate cardiomyocytes that comprise the heart tube are electrically connected by GAP junctions formed by innexin proteins in invertebrates. Thus, it is conceivable that the way these conserved proteins function within the mature heart to ensure a normal heartbeat has also evolved from a common evolutionary design that was in place prior to the invertebrate-vertebrate split.
In the following we review recent advances in elucidating the genetics of cardiac function and aging in Drosophila and propose that the control of the cardiac physiology and rhythmicity is conserved between in many ways vertebrates and invertebrates. As a consequence, the fly heart is a potentially useful genetic model not only for understanding congenital heart disease that causes developmental defects, but also for understanding functional abnormalities, such as cardiomyopathies and arrhythmias (Bier and Bodmer 2004).
Modulation of the heart rate
During larval stages, the Drosophila cardiovascular system is composed of a dorsal vessel containing 52 pairs of cardiomyocytes that display both endothelial and muscle cell characteristics. The cardiomyocytes form a tubular structure, flanked by pericardial cells. The organ can be divided into two distinct morphological and functional domains along the anterior-posterior (A/P) axis: the aorta, which constitutes the outflow tract, and the ‘heart’ itself, which is the only domain capable of spontaneous rhythmic activity (Figure 1). The heart, whose automatic myogenic activity generates the basic cardiac rhythm, ensures the pumping of the hemolymph in an open circulatory system. This relatively simple organization, coupled with sophisticated genetic tools available in Drosophila, provides a powerful model for elucidating the basic mechanisms of heart rhythm (pacemaking) and contractility. Mutations in a number of ion channels and transporters have been found to alter larval heart rate, including in SERCA, the Sarco-endoplasmic reticulum Ca2+-ATPase, and in the Ca2+-channel encoded by cacophony (Ray and Dowse 2005, Sanyal et al. 2006). Moreover, vertebrate homologs of potassium channels encoded by either-a-gogo (eag) and KCNQ are responsible for repolarization of the cardiac action potential (Kr/HERG and Ks/KCNQ; for review see Sanguinetti and Tristani-Firouzi, 2006) , and eag and KCNQ mutants in Drosophila also perturb proper heart function, further underlining the remarkable functional parallels in basic cardiac physiology between flies and humans (Johnson et al. 1998; Ocorr et al. 2007; for review see Bodmer et al. 2005). Investigation of ion channel expression, their cardiac-specific functions and detailed contractile properties of the Drosophila heart has become more readily possible in recent years, with the advent of new experimental techniques applied to this small organism.
Figure 1.
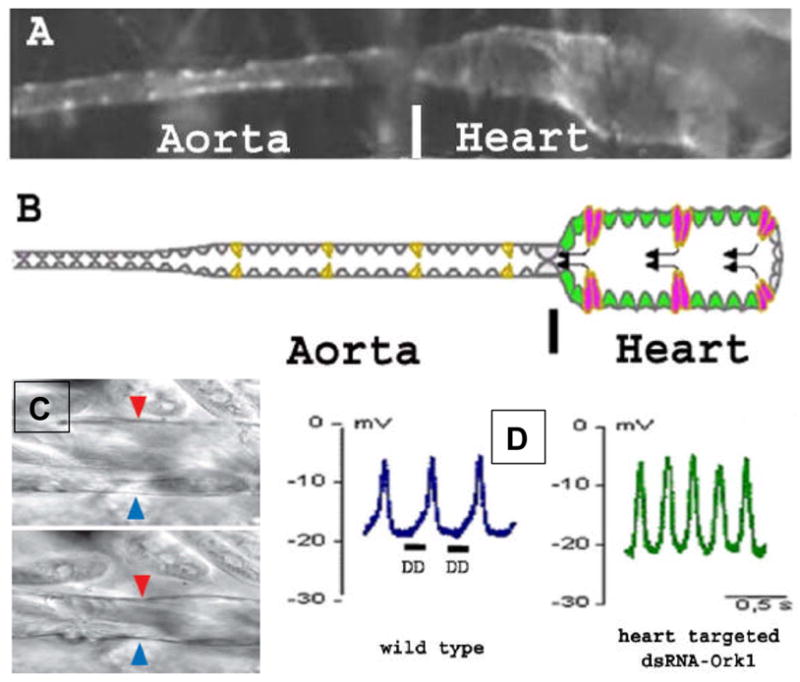
(A) Cardiac-specific expression of GFP (1029-Ga4/UAS-GFP) in semi-dissected Drosophila larvae outlines the morphology of the heart. Posterior is to the right. GFP allows monitoring the shape and function of the organ in living individuals. The thinner anterior portion is termed the aorta and the thicker posterior portion the heart proper, which displays automatic contraction activity. (B) Schematic drawing of the larval cardiac tube consisting of 104 cardiac myocytes. In the heart, cardiomyocytes expressing the orphan nuc lear receptor encoded by seven-up develop into functional ostia (in pink) which constitute the hemolymph inflow tracts. The heart’s tinman- expressing cardiomyocytes also express Ork1, HCN and NDAE1 and develop into autonomously beating cardiomyocytes (in green). Black arrows indicate the direction of the blood flow. (C) Micrograph of a segmental porti on of the heart in