Abstract
Since the functional state of a protein–protein interaction network depends on gene expression, a fundamental question is what relationships exist between protein interaction network and gene regulation. In particular, microRNAs have recently emerged as a major class of post-transcriptional regulators that influences a large proportion of genes in higher eukaryotes. Here we show that protein connectivity in the human protein–protein interaction network is positively correlated with the number of microRNA target-site types in the 3′ untranslated regions of the gene encoding the protein and that interacting proteins tend to share more microRNA target-site types than random pairs. Moreover, our results demonstrate that microRNA targeting propensity for genes in different biological processes can be largely explained by their protein connectivity. Finally, we show that for hub proteins, microRNA regulation complexity is negatively correlated with clustering coefficient, suggesting that microRNA regulation is more important to inter-modular hubs than to intramodular ones. Taken together, our study provides the first evidence for global correlation between microRNA repression and protein–protein interactions.
Keywords: hub protein, microRNA regulation, mRNA targeting propensity, protein–protein interaction network
INTRODUCTION
Interactions between proteins provide a mechanistic basis for most biological processes in an organism. Recent advances in biotechnology, such as high-throughput yeast two-hybrid screen, have allowed scientists to start building proteome-wide protein–protein interaction or “interactome” maps (Uetz et al. 2000; Ito et al. 2001; Giot et al. 2003; Li et al. 2004; LaCount et al. 2005; Rual et al. 2005; Stelzl et al. 2005). Conventionally, a protein–protein interaction map is represented as a static network, in which each node represents a protein and each edge represents a protein–protein interaction. In reality, the protein–protein interaction network (PPIN) is a dynamic entity: The functional state of the network depends on the expression of protein nodes (Han et al. 2004; Vidal 2005), which is intrinsically controlled by different regulatory mechanisms through time and space.
In higher eukaryotes, microRNAs (miRNAs) have emerged as an abundant class of regulatory genes that regulate protein expression (Lim et al. 2003; Ambros 2004; Bartel 2004). These ∼22 nucleotide (nt) endogenous noncoding RNAs can repress gene expression post-transcriptionally by binding to the 3′ untranslated regions (3′ UTRs) of their target messages. The critical region in a miRNA is the nucleotides 2–7 from the 5′ end, the so-called “seed” region, which usually requires perfect W–C base pairing to recognize target genes (Lewis et al. 2003; Brennecke et al. 2005). In the last several years, different methods have been developed to predict miRNA targets at the genome-wide level with a relatively high accuracy (Lewis et al. 2003; Stark et al. 2003; John et al. 2004; Kiriakidou et al. 2004; Rehmsmeier et al. 2004; Chan et al. 2005; Grun et al. 2005; Krek et al. 2005; Lewis et al. 2005; Rusinov et al. 2005; Lall et al. 2006). More recently, several studies have revealed a greater impact of miRNAs on gene expression and evolution than previously thought (Farh et al. 2005; Lim et al. 2005; Stark et al. 2005; Sood et al. 2006).
In the post-genomic era, a crucial task in molecular biology is to understand gene regulation in the context of biological networks. Recently, two studies about miRNA regulation of cellular networks have been published (Cui et al. 2006, 2007). However, the relationship between PPIN and miRNA regulation remains virtually unknown. Since proteins fulfill their functions largely through PPIN, it is important to study this relationship and understand how the dynamics of PPIN is influenced by miRNAs.
RESULTS
Global correlation between miRNA regulation and PPIN
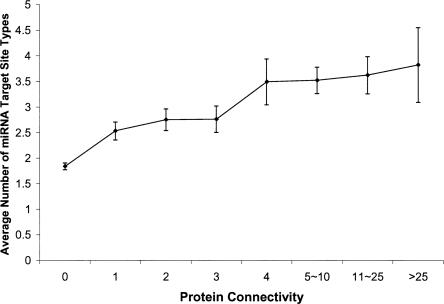
The most important property of a protein node in a PPIN is its connectivity, which is defined as the number of links the node has to other nodes. In general, this statistic quantitatively measures functional complexity of a protein node in the PPIN. To gain a global view of miRNA regulation on PPIN, we studied the relationship between protein connectivity and miRNA regulation complexity, using the largest available human PPIN data set (CCBS1-LCI) (Rual et al. 2005) and the miRNA target predictions from TargetScanS (Lewis et al. 2005), a leading program in the field. Interestingly, we found a highly significant positive correlation between the connectivity of a protein and the number of miRNA target-site types in the 3′ UTRs of the gene encoding the protein (Spearman's rank correlation, Rs = 0.30, P < 2 × 10−118, N = 6165). As shown in Figure 1, the average number of target-site types for a group of genes dramatically increases with their protein connectivity.
