Abstract
A number of highly regulated gene classes are regulated post-transcriptionally at the level of mRNA stability. A central feature in these mRNAs is the presence of A+U-rich elements (ARE) within their 3′ UTRs. Two ARE binding proteins, HuR and AUF1, are associated with mRNA stabilization and destabilization, respectively. Previous studies have demonstrated homomultimerization of each protein and the capacity to bind simultaneous or competitively to a single ARE. To investigate this possibility further, cell biological and biophysical approaches were undertaken. Protein–protein interaction was monitored by fluorescence resonance energy transfer (FRET) and by immunocytochemistry in live and fixed cells using fluorescently labeled CFP/YFP fusion proteins of HuR and p37AUF1. Strong nuclear FRET between HuR/HuR and AUF1/AUF1 homodimers as well as HuR/AUF1 heterodimers was observed. Treatment with the MAP kinase activator, anisomycin, which commonly stabilizes ARE-containing mRNAs, caused rapid nuclear to cytoplasmic shuttling of HuR. AUF1 also underwent shuttling, but on a longer time scale. After shuttling, HuR/HuR, AUF1/AUF1, and HuR/AUF1, FRET was also observed in the cytoplasm. In further studies, arsenite rapidly induced the formation of stress granules containing HuR and TIA-1 but not AUF1. The current studies demonstrate that two mRNA binding proteins, HuR and AUF1, are colocalized and are capable of functional interaction in both the nucleus and cytoplasm. FRET-based detection of AUF1/HuR interaction may serve as a basis of opening up new dimensions in delineating the functional interaction of mRNA binding proteins with RNA turnover.
Keywords: FRET, ARE, HuR, AUF1, protein–protein interaction
INTRODUCTION
The life cycle of an mRNA transcript from its genesis through its subsequent translation and ultimately to its degradation in the cytoplasm is a highly complex and tightly controlled biochemical process. One level of regulation is the differential interaction of numerous mRNA binding proteins that bind to a transcript and form a ribonucleoprotein complex (RNP) (Dreyfuss 1986; Parker and Song 2004; Moore 2005). Each mRNA transcript has several proteins that bind to its 5′-cap structure and to its 3′-untranslated region (3′-UTR), while other proteins bind to specific sequence motifs on the mature mRNA transcript and perform tasks such as delineating exon junctions, facilitating the translocation of the mRNA from the nucleus to the cytoplasm and to specific intracellular sites for translational silencing or for degradation (Kuersten and Goodwin 2005). Of particular interest is the class of A+U-rich (ARE) mRNA binding proteins involved in regulating mRNA turnover. Recent estimates indicate that 5%–8% of all genes (∼4000) contain AREs (Bakheet et al. 2006) and are thus subject to rapid decay rates (Chen et al. 1995; Yang et al. 2003).
A number of ARE binding proteins have been identified. Among these is HuR (Ma et al. 1996), a member of the ELAV/Hu family (Robinow and White 1988), which contains the closely related proteins, HuB, HuC, and HuD. Under unstimulated conditions, HuR is primarily localized to the nucleus, but its movement to the cytoplasm has been associated with stabilization of mRNA transcripts (Atasoy et al. 1998; Fan and Steitz 1998a,b). HuR itself has also been shown to be regulated at the protein level (Martinez-Chantar et al. 2006), with changes in expression correlating with a growth phenotype (Blaxall et al. 2000a; Gouble and Morello 2000) and with changes in phosphorylation affecting localization and function (Abdelmohsen et al. 2007; Doller et al. 2007). In contrast to HuR, a number of other proteins, e.g., TTP, KSRP, and BRF1, confer instability to ARE-containing mRNAs (Taylor and Blackshear 1995; Taylor et al. 1996; Carballo et al. 2000; Ming et al. 2001; Stoecklin et al. 2002; Gherzi et al. 2004; Lu and Schneider 2004; Raineri et al. 2004; Schmidlin et al. 2004; Linker et al. 2005; Ogilvie et al. 2005). Interestingly, the family of AUF1/hnRNPD mRNA binding proteins appears to be able to confer either stability or instability to target mRNAs, the effect being cell type and AUF1 isoform-dependent (Laroia and Schneider 2002). The AUF1 gene is expressed as four isoforms designated by their molecular weights as p37, p40, p42, and p45, which are generated by alternative splicing of a single pre-mRNA transcript (Wagner et al. 1998). Common to all four isoforms of AUF1 is an N-terminal, alanine-rich stretch of 28 amino acids putatively required for dimerization of the isoforms in solution and a C-terminal glutamine-rich domain required for high-affinity ARE binding (DeMaria et al. 1997). All four isoforms of AUF1 are predominantly nuclear (Sarkar et al. 2003a) and are able to shuttle between the nucleus and cytoplasm in response to environmental stimuli (Kawamura et al. 2002; Sarkar et al. 2003a; Suzuki et al. 2005).
In this report, we focus on the ARE binding proteins, HuR and AUF1, using a combined cell biological and biophysical approach. Previous reports describing HuR and AUF1 have relied on biochemical techniques that allow for the elucidation of protein–protein and protein–RNA interactions by immunoprecipitation and UV cross-linking studies. However, these biochemical techniques are unable to address the subcellular localization and interactions of ARE binding proteins and RNA in real time. Further, immunoprecipitation colocalization studies are inherently limited in terms of absolute resolution. In contrast, fluorescence microscopy and immunocytochemistry, when combined with fluorescence resonance energy transfer (FRET), allows for the detection of stably associated proteins in live cells in real time (Gordon et al. 1998; Vogel et al. 2006). Using these techniques, we were able to detect protein–protein interactions for p37AUF1 with itself, HuR with itself, and HuR with p37AUF1, in both the nucleus and cytoplasm in real time. We also show that the association of HuR with AUF1 is responsive to the intracellular environment and is subject to conditions of cellular stress such that the association between HuR and AUF1 can be disrupted resulting in the proteins being relocalized to different subcellular compartments within the cytoplasm.
RESULTS
Validation of FRET detection in DDT1-MF2 cells
Fluorescent resonance energy transfer is a powerful technique capable of identifying stable interactions between proteins both in fixed cells and in live cells in real time. To accurately measure FRET, the raw FRET signal is corrected for by subtracting the spectral bleed-through of each fluorescent protein into the FRET channel. A number of CFP and YFP constructs were individually transfected into DDT1-MF2 cells to measure the bleed-through of each fluorescent protein (FP) construct into the FRET channel. The calculated bleed-through number for the individual fluorophores was then used to verify the lack of FRET between noninteracting FP–protein constructs. The spectral bleed-through number empirical correction (see Materials and Methods) allowed for the determination of CFP and YFP values that produce a slight overcorrection (negative value) for FRET. Using this method ensures that the FRET signals detected between pairs of FP labeled proteins are a true indicator of protein interaction.
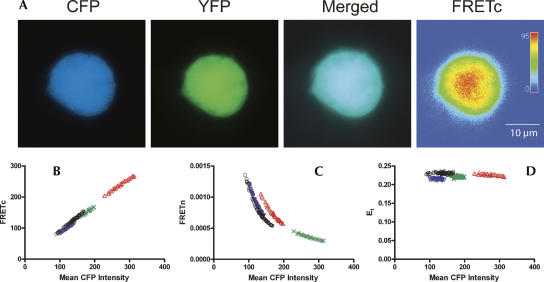
Empirically corrected CFP and YFP spectral bleed-through values were used to calculate FRET intensity using a tandem CFP:YFP construct (Galperin and Sorkin 2003), which served as a positive control. By definition, a one-to-one molar ratio of CFP to YFP yields the desired 1:10 signal intensity ratio of CFP:YFP. Figure 1 shows a representative image of a DDT1-MF2 smooth muscle cell transfected with the tandem CFP:YFP construct. Figure 1A depicts the individual channel signals as well as the merged CFP/YFP channel and FRETc signal. As expected, the tandem CFP:YFP protein is expressed uniformly throughout the cell. Due to the spherical nature of DDT1-MF2 cells, an architecture particularly evident in live cells, the FRETc image is of higher signal intensity at the center and of lower intensity at the cell's periphery. Mean CFP, YFP, and raw FRET intensity values collected from identically sized masks (1.7 μm in diameter) across the entire cell were used to calculate two different FRET indices, FRETc and FRETn, and the theoretical FRET efficiency, Et, (Fig. 1B–D). When FRETc is plotted versus CFP, a linear relationship independent of CFP intensity is observed. A linear relationship is also obtained if FRETc is plotted versus the mean YFP intensity (data not shown). Although FRETn can be a highly useful means of FRET correction, because of the nonlinear relationship between the abundance of donors or acceptors and FRETn (Vogel et al. 2006), we chose not to use this method of correction. Whereas FRETc and FRETn values are specific to each experimental setup, the FRET efficiency, E, allows direct comparison between experimental setups for the same cell type and fluorescent protein FRET pairs (Vogel et al. 2006). The theoretical FRET efficiency (percent) for the tandem CFP:YFP construct, which is the maximal FRET efficiency obtainable for this fluorescent protein pair in DDT1-MF2 cells, is 0.22 (Table 1). The FRET efficiencies for HuR/HuR, p37AUF1/p37AUF1, and HuR/p37AUF1, by definition, will be lower than the theoretical maximum obtained for the tandem CFP:YFP protein. Of the three interactions measured (see below), the HuR homodimer exhibited the highest FRET efficiency with E=0.11. The efficiency of the p37AUF1 homodimer was somewhat less with E=0.07. Finally, the HuR/p37AUF1 heterodimer exhibited the lowest efficiency with E=0.04 (Table 1).
FIGURE 1.