diastole (top) and systole (bottom). Arrowheads indicate the (outer) heart wall. (D) Action potentials (AP) recorded in larval automatic cardiomyocytes of a wildtype heart (left) or a heart with cardiac-specific dsOrk1 knockdown (right, 1029Gal4/UAS-ORK1-RNAi6.3). In the wild type, larval APs are characterized by a slow diastolic depolarization (DD) preceding the fast depolarization. As invertebrate cardiac APs in general, these relatively small APs are due to a relatively high potassiu m concentration of insect hemolymph (25mM). Upon cardiac ORK1 overexpression the heartbeat stops and the membrane potential is recorded at about -30mV, which drops to beyond -70mV in 5mK potassium concentration in artificial hemolymph (for details see Lalevée et al. 2006). In contrast, Ork1 knockdown seems to accelerate the DD phase (presumably due to a reduction in the outward K+ current), and thus the heart rate is increased (adapted from reference Lalvéee, et al, 2006).
The recording of intracellular action potentials in the larval heart with sharp glass microelectrodes has been achieved by using a floating electrode to record from the fly heart even as it contracts (Figures 1 C,D; Lalevée et al. 2006). Typical pacemaker action potentials were observed, with a slow diastolic depolarization preceding the fast depolarization which triggers contraction (Figure 1D). The size of the recorded action potential is typical for invertebrates, which tend to be relatively small (for details see legend of Fig. 1). The shape of the action potential is, however, reminiscent of that recorded in the mammalian sino-atrial node rather than in the ventricle. This type of action potential including the slow diastolic depolarization or pacemaker potential is recorded all along the heart portion of the fly’s dorsal vessel, suggesting that the larval population of cardiac myocytes may behave as both working and pacemaker cardiomyocytes.
To identify additional genes potentially involved in modulating cardiac activity, a screen for genes that are specifically expressed in the contractile myocytes of the heart but not in the aorta cardiomyocytes was carried out (L.P. and M. Sémériva, unpublished). Among them are the Na+-dependent bicarbonate anion exchanger 1 (NDAE1), the hyperpolarization cyclic nucleotide-gated channel (HCN, possibly generating the cardiac pacemaker Ih or If current), and the Outwardly Rectifying Potassium channel 1 (ORK1, a two-pore domain potassium (K2P) channel (Lalevée et al. 2006, Monier et al. 2005, Perrin et al. 2004).
Investigation of the role of ORK1 in cardiac physiology revealed that cardiac-specific inactivation of Ork1 leads to an increase in heart rate (Lalevée et al. 2006). By contrast, ORK1 overexpression in the heart abolishes the heartbeat. Interestingly, larvae without a heartbeat were alive but did not produce any adult flies, which suggests that regular heart contraction may not be absolutely necessary for larval survival. The waveform of action potentials recorded from ORK1-RNAi knockdown hearts (Figure 1C) suggests that the level of Ork1 activity sets the cardiac heartbeat frequency by controlling the duration of the slow diastolic depolarization phase. It is conceivable that lowering ORK1 activity diminishes an outward K+ current leading to an accelerated depolarization, while ORK1 overexpression induces K+ mediated hyperpolarization. These observations identify a new mechanism for cardiac rhythm control and suggest that K2P channels influence cardiac pacemaker activity. Since K2P channel activity is known to be regulated by many inputs, including neurotransmitters, hormones and anaesthetics, the discovery that ORK1 functions as a regulator of heart rate in Drosophila suggests that K2P channels may function similarly in vertebrates.
