Abstract
Early steps of embryo development are directed by maternal gene products and trace levels of zygotic gene activity in vertebrates. A major activation of zygotic transcription occurs together with degradation of maternal mRNAs during the midblastula transition in several vertebrate systems. How these processes are regulated in preparation for the onset of differentiation in the vertebrate embryo is mostly unknown. Here, we studied the function of TATA-binding protein (TBP) by knock down and DNA microarray analysis of gene expression in early embryo development. We show that a subset of polymerase II-transcribed genes with ontogenic stage-dependent regulation requires TBP for their zygotic activation. TBP is also required for limiting the activation of genes during development. We reveal that TBP plays an important role in the degradation of a specific subset of maternal mRNAs during late blastulation/early gastrulation, which involves targets of the miR-430 pathway. Hence, TBP acts as a specific regulator of the key processes underlying the transition from maternal to zygotic regulation of embryogenesis. These results implicate core promoter recognition as an additional level of differential gene regulation during development.
Keywords: maternal mRNA, MBT, TBP, transcription, zebrafish
Introduction
In most animal models, including Drosophila, Caenorhabditis elegans, zebrafish and Xenopus, the onset of zygotic gene activation is delayed until the midblastula transition (MBT). (Newport and Kirschner, 1982; Kimmel et al, 1995). Whereas there is no MBT in mammals, here zygotic gene activity is also delayed after fertilisation (Thompson et al, 1998). In the zebrafish blastula, the general delay in zygotic gene activity is followed by the sudden and broad activation of a large number of genes representing all main gene ontologies (Kane and Kimmel, 1993; Mathavan et al, 2005) leading to gastrulation. The activation of the zygotic genome is parallelled by an equally significant process, the differential degradation of maternally inherited mRNAs (Giraldez et al, 2005; Mathavan et al, 2005; De Renzis et al, 2007). Whereas little is known about the mechanisms of degradation of maternal mRNA, they are known to involve both transcription-dependent and -independent pathways (Bashirullah et al, 1999; Audic et al, 2001; Giraldez et al, 2005; Schier, 2007). Dynamic changes in expression of maternally and zygotically activated genes are observed during zygotic gene activation also in the mouse (Wang et al, 2004). Not all maternally inherited mRNAs degrade during early embryogenesis and many maternal mRNAs continue to influence embryo development until later developmental stages (Wagner et al, 2004; reviewed by Pelegri (2003)).
The initiation of zygotic transcription during MBT is believed to be regulated by a competition between chromatin and the assembly of the transcription machinery (Newport and Kirschner, 1982; Kimelman et al, 1987; Almouzni and Wolffe, 1995). The TATA-binding protein (TBP) has been implicated as a key regulator of transcription initiation in early embryo development in vertebrates (Veenstra et al, 2000; Müller et al, 2001; Martianov et al, 2002). TBP protein levels have been shown to be limiting for transcription before MBT and are dramatically upregulated at the initiation of zygotic transcription (Prioleau et al, 1994; Veenstra et al, 1999; Bártfai et al, 2004). TBP, together with TBP-associated factors (TAFs) are components of the TFIID complex, a key point at which activators can control transcription through the core promoter. Until recently, it was argued that TBP is required for the correct initiation of all RNA polymerase (Pol I, II and III)-mediated transcription in eukaryotes. However, recent reports have revealed the contrary: the composition of Pol II core promoter-binding complexes varies and is likely to represent a point of differential gene expression regulation (reviewed by Davidson (2003)). Consistently, whereas TBP is essential for early embryo development, it is not required for all Pol II transcription as demonstrated by studies on a small number of vertebrate genes (Veenstra et al, 2000; Müller et al, 2001; Martianov et al, 2002). The apparent redundancy of TBP in vertebrates is probably due to the function of TBP-like factors (TLF/TRF2) (Veenstra et al, 2000; Müller et al, 2001) and the recently described second set of TBP paralogue genes TBP2/TRF3 (Persengiev et al, 2003; Bártfai et al, 2004; Jallow et al, 2004). The functional requirement for different TBP family proteins in embryogenesis suggests specific nonoverlapping roles for these factors in regulating subsets of genes (Moore et al, 1999; Teichmann et al, 1999; Bártfai et al, 2004; Jallow et al, 2004).
Our objective was to investigate the transcriptional regulatory mechanisms that involve core promoter recognition proteins such as TBP in the whole organism. The transition of gene activity from maternally inherited mRNAs to zygotic gene expression provides an ideal model for the analysis of the control of transcription initiation (Newport and Kirschner, 1982). By using Morpholino (MO) knockdown and microarray expression profiling, we have addressed which genes require TBP for their activity and what is the function of TBP in regulating the transition from maternal to zygotic regulation during early vertebrate embryo development. We show that TBP is preferentially required for genes that exhibit dynamic changes in their expression during ontogeny. Furthermore, we provide evidence for a previously undocumented negative regulatory role of TBP in zygotic gene activation. Importantly, we also describe a novel biological function of TBP: a role in the degradation of a subset of maternal mRNAs after MBT.
Results
TBP regulates specifically a subset of mRNAs in the dome-stage embryo
In the early embryo, the steady-state levels of mRNA result from a dynamic process of gradual degradation of maternal mRNAs and the delayed initiation of zygotic gene expression at the MBT (Figure 1A). To investigate the role of TBP in regulating genes expressed in the early zebrafish embryo, we carried out a microarray analysis of 10 501 genes at the dome stage in embryos in which TBP function was blocked using MO antisense oligonucleotides as described previously (Müller et al, 2001). The dome stage occurs 1.3 h after the start of the global initiation of zygotic transcription at the MBT (Kane and Kimmel, 1993) and any TBP-dependent changes in gene activity detected at this stage are still expected to be mostly direct transcriptional effects. Loss of protein was confirmed by Western blot (Figure 1B). A total of 1927 genes from the 10 501 represented on the microarray were selected, having applied stringent but commonly used criteria (FDR cut off of 0.05) to eliminate potential false positives and false negatives from the analysis (see Materials and methods). Three distinct response groups of genes were identified: downregulated genes (⩽−2-fold change), genes with low variability in expression (values between >−2 and <+2-fold-change); upregulated genes (⩾+2-fold change) (Figure 1C and Supplementary Table I). The three groups thus identified were further validated by semiquantitative RT–PCR experiments; out of a total of 39 genes representing the above groups, 37 showed comparable activity to that observed in the microarray experiments (Supplementary Figure S1). The specificity of the effects detected was confirmed by microinjecting a second MO targeting TBP mRNA (TBP MO2), which resulted in comparable gene expression changes to the above TBP MO injection when analysed by RT–PCR of 28 genes (Supplementary Figure S2). Furthermore, the gene expression changes caused by TBP MO injection could be reverted by injecting a form of TBP mRNA that could not be targeted by the MO (Supplementary Figure S2).
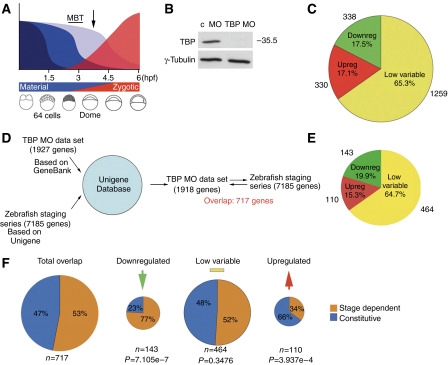
Figure 1.