FRET positive control and representative FRET calculations. A tandem vector construct of eCFP and eYFP was used as a positive control for the detection of FRET in DDT1-MF2 cells (Galperin et al. 2004). Slidebook software (v4.1.0.4) was used to obtain mean intensity values from identically sized areas sampled from the entire cell for CFP, YFP, and the raw FRET signal. These data were used to calculate FRETc, FRETn, and the FRET efficiency value, E, which are shown plotted vs. the mean CFP intensities. The maximal FRET efficiency (Et) obtained for CFP:YFP in DDT1-MF2 cells is 0.22±0.02. (A) DDT1-MF2 cell images. The CFP, YFP, and merged channel views show the uniform intracellular distribution of the CFP and YFP proteins throughout the cell. The FRETc image is shown in thermal gradient pseudocolor, where red indicates the strongest signal. The size scale bar (μm) applies to all images. (B) FRETc versus mean CFP intensity. (C) FRETn versus mean CFP intensity. (D) Et versus mean CFP intensity.
TABLE 1.
FRET efficiency for the YFP:CFP pair in DDT1-MF2 cells
Homo- and heterocomplex formation of HuR and AUF1 detected via FRET in live and fixed cells
It is generally accepted that both HuR and AUF1 can compete for binding to the same ARE on a specific RNA. More recently, it has been reported that HuR and AUF1 are capable of associating with the same target RNA on nonoverlapping ARE sites (Lal et al. 2004). This leads to the possibility of HuR and AUF1 binding to target mRNAs simultaneously and the possibility of direct protein–protein interaction for HuR and AUF1. Therefore, we wished to examine the potential for protein–protein interaction of HuR and p37AUF1 using FRET in both fixed cells and in live cells in real time.
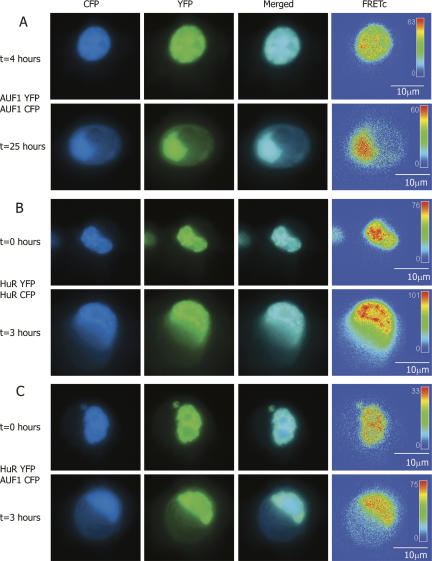
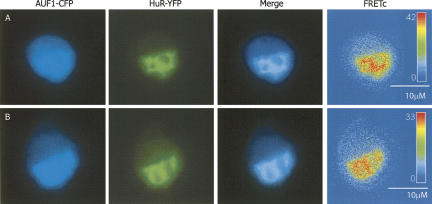
In previous studies, AUF1 has been shown to form homomultimers (DeMaria and Brewer 1996; DeMaria et al. 1997; Wilson et al. 1999). However, the multimeric state of AUF1 in the nucleus and cytoplasm has not been determined in vivo. Figure 2A shows the intracellular localization of p37AUF1 proteins labeled with either eYFP or eCFP and cotransfected into DDT1-MF2 cells. In unstimulated cells, and at times of <4 h, p37AUF1 protein was present almost exclusively in the nucleus. The formation of higher-order stable complexes between the two fluorescently labeled monomers of p37AUF1 is indicated by the strongly positive FRETc signal.
FIGURE 2.
FRET-based detection of p37AUF1/p37AUF1, HuR/HuR, and p37AUF1/HuR protein pair interactions in live cells. DDT1-MF2 cells were transiently transfected with pairs of complimentary, fluorescently labeled constructs of HuR and/or p37AUF1. Transfected cells were treated with anisomycin to stimulate nuclear/cytoplasmic shuttling of mRNA binding proteins. (A) p37AUF1-eYFP and p37AUF1-eCFP interaction. After 4 h of treatment with anisomycin, p37AUF1 remains localized to the nucleus (top row) and exhibits colocalization (merged channel column) detectable by FRETc, which is indicative of a stable homodimeric interaction (top row, FRETc column). After 25 h of anisomycin treatment, cytoplasmic shuttling of p37AUF is readily apparent (bottom row). The p37AUF1 present in the cytoplasm colocalizes, which is detectable by the merged channel view and the FRETc signal present in both the nucleus and cytoplasm. (B) HuR-eYFP and HuR-eCFP interaction. At time zero, HuR is present in the nucleus, and exhibits colocalization and a robust FRETc signal (top row). Anisomycin causes rapid shuttling of HuR to the cytoplasm, and a FRET-detectable, stable interaction between HuR is present in both the nucleus and cytoplasm (bottom row). (C) HuR-eYFP and p37AUF1-eCFP interaction. At time zero, p37AUF1 and HuR are present in the nucleus, colocalize, and display a FRETc signal indicative of a stable heteromeric interaction (top row). Treatment with anisomycin causes shuttling of both HuR and p37AUF1 into the cytoplasm. HuR and p37AUF1 colocalize in the cytoplasm and exhibit a FRET-detectable interaction in both the nuclear and cytoplasmic compartments. FRETc images are shown in thermal gradient pseudocolor. The scale bars in the FRETc image applies to all images in the same row.
Figure 2B shows a similar experiment for eYFP- and eCFP labeled HuR proteins. In unstimulated cells, both fluorescently labeled forms of HuR are localized to the nucleus. The strongly positive FRETc signal indicates that HuR is also forming a stable homomultimeric complex in the nucleus.
Given that both HuR and AUF1 may be part of a larger RNA mRNA binding protein complex, we wished to examine the possibility that there was direct protein–protein interaction between the two proteins in both nuclear and cytoplasmic compartments. Since HuR is typically found associated with the ribosomal and higher molecular weight polysome fractions, compartments that typically lack AUF1 (Pende et al. 1996; Lal et al. 2004), the possibility existed that the association of HuR and AUF1 might be location specific. Figure 2C shows a cell transiently cotransfected with both HuR-eYFP and p37AUF1-eCFP. In unstimulated cells, HuR and p37AUF1 colocalize to the nucleus. As indicated by the robust FRETc signal, HuR and p37AUF1 appear to form stable heterodimers.
To verify the specificity of these protein–protein interactions, additional FRET studies were performed with proteins not expected to interact with HuR. To this end, eCFP versions of the cytoplasmically expressed protein, PK-A R11A, and the nuclear and cytoplasmically localized protein, PH-PL-C, were analyzed. In each case, no evidence of FRET pairing with HuR-eYFP was evident (data not shown).
We have previously shown that anisomycin, a MAPK (p38, JNK) activator, increases the half-life of the β2-adrenergic receptor (AR) mRNA, an ARE-containing mRNA endogenously expressed in DDT1-MF2 cells (Headley et al. 2004). Thus, the β2-AR is similar to other ARE-containing mRNAs in that MAPK activation prolongs their half-lives (Mahtani et al. 2001; Dean et al. 2003). Further, the movement of HuR from nucleus to cytoplasm has been correlated with its ability to stabilize ARE-containing mRNAs (Atasoy et al. 1998). The anisomycin-dependent increase in the half-life of ARE-containing mRNAs could be due to increased binding of stabilizing factors, such as HuR, a decrease in the binding of destabilizing factors, such as AUF1, or a combination of both. Therefore, we wished to determine the effect of anisomycin on the intracellular localization of HuR and AUF1 in live cells.
To explore HuR shuttling at the level of higher-order associations, FRET was used to assess the interaction of HuR with itself once it has transited from nucleus to cytoplasm. As demonstrated in Figure 2B (bottom row), movement of HuR from the nucleus to the cytoplasm does not alter its ability to form homodimers. In contrast to the rapid shuttling observed for HuR, anisomycin-induced shuttling of p37AUF1 is a prolonged process that can take in excess of 24 h (Fig. 2A, bottom row). Like HuR, and as indicated by the FRETc signal, p37AUF1 maintains its ability to form homodimers in the cytoplasm. Similarly, anisomycin stimulated shuttling of AUF1 and HuR does not alter their heterodimeric association (Fig. 2C, bottom row).
These FRETc images indicate that HuR and AUF1 are in sufficient proximity to functionally associate in both the nucleus and the cytoplasm. Interestingly, the length of time required for anisomycin-induced shuttling of p37AUF1 appears greatly different when p37AUF1 is present as a complex with HuR (cf. Fig. 2C, bottom row, and Fig. 2A, bottom row). Although speculative, it is possible that the formation of a stable complex of p37AUF1 with HuR permits AUF1 to shuttle out of the nucleus on a shorter “HuR” time scale, when stimulated by anisomycin. This remains to be demonstrated.
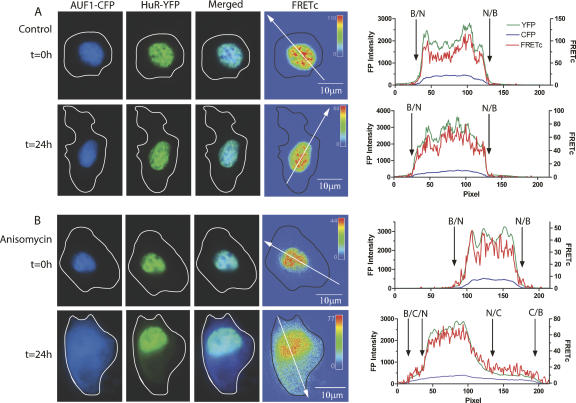
To verify that the images obtained for AUF1 and HuR in live cells could be recapitulated in fixed cells, a parallel set of experiments was performed. Figure 3, A and B, shows the intracellular localization of HuR-eYFP and p37AUF1-eCFP in fixed DDT-MF2 cells. Under control conditions, HuR and p37AUF1 remain colocalized within the nucleus, producing a high-intensity FRET signal. A graph depicting the intensity of each fluorescent protein and their associated FRET signals is presented to the right of each panel of images. The line intensity graph demonstrates clearly that in the control, untreated cells, there is no movement of HuR or p37AUF1 from the nucleus to the cytoplasm. However, following 24 h of anisomycin treatment, a substantial proportion of p37AUF1-eCFP and HuR-eYFP shuttled from nucleus to cytoplasm. A FRETc signal is present in the nucleus and throughout the entire cytoplasm with the highest signal intensity being found closest to the nucleus. Whether the decrease in FRET signal intensity further away from the nucleus is due to dissociation of HuR from p37AUF1 or due merely to lower concentrations of HuR/p37AUF1 complexes is not known. As has been noted previously, both HuR and p37AUF1 are excluded from the nucleoli of the nucleus (Fan and Steitz 1998a; Fawal et al. 2006). This can be seen clearly from the line graphs, which indicate that the concentration of both HuR and p37AUF1 in the nucleus is nonhomogeneous. The concordance of the results presented in Figures 3 and 2 demonstrates that data obtained from fixed cells can be an accurate representation of the live cell conditions in DDT1-MF2 cells.