Even though the larval heart is a relatively simple organ, its myocardial cells in Drosophila display several hallmarks of pacemaker cardiomyocytes, suggesting that this bug may indeed serve as a useful model for genetic analysis of heart function, including cardiac pacemaking and regulation of heart rate. In addition, while vertebrate genes are usually present in multiple copies, most Drosophila genes exist as single copies. This lack of genomic redundancy allows gene function to be rigorously assessed. For example, HCN channels are encoded by four different genes in mammals, whereas in Drosophila, there is only one HCN gene, expressed specifically in the heart portion of the dorsal vessel (Monier et al. 2005). The fact that normal heart function is not essential for larval viability provides an enormous advantage for analysis of severe abnormalities and for conducting large scale genetic screens to identify new genes involved in cardiac function.
Modulation of the heart rhythm
New methods for investigating the heart function in organisms that are too small for conventional heart analysis have recently been developed that promise to dramatically expand the possibilities for screening mutants and for detailed analysis of the cardiac contractility in Drosophila. One method uses a non-contact echography, optical coherence tomography (OCT), to image the heart contractions in an ultrasound- like fashion visualizing the dynamics of the heart lumen (Wolf et al. 2006). Although somewhat limited in its spatial resolution, this non-invasive method seems to be applicable for high throughput screening and to investigate ‘dilated heart’ phenotypes. Interestingly, troponin I and tropomyosin mutants or flies with cardiac overexpression of a mutant cardiomyopathic form of sarcoglycan exhibited systolic dysfunction phenotypes reminiscent of the corresponding dilated cardiomyopathy syndromes in humans (Wolf et al. 2006).
For more detailed analysis of the cardiac contractions it is necessary to surgically expose the fly’s heart in order to make it accessible for high-speed, high-resolution video microscopy (Figure 2A). A semi-intact fly preparation was recently developed, which is well-suited to study cardiac arrhythmia, as well as other parameters of cardiac contractions (Ocorr et al. 2007). Movies of exposed heart tube preparations bathed in oxygenated, physiological saline, allow monitoring the movement of the edges of the heart at various locations along the heart tube using specialized software (developed by M. Fink). To do this a specified region of one pixel width spanning the diameter of the heart is electronically “cut” out of each frame of the movie (Figure 2A) by the softw are and aligned horizontally, creating an image montage that describes the movement of the heart walls over time (Figure 2B). The resulting image is similar to the M-mode of cardiac ultrasound measurements in humans. Because these movies are taken at speeds up to 200 frames per second, the M-modes are able to track even very rapid movements during contraction. Remarkably, as the fly ages the rhythmicity of its heartbeat is less and less regular with increased episodes of arrhythmias [Figure 2B; (Ocorr et al. 2007) ]. The observed changes in cardiac rhythmicity with age are reminiscent of the increased incidence of atrial fibrillation and heart failure in the aging human population [reviewed in (Lakatta and Levy 2003)]. Thus, the regular cardiac rhythm not only declines with age in humans but also in flies, suggesting that the heart in Drosophila could serve as a useful genetic model. This semi-intact fly preparation has also been used to measure tension generated by the cardiac muscle using carbon fibers (Ocorr et al. 2007).
Figure 2.