TBP is selectively required for both activation and repression of genes in the early zebrafish embryo. (A) Schematic diagram of the dynamics of mRNA degradation and zygotic gene activation during the MBT in the zebrafish embryo. Shades of blue indicate differential degradation of maternal mRNAs before the MBT. The red curve indicates the dynamics of zygotic gene activation. Time after fertilisation is indicated in hpf. Schematic drawing of respective stages of embryo development are shown below. The arrow indicates time point for collection of embryos for microarray analysis. (B) Western blot analysis of TBP protein levels in dome-stage zebrafish embryos injected with TBP (MOTBP MO) and control MO antisense oligonucleotides. (C) Pie chart diagram summary of expression profiling data of 1927 probes from microarray experiments carried out in dome-stage zebrafish embryos. (D) Schematic diagram of the protocol for identification of the intersection of genes analysed for TBP dependence among genes analysed for their expression dynamics during zebrafish development via the Unigene database. (E) Pie chart diagram of the proportion of genes found in the three response groups following TBP knockdown and overlap with the stage-dependent expression microarray. (F) Pie chart diagrams showing the distribution of constitutive and stage-specific genes among the total and the three response groups of genes in TBP morphant embryos. Numbers below the charts indicate the number of overlapping genes between the two data sets compared and χ2 analysis of the gene distributions.
Within the 1927 genes that were used for this analysis, a large proportion of genes expressed in the dome-stage embryo (65.3%) showed no significant difference in signal strength between TBP MO- and control MO (c MO)-injected embryos, indicating that their steady-state mRNA levels are independent of TBP function. A smaller group of genes showed a significant reduction of expression demonstrating that these genes require TBP directly or indirectly for their activation (17.5%). A similar number of genes (17.1%) showed increased levels of transcripts in TBP MO injected embryos, suggesting that TBP is required for controlling their steady-state levels by reducing their transcription and/or by enhancing mRNA degradation.
Most TBP-activated genes are dynamically regulated during zebrafish ontogeny
To characterise further the genes affected by loss of TBP function, we tested whether genes in the three response groups described above showed discrete expression dynamics during zebrafish ontogeny. To this end, we compared the data set described above to an ontogenic stage-dependent expression profiling experiment on the zebrafish transcriptome (Konantz M, Otto G-W, Weller C, Saric M, Geisler R. Microarray analysis of gene expression in zebrafish development, manuscript in preparation). The two gene sets share 717 genes (Figure 1D and E) and the proportions of the three TBP morphant response groups among these 717 genes are similar to those of the total TBP microarray data set (Figure 1C and E).
Meta-analysis of the ontogenic stage-dependent gene expression array was carried out to define two classes of genes. Genes showing stage specific peaks of expression activity during zebrafish ontogeny were classified as ‘stage-dependent' and genes that showed no significant variation in gene expression during ontogeny were considered as constitutively active genes (Supplementary Figure S3, see Materials and methods). Nearly half of the 717 genes (46.9%) that overlap between the two microarray data sets were shown to be constitutively expressed genes. The remaining genes belonged to the stage-dependent class (53.1%) showing dynamic activity during zebrafish ontogeny (Figure 1F and Supplementary Figure S3).
Applying the ‘constitutive versus stage-dependent' classification to the TBP microarray gene response groups revealed that genes that require TBP for their activation were predominantly stage-dependent (77%, Figure 1F), whereas upregulated genes in TBP morphants showed the opposite tendency. The low-variable group of genes did not show a bias to either stage-dependent or constitutively expressed genes. These results indicate that TBP-dependent activation tends to be a property of genes that show dynamic activity during ontogeny. Moreover, TBP tends to negatively regulate steady-state levels of constitutively active genes.
TBP dependence of transcription from isolated zebrafish promoters
TBP could influence steady-state levels of mRNA in zebrafish embryos both through transcriptional as well as post-transcriptional processes. To address the former, we tested 23 GFP constructs using promoters of zebrafish genes expressed at the sphere/dome-stage and representing various gene ontology classes (O'Boyle et al, 2007). TBP-dependent promoter activation was evident for seven promoters, including the otx1 gene promoter (Figure 2A and Supplementary Figure S4 and Supplementary Table II). This result is consistent with the proposed role of TBP in activating zygotic transcription of many genes during development. On the other hand, 12 promoters, including the apoeb gene promoter, did not show significant changes of activity upon loss of TBP function (Figure 2A and Supplementary Figure S4). TBP independence of apoeb transcription was further confirmed by its mRNA levels (Supplementary Figure S1) and the utilisation of its TSS (data not shown) in TBP morphants. No correlation was found between known promoter motifs (such as TATA boxes, CpG islands, etc.) and TBP response (data not shown).
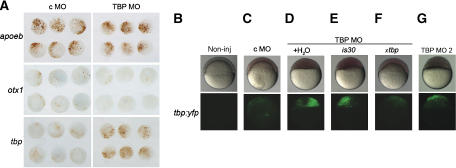
Figure 2.
TBP is required for both activation as well as repression of zebrafish promoters. (A) Representative samples of whole-mount immunochemical staining of embryos injected with promoter:gfp constructs (view on animal pole) with brown staining indicating mosaic pattern of GFP activity. (B–F) Rescue of the TBP morphant phenotype by overexpression of recombinant TBP. (B) Noninjected embryos, (C–G) Injection of tbp:yfp reporter construct carried out together with MO oligonucleotides as indicated above the images. Injected embryos were split into separate batches and exposed to a subsequent injection of water, tbp or is30 mRNA, as indicated below the horizontal line (D–F). Lateral views of 7 hpf embryos in bright field (top) or fluorescence views under YFP (bottom).
Several promoters (4 out of 23) showed a clear increase of promoter activity upon loss of TBP, including the 1.4-kb promoter of the tbp gene (Figure 2A and Supplementary Figure S4). This finding suggests negative regulatory role of TBP on the tbp gene promoter and is in line with the inverse correlation between tbp mRNA and TBP protein levels at the late blastula and early gastrula stages (Bártfai et al, 2004; Supplementary Figure S4D). Co-injection of a synthetic TBP MO-resistant Xenopus (x) tbp mRNA, but not of bacterial IS30 transposase control mRNA rescued the epiboly movements of the animal cap (Figure 2C–F, bright field view) and tbp:yfp activity (Figure 2C–F, fluorescence views). Finally, the injection of TBP MO2 resulted in comparable effects to TBP MO both in blocking epiboly movements and in the increased activity of the tbp:yfp promoter construct (Figure 2G). These results demonstrate that the specific loss of TBP protein is the reason for the observed upregulation of the tbp promoter in TBP MO-injected embryos.
TBP is required for degradation of a large number of maternal mRNAs
It is known that degradation of many maternal mRNAs involves zygotic transcription-dependent mechanisms, which may be specifically regulated by TBP. Thus, the steady-state levels of maternal mRNAs may appear increased in TBP morphants. To test if the inhibition of the degradation of maternal mRNA occurs in TBP morphants, we searched for maternally expressed genes in the TBP morphant microarray gene sets.