FIGURE 1.
A positive correlation between protein connectivity and number of miRNA target-site types. Error bars indicate the standard deviations from the mean values.
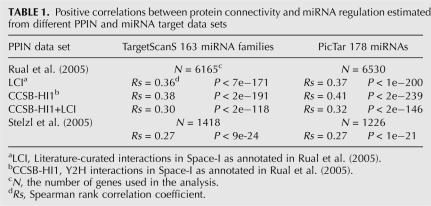
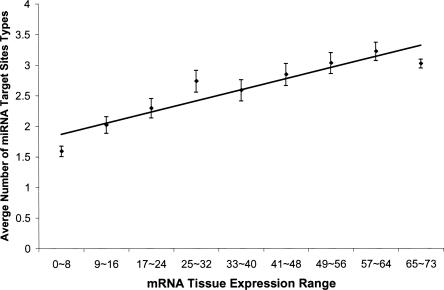
In view of considerable noise in current protein–protein interaction data and miRNA target prediction, we carried out further analyses to examine the validity of the above correlation. First, to see whether the observation is robust for different data sets, we performed the same analysis on four different PPIN data sets (Rual et al. 2005; Stelzl et al. 2005) using two different sets of miRNA target predictions (TargetScanS and PicTar) (Krek et al. 2005; Lewis et al. 2005). As shown in Table 1, all the analyses gave very consistent results, supporting a strong positive correlation between the number of miRNA target-site types and protein connectivity. Second, because the miRNA target prediction programs we used strongly rely on the length and sequence conservation of 3′ UTRs, it is important to rule out the possibility that the above correlation is a side effect of variations in 3′ UTR length and conservation across genes in the PPIN. Through the analyses of 3′ UTRs, we found that the increasing tendency of number of miRNA target-site types with protein connectivity cannot be explained by the corresponding variations in 3′ UTR length and conservation (Supplemental Fig. S1). Finally, using independent mRNA expression data, we studied the relationship between mRNA tissue expression range and miRNA regulation complexity, since tissue expression range can be regarded as another index for functional complexity of a gene. For this purpose, we extracted mRNA expression profiles from the Human Expression Atlas (Su et al. 2004) and calculated the number of tissues in which a gene is expressed. Again, we found a highly significant positive correlation between mRNA tissue expression range and the number of miRNA target-site types (Spearman's rank correlation, Rs = 0.21, P < 2 × 10−140, N = 14,285). In fact, for a group of proteins with a similar tissue expression range (x), the average number of target-site types (y) in the 3′ UTRs of their coding genes can be quantitatively predicted (y = 0.0225x + 1.774, R = 0.93, P < 2 × 10−3) (Fig. 2). This observation is also consistent with a very recent study showing a similar trend in fly and mouse (Yu et al. 2007). Taken together, our results indicate that proteins with more interacting partners tend to be regulated by more miRNAs.
TABLE 1.
Positive correlations between protein connectivity and miRNA regulation estimated from different PPIN and miRNA target data sets
FIGURE 2.
A positive correlation between mRNA expression range and number of miRNA target-site types. The overall tendency is shown with the line from a least squares regression, y = 0.0225x + 1.774, R = 0.93, P < 2 × 10−3, where y is the average number of miRNA target-site types and x is the number of mRNA expressed tissues, which ranges from 0 to 73. Error bars indicate the standard deviations from the mean values.