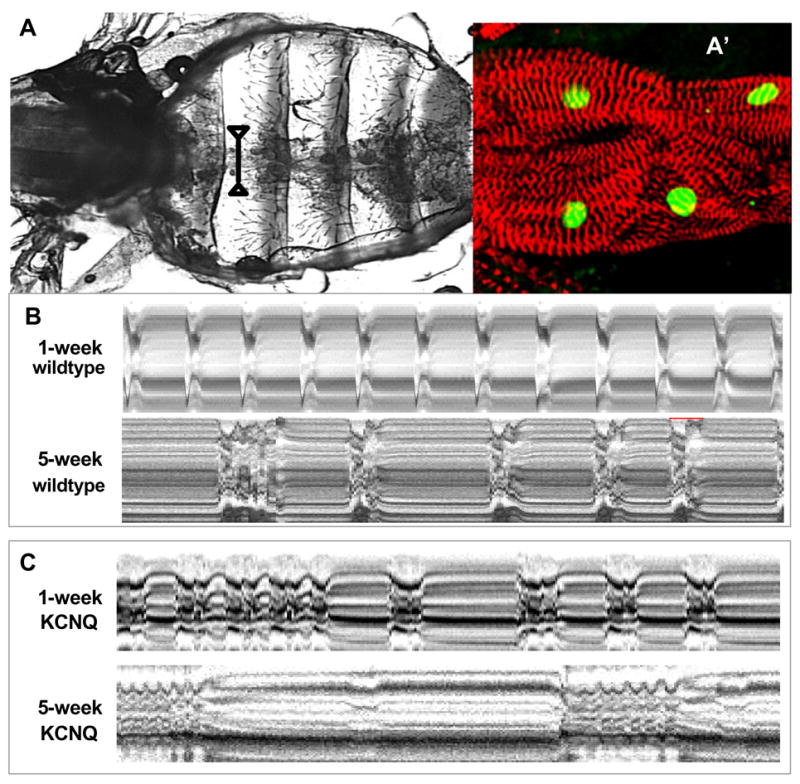
(A) Semi-intact heart preparation where the abdominal heart is exposed by removing the head, ventral thorax, gut, and fat bodies. The exposed heart can beat for hours in oxygenated, artificial hemolymph. The line between arrow heads indicates the 1 pixel-wide portion of the movie frame used to generate the M-mode traces shown in B and C. (A’) A portion of the heart around the region indicated in (A) is double-stained for actinin (red) labeling Z-lines of myofibers within cardiomyocytes and for neuromancer (H15-lacZ, green) labeling cardiomyocyte nuclei). (B) M-mode traces prepared from high speed movies of dissected flies. A 1 pixel-wide region with both edges of the heart tube is defined in a single movie frame. Same regions are electronically cut from all of the frames in the movie and aligned horizontally to produce the trace. Arrhythmic heart beats are evident in KCNQ mutants as early as 1 week of age. The incidence and severity of arrhythmias increase with age in all flies but is consistently more in KCNQ mutants than in wildt-ype (wt). For quantitative analysis, see Ocorr et al. (2007).
Mutations in human KCNQ genes, encoding the pore-forming subunits underlying Ks I , cause type 1 long QT syndrome (LQT1) and are associated with an increased risk for torsades des pointes (TdP) arrhythmias and sudden death (Jentsch 2000, Towbin 2004, Sanguinetti and Tristani-Firouzi 2006). There is only one copy of the KCNQ gene found in Drosophila genome (Wen et al. 2005) and deletion mutants of the single Drosophila KCNQ gene are viable but show a 3-fold higher than normal susceptibility to pacing-induced heart failure [see below; and (Ocorr et al. 2007)]. Recent investigation of the heart rhythm of KCNQ mutants revealed that Drosophila, the only invertebrate genetic model organism with a heart (Bier and Bodmer 2004), may also be a surprisingly good model for studying the genetics of arrhythmias, as a result of both mutations and aging. High-speed video recording and display of heart wall movements in M-mode show episodes of prolonged heart contraction and fibrillation in KCNQ mutants, and this phenotype gets progressively more severe as the flies age [Figure 2C; (Ocorr et al. 2007)]. The fibrillations observed in mutant flies are likely due to delayed relaxation of the myocardium, as revealed by increases in the duration of phasic contractions, extracellular field potentials and systolic tension. In addition to the KCNQ channel, mutations in the human ether-a-gogo related gene (HERG), which encode subunits of channels underlying the cardiac IKr current, also affect human heart function and lead to the LQT2 and most drug-induced LQT syndromes. Remarkably, mutants or heart-specific knockdown of HERG-related fly genes als o exhibit a significantly elevated frequency of arrhythmias (K.O. and R.B., unpublished). These findings suggest that flies can develop congenital and age-dependent arrhythmias and the underlying causes may be similar to those that produce arrhythmias in mammals.