We classified genes as being maternal or zygotic through another microarray experiment utilizing mRNA pre- and post-MBT; those showing a decrease of mRNA levels from pre- to post-MBT (MBT down) were classified as prevalently maternal and vice versa (MBT up) for prevalently zygotic ones (Supplementary Tables III and IV). We then compared this experiment with the TBP morphants data set, which resulted in an overlap of 131 genes (Supplementary Tables III and IV). The overlap showed that maternal mRNAs were enriched among the upregulated genes of TBP morphants (Figure 3A, MBT down) and the inverse was observed for zygotic mRNAs (Figure 3A, MBT up). A side by side hierarchical clustering analysis of gene activity fold changes in the MBT experiment versus the TBP MO experiment demonstrates further the inverse correlation between the levels of mRNAs before or after MBT as compared to mRNA levels in TBP morphants versus controls (Figure 3B).
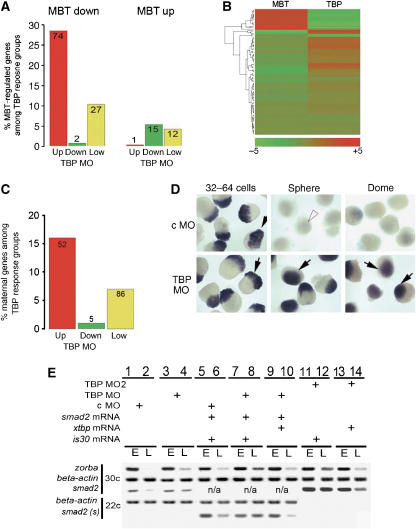
Figure 3.
A programme of maternal mRNA degradation requires TBP function. (A) Distribution of TBP MO response genes given as percentage of the genes expressed primarily at pre- or post-MBT stages. The number of overlapping genes between the microarray data sets is shown in the columns. Genes showing a higher abundance in pre-MBT stages (primarily maternal) are termed MBT down, genes showing a higher abundance in post-MBT stages (primarily zygotic) are termed MBT up. (B) Hierarchical clustering of genes based on their fold change values derived from the intersection of two microarray experiments. In the left column, the change of gene activities at post-MBT stages is shown in comparison to pre MBT (MBT). On the right, the response of the same genes is shown upon TBP knockdown (TBP). The fold change values for each gene are demonstrated as lines with colour intensities where red indicates higher expression. (C) Distribution of maternally expressed genes showing highest accumulation in the unfertilised egg (Mathavan et al, 2005) among significantly regulated genes present on the TBP-knockdown microarray given as percentages of the respective TBP response groups. The number of overlapping genes are given in the respective bars. (D) The expression patterns and levels of the maternally expressed zorba gene before (64 cells) and after MBT (sphere, dome). WISH of randomly oriented groups of embryos with DIG-labelled riboprobes against zorba are shown in MO injected embryos. Dark blue/purple staining in the animal pole (arrows) indicate specific gene activity in individual embryos. Arrowheads indicate loss of zorba mRNA signal. (E) RT–PCR analysis of selected maternally expressed genes before MBT and after MBT. Injection of MOs xtbp or is30 transposase mRNAs are indicated by ‘+' symbol. Abbreviations, hpf, hours post fertilisation; up, upregulated; down, downregulated; low var, low variable expression in TBP MO injected embryos; c, cycle number of PCR reactions; s, synthetic mRNA; E, early embryos before MBT; L, late embryos after MBT.
We further verified our findings by intersecting the TBP MO microarray experiment with an independent set of 622 maternal mRNAs (Mathavan et al, 2005), which resulted in an overlap of 143 genes (Supplementary Table V). As shown in Figure 3C, maternally inherited transcripts were significantly (P-value 1.043e−11) enriched among mRNAs upregulated in TBP morphants and underrepresented in the downregulated gene set (P-value 3.483e−5). Together, these results suggest that the upregulation of genes observed in TBP morphants could be in large part due to the specific loss of degradation of many maternal mRNAs.
Identification of TBP-dependent maternal transcripts
To validate the predicted involvement of TBP in the degradation of maternal mRNAs, we investigated the fate of individual maternal mRNAs. Zorba is a maternally expressed gene (Bally-Cuif et al, 1998), which is upregulated 2.51-fold in TBP MO embryos. We analysed the expression of zorba in wild-type and TBP-morphant embryos at regular intervals for the first 6 h of development by whole-mount in situ hybridisation (WISH). We found high levels of zorba expression in fertilised wild-type eggs and early embryos before MBT, followed by a sharp decrease soon after the MBT, followed by a slight increase at the dome stage (Figure 3D). In contrast, in TBP morphant embryos, zorba mRNA levels showed similar levels throughout early development, consistent with the assumption that degradation of maternal mRNA was impaired. We verified that the lack of degradation of zorba mRNA in TBP morphants was not due to a general delay in embryo development by observing the expression of two zygotically expressed genes: the TBP-independent gene no tail (Schulte-Merker et al, 1994), which correctly initiated transcription after the sphere stage in TBP morphants (Supplementary Figure S5A); and the TBP-dependent goosecoid (Schulte-Merker et al, 1994), whose activity was lost in TBP morphants (Supplementary Figure S5B). These results suggest efficient depletion of TBP at dome stage. To further verify the defect in maternal mRNA degradation, RT–PCR analysis was carried out on several maternally expressed genes that showed upregulation in TBP morphants. Zorba and smad2 (expressed both maternally and zygotically; Muller et al, 1999) showed elevated levels at dome stage in comparison to c MO-injected embryos, suggesting loss of degradation of maternal mRNA, as opposed to the control gene β-actin, which did not show a change in its steady-state levels (Figure 3E, lanes 1–4). RT–PCR analysis of zygotic genes was also carried out; the TBP-independent ntl showed no change in its mRNA levels, whereas zygotic activity of gsc dropped (data not shown) as shown previously by WISH.
To test directly the fate of mRNAs deposited in the egg, we utilised a synthetic smad2 mRNA microinjected into the fertilised eggs (Figure 3E, smad2 (s)). This mRNA could be readily distinguished from endogenous smad2 by reducing the cycles in the RT–PCR reaction (Figure 3E, compare lanes 1–2 to 5–6 of smad2 (s)). Microinjected smad2 mRNA was more efficiently degraded in c MO- than in TBP MO-injected embryos (Figure 3E, compare lanes 6 and 8) and similar results were obtained by WISH (data not shown). Thus, the apparent increase of smad2 mRNA levels in TBP morphants is not due to premature activation of zygotic smad2 expression, but due to the loss of degradation of smad2 mRNAs.
To verify the specificity of the maternal mRNA degradation phenotype to loss of TBP protein function, the ability of a MO-insensitive TBP mRNA to rescue the phenotype in TBP MO-injected embryos was tested. TBP MO and smad2 (s) co-injected embryos were split after injection and separate batches were injected for a second time either by xtbp mRNA or is30 mRNA. Expression of recombinant TBP resulted in increase of degradation of zorba (Figure 3E, compare lanes 7, 8 with 9, 10) as well as that of microinjected synthetic smad2 mRNA. In contrast, is30 tpase control mRNA did not result in rescue of the degradation phenotype of TBP morphants (Figure 3E, compare lanes 8 and 10). These results demonstrate that the effect of TBP MO on maternal mRNA degradation is directly attributable to the loss of TBP protein function.