FIGURE 3.
Anisomycin-induced shuttling of HuR and p37AUF1 in fixed cells. DDT1-MF2 cells were transiently transfected with HuR-eYFP and p37AUF1-eCFP and treated with anisomycin for up to 24 h. HuR-eYFP and p37AUF1-eCFP are present in the nucleus of both the vehicle- and anisomycin treated cells at time zero (A,B, t=0 h, CFP and YFP columns). The merged CFP:YFP image shows that both proteins colocalize to the nucleus but also exhibit a non-identical nuclear distribution (see graphs in right panel). The FRETc image indicates that HuR and p37AUF1 are in sufficiently close proximity in the nucleus to be of functional significance (A,B, t=0 h, FRETc column). After 24 h of vehicle treatment, HuR and p37AUF1 remain in the nucleus (A, t=24 h). After 24 h of anisomycin treatment, shuttling of HuR and p37AUF1 from nucleus to cytoplasm is readily apparent (B, t=24 h). The FRETc image indicates the continued stable interaction of HuR and p37AUF1 in the nucleus and that the proteins retain this interaction in the cytoplasm. (Free-form line) Cell boundary for each image. The intensity for each image in the FRETc column is represented in a thermal gradient pseudocolor. The scale bar in the FRETc image applies to the entire row. (Graphs) The CFP and YFP intensities (left axis) and calculated FRETc (right axis) as described by the white directional arrow in the FRETc panel. (Black arrows) Location of the nuclear/cytoplasmic/background transition, (N) nucleus, (C) cytoplasm, (B) background.
Detection of endogenously expressed HuR and AUF1
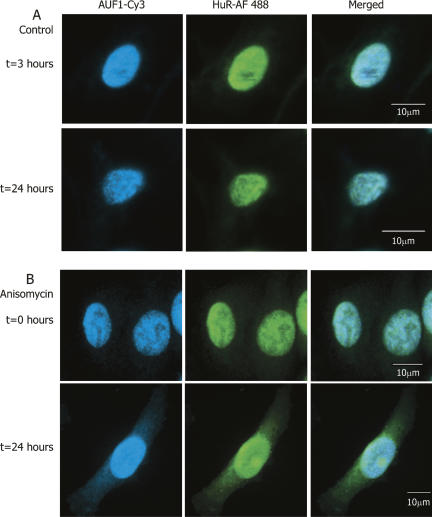
To ensure that the results observed for ectopic expression of fluorescently tagged HuR and p37AUF1 are not artifactual, immunocytochemistry was employed to detect endogenously expressed HuR and AUF1. These data, represented in Figure 4, reveal that under both unstimulated and anisomycin-stimulated conditions, endogenous HuR and AUF1 exhibit a highly similar intracellular localization to that of ectopically expressed, fluorescently tagged proteins. In general, the immunostained cells show a somewhat greater amount of HuR and AUF1 present in the cytoplasm at “t=0” than do transiently transfected cells. It is possible that this result is due to the relative ability of the antibody to detect cytoplasmic HuR. AUF1 is also detected to a greater extent at time zero in both control and anisomycin-treated cells. It is possible that this may be due to the ability of the antibody to detect all four isoforms of AUF1, whereas in overexpression studies only p37AUF1 was examined.
FIGURE 4.
Colocalization of endogenously expressed HuR and AUF1. Immunocytochemistry was used to detect the intracellular location of endogenously expressed HuR and AUF1 proteins in DDT1-MF2 cells. (A) In control cells, endogenous HuR (Alexa Fluor 488 conjugated 2° antibody) and AUF1 (Cy3 conjugated 2° antibody) are expressed almost exclusively in the nucleus and are shown to colocalize. (B) Anisomycin treatment induces shuttling of endogenous AUF1 and HuR from the nucleus to the cytoplasm (cf. t=0 h and t=24 h). The merged view shows the colocalization of HuR and AUF1 in the nucleus and the cytoplasm after 24 h of anisomycin treatment. The scale bars in the merged view apply to all images in the same row.
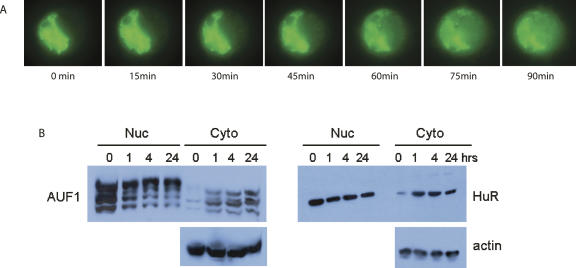
To examine in greater detail the time frame of HuR nuclear/cytoplasmic shuttling, a series of time-lapse images of a single live cell transiently transfected with HuR-eYFP was examined (Fig. 5A). Upon treatment of anisomycin, HuR moves rapidly from its primarily nuclear localization to a high concentration in the cytoplasm. To verify this result biochemically, Western blots of HuR and AUF1 were performed on both cytosolic and nuclear fractions from anisomycin-treated DDT1-MF2 cells. Figure 5B demonstrates that anisomycin induces marked shuttling of both HuR and AUF1 from nucleus to cytoplasm with notable detection of HuR protein by 1 h. In contrast, the time course of AUF1 accumulation in the cytoplasm was somewhat slower and proportionately less robust. These results confirm that stabilization of ARE-containing transcripts by MAPK activation temporally coincides with the increased presence of HuR in the cytoplasm.
FIGURE 5.
MAPK-induced nuclear/cytoplasmic shuttling of ARE binding proteins. (A) DDT1-MF2 cells were transiently transfected with HuR-eYFP. Anisomycin was added to the cells to stimulate nuclear to cytoplasmic shuttling. A single cell was tracked for the time course indicated. eYFP-labeled HuR rapidly shuttles from the nucleus to the cytoplasm. Cells were maintained at 37°C, 5% CO2 for the duration of the experiment. (B) Following treatment with anisomycin (1–24 h), DDT1-MF2 cells were harvested and nuclear and cytosolic fractions isolated. Immunodetection of endogenous AUF1 (p37, p40, p42, p45) and HuR was performed by Western blotting. For normalization, blots were stripped and reprobed for actin protein.
HuR, not AUF1, is present in stress granules
The formation of nuclear and cytoplasmic complexes of HuR and AUF1 shown above builds significantly on previous reports indicating the presence of HuR and AUF1 in the cytoplasm. Further, previous reports have shown HuR to be present on polysomes in cytoplasmic cell fractions, whereas AUF1 was not found in these same fractions (Lal et al. 2004). However, data are emerging indicating that ARE mRNA binding protein complexes are both large and dynamic (Tenenbaum et al. 2002; Mazan-Mamczarz et al. 2003; Lal et al. 2004; Keene and Lager 2005). Under conditions of stress, discrete cytoplasmic entities composed of high concentrations of proteins and RNA have been identified (Abraham et al. 2002; Kedersha and Anderson 2002; Cougot et al. 2004; Brengues et al. 2005; Kedersha et al. 2005; Anderson and Kedersha 2006). These “cytoplasmic” or “stress” granules have been shown to be composed of proteins involved in mRNA translation initiation, translation control, and mRNA decay (Newbury et al. 2006).
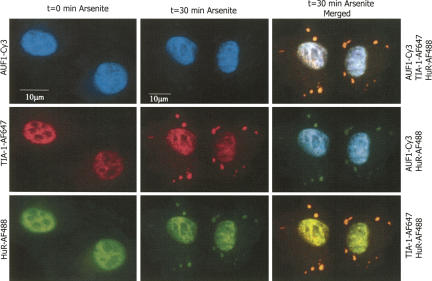
We were interested in determining the localization and interaction of HuR with AUF1 during times of cellular stress. Figure 6 shows immunocytochemical detection of several mRNA binding proteins in DDT1-MF2 cells in response to sodium arsenite. TIA-1 was used as a marker of stress granules (Kedersha et al. 1999, 2000, 2005). The left-hand column of Figure 6 shows the individual channel data at time zero prior to arsenite treatment. As noted previously, endogenous AUF1 and HuR are present mainly in the nucleus, whereas TIA-1 is present in both the nucleus and to some degree in the cytoplasm. In the absence of arsenite treatment, no stress granules are detected (left column). However, 30 min after arsenite treatment, TIA-1-positive stress granules are readily detected (Fig. 6, center column). Arsenite-induced stress also causes HuR to move into stress granules, and the colocalization of HuR and TIA-1 to the stress granules is readily apparent. The observed stress-induced movement of HuR to cytoplasmic granules is consistent with previous results using other types of stressors (Kedersha et al. 2000, 2005; Kedersha and Anderson 2002). In contrast, arsenite treatment had no obvious effect on the intracellular localization of AUF1.
FIGURE 6.
HuR, but not AUF1, is present in arsenite-induced stress granules. Sodium arsenite was used to induce stress granule formation in DDT1-MF2 cells. TIA-1 was used as a marker for stress granule formation. Immunocytochemistry employing triple antibody staining shows that in untreated cells, HuR, TIA-1 and AUF1 are all localized in the nucleus with a lesser amount of TIA-1 and HuR present in the cytoplasm (left column, t=0 min Arsenite). Rapid formation of stress granules containing HuR and TIA-1 occurs after 30 min of arsenite treatment, (center column, t=30 min Arsenite; right column, merged views). Sodium arsenite did not affect the cellular localization of AUF1, (center column, t=30 min Arsenite; right column, merged views). The scale bar in the left column applies to all images in this column. The scale bar in the center column applies to all images in this and the right column.