Transcription factors in cardiac function
Defects in a number of transcription factor genes required for specification and differentiation of diverse cardiac lineages have been associated with human congenital heart disease (CHD) . Recent evidence suggests that these cardiogenic transcription factors are also involved in causing heart function defects that are manifest primarily in adults. For example, null mutants in the fly’s cardiac determinant tinman give rise to severe developmental defects and lack most dorsal mesodermal derivatives, including the cardiac primordium. In contrast, mouse knockout of the tinman homolog Nkx2–5 or double knockout of Nkx2–5/2–6 develop almost normally until the linear heart tube stage (Lyons et al. 1995, Tanaka et al. 1999, 2001). This apparent discrepancy between flies and mammals was recently resolved by investigating the specific requirement of tinman only within the cardiac progenitors, as opposed to its role in the entire early mesoderm. These cardiac progenitors are the equivalent of the mammalian cardiac crescent that expresses the cardiac Nkx genes and gives rise to myocardium. In order to remove the heart progenitor -specific function of tinman in Drosophila, the enhancer responsible for ‘late’ cardiac-only tinman expression was selectively eliminated in a genomic rescue construct in a tinman null mutant background. These ‘cardiac tinman’-deficient flies are viable and do form a heart tube initially but have a reduced lifespan, and importantly, exhibit severely compromised adult cardiac morphology and function (Zaffran et al. 2006). This suggests that in addition to its cardiogenic role in the early mesoderm, tinman is also required for proper cardiac morphogenesis and differentiation, as is the case in mammals, again underlying the unified principles of organogenesis. Further supporting this notion is the finding that haploinsufficient human and late knockout mouse mutations in Nkx2–5 cause atrial-septal defects during development, and in addition cause disruption of the conduction system in the adult heart (Schott et al. 1998; Pashmforoush et al. 2004).
A future challenge will be the exploration of polygenic modifiers of these CHD syndromes that apparently influence dramatically the expression of the heart disease phenotype. Drosophila is particularly powerful as a genetic animal model to investigate genetic interactions, and is ideally suited to test the influence of polygenic variants relevant to disease genes. Indeed, there seems to be an emerging genetic network involving homeobox, T-box and GATA factor genes that is responsible in establishing or maintaining proper cardiac function (L.Q. and R.B., unpublished), in addition to their established role in development. It is possible that similar interactions also exist in the mammalian adult heart.
Cardiac aging
Cardiac aging can be regarded as a progressive decline in heart function that is intrinsic to the organ itself and that correlates with the age of the organism. Most elderly adults are much more concerned about preserving their functionality later in life rather than simply extending their lifespan (Phelan et al. 2004). For this reason the study of functional senescence of the heart as distinguished from its effect on global mortality becomes important. Some genes that specifically influence the functional status of the organ may not affect organismal survival. It will be interesting to distinguish between those processes that regulate lifespan from those that control age-dependent decline in individual organ systems.
One of the most notable phenotypes of cardiac senescence in humans and rodents is the reduction in maximum heart rate that can be achieved under stress (Lakatta 2001). This phenomenon of impaired cardiovascular acceleration during stress is a biomarker of mammalian cardiac aging, but its genetic basis is not well-determined, partly owing to the complex interactions between genes, age, disease and lifestyle (Lakatta 2001). An initial understanding of the interactions may only be possible by examining cardiac aging using simpler genetic animal models.
In this regard, Drosophila may be uniquely situated for studies of cardiac aging, since it is the only one among other simple genetic models that possesses a fluid pumping heart (Bier and Bodmer 2004). Because the survival of the organism is not as tightly coupled to heart function as it is in vertebrates it will be possible to investigate this organ’s senescence in relative independence of organismal senescence. Moreover, the maximal heart rate of Drosophila under stress conditions, such as elevated ambient temperature and external electrical pacing, is also significantly and reproducibly reduced with aging, analogous to observations in elderly humans (Paternostro et al. 2001, Wessells et al. 2004). Thus, Drosophila with its versatile genetic tools can also be exploited for identifying and studying genes involved in cardiac functional senescence. Again, the insights from these studies are likely to significantly impact the field of human cardiac pathology and aging.