TBP regulates a zygotic transcription-dependent mRNA degradation process
Little is known about the mechanisms of maternal mRNA degradation in zebrafish, however, it is likely to involve several maternal as well as zygotic transcription-dependent mechanisms. Not all maternal mRNAs were degraded in TBP morphants (Figure 3A and C). This may be due to different regulatory mechanisms acting in parallel during maternal mRNA degradation. To investigate this further, we verified the kinetics of mRNA degradation by exploiting a published microarray data set on maternal mRNAs (Mathavan et al, 2005) and compared it to our TBP morphant data set. We established three classes of mRNAs based on the time of their degradation (see Figure 4A and B and Supplementary Table X): a ‘fast' group of mRNAs, which degrade transcription independently or in a transcription dependent manner immediately after initiation of zygotic transcription; a ‘medium' group which is mostly degraded after MBT by early gastrula stage; and a ‘late' group degraded during neurulation and somitogenesis. The comparison of these groups with the TBP-morphant experiment (Figure 4C) showed that maternal mRNAs upregulated in TBP morphants follow the pattern of expression dynamics of the ‘medium' group (P-value=2.205e−06). TBP-dependent maternal mRNAs showed minimal degradation until MBT (3 h post-fertilisation (hpf)) and accelerated degradation by early gastrulation (4.5 hpf), suggesting that zygotic transcription-dependent mechanisms are involved in their degradation.
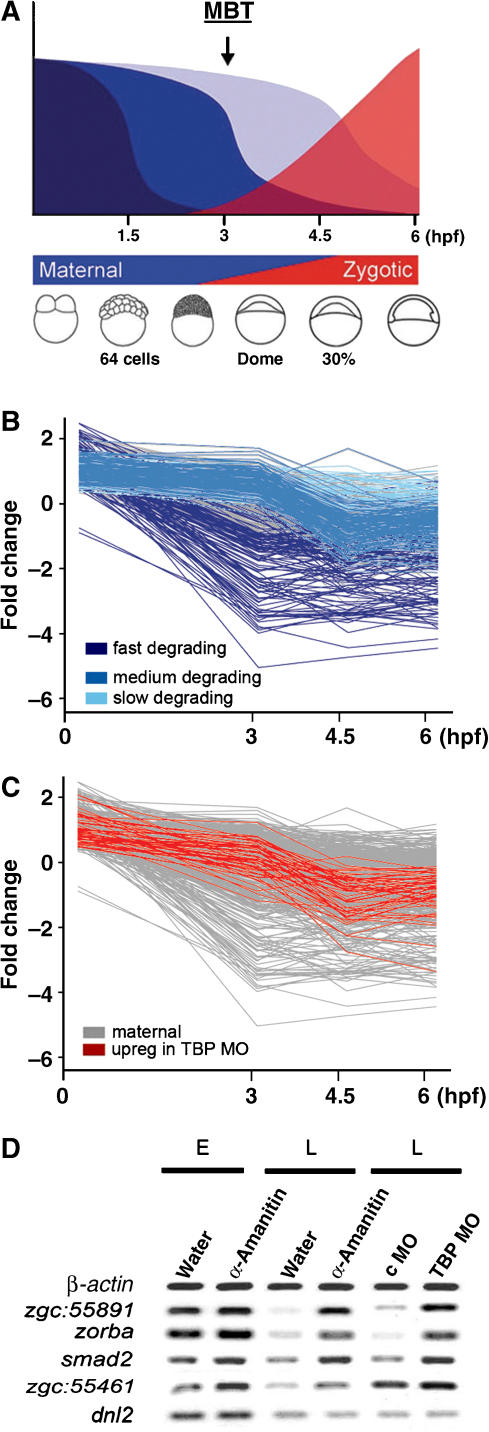
Figure 4.
TBP is required for the degradation of a subset of maternal mRNAs. (A) Schematic representation of early gene activities of the zebrafish embryo as shown in Figure 1A. (B) Degradation pattern of maternally accumulated genes during early zebrafish development until gastrulation (Mathavan et al, 2005). The ‘fast' group of mRNAs, degraded before and immediately after MBT, is shown in dark blue), the ‘medium' group, degraded after transcription starts at MBT, is shown in medium blue and the ‘late' group, degraded during neurulation and somitogenesis, is shown in light blue. (C) Degradation pattern of genes upregulated in TBP Morphant embryos (red) in comparison to the degradation dynamics of all maternal genes (grey). (D) Degradation of maternal RNA in control and α-amanitin-injected embryos as compared to c MO- and TBP MO-injected embryos before MBT and after MBT. Abbreviations, as in Figure 3.
These results suggest that TBP is only acting on a subset of mRNAs and that these mRNAs require transcription for their degradation. To test the transcriptional requirement for degradation of maternal mRNAs, we treated embryos with α-amanitin at a concentration that inhibits Pol II activity (Müller et al, 2001) and carried out RT–PCR analysis of gene expression. High levels of zorba, zgc:55891 and smad2 mRNAs were retained after MBT in α-amanitin-injected embryos similarly to TBP morphants (Figure 4D), demonstrating that these mRNAs require transcription and TBP for their degradation. These results taken together with the results obtained using synthetic smad2 mRNAs (Figure 3E) suggest that the sustained levels of maternal mRNAs in TBP morphants are due to the inhibition of maternal mRNA degradation rather than ectopic activation of zygotic transcription of the respective genes. Not all maternal mRNAs require zygotic transcription (and TBP) for their degradation as confirmed by the fact that degradation of the dnl2 gene is unaffected by α-amanitin and TBP MO (Figure 4D) and its degradation is primarily mediated by mechanisms acting before MBT (Figure 4C). Thus, TBP appears to function within a transcription dependent mechanism directing the degradation of a subset of maternal mRNAs eliminated in a tight time window after MBT during early gastrulation.
Degradation of maternal mRNA by the miR-430 microRNA is specifically affected in TBP morphants
Recently, a novel mechanism for maternal mRNA degradation has been described, which involves the zygotically transcribed miR-430 microRNA (Giraldez et al, 2006). Importantly, the maternally inherited zorba and smad2 transcripts have been shown to be targets of miR-430 regulation (Giraldez et al, 2006), suggesting a potential link between TBP and miR-430 function in mRNA degradation. Therefore, we explored the relationship between miR-430- and TBP-dependent mRNA degradation mechanisms.
We first addressed the question whether TBP-dependent maternally inherited transcripts represent targets of miR-430-mediated mRNA degradation in general. We compared the overlap between experimentally verified miR-430 target genes (Giraldez et al, 2006) and our TBP morphant microarray gene sets (see Supplementary Tables XI, XII and Materials and methods). As shown in Figure 5A, a significant enrichment of miR-430 target genes among the upregulated genes of TBP morphants was observed (P=0.002074). The proportion of miR-430 target genes was found to be higher among TBP-upregulated genes (20%) than among maternal genes in general (14%, P=0.0276). This difference is significant also after 100 randomisation experiments in which we randomly selected 100 maternal genes (15% s.d.=3, P=0.0507). This suggests that the enrichment of miR-430 targets among the upregulated genes of TBP morphants is not simply reflecting the high proportion of maternal genes among miR-430 targets and upregulated genes in TBP morphants.
Figure 5.