Given the positive correlation between protein connectivity and the number of miRNA target-site types, one would expect that interacting proteins tend to share more miRNA target-site types than a randomly chosen protein pair, because across the whole PPIN, proteins with a higher connectivity have, on average, more target-site types, and by definition, are involved in more interactions than proteins with a lower connectivity. With this in mind, we investigated whether interacting proteins tend to be coregulated by more miRNAs, compared with the situation where their target-site patterns are randomized. For this purpose, we constructed a Gene-by-TargetSite matrix for genes encoding protein nodes in the PPIN, which contains the information about whether a target site of a given miRNA family is present in a given node. Then we introduced two indexes: (1) the number of protein interacting pairs that share at least one type of target site and (2) the total number of the target-site types shared by interacting partners. To estimate their statistical significance, we generated 10,000 control Gene-by-TargetSite matrices by randomly shuffling the miRNA-TargetSite profile for each gene. We found that, compared with random expectation, both the number of interacting pairs that share at least one type of target site (observation 451 versus expectation 314, Z-score = 9.9, P < 10−24) and the total number of the target-site types shared by interacting partners (observation 698 versus expectation 409, Z-score = 15.4, P < 10−54) are significantly higher. These results suggest a role of miRNA regulation on the coordination of interacting proteins in PPIN.
Biological basis of miRNA targeting propensity in PPIN
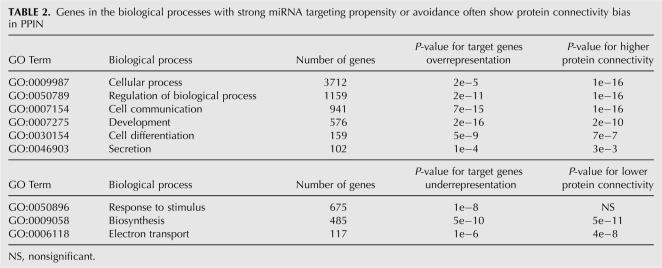
It has been reported that miRNAs have different propensities to target messages in different biological processes or functional categories (Bartel 2004; Lewis et al. 2005; Stark et al. 2005). For example, in animals, miRNA target genes have a strong propensity to be involved in regulatory and development processes, whereas genes in biosynthesis tend to avoid miRNA regulation. However, the basis of miRNA targeting propensity at the system level remains unclear. To address this question, we first used GO term analysis to identify the biological processes where genes strongly favor or avoid miRNA targeting. Then we studied the connectivity distributions of genes in these different processes. As shown in Table 2, genes in the biological processes with strong miRNA targeting propensity have a significantly higher connectivity than other genes, whereas genes in the processes with strong target avoidance tend to have fewer interacting partners. The results suggest that miRNA targeting propensity can be largely explained by the functional behavior of proteins in PPIN.
TABLE 2.
Genes in the biological processes with strong miRNA targeting propensity or avoidance often show protein connectivity bias in PPIN
miRNA regulation of hub proteins
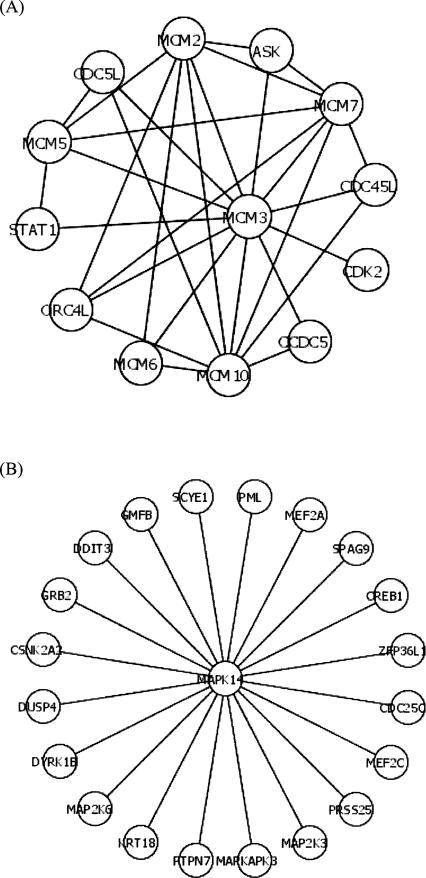
Highly connected proteins, i.e., hub proteins, play a dominant role in maintaining the functionality of the PPIN (Jeong et al. 2001; Barabasi and Oltvai 2004). For hub proteins, besides connectivity, clustering coefficient is another important characteristic, which is defined as the ratio of the number of existing links among a node's neighbors and the maximum possible number of links among them. (In contrast, for poorly connected nodes, clustering coefficient is much less informative. For instance, nodes with “0” or “1” connectivity have a “0” clustering coefficient by definition.) In general, hub proteins with a high clustering coefficient are likely to be intramodular hubs, while hub proteins with a low clustering coefficient tend to be intermodular hubs that coordinate different functional modules. In this study, we focused on the top 5% of protein nodes with the highest connectivity (≥8) and used a multiple variable regression analysis to evaluate the relative impacts of connectivity and clustering coefficient on miRNA regulation complexity of these genes. We found that the number of miRNA target-site types of a gene encoding a hub protein is significantly negatively correlated with clustering coefficient (N = 340, F-statistics = 5.2, P < 0.02). To illustrate this point clearly, two contrasting examples are shown in Figure 3: As an intramodular hub, MCM3, which is a subunit of the prereplication complex, contains no miRNA target sites; whereas, as an intermodular hub, MAPK14, which is involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation, and development, contains 11 types of miRNA target sites. This result suggests that different types of hub proteins have different miRNA targeting propensity: regulation by miRNAs is more important in the behavior of intermodular hubs than in that of intramodular ones.