Several genes and pathways have already been identified that play a crucial role in fly cardiac aging. The Drosophila SUR gene (dSUR) encodes a subunit of the ATP- sensitive potassium channel (KATP). which plays important roles in various cellular processes by coupling cell metabolism to electrical activity (Akasaka et al. 2006, Seino 1999). The expression of dSUR in the heart is dramatically diminished in the aging heart, and RNAi-mediated knockdown of dSUR in young fly hearts phenocopies aged hearts under conditions of pacing-induced stress (Akasaka et al. 2006). These results provide evidence for a role of dSUR in protecting against declining performance during cardiac aging. Mutations in human SUR2, the mammalian homolog of dSUR, result in cardiomyopathy by compromising K ATP channel function (Bienengraeber et al. 2004), and it is possible that a reduction in K ATP channel activity could cause membrane electrical instability, especially in older hearts (Akasaka et al. 2006). This suggests that similar to the fly’s dSUR, human SUR2 may also protect against cardiac senescence.
The insulin/insulin-like growth factor (IGF) signaling is a well-established genetic pathway that regulates longevity (Kenyon 2001, Tatar et al. 2003). Drosophila mutants of insulin-like receptor (InR) and chico (encoding the insulin receptor substrate) extend the lifespan of the organism (Clancy et al. 2001, Tatar et al. 2001) as well as protect their hearts from two age-related phenomena: decreases in resting heart rate and increases in heart failure resulting from pacing-induced stress (Wessells et al. 2004). Additionally, interfering with InR signaling exclusively in the heart, by overexpressing the phosphatase dPTEN (a negative regulator of insulin/IGF signaling) or the forkhead transcription factor dFOXO (a negatively-regulated target of insulin/IGF signaling), prevents the decline in cardiac fitness with age (Wessells et al. 2004). These data indicate that insulin-like signaling is involved in both systemic as well as cardiac -specific senescence. Moreover, the ablation of insulin-producing cells (IPCs) in flies also slows demographic aging and reduces age-dependent heart failure (Wessells et al. 2004), indicating that both a reduction of insulin receptor signaling and circulating insulin levels influence organismal aging and age-related cardiac susceptibility to pacing stress. This is consistent with the observations that overexpressing dFOXO in the adult fat body results in long-lived flies (Giannakou et al. 2004, Hwangbo et al. 2004), whereas cardiac-specific overexpression of InR and dPTEN do not seem to affect the overall lifespan of the animal (Wessells, et al., 2004). Lifespan-altering manipulation of insulin/IGF signaling specifically in the fat body of the head also decreases insulin production from the IPCs, thus affecting longevity and possibly organ senescence indirectly, which is suggestive of a complex coordination of aging with organ-autonomous endpoint effects (Hwangbo et al. 2004, Wessells et al. 2004).
Interestingly, upregulating the expression level of dFOXO in the adult fat body protects against the oxidative stressor paraquat (Hwangbo et al. 2004), raising the possibility that insulin-like signaling could, at least in part, delay cardiac performance senescence via reducing oxidative stress damage. However, in the chico mutants with extended lifespan (Clancy et al., 2001) and lowered cardiac aging (Wessells et al. 2004), there seems to be no alteration in their resistance to oxidative stress (Clancy et al. 2001). Thus, the contribution of oxidative damage to heart functional aging remains to be further examined.
Another example of how alteration in energy homeostasis could be coupled to aging and organ senescence is illustrated by manipulations of the Drosophila Target of Rapamycin (dTOR) pathway. In addition to its well-studied role in nutrient sensing and cellular growth in response to insulin/IGF and PI3K, TOR signaling is also involved in global lifespan regulation in model organisms. A recent study further showed that lowering TOR activity in Drosophila prevents age-dependent functional decline of heart performance, and this could be attributed to a reallocation of energy stores preferentially for the regulation of “long term” responses such as lifespan and organ maintenance (Luong et al., 2006). Collectively, these findings support the notion that insulin/TOR signaling plays an important role in modulating organismal and cardiac aging, and it remains to be determined whether TOR regulates cardiac functional senescence via its known downstream effectors such as S6K and 4E-BP, or through some other novel factors. Given the high degree of parallel mechanisms controlling organismal aging and lifespan, it is likely that the mechanisms underlying cardiac functional aging in flies are also conserved, and will have significant implications on mammalian heart aging and physiology.