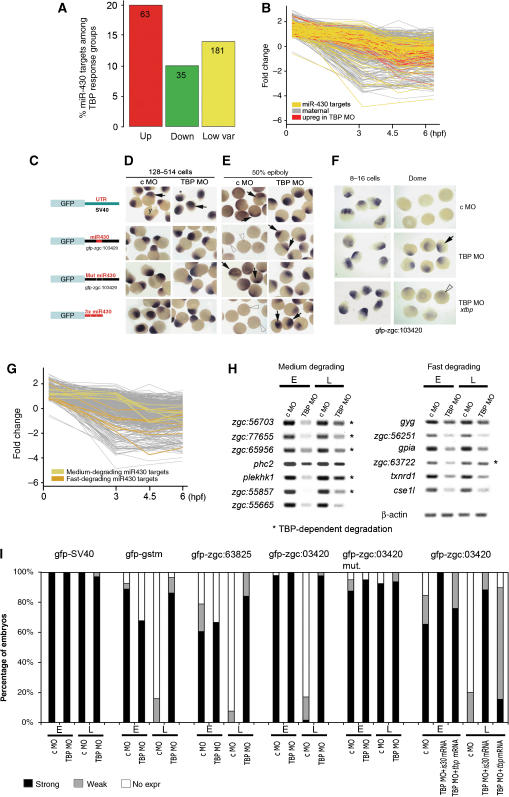
TBP is required for miR-430-dependent maternal mRNA degradation. (A) Distribution of miR-430 target genes among the different response groups of genes regulated in TBP Morphant embryos is shown as percentage of the respective TBP MO response groups. The number of overlapping genes between the two microarray data sets compared is indicated in the columns. (B) Degradation dynamics of miR-430 target maternal mRNAs during early zebrafish development (yellow) in comparison to the maternal mRNAs upregulated in TBP Morphants (red) and all maternal mRNAs (grey). (C–F) MiR-430 target mRNAs are degraded by a TBP-dependent mechanism. Synthetic mRNAs injected into zebrafish embryos are shown (A). Microinjected GFP mRNAs are detected by WISH (arrows) in randomly oriented representative groups of early embryos before MBT (D) and several hours after MBT). (F) Injection of TBP mRNA rescues the mRNA degradation phenotype. Detection of microinjected synthetic gfp-zgc:103420 mRNA by WISH using a gfp antisense probe. Embryos are shown at the indicated stages in random orientation. (G) Degradation dynamics of maternal miR-430 target genes. Representative examples of ‘fast'- and ‘medium'-degrading mRNAs used in RT–PCR analyses (H) are shown. (H) RT–PCR analysis of the mRNAs at 16-cell stage and at 30% epiboly stage in embryos injected with c MO or TBP MO. (I) Degradation of synthetic mRNAs injected in zebrafish embryos. Embryos were injected with mRNAs indicated above the bar chart. Percentage of embryos with different signal levels after in situ hybridisation using a gfp probe are shown. The numbers of embryos injected are shown in Supplementary Table III. Abbreviations are as in Figure 3.
Subsequently, we have checked the degradation patterns of miR-430 target genes by analysing the overlap between the maternal genes with known degradation kinetics (Mathavan et al, 2005) and miR-430 target genes (Supplementary Table VI). The degradation kinetics of miR-430 target genes largely but not exclusively overlap with that of the maternal genes degraded by TBP-dependent mechanisms (Figure 5B). Taken together, these results indicate a correlation between miR-430- and TBP-dependent mRNA degradation.
We next tested whether miR-430-dependent mRNA degradation requires TBP function. Embryos were injected at the zygote stage with a combination of synthetic mRNAs containing UTR sequences with or without miR-430 target sites (Giraldez et al, 2006) together with TBP MO or c MO, and mRNA distribution was detected by WISH before and after the MBT. A synthetic gfp mRNA containing the 3′ UTR region from SV40 that lacks miR-430 target sequences was not degraded until the 50% epiboly stage (Figure 5D, E and I and Supplementary Table VIII), suggesting that they are not degraded by the miR-430 pathway. In contrast, gfp mRNA linked to the UTR from the zgc:103420 gene containing a miR-430 target site is degraded by the 50% epiboly stage in c MO-injected embryos, but not in TBP MO-injected embryos (Figure 5C–E and I). Similar results were obtained with gfp mRNA fused to the 3′ UTR sequences of the zgc:63825 and gstm genes containing miR-430 target sites or when only the miR430 target site sequences were added to gfp (Figure 5C–E and I). These results suggest that TBP is required for miR-430-dependent degradation of several mRNAs. Upon injection of a transcript containing a mutated 3′ UTR sequence of zgc:103420 lacking the miR-430 target site, the mRNA became insensitive to degradation until the 50% epiboly stage (Figure 5C–E and I), indicating that the degradation of these synthetic mRNAs are indeed miR-430-dependent. The defects in miR-430-dependent degradation of mRNA in TBP morphants was also rescued by overexpression of recombinant TBP. Co-injection of synthetic tbp mRNA with gfp-zgc:103420 and TBP MO resulted in reversal of the mRNA degradation phenotype of TBP morphants, indicating that the mRNA degradation effects relate directly to loss of TBP (Figure 5F and I and Supplementary Table VIII). As miRNA genes are known to be transcribed by polymerase II (Lee et al, 2004) miR-430 may be a candidate target of TBP-dependent transcription regulation. However, miR-430 expression in TBP morphants is unaffected (Supplementary Figure S5D), suggesting that TBP functions downstream of miR-430 production in the mRNA degradation process. We investigated further whether TBP is involved in general microRNA function or the mir-430 pathway specifically. Thus, we co-injected miR-430 and miR-1 with their respective target mRNAs (Giraldez et al, 2006) with TBP and c MO. miR-430-mediated mRNA degradation was blocked in TBP morphants, as opposed to that by miR-1, confirming the specificity of TBP function to a subset of miRNA-dependent processes (Supplementary Figure S6).
Given the tight temporal control of TBP-dependent mRNA degradation, we hypothesised that those miR-430 target mRNAs are degraded in a TBP-dependent manner, which are eliminated at a ‘medium' rate during late blastulation/early gastrulation. To test this hypothesis, we utilised the overlap between miR-430 target genes and maternal genes and identified those, which show either medium or fast degradation (Figure 5G). Then we tested both fast- and medium-degrading miR430 target genes for TBP dependence of their degradation. The results indicate that medium-degrading mRNAs are more likely to be TBP-dependent (increased accumulation in TBP MO injected embryos) than fast-degrading mRNAs (5 of 7 versus 1 of 6, respectively, Figure 5H). Taken together, our results indicate that TBP is specifically required for the degradation of a subset of miR-430-dependent mRNAs that are eliminated at late blastula/early gastrula stages.
Discussion
In summary, we have demonstrated a differential requirement for TBP in the regulation of mRNA levels in the early zebrafish embryo and pinpointed three levels of regulatory activities associated with TBP function. The approach to block TBP function by MO oligonucleotides resulted in the efficient depletion of TBP in the embryo before the MBT, and thus allowed the detection of the earliest effects of loss of TBP protein on zygotic transcription and associated maternal degradation processes. The extensive use of a second TBP targeting MO and rescue experiments with a MO-insensitive recombinant TBP in this study provided strong experimental verification of the specificity of our key findings to loss of TBP function.