AUF1 and HuR are able to bind to a target ARE in an additive manner
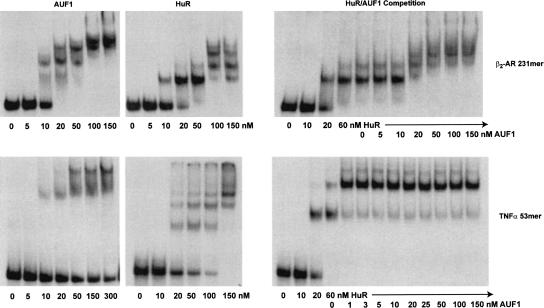
To examine the interaction of AUF1 and HuR biochemically, nondenaturing gel shift assays were used to determine whether or not HuR and p37AUF1 proteins can bind to target AREs and whether they can compete for a single ARE or bind to it additively. First, we demonstrate that purified recombinant GST-HuR and GST-p37AUF1 bind individually to the relatively simple 53-nucleotide (nt) ARE from TNFα mRNA (Fig. 7, top), and to the more complex ARE 231 nucleotides from the β2-AR mRNA (Fig. 7, bottom). As has been shown previously, the banding pattern of the gel shift assays indicates that the size and composition of the protein–mRNA complex formed is dependent on the amount of protein present (Wilson et al. 1999), as both HuR and p37AUF1 are able to form multiple complexes with a single ARE. Competition of p37AUF1 with a preformed HuR–mRNA complex had the potential to yield several possible results. If p37AUF1 displaced HuR, then no substantial change in the relative migration of bands should be observed, as HuR and p37AUF1 are relatively close in molecular weight. However, if p37AUF1 is able to bind to a preformed HuR–mRNA complex, then larger, more slowly migrating bands should form. Figure 7 shows that for both TNFα and the β2-AR AREs, the addition of GST-p37AUF1 (or His6-p37AUF1, data not shown) leads to larger protein–mRNA complexes, indicating that both HuR and p37AUF1 are complexing with a single RNA. Although the data do not directly support the conclusion that the two proteins are competing for individual binding sites on the ARE, the resolution of a gel shift assay with a large complex ARE is likely insufficient to eliminate this possibility.
FIGURE 7.
Competitive binding of p37AUF1 and HuR to 3′ -UTR ARE. Nondenaturing gel-shift assays were performed to assess the competitive interaction of purified recombinant p37AUF1 and HuR proteins for the TNFα 53 nt ARE and the hamster β2-AR 231 nt ARE. 32P-UTP labeled RNAs encoding the hamster β2-AR 231 nt ARE or TNFα 53-nucleotide AREs were incubated with either p37AUF1 alone (left), HuR alone (center), or HuR in competition with p37AUF1 (right). Holding the concentration of HuR at ∼3×Kd (60 nM), increasing amounts of purified recombinant GST-p37AUF1 were added in an attempt to displace HuR binding.
Competition between p37AUF1 and HuR for the TNFα demonstrates that p37AUF1 is able to bind to the ARE simultaneously with HuR and form a single complex. Due to the size of this mRNA molecule, 53 nt, the possibility exists for one stable nucleoprotein complex. This result, however, is not able to distinguish between the possible order and location of binding of HuR and p37AUF1 to the ARE. For the β2-AR ARE, a greater number of combinations of protein and mRNA are possible. However, only two ribonuclear protein complexes are formed at the highest protein concentrations. These results clearly demonstrate that HuR and AUF1 can bind additively to the AREs tested and presumably have the potential to do so on other ARE containing RNAs.
RNA dependence of the ARE binding protein interaction
To address whether the stable, cytoplasmic interaction of HuR and p37AUF1 that we have shown in Figures 2 and 3 is RNA dependent, DDT1-MF2 cells were transiently transfected with HuR-eYFP and p37AUF1-eCFP, incubated for 4 h with anisomycin, and treated with ribonuclease A prior to fixation (Maison et al. 2002). Figure 8 demonstrates that there is no qualitative difference in the HuR/p37AUF1 interaction, as indicated by the FRETc signal, between mock treated and cells treated with ribonuclease A. To further verify this result, the FRET efficiency value, E, was calculated for the HuR/p37AUF1 interaction in mock- versus ribonuclease A treated cells. The E value, calculated from a sampling of >100 individual cells, was 0.04, a value identical to that found for the HuR/p37AUF1 heterodimer in live and fixed cells. This result supports the conclusion that at least in the whole cell context, RNA binding is not an absolute requirement for the AUF1/HuR protein–protein interaction.
FIGURE 8.
RNA dependence of HuR/p37AUF1 heterodimer. DDT1-MF2 cells were transiently transfected with HuR-eYFP and p37AUF1-eCFP. Anisomycin was added to the cells for 4 h to stimulate nuclear to cytoplasmic shuttling. Following permeabilization with Triton X-100, cells were either treated with ribonuclease A or mock treated prior to fixation. Both the mock- and ribonuclease-treated cells shows cytoplasmic localization of HuR and AUF1 (CFP and YFP channels), colocalization of HuR and AUF1 (merged view), and the presence of a FRETc signal in both the nucleus and cytoplasm. (A) Mock treated DDT1-MF2 cells, (B) ribonuclease treated cells.
Potential ARE binding protein interactions
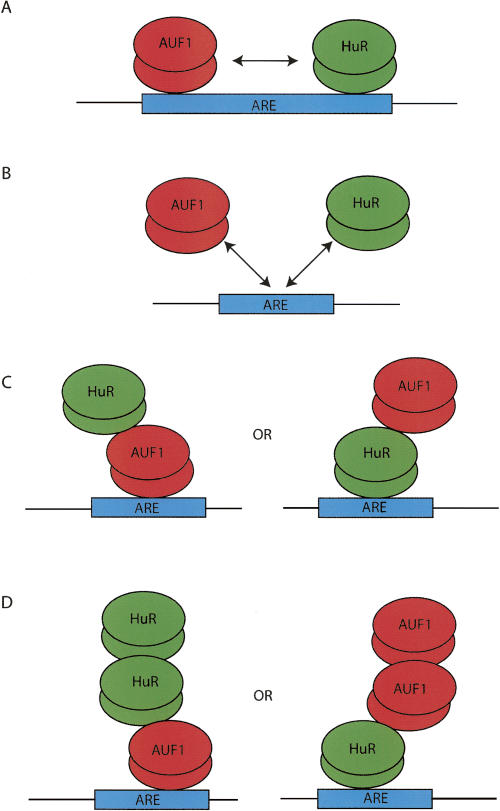
Figure 9 is a model illustrating potential binding interactions between AUF1 and HuR and a target ARE. As stated above, the results of the gel-shift competition assays depicted in Figure 7 do not support scenario B (mutually exclusive binding); however, this finding may simply be due to issues of resolution. In fact, data published by Lal et al. (2004) support the ability of AUF1 and HuR to directly compete for binding but only at select AREs. For both the TNFα 53mer and the β2-AR 231mer AREs, the data also do not permit us to distinguish between scenarios A and C (Fig. 9). What is apparent, however, is that a small amount of a higher molecular weight complex is formed in both competition experiments. Thus, scenario D may occur if, after the binding sites on the ARE are saturated, proteins are then able to bind to themselves to form even larger mRNA–protein–protein complexes. This result has previously been shown for AUF1 alone, forming a dimer in solution but a hexamer when bound to an ARE (DeMaria and Brewer 1996; DeMaria et al. 1997; Lal et al. 2004).
FIGURE 9.
Model of potential interactions of HuR and AUF1. AUF1 has been shown to form homomultimers in solution (DeMaria et al. 1997) as well as when bound to an ARE (Fig. 2A; Wilson et al. 1999; Blaxall et al. 2000b, 2002). HuR has also been shown to be able to form dimers (Fig. 2B; Soller and White 2005). AUF1 and HuR have also been shown to bind to mRNA concurrently and in a competitive manner (Lal et al. 2004). Based on these results and those presented in Figure 6, potential protein–protein and protein–RNA interactions of HuR and AUF1 are presented.
DISCUSSION
In this manuscript, immunocytochemistry and immunofluorescence techniques, including live-cell FRET, were used to demonstrate that two ARE binding proteins, HuR and AUF1/hnRNP D, are capable of intracellular interaction. Although biochemical methods have previously demonstrated AUF1/AUF1 and HuR/HuR interactions, the results herein demonstrate for the first time that interactions between these two proteins are detectable in live cells. The advantage of FRET over immunoprecipitation and colocalization experiments using standard confocal microscopy is that for FRET to occur, the proximity of proteins is such that it is sufficient for true functional interaction.
FRET is a powerful biophysical technique that is increasingly being used to visualize cellular processes in vivo (Galperin et al. 2004; Vogel et al. 2006). Intrinsically, FRET is dependent on both the absolute distance between, and the correct orientation of, two fluorophores that constitute defined FRET pairs (Forster 1948). The strength of the FRET signal obtained from a pair of interacting, fluorescently labeled proteins is also dependent on the FRET efficiency, E, which is the maximum energy transfer from the FRET donor to acceptor.