Acknowledgments
We thank Natalie Lalevée for helpful discussions and kindly providing the pictures for the electrophysiology traces in Figure 1. This work was supported by the American Heart Association (L.Q.) and by NHLBI and NIA of the National Institutes of Health (R.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akasaka T, Klinedinst S, Ocorr K, Bustamante EL, Kim SK, Bodmer R. The ATP-sensitive potassium (KATP) channel-encoded dSUR gene is required for Drosophila heart function and is regulated by tinman. Proc Natl Acad Sci U S A. 2006;32:11999–2004. doi: 10.1073/pnas.0603098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu N, Frasch M. Tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7B:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- Bienengraeber M, Olson TM, Selivanov VA, Kathmann EC, O'Cochlain F, Gao F, et al. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat Genet. 2004;4:382–387. doi: 10.1038/ng1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E, Bodmer R. Drosophila, an emerging model for cardiac disease. Gene. 2004;342:1–11. doi: 10.1016/j.gene.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:1301–1306. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- Bodmer R. Heart development in Drosophila and its relationship to vertebrate systems. Trends in Cardiovascular Medicine. 1995;5:21–27. doi: 10.1016/1050-1738(94)00032-Q. [DOI] [PubMed] [Google Scholar]
- Bodmer R, Jan LY, Jan YN. A new homeobox-containing gene, msh-2, is transiently expressed early during mesoderm formation of Drosophila. Development. 1990;3:661–9. doi: 10.1242/dev.110.3.661. [DOI] [PubMed] [Google Scholar]
- Bodmer R, Wessells RJ, Johnson EC, Dowse H. Heart development and function. In: Gilbert LI, Iatrou K, Gill S, editors. Comprehensive Molecular Insect Science. 1–7. Vol. 2. Elsevier; 2005. pp. 199–250. [Google Scholar]
- Chien KR, Olson EN. Converging pathways and principles in heart development and disease: CV@CSH. Cell. 2002;2:153–62. doi: 10.1016/s0092-8674(02)00834-6. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;5514:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol. 2002;1:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Junger MA, Hafen E, Leevers SJ, Partridge L. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;5682:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- Harvey RP. NK-2 homeobox genes and heart development. Dev Biol. 1996;2:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- Hwangbo DS, Gersham B, Tu MP, Palmer M, Tatar M. Regulation of aging and neuronal insulin transcription by dFOXO in fat body of adult Drosophila. Nature. 2004;480:652–656. [Google Scholar]
- Jentsch TJ. Neuronal KCNQ potassium channels: physiology and role in disease. Nat Rev Neurosci. 2000;1:21–30. doi: 10.1038/35036198. [DOI] [PubMed] [Google Scholar]
- Johnson E, Ringo J, Bray N, Dowse H. Genetic and pharmacological identification of ion channels central to the Drosophila cardiac pacemaker. J Neurogenet. 1998;12:1–24. doi: 10.3109/01677069809108552. [DOI] [PubMed] [Google Scholar]
- Kenyon C. A conserved regulatory system for aging. Cell. 2001;2:165–168. doi: 10.1016/s0092-8674(01)00306-3. [DOI] [PubMed] [Google Scholar]
- Lakatta EG. Heart aging: a fly in the ointment? Circ Res. 2001;10:984–986. doi: 10.1161/hh1001.091963. [DOI] [PubMed] [Google Scholar]
- Lakatta EG, Levy D. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: Part II: the aging heart in health: links to heart disease. Circulation. 2003;2:346–354. doi: 10.1161/01.cir.0000048893.62841.f7. [DOI] [PubMed] [Google Scholar]
- Lalevee N, Monier B, Senatore S, Perrin L, Semeriva M. Control of cardiac rhythm by ORK1, a Drosophila two-pore domain potassium channel. Curr Biol. 2006;15:1502–1508. doi: 10.1016/j.cub.2006.05.064. [DOI] [PubMed] [Google Scholar]
- Luong N, Davies C, Wessells R, Graham S, King M-T, Veech R, et al. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metabolism. 2006;4:133–142. doi: 10.1016/j.cmet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev. 1995;13:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Monier B, Astier M, Semeriva M, Perrin L. Steroid-dependent modification of Hox function drives myocyte reprogramming in the Drosophila heart. Development. 2005;23:5283–5293. doi: 10.1242/dev.02091. [DOI] [PubMed] [Google Scholar]