Redundant and specific function of TBP in the activation of subsets of genes at MBT
In this study of the transcriptome of the early zebrafish embryo, we have found that the expression of most genes remains weakly or not affected in TBP morphants. It is tempting to speculate that the TBP paralogue TBP2/TRF3, which has similar DNA-binding properties to TBP and is expressed at high levels in ovaries (Bártfai et al, 2004) may contribute to the control of steady-state levels of mRNAs before gastrulation, thus complementing TBP function. However, due to the presence of maternally inherited TBP2 protein in the early zebrafish embryo, MO knockdown of TBP2 is inefficient before gastrulation (Bártfai et al, 2004) and new ways of interfering with TBP2 function in the oocyte will have to be developed to address TBP2 function in the early zebrafish embryo. Another TBP-related factor (TLF/TLP/TRF2) has also been shown to affect transcription regulation (Veenstra et al, 2000; Müller et al, 2001). Together, these alternative transcription initiation mechanisms may explain the large number of unaffected gene activities in TBP morphants and emphasise the need to define the boundaries of TBP-regulatory mechanisms.
The differential regulation of genes during early development by TBP raises the question of which genes are specifically regulated by TBP and what promoter properties do they possess. Among genes requiring TBP for their activation, there was an enrichment for genes with stage-dependent activity during ontogeny. This observation suggests that TBP is required for genes that are expressed in a tightly regulated manner and demonstrates an important regulatory role for TBP as a core promoter-binding factor during ontogeny. Does the correlation between TBP function and the type of genes regulated by TBP mean a correlation between TBP dependence and core promoter motif composition? The recent genome-wide analysis of mouse and human promoters revealed that the binding site for TBP, known as the TATA box, is more likely to be present in genes associated with tightly regulated transcription and tissue specificity (Carninci et al, 2006). Our results, which show that ontogenic stage-dependent genes preferentially require TBP, are in line with the observations in mammals. However, our failure to detect a direct correlation between the presence of a TATA box and TBP dependence of gene activation in zebrafish is possibly due to the small set of genes available for this analysis. The small number of promoters analysed combined with the very low number of TATA boxes encountered in zebrafish promoters (for less than 10% of genes verified, unpublished data) is consistent with findings in mammals (Carninci et al, 2006) and hampers the establishment of statistically significant correlations.
TBP limits certain gene expression activities in the zebrafish embryo
The striking presence of upregulated genes in TBP morphants could be attributed to two distinct mechanisms: the direct or indirect negative regulatory role of TBP on zygotic transcription as well as the block of maternal mRNA degradation. In this paper, we show evidence for both mechanisms, thus revealing the complexity of regulatory roles played by TBP during the maternal to zygotic transition in zebrafish.
The observed negative regulatory role of TBP in zygotic gene activation is consistent with recent observations made in cultured cells. Inverse correlation between TBP occupancy and gene activity has been reported recently in Drosophila cells (Lebedeva et al, 2005). Moreover, TBP was found to inhibit transcription of the NF1 promoter in transfection experiments (Chong et al, 2005). Thus, published biochemical evidence (Chong et al, 2005) together with our observations on the biological roles of TBP together make it feasible to hypothesise that TBP may repress promoters directly during embryogenesis. To test this possibility, as well as to assess the direct effects of TBP in early development, will require the establishment of assays of promoter occupancy by TBP in vivo during zebrafish embryonic development.
The mRNA degradation machinery active during maternal to zygotic transition requires TBP function
The temporally regulated degradation of maternal mRNAs at and after the MBT is part of a general mechanism to regulate the maternal to zygotic transition and is required for normal development of the zebrafish embryo (Giraldez et al, 2006; Schier, 2007). We showed here that TBP is required for a zygotic developmental programme, which facilitates the regulated degradation of a subset of maternally deposited mRNAs during late blastula/early gastrula stages in the zebrafish. This finding is one of the few currently available explanations for the interplay between zygotic transcription and mRNA degradation during early embryogenesis (De Renzis et al, 2007).
Where does TBP function in maternal mRNA degradation? The most likely scenario is that TBP is required for the expression of a zygotic gene product, which acts in the mRNA degradation mechanism. The components of the mRNA degradation pathway(s) in the embryo that may be affected by TBP function are largely unknown. However, our analysis of the temporal regulation of maternal mRNA degradation allowed us to characterise the set of mRNAs that require TBP for their elimination and provides useful tools for future research into the molecular mechanisms involved in the control of mRNA degradation by transcription-dependent mechanisms. We concluded that TBP-dependent mRNAs are specifically degraded after the MBT but before the end of gastrulation in a transcription-dependent manner. The zygotically expressed miR-430 becomes active at the MBT and was recently shown to regulate the degradation of many mRNAs in zebrafish (Giraldez et al, 2006). In our study, TBP was found to affect miR-430 target mRNAs but only a subset of them. Importantly, we were able to pinpoint the set of miR-430-dependent maternal mRNAs degraded during late blastula early gastrula that require TBP. Thus, TBP may act on yet unidentified components of a pathway feeding into or functioning genetically downstream to the miR-430-dependent maternal mRNA degradation pathway. The pursuit for other factors in this pathway, including other potential TBP-dependent miRNAs, remains an urgent subject for future investigation. It will be important to address whether the restricted role TBP plays in the degradation of a subset of mRNAs can be associated with a particular developmental programme such as the various aspects of spatial and temporal control of gene activities during gastrulation. Another interesting question is whether TBP-dependent degradation of maternal mRNAs is a general feature of vertebrate development. Whereas it is premature to make direct comparisons with other vertebrates, there are indications for similarities in mRNA degradation dynamics. For example, smad2 is a maternally expressed gene in the mouse with a similar early degradation pattern to that described here for zebrafish (Wang et al, 2004).
The role of TBP in regulating specific subsets of genes during early embryo development demonstrates that promoter recognition proteins, once considered as constitutive components of transcription initiation, play specialised roles in differential gene expression regulation and may explain the diversity of core promoters (Butler and Kadonaga, 2002). Our findings support the model that promoter recognition by general transcription factors represents an additional regulatory level in transcription, which may provide flexibility for the cells of the animal to respond to signals at the core promoter level during ontogeny.
Materials and methods
Embryo injection experiments
Wild-type embryos (Tubingen AB) were collected after fertilisation and dechorionated by pronase treatment as described previously (Westerfield, 1995). The TBP-specific MO (TBP MO) used was described previously (Müller et al, 2001). TBP2 MO was CAAAAGACGTAAACGATAATTCGCA. Embryos were injected also with a c MO with five mismatches relative to the TBP-specific MO (GACGTACGCTGTTCTTCTCCTCGAT) or a standard c MO (CCTCTTACCTCAGTTACAATTTATA) provided by Gene-tools LLC (Phylomath, USA). MOs were injected into the cytoplasm of zebrafish embryos at the one cell stage. A total of 1200 embryos were collected for microarray analysis at the dome stage. Embryos were co-injected with combinations of MOs and mRNAs for analysis of gene expression. For synthetic tbp mRNA production, the pCS2+xcTBP was used containing as a XhoI–XbaI fragment of the biologically active core domain (aa 104–297) (Schmidt et al, 2003) of the Xenopus laevis TBP (Veenstra et al, 1999). The pFOL876 plasmid in a pCS2+ vector with an Escherichia coli IS30 transposase gene was used to make control mRNA (Szabo et al, 2003). Both mRNAs were injected at 100 ng/ml concentration. mRNAs were produced by in vitro transcription of linearised plasmids using the mMESSAGE mMACHINE Kit (Ambion, UK). Plasmids for the production of synthetic gfp mRNAs with various miR-430 target sequences were described by Giraldez et al (2006). In co-injection experiments, MOs were co-injected with 20 ng/ml gfp-UTR mRNAs followed by injection with tbp or is30 tpase mRNAs.