To attempt to quantify the relative efficiency of FRET between the ARE binding proteins p37AUF1 and HuR, the maximum theoretical FRET efficiency, Et, was first obtained by using a tandem CFP:YFP protein construct (Galperin and Sorkin 2003). Under these conditions, the maximum efficiency obtained in our live cell system was 22%. In the same system, HuR homodimers, p37AUF1 homodimers, and p37AUF1/HuR heterodimers yielded Ed values of 11%, 7%, and 4% respectively, Table 1. These Ed values are in the range measured for other protein pairs in other cellular systems (Galperin et al. 2004). The calculated Ed values for HuR and p37AUF1 show that both HuR and p37AUF1 have the potential to form strong homodimers in the cell (Figs. 2, 3). The lesser Ed for the HuR/AUF1 heterodimer may be due to any number of factors. One possibility is that the formation of HuR and p37AUF1 homodimers sequesters HuR and p37AUF1, thereby preventing the formation of p37AUF1/HuR heterodimers. If so, this would result in an artifactual reduction in the FRET signal. It is also possible that mRNA-dependent binding of HuR and p37AUF1 imposes steric factors onto the respective proteins such that the protein–RNA interaction(s) precludes or diminishes the FRET detectable interactions. Alternatively, and perhaps most simply, the possibility exists that the heteromeric interactions between p37AUF1 and HuR are weaker than the respective homomeric interactions. In support of this later interpretation is the fact that Ed is independent of the concentration(s) of fluors (and thus is likely not an artifact of differential expression), and the fact that FRET efficiency can under certain circumstances be correlated directly with the affinity of protein–protein interaction (Chilibeck et al. 2006).
Results obtained from nondenaturing electrophoresis experiments (Fig. 7) indicate that biochemical associations between HuR and p37AUF1 are indeed observable. For both p37AUF1 and HuR, binding either to the TNFα 53mer ARE or the β2-AR 231mer ARE, homomeric higher-order structures are observed. These results are concordant with our previous observations and with those of others (Wilson et al. 1999; Blaxall et al. 2000b; Blaxall et al. 2002). In competitive binding experiments where p37AUF1 is added to mRNA pre-bound to HuR, rather than displacing HuR from either ARE, p37AUF1 appears to further shift the mRNA–protein complex. These data provide strong evidence that for both a simple ARE, such as that of TNFα 53mer, or for a long, complex ARE, that of the β2-AR 231mer, co-residence of both proteins on an ARE can occur. What nondenaturing gel shift assays cannot reveal is the RNA dependence of these protein–protein interactions. In this regard, it is well established that AUF1 can form RNA-independent homomultimers (DeMaria et al. 1997; Wilson et al. 1999). HuR also appears to homomultimerize (Soller and White 2005), the RNA dependence of that interaction being unknown. Conversely, coexistence of HuR and AUF1 on AREs appears to be dependent on the specific ARE. What remained unclear was the RNA dependence of these interactions. As demonstrated by Lal et al. (2004), coimmunoprecipitation of HuR and AUF1 does not occur when the protein–RNA complex is subjected to RNase pretreatment. In contrast, in the current manuscript, we demonstrate that HuR/AUF1 interaction (Fig. 8) can be detected by FRET even after RNase treatment. These findings are not incongruous as the preparations and conditions are entirely different, one being in the whole cell context and the other being in a purified biochemical preparation. Regardless, the dependence or independence of protein–protein interaction on RNA binding in no way diminishes or precludes the possibility of functionally relevant interactions between the two proteins, both direct or indirect, e.g., by RNA remodeling affecting the binding of one or the other protein (Wilson et al. 2001a,b).
As detailed in Figure 9, there are several ways in which AUF1 and HuR might compete for binding to, or co-reside on, a single ARE. Each of the AREs examined herein, as well as other AREs we have examined previously (Blaxall et al. 2000b, 2002), are capable of binding both AUF1 and HuR with high affinity. However, certain AREs appear to bind only AUF1, others bind only HuR, whereas still others bind either AUF1 or HuR (Lal et al. 2004). Other studies have clearly demonstrated that binding of HuR to the AREs of a broad spectrum of mRNAs is associated with their stabilization (Fan and Steitz 1998b; Peng et al. 1998; Dean et al. 2001; Figueroa et al. 2003; Raineri et al. 2004). Although somewhat less consistent, and perhaps splice-form dependent, there is a general association between AUF1 binding and mRNA destabilization (Buzby et al. 1996; Sirenko et al. 1997; Laroia et al. 1999; Sarkar et al. 2003b). What remains to be examined, especially in the case of an mRNA containing a complex ARE, is the functional significance of more than one protein simultaneously occupying the same ARE or the same mRNA on sequential AREs.
Regulation of the life cycle of an mRNA molecule is a complex process dependent on a number of variables. One level of regulation is that of the abundance of the mRNA binding proteins themselves. To this end, the functional, intracellular concentrations of HuR and AUF1 are regulated by multiple factors including external stimuli, the cell cycle, and pathophysiological processes such as cellular hypertrophy or hyperplasia (Pende et al. 1996; Lafon et al. 1998; Blaxall et al. 2000a; Gouble and Morello 2000; Wang et al. 2000a). A second level of regulation is that of the intracellular localization of the mRNA binding protein and/or the mRNA. There is well-documented evidence both in the literature and presented herein that AUF1 and HuR undergo nuclear/cytoplasmic shuttling.
One major regulatory pathway for both the binding and function of AUF1 is phosphorylation. As demonstrated by Wilson and colleagues (2003a), phosphorylation of AUF1 can markedly affect its affinity for an ARE. Phosphorylation of AUF1 can also induce structural remodeling of the ARE (Wilson et al. 2003a,b; Fialcowitz et al. 2005). Further, the peptidyl–prolyl isomerase, Pin1, binds to AUF1 (Shen et al. 2005). Isomerization by Pin1 could potentially alter the binding affinity of AUF1 by inducing conformational changes in phosphorylated forms of the protein. Like AUF1, and other mRNA-destabilizing proteins including TTP, BRF1, and KSRP, there is recent evidence to date that HuR is also a phosphoprotein. In the case of HuR, induction of cytoplasmic shuttling and increased binding to COX-2 mRNA has recently been linked to phosphorylation by PK-Cα (Doller et al. 2007). HuR has also been shown to be phosphorylated by the cell cycle checkpoint kinase Chk2 (Abdelmohsen et al. 2007). However, phosphorylation by Chk2 does not appear to effect HuR shuttling. Given the impact of phosphorylation state on the binding of AUF1 to an mRNA, it is reasonable to conclude that this mode of regulation might impact the interaction of AUF1 and HuR.
The persistent association of HuR and/or AUF1 with a target mRNA throughout the life of an mRNA in the cell has yet to be definitively tested. HuR has been associated with the stabilization of mRNA transcripts in the cytoplasm, and studies have been conducted on the movement of HuR from the nucleus to the cytoplasm (Atasoy et al. 1998; Fan and Steitz 1998a,b; Seko et al. 2004). However, it is not clear whether HuR binds to a target mRNA and remains bound to a particular mRNA transcript, not only as it is translocated from the nucleus to the cytoplasm, but also through its lifespan in the cytoplasm. This idea can be extended to include whether or not AUF1 also binds to one mRNA transcript for its lifespan and whether or not combinations of other destabilizing and stabilizing RBPs do so as well.
Anisomycin, by virtue of its ability to activate MAPKs, stabilizes ARE-containing mRNAs (Winzen et al. 1999; Mahtani et al. 2001; Neininger et al. 2002; Dean et al. 2003; Stoecklin et al. 2004; Brook et al. 2006; Martinez-Chantar et al. 2006), and causes rapid shuttling of HuR from nucleus to cytoplasm. Figure 5A demonstrates that the time course of shuttling of HuR (30 min) coincides with previous data on the stabilization of ARE-containing mRNAs by MAPK activation (Headley et al. 2004). Conversely, short-term exposure to anisomycin has a more modest effect on the localization of AUF1. These observations lead to the question of how RNPs move from the nucleus to the cytoplasm. The nuclear pore complex (NPC) allows passive diffusion of protein complexes under 60 kDa; larger molecules require assistance for transport. Monomeric HuR and AUF1 are between 30 and 45 kDa. However, when they are bound to an ARE as multimers, the size of an ABP-bound RNA complex necessitates assistance for translocation from the nucleus. Studies detailing the nucleocytoplasmic shuttling of HuR and AUF1 demonstrate that two different mechanisms appear to be utilized for the shuttling of each of these proteins. HuR has been shown to use the CRM1 assisted method of shuttling (Gallouzi et al. 2001; Kimura et al. 2004; Higashino et al. 2005; Cherradi et al. 2006; Fries et al. 2006; Prechtel et al. 2006). The exact mechanism of AUF1 nucleocytoplasmic shuttling has yet to be elucidated; however, AUF1 shuttling appears to be carrier-mediated but not CRM1 associated (Sarkar et al. 2003a).
One condition under which the protein composition bound to a particular mRNA can be altered is that of cellular stress. Upon stress, such as that engendered by treating cells with arsenite, specific, well-defined granules appear. These so-called stress granules (SGs) are known to be protein- and mRNA-dense foci. The mRNA transcripts present in these SGs are those present in the cytoplasm at the onset of the stress conditions and by one mechanism or another are transported into these structures. An example of this would be the TNFα mRNA, which appears to be localized to SGs when associated with the ARE binding protein, tristetraprolin (TTP) (Kedersha et al. 2005; Lian et al. 2006). Other proteins commonly found in stress granules are those involved in RNA deadenylation and in translational silencing (Kedersha et al. 2005). In our model system, we have demonstrated that HuR is present and colocalizes with TIA-1 in stress granules. In contrast, AUF1 does not appear to colocalize to SGs, at least not under conditions of arsenite induction. This result is fully consistent with results obtained in other cell types (Newbury et al. 2006; Eulalio et al. 2007). As stress granules are an accumulation of cytoplasmic mRNA putatively not required during the stress response and which are stabilized and held in translational arrest (Kedersha et al. 2000, 2005; Kedersha and Anderson 2002), the presence of a stabilizing ARE binding protein, such as HuR, would be fully expected. The lack of AUF1 in stress granules implies either that (1) a cellular mechanism exists for removing AUF1 from an mRNA transcript prior to localization to a stress granule, (2) that mRNA transcripts with AUF1 bound to them are excluded from stress granules and remain present in the cytoplasm at times of stress, or (3) AUF1-bound mRNA would be more rapidly degraded under these conditions with AUF1 returning to the nucleus. The results presented here show that sodium arsenite-induced stress does not alter the cellular AUF1 localization. However, the data presented here do not address the fate of cellular AUF1-bound mRNA transcripts at the onset of a stress response.