- Ocorr K, Reeves N, Wessells R, Fink M, Chen H-SV, Akasaka T, et al. KNCQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. PNAS. 2007;103:3943–3948. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. A decade of discoveries in cardiac biology. Nat Med. 2004;5:467–474. doi: 10.1038/nm0504-467. [DOI] [PubMed] [Google Scholar]
- Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, et al. Nkx2–5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- Paternostro G, Vignola C, Bartsch DU, Omens JH, McCulloch AD, Reed JC. Age-associated cardiac dysfunction in Drosophila melanogaster. Circ Res. 2001;10:1053–1058. doi: 10.1161/hh1001.090857. [DOI] [PubMed] [Google Scholar]
- Perrin L, Monier B, Ponz ielli R, Astier M, Semeriva M. Drosophila cardiac tube organogenesis requires multiple phases of Hox activity. Dev Biol. 2004;2:419–431. doi: 10.1016/j.ydbio.2004.04.036. [DOI] [PubMed] [Google Scholar]
- Phelan EA, Anderson LA, LaCroix AZ, Larson EB. Older adults' views of "successful aging"--how do they compare with researchers' definitions? J Am Geriatr Soc. 2004;2:211–216. doi: 10.1111/j.1532-5415.2004.52056.x. [DOI] [PubMed] [Google Scholar]
- Prall OW, Elliott DA, Harvey RP. Developmental paradigms in heart disease: insights from tinman. Ann Med. 2002;3:148–156. [PubMed] [Google Scholar]
- Ray VM, Dowse HB. Mutations in and deletions of the Ca2+ channel-encoding gene cacophony, which affect courtship song in Drosophila, have novel effects on heartbeating. J Neurogenet. 2005;1:39–56. doi: 10.1080/01677060590953066. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Jennings T, Dowse H, Ramaswami M. Conditional mutations in SERCA, the Sarco-endoplasmic reticulum Ca(2+)-ATPase, alter heart rate and rhythmicity in Drosophila. J Comp Physiol [B] 2006;3:253–263. doi: 10.1007/s00360-005-0046-7. [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, et al. Congenital heart disease caused by mutations in the transcription factor NKX2–5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440:463–469. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- Seidman CE, Seidman JG. The coming of age of cardiovascular science. Cold Spring Harb Symp Quant Biol. 2002;67:543–550. doi: 10.1101/sqb.2002.67.543. [DOI] [PubMed] [Google Scholar]
- Seino S. ATP-sens itive potassium channels: a model of heteromultimeric potassium channel/receptor assemblies. Annu Rev Physiol. 1999;61:337–362. doi: 10.1146/annurev.physiol.61.1.337. [DOI] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;6:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Schinke M, Liao HS, Yamasaki N, Izumo S. Nkx2.5 and Nkx2.6, homologs of Drosophila tinman, are required for development of the pharynx. Mol Cell Biol. 2001;13:4391–4398. doi: 10.1128/MCB.21.13.4391-4398.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;5514:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Towbin JA. Molecular genetic basis of sudden cardiac death. Pediatr Clin North Am. 2004;5:1229–1255. doi: 10.1016/j.pcl.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Wen H, Weiger TM, Ferguson TS, Shahidullah M, Scott SS, Levitan IB. A Drosophila KCNQ channel essential for early embryonic development. J Neurosci. 2005;44:10147–10156. doi: 10.1523/JNEUROSCI.3086-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessells RJ, Fitz gerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nat Genet. 2004;12:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- Wolf MJ, Amrein H, Izatt JA, Choma MA, Reedy MC, Rockman HA. Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci U S A. 2006;5:1394–1399. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran S, Frasch M. Early signals in cardiac development. Circ Res. 2002;6:457–469. doi: 10.1161/01.res.0000034152.74523.a8. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Reim I, Qian L, Lo PC, Bodmer R, Frasch M. Cardioblast-intrinsic Tinman activity controls proper diversification and differentiation of myocardial cells in Drosophila. Development. 2006;20:4073–4083. doi: 10.1242/dev.02586. [DOI] [PubMed] [Google Scholar]