FIGURE 3.
miRNA regulation of different types of hub proteins. (A) An example of intra-modular hub. MCM3, which is a subunit of the pre-replication complex, contains no miRNA target sites. (B) An example of inter-modular hub. MAPK14, which is involved in a wide variety of cellular processes, contains 11 types of miRNA target sites. The interactions among proteins are shown as lines.
DISCUSSION
Our study reveals a positive correlation between protein connectivity and miRNA regulation complexity; namely, for proteins with more interacting partners, their genes tend to be regulated by more miRNA types. This may reflect a general connection between functional complexity of proteins and miRNA regulation of their coding genes. It has been suggested that miRNA can influence protein production of target genes in a qualitative or quantitative way (Bartel 2004). For some target genes, the so-called “switch targets,” miRNAs reduce their protein expression to a negligible level in certain cell types where they should not be expressed. In this case, miRNA regulation is equivalent to a switch between “on” and “off.” For other messages called “tuning targets,” miRNAs can adjust their protein output to allow customized expression in different cell types (Bartel 2004). From either point of view, proteins with more interacting partners would be under stronger selective pressure to be associated with more complex miRNA regulation: (1) without corresponding miRNA repression, expression of a protein with more interacting partners at undesirable time or place may lead to a more severe fitness effect, because it has a potential to interfere in more interactions; (2) genes with many interactions may require a more tight control of protein production at the post-transcriptional level, thereby exerting protein interactions accurately and efficiently. Consistently, our results also suggest that intermodular hub proteins are more likely to be under miRNA regulation than intramodular hubs. Again, this can be attributed to the functional difference between these two types of hub proteins: intramodular hub proteins interact with most of their partners simultaneously, while intermodular hubs tend to bind different partners at different time and place. Therefore, intramodular hubs would have relatively less pressure to achieve expression diversity. Moreover, our study offers a new perspective to understand functional bias of miRNA targets. Previously known miRNA targeting propensity for genes in different biological processes can be largely explained by their connectivity, which highlights the role of PPIN as a platform to understand gene regulation.
Analyzing miRNA targets in the context of PPIN also provides important insights into how the dynamics of a biological system can be efficiently controlled. In a higher eukaryotic cell, first transcriptional regulation determines mRNA expression specificity and level, then miRNAs, as “micromanagers of gene expression” (Bartel and Chen 2004), regulate protein output, and finally proteins fulfill their functions through the interaction network. During this process, “output” macromolecules from a given step immediately become the inputs in the next step. As demonstrated in present and previous studies (Cui et al. 2007; Yu et al. 2007), large-scale coordination exists at every step: Broadly expressed mRNAs tend to be regulated by more miRNA types; broadly expressed miRNAs tend to regulate more target genes (Supplemental Materials); and proteins with more interacting partners tend to be associated with more extensive miRNA regulation. Moreover, we found the genes of two interacting proteins tend to be under similar miRNA regulation, which parallels the previous finding that genes encoding interacting proteins tend to have similar mRNA expression profiles (Ge et al. 2001; Li et al. 2004; Rual et al. 2005; Wuchty et al. 2006). Via such global or local coordination, a biological system can efficiently control various functional states and improve the performance of each component.