In conclusion, the results presented herein demonstrate that two ARE binding proteins with potentially opposite functional effects on mRNA stability can co-associate in mRNA–protein complexes. These interactions occur in both the nucleus and cytoplasmic compartments. What remain to be determined are the functional consequences of this interaction. An extension of the current findings is that the use of FRET to detect these interactions opens up distinct new possibilities in the ability to detect the interactions of multiple individual mRNA binding proteins in live cells in real time.
MATERIALS AND METHODS
Vector constructs
An 860 nt HindIII/ApaI fragment of myc-tagged p37AUF1 was subcloned into either pEYFP.C1 or pECFP.C1 (Clonetech), yielding the p37AUF1-eYFP and p37AUF1-eCFP vector constructs. Similarly, a HindIII/EcoRI fragment of myc-tagged HuR was subcloned into either pEYFP.C1 or pECFP.C1, yielding the HuR-eYFP and HuR-eCFP vector constructs. In all constructs the fluorophor is N-terminal to the protein of interest.
Cell culture
DDT1-MF2 hamster smooth muscle cells were grown in 100 mm dishes at 37°C, 5% CO2 in Dubelcco's Modified Eagle Medium (Invitrogen) supplemented with 5% Fetal Bovine Serum (Hyclone) and 0.1% penicillin/streptomycin solution (Invitrogen). For transient transfections, cells were grown on either 18-mm or 25-mm glass coverslips to ∼80% confluence. For immunocytochemistry, cells were grown on 18-mm glass coverslips to ∼60% confluence. Where indicated, cells were treated with anisomycin (75 nM in dimethyl sulphoxide) or sodium arsenite (0.5 mM).
Transient transfection
Vectors were transfected into DDT1-MF2 cells using FuGene 6 (Roche) following the manufacturer's protocol. The amount of plasmid DNA transfected was adjusted empirically to yield the desired 1:1 molar ratio of the CFP protein construct to the YFP protein construct.
Immunocytochemistry
DDT1-MF2 cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences), permeabilized with 0.2% Triton X-100, treated with 0.1% sodium borohydride (Sigma), and blocked with 10% horse serum in 1% BSA. Cells were incubated overnight at 4°C with 1°Ab and with 2°Ab for 1 h at room temperature in the dark. Coverslips were mounted on glass slides using a 12.5% solution of Mowiol (Calbiochem) in 50% glycerol. Detection of HuR was via a 1:400 dilution of monoclonal 1°Ab (Santa Cruz Biotechnology) and a 1:2000 dilution of 2°Ab labeled with Alexa Fluor 488 (Molecular Probes); detection of AUF1 was via a 1:500 dilution of a polyclonal 1°Ab (Upstate), which recognizes all four isoforms of AUF1, and a 1:800 dilution of 2°Ab labeled with Cy3 (Jackson ImmunoResearch); detection of TIA-1 was via a 1:500 dilution of polyclonal 1°Ab (Santa Cruz Biotechnology) and a 1:400 dilution of 2°Ab labeled with Alexa Fluor 647 (Molecular Probes).
Subcellular fractionation
Cytoplasmic and nuclear fractions were produced as described by Wang et al. (2000b). Briefly, cells were rinsed with cold phosphate buffered saline followed by incubation on ice in hypotonic buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2) containing protease inhibitors (leupeptine [1 μg/mL], aprotinin [1 μg/mL], and 0.5 mM PMSF). Cells were lysed by the addition of buffer A with protease inhibitors containing 2.5% Nonidet P-40. Lysed cells were centrifuged (3500 rpm for 4 min) to pellet nuclei. The resultant supernatant was isolated, subjected to five freeze–thaw cycles, and recentrifuged. Cytosolic fractions were prepared by centrifugation of the second supernatant (14,000 rpm for 60 min at 4°C). Nuclear pellets were incubated on ice in extraction buffer C (20 mM HEPES [pH 7.9], 0.45 M NaCl, 1 mM EDTA) with protease inhibitors followed by centrifugation (14,000 rpm for 10 min at 4°C).
Western blotting
Endogenous HuR and AUF1 proteins were detected as described previously (Blaxall et al. 2000a). Briefly, AUF1 and HuR proteins were detected using polyclonal 1° antibodies (1:1000, Upstate) and detected by a GAR 2° antibody (1:2000, Jackson ImmunoReserearch). For normalization, blots were stripped and reprobed with a mouse monoclonal anti-actin antibody (1:1000) with detection by a GAM 2° antibody (1:4000, Jackson ImmunoResearch).
Cell fixation
DDT1-MF2 cells were fixed with 3.4% formaldehyde. The coverslips were mounted on glass slides using a 12.5% solution of Mowiol (Calbiochem) in 50% glycerol.
Microscopy
Images of live and fixed cells were captured at 100× magnification under oil immersion, in 2×2 binning mode using a Nikon Eclipse TE3000 fluorescence microscope running Slidebook software (3I, version 4.0.1.43), or a Zeiss Axiovert 200M fluorescence microscope, fitted with an atmosphere controlled chamber, running Slidebook software, (3I, version 4.1.0.12). All images were collected using the following criteria: integration time >25 msec, channel intensity <4000. All images shown are background corrected by subtracting the intensity of a cell devoid area from each individual image. For images containing CFP- and YFP-labeled proteins, integration time was between 25 and 250 msec. Cells containing CFP- and YFP-labeled proteins were viewed via the CFP channel to prevent photobleaching of YFP. All images presented have a signal intensity >100 after background correction. FRETc images were produced by Slidebook using spectral bleed-through numbers determined experimentally. FRETc data was calculated using the raw intensity values obtained from Slidebook.
Spectral bleed-through number calculations
The following constructs were used to determine the average CFP spectral bleed-through number: eCFP vector, p37AUF1-eCFP, HuR-eCFP, TTP-eCFP and KSRP-eCFP. To determine the YFP spectral bleed-through number, the following vector constructs were used: eYFP vector, HuR-eYFP, and BRF1-eYFP. As a further proof of the bleed-through numbers, transient transfection of non-FRET pairs was used for empirical correction of the spectral bleed-through number; eCFP vector and eYFP vector; HuR-eYFP and eCFP vector; p37AUF1-eCFP and eYFP vector; HuR-eYFP with both an unrelated cytoplasmically localized protein (PKA R11A) and an unrelated nuclear and cytoplasmically localized protein (PH-PLC), labeled with CFP. Both unrelated protein vector constructs were kindly provided by Dr. Mark Dell'Acqua, University of Colorado.
Calculation of FRETc, FRETn, and FRET efficiency E:
To calculate FRETc, FRETn, and the FRET efficiency, E, the following equations were used:
FRETc is the corrected FRET value, raw FRET is the uncorrected FRET intensity, D is the donor fluorophore intensity, X is the donor fluorophore spectral bleed-through number, A is the acceptor fluorophore intensity, and Y is the acceptor fluorophore spectral bleed-through number.
FRETn is the normalized FRET value, FRETc is the corrected FRET value, D is the donor fluorophore intensity, and A is the acceptor fluorophore intensity.
E is the FRET efficiency, FRETc is the corrected FRET value, D is the donor fluorophore intensity, and G is a constant determined for the specific filter set used (Gordon et al. 1998; Galperin et al. 2004). To calculate the FRETc, FRETn, and E calculations, the following criteria were used: YFP:CFP ratio between 9 and 11, CFP intensity >100.
Production of ARE-containing mRNAs
PCR amplification of a 231 nt section of the 3′ -UTR of the hamster β2-AR gene using the following primers (5′-CCGCTCGAGCAGGCTTT; 3′-GATCTCTAGATTTAAGTT, both shown in the 5′-3′ direction) was cloned into the pCDNA3.1 zeo+ vector. This construct was used to make the 231 nt ARE containing mRNA fragment utilized for the nondenaturing gel shift assays. The production of the TNFα 53 nt mRNA was as described (Stoecklin et al. 2001).
Production of recombinant GST-p37AUF1 protein
The coding region of p37AUF1 (Pende et al. 1996) was inserted into a pGEX-2T plasmid (Amersham Biosciences). The resulting pGEX-2T-AUF1 plasmid was transformed into Escherichia coli BL21 cells, and protein expression was induced with 1 mM IPTG at 37°C for 3–4 h. Cells were centrifuged at 4000 rpm for 10 min, 4°C, and resuspended in 5 mL of Buffer A (50 mM Tris pH 8.0, 200 mM NaCl, 1 mM EDTA). When fully resuspended, 500 μL of 10 mg/mL lysozyme was added. The cell suspension was incubated on ice for 20 min followed by centrifugation at 13,000 rpm, 30 min, 4°C. The supernatant was incubated with 1 mL of glutathione sepharose beads slurry (50% slurry) for 1 h at 40°C with agitation. The beads were washed twice with Buffer B (50 mM Tris-HCl pH 8, 200 mM NaCl, 1 mM EDTA, and 1% Triton X-100), and fractions were eluted with Elution Buffer (50 mM Tris-HCl [pH 8], 5 mM Glutathione). Human alpha-lactalbumin (Sigma) was added to a final concentration of 100 μg/mL prior to storage at −80°C. Production of recombinant GST-HuR and His6-p37AUF1 has been described previously (Blaxall et al. 2002).
Nondenaturing gel shift assays
Briefly, 1 fmol of 32P-labeled, in vitro transcribed RNA (∼5000 CPM) (Pende et al. 1996) was incubated with increasing concentrations of purified recombinant protein in a final volume of 10 μL of binding buffer (10 mM Tris-HCl [pH 7.5], 5 mM magnesium acetate, 100 mM potassium acetate, 2 mM DTT, 0.1 μg/μL of BSA, 10 units of RNasin [Promega], 0.2 mg/mL yeast tRNA, 5 μg/μL heparin, and 0.1 μg/μL poly[C]) for 10 min on ice. Complexes were resolved by electrophoresis through nondenaturing, 5%–6%, polyacrylamide gels and visualized by autoradiography.