Twenty-three promoter constructs were made which contained 1–2 kb upstream and 30–300 bp downstream sequence around the predicted (9 promoters) or 5′RACE verified (14 promoters) transcriptional start site (TSS) linked to a GFP reporter. Promoter fragments were amplified from genomic DNA using TripleMaster Enzyme Mix (Eppendorf, Germany) and cloned into the pGlow-TOPO vector (Invitrogen, Germany). The promoter constructs were injected into the cytoplasm of one-cell stage embryos. At 50% epiboly, embryos were fixed overnight in 4% PFA. TBP-dependence analysis of promoters was carried out by co-injecting MOs in a concentration of 1 mM with 100 ng/μl of mRNA. For recombinant TBP rescue experiments, the tbp promoter was PCR-amplified and subcloned into the pCS2+yfp replacing the CMV promoter by using SalI and NcoI sites. Yellow fluorescence protein was visualised by using a Leica MZ16F fluorescent stereo microscope and digital image recording. Western blotting with the 3G3 antibody against TBP was carried out as described previously (Müller et al, 2001).
Whole-mount in situ hybridisation and immunostaining
Fixed embryos were incubated overnight at 4°C with wild-type GFP (1:500) (Torrey Pines Biolabs, USA) or cycle3 GFP antibodies (1:200) (Invitrogen, Germany). Embryos were incubated with goat-anti-rabbit HRP secondary antibody in PBT (1:500) (DakoCytomation, Denmark) for detection by diaminobenzidine solution according to manufacturer's instruction (Vector, USA). An LNA oligonucleotide probe was used in WISH to detect miR-430 miRNA as described previously (Giraldez et al, 2006). WISHs were carried out with standard protocols (Westerfield, 1995).
RT–PCR analysis of maternal mRNA degradation
Analysis of a selected set of genes by RT–PCR was carried out using 50 embryos for each treatment group. Microinjected and wild-type embryos were collected at the 512-cell stage and shield stage. Total RNA was extracted using Trizol (Invitrogen) following manufacturer instructions. A 1 μg portion of total RNA was reverse transcribed (M-MLV Reverse Transcriptase, Promega, Germany) and PCR was carried out from several targets using the oligonucleotide primers specified in Supplementary Table IX. PCR products were separated in a 2% agarose gel.
Gene identification and statistical analysis of EST microarray data
Three developmental stages around MBT were analysed on microarrays containing the primary set of 16 177 oligos (16 k set). RNA was obtained from freshly laid eggs (zygotic target), 1k-stage embryos (1k target) and 30% epiboly embryos (30% epiboly target). The following hybridisations were performed with dye swap each: zygotic versus 1k, zygotic versus 30% epiboly and 1k versus 30% epiboly. Microarray chips representing 10 501 nonredundant Genbank ESTs of the primary 16 416 set of 65-mer oligonucleotides were used in the subsequent hybridisations. Two independent hybridisations were carried out for three biological repeats of treatment groups (TBP MO- versus c MO-injected embryos), resulting in a maximum of 12 data points per gene. The reliability of the fold changes was assessed using a regularised t-test (Baldi and Long, 2001) and the adjustment of P-values to control the false discovery rate (FDR) (Benjamini and Hochberg, 1995) selecting genes with FDR smaller than 0.05 (minimum 5 data points). This cut-off value of FDR is the minimal widely used (Shi et al, 2006) and results in a good balance of quality versus quantity in the selected gene lists.
Annotation of ESTs of the TBP microarray, in relation to the stage-dependence array and to the zebrafish genome
Several meta-analyses of different, unrelated microarray experiments were carried out as has been described previously (Cavallo et al, 2005). Gene sets of the TBP morphant microarray were compared with existing gene sets (Mathavan et al, 2005) generated on the same platform (Compugen microarray), with the experiment GSE4201, http://www.ncbi.nlm.nih.gov/projects/geo (Giraldez et al, 2006) and the experiment E-TABM-33 from http://www.ebi.ac.uk/arrayexpress (Konantz M, Otto G-W, Weller C, Saric M, Geisler R. Microarray analysis of gene expression in zebrafish development, manuscript in preparation) executed on the Affymetrix platform (See Supplementary data for details). The outdated gene annotation by Compugene (Mathavan et al, 2005) was updated by converting all Entrez Nucleotide identifiers of the probes to their respective Unigene identifiers using a custom Perl script and release 91 of Unigene. The heatmap and hierarchical clustering of microarray experiments was created using the R programming language version 2.2.0. The clustering of fold changes is based on the ‘complete linkage' method provided by R. See http://cran.r-project.org/ for details.
Degradation pattern of maternal transcripts
Maternal genes present in the TBP-knockdown and miR-430 targets gene sets (Giraldez et al, 2006) were identified by utilizing an existing data set of transcripts accumulated in the unfertilised egg (Mathavan et al, 2005). The variation of steady-state mRNA levels over developmental time was determined by comparing fold changes as described in (Mathavan et al, 2005) and was visualised by plotting their values in the unfertilised egg and at 3, 4.5 and 6 hpf). For a clustering of maternal mRNAs into fast-, medium- and slow-degrading subgroups the following criteria were used: fast-degrading, >200% decrease of fold change from 0 to 3 hpf; medium–degrading, <100% from 0 to 3 hpf, >100% from 3 to 4.5 hpf; slow-degrading, <50% decrease of fold change till 4.5 hpf).
Identification of miR-430 targets among the genes of the TBP microarray
We downloaded the raw data of the experiments (Giraldez et al, 2006; wild type, MZ Dicer and MZ Dicer miR-430-injected) by the GEO database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4201). The probe summaries were generated by using RMA from the BioConductor collection of packages. The Limma package was used to perform the statistical analysis and select differentially expressed genes. The following comparisons were made: MZ Dicer versus wild type; MZ Dicer versus MZ Dicer miR-430–injected; wild type versus MZ Dicer miR-430-injected. We selected as differentially expressed genes all the genes resulting in at least one comparison with a corrected P-value ⩽0.05. All the genes significantly upregulated in MZ DICER compared to both wild type and MZ DICER miR-430-injected were considered as potential miR-430 target (1650 probes).
Supplementary Material
Supplementary Figures
Supplementary Table I
Supplementary Table II
Supplementary Table III
Supplementary Table IV
Supplementary Table V
Supplementary Table VI
Supplementary Table VII
Supplementary Table VIII
Supplementary Table IX
Supplementary Table X
Supplementary Table XI
Supplementary Table XII
Acknowledgments
We thank L Tora for TBP antibodies, discussions and critical reading of this manuscript. We also thank NS Foulkes and M-E Torres Padilla for critical comments and R Caogero and F McNish for their advice. We acknowledge S Schindler and N Borel for technical assistance and T Dalmay and T Rathjen for their help in detecting miR-430 in embryos. We thank A Giraldez and A Schier for sharing GFP constructs containing miR-430 targets, and L Byrnes, R Bree, S Goyle and R Geisler for sharing unpublished data. This work was supported by funds from the DFG (MU 1768/2) to FM and by the EU (contract 511990) to FM and ES and the BMBF to FM and FO and an ESF travel grant to MF.