As to the basis of our study, the large-scale protein–protein interaction and miRNA target prediction data we used have proved to be useful for gaining biological knowledge (Farh et al. 2005; Rual et al. 2005; Sood et al. 2006). Although these data sets are far from being complete and may contain considerable noise, it is unlikely that incompleteness or noise can totally distort our results. As to the incompleteness of PPIN, a widely accepted assumption in the field is that, as long as the PPIN is neither strongly biased nor too small, the node-related statistics in general (i.e., connectivity and clustering coefficient) should correlate well with the ones in the complete version. For false positives in PPIN, as can be seen from consistent results across various data sets, this factor appears to have no serious effects on our analysis. In fact, the strength of large-scale data mining lies on reducing noise effect and identifying general trends that would not have been discovered otherwise. A similar comment applies to miRNA target prediction data: although there is some uncertainty in predicting miRNA targets for individual genes, the overall signal inferred from thousands of genes would be strong enough to reflect real biology. Finally, we note that our study may significantly contribute to both PPIN and miRNA research: On the one hand, by incorporating miRNA targeting data, our results make important steps to reveal the dynamic properties and organizational principles of the human PPIN; on the other hand, this study also highlights the potential to improve current miRNA target prediction by adding protein–protein interaction data.
MATERIALS AND METHODS
Analysis on miRNA regulation complexity and protein connectivity in PPIN
We obtained protein–protein interaction data from Rual et al. (2005). The authors used a stringent, high-throughput yeast two-hybrid (Y2H) system to identify binary protein–protein interactions in a “search space” (Space-I) defined by a ∼7200 × 7200 matrix of human protein-coding genes. Combining 2754 Y2H interactions (CCSB-HI1) with 4067 literature-curated interactions (LCI), they presented a PPIN containing 6726 edges (CCSB-HI1+LCI). In the analysis, self-binding protein interactions were excluded. We downloaded miRNA target gene predictions from the TargetScanS Web server (version 3.1) (http://www.targetscan.org/), which included miRNA target prediction for ∼17,000 genes against 163 conserved miRNA families (corresponding to 238 miRNAs). In our analysis, we focused on those conserved miRNA target sites (across human, mouse, rat, and dog), because (1) they have been conserved in the long history of eutherian evolution and may represent the most important target sites, (2) such a requirement gives a relatively high signal-to-noise ratio (Lewis et al. 2005), and (3) such a requirement largely reduces the distraction of “neutral targets” (Bartel and Chen 2004), which are under no selective pressure and are very likely to be nonconserved. In the analysis, we used 6165 genes within the intersection between miRNA target prediction and PPIN data sets. Since the underlying distributions of protein connectivity and the number of miRNA target-site types were not normal, we used Spearman's rank correlation test to determine whether there is a monotonic relation between two variables.
To test the robustness of our results, we performed the analysis using additional PPIN and miRNA target data sets. (1) We used CCSB-HI1 and LCI protein–protein data separately. These two data sets have different merits and drawbacks: CCSB-HI1 is not biased toward any particular biological interest, while LCI is largely free of technical false positives. Our results appear not to be influenced by these potential confounding factors. (2) We used independent protein–protein interaction data from Stelzl et al. (2005), which presents a PPIN of 1705 nodes using Y2H. (3) We used another set of miRNA targets from PicTar, another leading prediction program. We obtained PicTar targets from the UCSC Genome Browser, which covered target prediction for ∼15,000 genes (after converting RefSeq transcripts into genes) against 178 miRNAs. We found very consistent results as using the TargetScanS predictions. Through the rest of this study, we focused on CCSB-HI1+LCI PPIN and TargetScanS miRNA target data sets, since they are much more extensive.