In vivo RNA digestion
DDT1-MF2 cells grown on 18 mm glass coverslips were transiently transfected as above. Cells were permeabilized with 0.01% Triton X-100, treated with 1 mg/mL ribonuclease A (Sigma) for 10 min at room temperature, and fixed with 4% paraformaldehyde (Maison et al. 2002). Coverslips were mounted on glass slides using a 12.5% solution of Mowiol in 50% glycerol.
ACKNOWLEDGMENTS
We thank Dr. Georg Stoecklin for providing the TNFα probe template and Dr. Mark Dell'Acqua for the PKA(R11A) and PH-PLC CFP labeled constructs. We would also like to thank Drs. Alexander Sorkin, Emilia Galperin, and Mark Dell'Acqua for many helpful discussions pertaining to FRET microscopy.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.501707.
REFERENCES
- Abdelmohsen, K., Pullmann R., Jr, Lal, A., Kim, H.H., Galban, S., Yang, X., Blethrow, J.D., Walker, M., Shubert, J., Gillespie, D.A., et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham, W.T., Gilbert, E.M., Lowes, B.D., Minobe, W.A., Larrabee, P., Roden, R.L., Dutcher, D., Sederberg, J., Lindenfeld, J.A., Wolfel, E.E., et al. Coordinate changes in Myosin heavy chain isoform gene expression are selectively associated with alterations in dilated cardiomyopathy phenotype. Mol. Med. 2002;8:750–760. [PMC free article] [PubMed] [Google Scholar]
- Anderson, P., Kedersha, N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy, U., Watson, J., Patel, D., Keene, J.D. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J. Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- Bakheet, T., Williams, B.R., Khabar, K.S. ARED 3.0: The large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxall, B.C., Dwyer-Nield, L.D., Bauer, A.K., Bohlmeyer, T.J., Malkinson, A.M., Port, J.D. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol. Carcinog. 2000a;28:76–83. [PubMed] [Google Scholar]
- Blaxall, B.C., Pellett, A.C., Wu, S.C., Pende, A., Port, J.D. Purification and characterization of β-adrenergic receptor mRNA-binding proteins. J. Biol. Chem. 2000b;275:4290–4297. doi: 10.1074/jbc.275.6.4290. [DOI] [PubMed] [Google Scholar]
- Blaxall, B.C., Pende, A., Wu, S.C., Port, J.D. Correlation between intrinsic mRNA stability and the affinity of AUF1 (hnRNP D) and HuR for A+U-rich mRNAs. Mol. Cell. Biochem. 2002;232:1–11. doi: 10.1023/a:1014819016552. [DOI] [PubMed] [Google Scholar]
- Brengues, M., Teixeira, D., Parker, R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook, M., Tchen, C.R., Santalucia, T., McIlrath, J., Arthur, J.S., Saklatvala, J., Clark, A.R. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol. Cell. Biol. 2006;26:2408–2418. doi: 10.1128/MCB.26.6.2408-2418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzby, J.S., Lee, S.M., Van Winkle, P., DeMaria, C.T., Brewer, G., Cairo, M.S. Increased granulocyte-macrophage colony-stimulating factor mRNA instability in cord versus adult mononuclear cells is translation dependent and associated with increased levels of A+U-rich element binding factor. Blood. 1996;88:2889–2897. [PubMed] [Google Scholar]
- Carballo, E., Lai, W.S., Blackshear, P.J. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- Chen, C.-Y., Xu, N., Shyu, A.-B. mRNA decay is mediated by two distinct AU-rich elements from the c-fos and granulocyte-macrophage colony-stimulating factor transcripts: Different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherradi, N., Lejczak, C., Desroches-Castan, A., Feige, J.J. Antagonistic functions of tetradecanoyl phorbol acetate-inducible-sequence 11b and HuR in the hormonal regulation of vascular endothelial growth factor messenger ribonucleic acid stability by adrenocorticotropin. Mol. Endocrinol. 2006;20:916–930. doi: 10.1210/me.2005-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilibeck, K.A., Wu, T., Liang, C., Schellenberg, M.J., Gesner, E.M., Lynch, J.M., MacMillan, A.M. FRET analysis of in vivo dimerization by RNA-editing enzymes. J. Biol. Chem. 2006;281:16530–16535. doi: 10.1074/jbc.M511831200. [DOI] [PubMed] [Google Scholar]
- Cougot, N., Babajko, S., Seraphin, B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, J.L., Wait, R., Mahtani, K.R., Sully, G., Clark, A.R., Saklatvala, J. The 3′-untranslated region of tumor necrosis factor α mRNA is a target of the mRNA-stabilizing factor HuR. Mol. Cell. Biol. 2001;21:721–730. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, J.L.E., Sarsfield, S.J., Tsounakou, E., Saklatvala, J. p38 mitogen-activated protein kinase stabilizes mRNAs that contain cyclooxygenase-2 and tumor necrosis factor AU-rich elements by inhibiting deadenylation. J. Biol. Chem. 2003;278:39470–39476. doi: 10.1074/jbc.M306345200. [DOI] [PubMed] [Google Scholar]
- DeMaria, C.T., Brewer, G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- DeMaria, C.T., Sun, Y., Long, L., Wagner, B.J., Brewer, G. Structural determinants in AUF1 required for high affinity binding to A+U-rich elements. J. Biol. Chem. 1997;272:27635–27643. doi: 10.1074/jbc.272.44.27635. [DOI] [PubMed] [Google Scholar]
- Doller, A., Huwiler, A., Muller, R., Radeke, H.H., Pfeilschifter, J., Eberhardt, W. PKCα-dependent phosphorylation of the mRNA stabilizing factor HuR: Implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell. 2007;18:2137–2148. doi: 10.1091/mbc.E06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss, G. Structure and function of nuclear and cytoplasmic ribonucleoprotein particles. Annu. Rev. Cell Biol. 1986;2:459–498. doi: 10.1146/annurev.cb.02.110186.002331. [DOI] [PubMed] [Google Scholar]
- Eulalio, A., Behm-Ansmant, I., Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Fan, X.C., Steitz, J.A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. 1998a;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, X.C., Steitz, J.A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998b;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawal, M., Armstrong, F., Ollier, S., Dupont, H., Touriol, C., Monsarrat, B., Delsol, G., Payrastre, B., Morello, D. A “liaison dangereuse” between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood. 2006;108:2780–2788. doi: 10.1182/blood-2006-04-014902. [DOI] [PubMed] [Google Scholar]
- Fialcowitz, E.J., Brewer, B.Y., Keenan, B.P., Wilson, G.M. A hairpin-like structure within an AU-rich mRNA-destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics. J. Biol. Chem. 2005;280:22406–22417. doi: 10.1074/jbc.M500618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa, A., Cuadrado, A., Fan, J., Atasoy, U., Muscat, G.E., Munoz-Canoves, P., Gorospe, M., Munoz, A. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol. Cell. Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster, T. Zwischenmolekulare energiewanderung und fluoreszenz. Ann. Phys. 1948;2:57–75. [Google Scholar]
- Fries, B., Heukeshoven, J., Hauber, I., Gruttner, C., Stocking, C., Kehlenbach, R.H., Hauber, J., Chemnitz, J. Analysis of nucleocytoplasmic trafficking of the Hur ligand APRIL and its influence on CD83 expression. J. Biol. Chem. 2006;282:4504–4515. doi: 10.1074/jbc.M608849200. [DOI] [PubMed] [Google Scholar]
- Gallouzi, I.E., Brennan, C.M., Steitz, J.A. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA. 2001;7:1348–1361. doi: 10.1017/s1355838201016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin, E., Sorkin, A. Visualization of Rab5 activity in living cells by FRET microscopy and influence of plasma-membrane-targeted RAB5 on clathrin-dependent endocytosis. J. Cell Sci. 2003;116:4799–4810. doi: 10.1242/jcs.00801. [DOI] [PubMed] [Google Scholar]
- Galperin, E., Verkhusha, V.V., Sorkin, A. Three-chromophore FRET microscopy to analyze multiprotein interactions in living cells. Nat. Methods. 2004;1:209–217. doi: 10.1038/nmeth720. [DOI] [PubMed] [Google Scholar]
- Gherzi, R., Lee, K.Y., Briata, P., Wegmuller, D., Moroni, C., Karin, M., Chen, C.Y. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Gordon, G.W., Berry, G., Liang, X.H., Levine, B., Herman, B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouble, A., Morello, D. Synchronous and regulated expression of two AU-binding proteins, AUF1 and HuR, throughout murine development. Oncogene. 2000;19:5377–5384. doi: 10.1038/sj.onc.1203910. [DOI] [PubMed] [Google Scholar]
- Headley, V., Tanveer, R., Greene, S.M., Zweifach, A., Port, J.D. Reciprocal regulation of β-adrenergic receptor mRNA stability by mitogen activated protein kinase activation and inhibition. Mol. Cell. Biochem. 2004;285:109–119. doi: 10.1023/b:mcbi.0000012841.03400.42. [DOI] [PubMed] [Google Scholar]
- Higashino, F., Aoyagi, M., Takahashi, A., Ishino, M., Taoka, M., Isobe, T., Kobayashi, M., Totsuka, Y., Kohgo, T., Shindoh, M. Adenovirus E4orf6 targets pp32/LANP to control the fate of ARE-containing mRNAs by perturbing the CRM1-dependent mechanism. J. Cell Biol. 2005;170:15–20. doi: 10.1083/jcb.200405112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura, H., Tomozoe, Y., Akagi, T., Kamei, D., Ochiai, M., Yamada, M. Identification of the nucleocytoplasmic shuttling sequence of heterogeneous nuclear ribonucleoprotein D-like protein JKTBP and its interaction with mRNA. J. Biol. Chem. 2002;277:2732–2739. doi: 10.1074/jbc.M108477200. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., Anderson, P. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- Kedersha, N.L., Gupta, M., Li, W., Miller, I., Anderson, P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 α to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., Cho, M.R., Li, W., Yacono, P.W., Chen, S., Gilks, N., Golan, D.E., Anderson, P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fitzler, M.J., Scheuner, D., Kaufman, R.J., Golan, D.E., Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, J.D., Lager, P.J. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 2005;13:327–337. doi: 10.1007/s10577-005-0848-1. [DOI] [PubMed] [Google Scholar]