References
- Almouzni G, Wolffe AP (1995) Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. EMBO J 14: 1752–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic Y, Anderson C, Bhatty R, Hartley RS (2001) Zygotic regulation of maternal cyclin A1 and B2 mRNAs. Mol Cell Biol 21: 1662–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, Long AD (2001) A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519 [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L, Schatz WJ, Ho RK (1998) Characterization of the zebrafish Orb/CPEB-related RNA binding protein and localization of maternal components in the zebrafish oocyte. Mech Dev 77: 31–47 [DOI] [PubMed] [Google Scholar]
- Bártfai R, Balduf C, Hilton T, Rathmann Y, Hadzhiev Y, Tora L, Orbán L, Müller F (2004) TBP2, a vertebrate-specific member of the TBP family is required in embryonic development of zebrafish. Curr Biol 14: 593–598 [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, Fu W, Hamilton JK, Etkin LD, Lipshitz HD (1999) Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J 18: 2610–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 136–143 [Google Scholar]
- Butler JE, Kadonaga JT (2002) The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev 16: 2583–2592 [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, Forrest AR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y et al. (2006) Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635 [DOI] [PubMed] [Google Scholar]
- Cavallo F, Astolfi A, Iezzi M, Cordero F, Lollini PL, Forni G, Calogero R (2005) An integrated approach of immunogenomics and bioinformatics to identify new tumor associated antigens (TAA) for mammary cancer immunological prevention. BMC Bioinformatics 6 (Suppl 4): S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JA, Moran MM, Teichmann M, Kaczmarek JS, Roeder R, Clapham DE (2005) TATA-binding protein (TBP)-like factor (TLF) is a functional regulator of transcription: reciprocal regulation of the neurofibromatosis type 1 and c-fos genes by TLF/TRF2 and TBP. Mol Cell Biol 25: 2632–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson I (2003) The genetics of TBP and TBP-related factors. Trends Biochem Sci 28: 391–398 [DOI] [PubMed] [Google Scholar]
- De Renzis S, Elemento O, Tavazoie S, Wieschaus EF (2007) Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol 5: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF (2005) MicroRNAs regulate brain morphogenesis in zebrafish. Science 308: 833–838 [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF (2006) Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ (2004) Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci USA 101: 13525–13530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB (1993) The zebrafish midblastula transition. Development 119: 447–456 [DOI] [PubMed] [Google Scholar]
- Kimelman D, Kirschner M, Scherson T (1987) The events of the midblastula transition in Xenopus are regulated by changes in the cell-cycle. Cell 48: 399–407 [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310 [DOI] [PubMed] [Google Scholar]
- Lebedeva LA, Nabirochkina EN, Kurshakova MM, Robert F, Krasnov AN, Evgen'ev MB, Kadonaga JT, Georgieva SG, Tora L (2005) Occupancy of the Drosophila hsp70 promoter by a subset of basal transcription factors diminishes upon transcriptional activation. Proc Natl Acad Sci USA 102: 18087–18092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Viville S, Davidson I (2002) RNA polymerase II transcription in murine cells lacking the TATA binding protein. Science 298: 1036–1039 [DOI] [PubMed] [Google Scholar]
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, Ruan Y, Korzh V, Gong Z, Liu ET, Lufkin T (2005) Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet 1: 260–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PA, Ozer J, Salunek M, Jan G, Zerby D, Campbell S, Lieberman PM (1999) A human TATA binding protein-related protein with altered DNA binding specificity inhibits transcription from multiple promoters and activators. Mol Cell Biol 19: 7610–7620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F, Blader P, Rastegar S, Fischer N, Knochel W, Strahle U (1999) Characterization of zebrafish smad1, smad2 and smad5: the amino-terminus of smad1 and smad5 is required for specific function in the embryo. Mech Dev 88: 73–88 [DOI] [PubMed] [Google Scholar]
- Müller F, Lakatos L, Dantonel J, Strahle U, Tora L (2001) TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr Biol 11: 282–287 [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M (1982) A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 30: 687–696 [DOI] [PubMed] [Google Scholar]
- O'Boyle S, Bree RT, McLoughlin S, Grealy M, Byrnes L (2007) Identification of zygotic genes expressed at the midblastula transition in zebrafish. Biochem Biophys Res Commun 358: 462–468 [DOI] [PubMed] [Google Scholar]
- Pelegri F (2003) Maternal factors in zebrafish development. Dev Dyn 228: 535–554 [DOI] [PubMed] [Google Scholar]
- Persengiev SP, Zhu X, Dixit BL, Maston GA, Kittler EL, Green MR (2003) TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc Natl Acad Sci USA 100: 14887–14891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau MN, Huet J, Sentenac A, Mechali M (1994) Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell 77: 439–449 [DOI] [PubMed] [Google Scholar]
- Schier AF (2007) The maternal-zygotic transition: death and birth of RNAs. Science 316: 406–407 [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Bondareva AA, Radke JR, Capecchi MR (2003) Fundamental cellular processes do not require vertebrate-specific sequences within the TATA-binding protein. J Biol Chem 278: 6168–6174 [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, Hammerschmidt M, Beuchle D, Cho KW, De Robertis EM, Nusslein-Volhard C (1994) Expression of zebrafish goosecoid and no tail gene products in wild-type and mutant no tail embryos. Development 120: 843–852 [DOI] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM et al. (2006) The MicroArray quality control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol 24: 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo M, Muller F, Kiss J, Balduf C, Strahle U, Olasz F (2003) Transposition and targeting of the prokaryotic mobile element IS30 in zebrafish. FEBS Lett 550: 46–50 [DOI] [PubMed] [Google Scholar]
- Teichmann M, Wang Z, Martinez E, Tjernberg A, Zhang D, Vollmer F, Chait BT, Roeder RG (1999) Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc Natl Acad Sci USA 96: 13720–13725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EM, Legouy E, Renard JP (1998) Mouse embryos do not wait for the MBT: chromatin and RNA polymerase remodeling in genome activation at the onset of development. Dev Genet 22: 31–42 [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Destree OH, Wolffe AP (1999) Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Mol Cell Biol 19: 7972–7982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, Weeks DL, Wolffe AP (2000) Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science 290: 2312–2315 [DOI] [PubMed] [Google Scholar]
- Wagner DS, Dosch R, Mintzer KA, Wiemelt AP, Mullins MC (2004) Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Dev Cell 6: 781–790 [DOI] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M (2004) A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell 6: 133–144 [DOI] [PubMed] [Google Scholar]
- Westerfield M (1995) The zebrafish book. Eugene: University of Oregon Press [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table I
Supplementary Table II
Supplementary Table III
Supplementary Table IV
Supplementary Table V
Supplementary Table VI
Supplementary Table VII
Supplementary Table VIII
Supplementary Table IX
Supplementary Table X
Supplementary Table XI
Supplementary Table XII