For mRNA expression range analysis, we used the second version of the Gene Expression Atlas (Su et al. 2004), which contained the mRNA expression patterns of human genes across 73 normal tissues. We downloaded the data and its annotation from the NCBI GEO database and calculated mRNA expression level as described elsewhere (Farh et al. 2005). We used an AD value of 200 as the threshold calling a gene “expressed in a given tissue,” because a value below 200 largely reflects background noise (Su et al. 2002; Yang et al. 2005). In the analysis, we used 14,285 genes within the intersection between mRNA expression and miRNA TargetScanS prediction data sets.
To find whether interacting proteins tend to share more target-site types, we first constructed a 6165 × 163 Gene-by-TargetSite matrix: for a gene and a miRNA family, if the gene contained the miRNA target site, it was marked “1,” otherwise, “0.” Then we generated 10,000 control Gene-by-TargetSite matrices by randomly shuffling the miRNA-TargetSite profile for each gene. This shuffling allowed us to consider target-site patterns separately from the number of target-site types, because the latter was maintained constant for each gene in the randomization process. For each index, the probability of the observed value was derived from the empirical background distribution.
GO term analysis
To study miRNA targeting propensity in the PPIN, we used the GO Term Mapper Web server at Princeton University (http://go.princeton.edu/cgi-bin/GOTermMapper). Among the 6165 genes, 4813 genes were annotated into different biological processes by the default GOA slim file (a list of general GO terms). For each biological process, we performed a Wilcoxon rank test to determine whether the number of miRNA target-site types of genes in the process is significantly different than the remaining genes. To count the effect of multiple testing, 0.002 was used to call a biological process with strong miRNA targeting propensity (or avoidance), which is equivalent to the conventional threshold (0.05) after Bonferroni correction. Then for each of the identified nine slim GO terms, we used a Wilcoxon rank test (one-tailed) to test whether there is a corresponding variation (significantly higher or lower) in terms of protein connectivity of genes in the biological process.
Multiple variable regression analysis of hub proteins
The clustering coefficient of a node i is calculated as Ci = 2ni/k(k−1), where n is the number of links connecting the k neighbors of node i to each other. To study the impacts of connectivity and clustering coefficient on miRNA regulation of hub proteins, for 340 hub proteins (connectivity ≥8), we performed a multiple variable linear regression analysis among the number of miRNA target-site types (y), protein connectivity (x 1), and clustering coefficient (x 2). The forward selection algorithm was used to choose variable(s) based on Akaike Information Criterion (the lower bound was set to contain no variable, and the upper bound was set to contain both variables). F-test was used to determine the statistical significance of a correlation, which is known to be robust for non-normal distributions.
SUPPLEMENTAL DATA
Supplemental material can be found at http://pondside.uchicago.edu/∼lilab/downloads/Liang&Li_RNA_suppl.pdf.
ACKNOWLEDGMENTS
We thank Henry Lu, Matt Saunders, Lingling Chen, and Ay Prachumwat for suggestions. We also thank Nikolaus Rajewsky and Dominic Gruen for their help in obtaining PicTar miRNA target prediction data. This work was supported by NIH grants to W.-H.L.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.634607.
REFERENCES
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Barabasi, A.L., Oltvai, Z.N. Network biology: Understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P., Chen, C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Stark, A., Russell, R.B., Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, C.S., Elemento, O., Tavazoie, S. Revealing posttranscriptional regulatory elements through network-level conservation. PLoS Comput. Biol. 2005;1:e69. doi: 10.1371/journal.pcbi.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Q., Yu, Z., Purisima, E.O., Wang, E. Principles of microRNA regulation of a human cellular signaling network. Mol. Syst. Biol. 2006;2:46. doi: 10.1038/msb4100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Q., Yu, Z., Pan, Y., Purisima, E.O., Wang, E. MicroRNAs preferentially target the genes with high transcriptional regulation complexity. Biochem. Biophys. Res. Commun. 2007;352:733–738. doi: 10.1016/j.bbrc.2006.11.080. [DOI] [PubMed] [Google Scholar]
- Farh, K.K., Grimson, A., Jan, C., Lewis, B.P., Johnston, W.K., Lim, L.P., Burge, C.B., Bartel, D.P. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Ge, H., Liu, Z., Church, G.M., Vidal, M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae . Nat. Genet. 2001;29:482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- Giot, L., Bader, J.S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y.L., Ooi, C.E., Godwin, B., Vitols, E., et al. A protein interaction map of Drosophila melanogaster . Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Grun, D., Wang, Y.L., Langenberger, D., Gunsalus, K.C., Rajewsky, N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J.D., Bertin, N., Hao, T., Goldberg, D.S., Berriz, G.F., Zhang, L.V., Dupuy, D., Walhout, A.J., Cusick, M.E., Roth, F.P., et al. Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., Sakaki, Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H., Mason, S.P., Barabasi, A.L., Oltvai, Z.N. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- John, B., Enright, A.J., Aravin, A., Tuschl, T., Sander, C., Marks, D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou, M., Nelson, P.T., Kouranov, A., Fitziev, P., Bouyioukos, C., Mourelatos, Z., Hatzigeorgiou, A. A combined computational–experimental approach predicts human microRNA targets. Genes & Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek, A., Grun, D., Poy, M.N., Wolf, R., Rosenberg, L., Epstein, E.J., MacMenamin, P., da Piedade, I., Gunsalus, K.C., Stoffel, M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- LaCount, D.J., Vignali, M., Chettier, R., Phansalkar, A., Bell, R., Hesselberth, J.R., Schoenfeld, L.W., Ota, I., Sahasrabudhe, S., Kurschner, C., et al. A protein interaction network of the malaria parasite Plasmodium falciparum . Nature. 2005;438:103–107. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- Lall, S., Grun, D., Krek, A., Chen, K., Wang, Y.L., Dewey, C.N., Sood, P., Colombo, T., Bray, N., Macmenamin, P., et al. A genome-wide map of conserved microRNA targets in C. elegans . Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., Burge, C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Burge, C.B., Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li, S., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D., Chesneau, A., Hao, T., et al. A map of the interactome network of the metazoan C. elegans . Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, L.P., Glasner, M.E., Yekta, S., Burge, C.B., Bartel, D.P. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier, M., Steffen, P., Hochsmann, M., Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual, J.F., Venkatesan, K., Hao, T., Hirozane-Kishikawa, T., Dricot, A., Li, N., Berriz, G.F., Gibbons, F.D., Dreze, M., Ayivi-Guedehoussou, N., et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Rusinov, V., Baev, V., Minkov, I.N., Tabler, M. MicroInspector: A web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33:W696–W700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood, P., Krek, A., Zavolan, M., Macino, G., Rajewsky, N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Russell, R.B., Cohen, S.M. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:e60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Bushati, N., Russell, R.B., Cohen, S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′-UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stelzl, U., Worm, U., Lalowski, M., Haenig, C., Brembeck, F.H., Goehler, H., Stroedicke, M., Zenkner, M., Schoenherr, A., Koeppen, S., et al. A human protein–protein interaction network: A resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Su, A.I., Cooke, M.P., Ching, K.A., Hakak, Y., Walker, J.R., Wiltshire, T., Orth, A.P., Vega, R.G., Sapinoso, L.M., Moqrich, A., et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, A.I., Wiltshire, T., Batalov, S., Lapp, H., Ching, K.A., Block, D., Zhang, J., Soden, R., Hayakawa, M., Kreiman, G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae . Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Vidal, M. Interactome modeling. FEBS Lett. 2005;579:1834–1838. doi: 10.1016/j.febslet.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Wuchty, S., Barabási, A.-L., Ferdig, M.T. Stable evolutionary signal in a yeast protein interaction network. BMC Evol. Biol. 2006;6:8. doi: 10.1186/1471-2148-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Su, A.I., Li, W.H. Gene expression evolves faster in narrowly than in broadly expressed mammalian genes. Mol. Biol. Evol. 2005;22:2113–2118. doi: 10.1093/molbev/msi206. [DOI] [PubMed] [Google Scholar]
- Yu, Z., Jian, Z., Shen, S.-H., Purisima, E., Wang, E. Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res. 2007;35:152–164. doi: 10.1093/nar/gkl1032. [DOI] [PMC free article] [PubMed] [Google Scholar]