- Kimura, T., Hashimoto, I., Nagase, T., Fujisawa, J. CRM1-dependent, but not ARE-mediated, nuclear export of IFN-α1 mRNA. J. Cell Sci. 2004;117:2259–2270. doi: 10.1242/jcs.01076. [DOI] [PubMed] [Google Scholar]
- Kuersten, S., Goodwin, E.B. Linking nuclear mRNP assembly and cytoplasmic destiny. Biol. Cell. 2005;97:469–478. doi: 10.1042/BC20040106. [DOI] [PubMed] [Google Scholar]
- Lafon, I., Carballes, F., Brewer, G., Poiret, M., Morello, D. Developmental expression of AUF1 and HuR, two c-myc mRNA binding proteins. Oncogene. 1998;16:3413–3421. doi: 10.1038/sj.onc.1201895. [DOI] [PubMed] [Google Scholar]
- Lal, A., Mazan-Mamczarz, K., Kawai, T., Yang, X., Martindale, J.L., Gorospe, M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia, G., Schneider, R.J. Alternate exon insertion controls selective ubiquitination and degradation of different AUF1 protein isoforms. Nucleic Acids Res. 2002;30:3052–3058. doi: 10.1093/nar/gkf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia, G., Cuesta, R., Brewer, G., Schneider, R.J. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- Lian, S., Jakymiw, A., Eystathioy, T., Hamel, J.C., Fritzler, M.J., Chan, E.K. GW bodies, microRNAs and the cell cycle. Cell Cycle. 2006;5:242–245. doi: 10.4161/cc.5.3.2410. [DOI] [PubMed] [Google Scholar]
- Linker, K., Pautz, A., Fechir, M., Hubrich, T., Greeve, J., Kleinert, H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4827. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J.Y., Schneider, R.J. Tissue distribution of AU-rich mRNA binding proteins involved in regulation of mRNA decay. J. Biol. Chem. 2004;279:12974–12979. doi: 10.1074/jbc.M310433200. [DOI] [PubMed] [Google Scholar]
- Ma, W.-J., Cheng, S., Campbell, C., Wright, A., Furneaux, H. Cloning and characterization of HuR, a ubiquitously expressed elav-like protein. J. Biol. Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- Mahtani, K.R., Brook, M., Dean, J.L., Sully, G., Saklatvala, J., Clark, A.R. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor α mRNA stability. Mol. Cell. Biol. 2001;21:6461–6469. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison, C., Bailly, D., Peters, A.H., Quivy, J.P., Roche, D., Taddei, A., Lachner, M., Jenuwein, T., Almouzni, G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Martinez-Chantar, M.L., Vazquez-Chantada, M., Garnacho, M., Latasa, M.U., Varela-Rey, M., Dotor, J., Santamaria, M., Martinez-Cruz, L.A., Parada, L.A., Lu, S.C., et al. S-adenosylmethionine regulates cytoplasmic HuR via AMP-activated kinase. Gastroenterology. 2006;131:223–232. doi: 10.1053/j.gastro.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz, K., Galban, S., Lopez de Silanes, I., Martindale, J.L., Atasoy, U., Keene, J.D., Gorospe, M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming, X.F., Stoecklin, G., Lu, M., Looser, R., Moroni, C. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol. Cell. Biol. 2001;21:5778–5789. doi: 10.1128/MCB.21.17.5778-5789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J. From birth to death: The complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Neininger, A., Kontoyiannis, D., Kotlyarov, A., Winzen, R., Eckert, R., Volk, H.-D., Holtmann, H., Kollias, G., Gaestel, M. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 2002;277:3065–3068. doi: 10.1074/jbc.C100685200. [DOI] [PubMed] [Google Scholar]
- Newbury, S.F., Muhlemann, O., Stoecklin, G. Turnover in the Alps: An mRNA perspective. Workshops on mechanisms and regulation of mRNA turnover. EMBO Rep. 2006;7:143–148. doi: 10.1038/sj.embor.7400628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie, R.L., Abelson, M., Hau, H.H., Vlasova, I., Blackshear, P.J., Bohjanen, P.R. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- Parker, R., Song, H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Pende, A., Tremmel, K.D., DeMaria, C.T., Blaxall, B.C., Minobe, W., Sherman, J.A., Bisognano, J., Bristow, M.R., Brewer, G., Port, J.D. Regulation of the mRNA-binding protein AUF1 by activation of the β-adrenergic receptor signal transduction pathway. J. Biol. Chem. 1996;271:8493–8501. doi: 10.1074/jbc.271.14.8493. [DOI] [PubMed] [Google Scholar]
- Peng, S.S., Chen, C.Y., Xu, N., Shyu, A.B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prechtel, A.T., Chemnitz, J., Schirmer, S., Ehlers, C., Langbein-Detsch, I., Stulke, J., Dabauvalle, M.C., Kehlenbach, R.H., Hauber, J. Expression of CD83 is regulated by HuR via a novel cis-active coding region RNA element. J. Biol. Chem. 2006;281:10912–10925. doi: 10.1074/jbc.M510306200. [DOI] [PubMed] [Google Scholar]
- Raineri, I., Wegmueller, D., Gross, B., Certa, U., Moroni, C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow, S., White, K. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev. Biol. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- Sarkar, B., Lu, J.Y., Schneider, R.J. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 2003a;278:20700–20707. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- Sarkar, B., Xi, Q., He, C., Schneider, R.J. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol. Cell. Biol. 2003b;23:6685–6693. doi: 10.1128/MCB.23.18.6685-6693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidlin, M., Lu, M., Leuenberger, S.A., Stoecklin, G., Mallaun, M., Gross, B., Gherzi, R., Hess, D., Hemmings, B.A., Moroni, C. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 2004;23:4760–4769. doi: 10.1038/sj.emboj.7600477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seko, Y., Azmi, H., Fariss, R., Ragheb, J.A. Selective cytoplasmic translocation of HuR and site-specific binding to the interleukin-2 mRNA are not sufficient for CD28-mediated stabilization of the mRNA. J. Biol. Chem. 2004;279:33359–33367. doi: 10.1074/jbc.M312306200. [DOI] [PubMed] [Google Scholar]
- Shen, Z.J., Esnault, S., Malter, J.S. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat. Immunol. 2005;6:1280–1287. doi: 10.1038/ni1266. [DOI] [PubMed] [Google Scholar]
- Sirenko, O.I., Lofquist, A.K., DeMaria, C.T., Morris, J.S., Brewer, G., Haskill, J.S. Adhesion-dependent regulation of an A+U-rich element-binding activity associated with AUF1. Mol. Cell. Biol. 1997;17:3898–3906. doi: 10.1128/mcb.17.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller, M., White, K. ELAV multimerizes on conserved AU4-6 motifs important for ewg splicing regulation. Mol. Cell. Biol. 2005;25:7580–7591. doi: 10.1128/MCB.25.17.7580-7591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin, G., Stoeckle, P., Lu, M., Muehlemann, O., Moroni, C. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA. 2001;7:1578–1588. [PMC free article] [PubMed] [Google Scholar]
- Stoecklin, G., Colombi, M., Raineri, I., Leuenberger, S., Mallaun, M., Schmidlin, M., Gross, B., Lu, M., Kitamura, T., Moroni, C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21:4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin, G., Stubbs, T., Kedersha, N., Wax, S., Rigby, W.F., Blackwell, T.K., Anderson, P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, M., Iijima, M., Nishimura, A., Tomozoe, Y., Kamei, D., Yamada, M. Two separate regions essential for nuclear import of the hnRNP D nucleocytoplasmic shuttling sequence. FEBS J. 2005;272:3975–3987. doi: 10.1111/j.1742-4658.2005.04820.x. [DOI] [PubMed] [Google Scholar]
- Taylor, G.A., Blackshear, P.J. Zinc inhibits turnover of labile mRNAs in intact cells. J. Cell. Physiol. 1995;162:378–387. doi: 10.1002/jcp.1041620310. [DOI] [PubMed] [Google Scholar]
- Taylor, G.A., Carballo, E., Lee, D.M., Lai, W.S., Thompson, M.J., Patel, D.D., Schenkman, D.I., Gilkeson, G.S., Broxmeyer, H.E., Haynes, B.F., et al. A pathogenetic role for TNF α in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- Tenenbaum, S.A., Lager, P.J., Carson, C.C., Keene, J.D. Ribonomics: Identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- Vogel, S.S., Thaler, C., Koushik, S.V. Fanciful FRET. Sci. STKE. 2006;2006:re2. doi: 10.1126/stke.3312006re2. [DOI] [PubMed] [Google Scholar]
- Wagner, B.J., DeMaria, C.T., Sun, Y., Wilson, G.M., Brewer, G. Structure and genomic organization of the human AUF1 gene: Alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- Wang, W., Caldwell, M.C., Lin, S., Furneaux, H., Gorospe, M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000a;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Furneaux, H., Cheng, H., Caldwell, M.C., Hutter, D., Liu, Y., Holbrook, N., Gorospe, M. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 2000b;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, G.M., Sun, Y., Lu, H., Brewer, G. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J. Biol. Chem. 1999;274:33374–33381. doi: 10.1074/jbc.274.47.33374. [DOI] [PubMed] [Google Scholar]
- Wilson, G.M., Sutphen, K., Chuang, K., Brewer, G. Folding of A+U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J. Biol. Chem. 2001a;276:8695–8704. doi: 10.1074/jbc.M009848200. [DOI] [PubMed] [Google Scholar]
- Wilson, G.M., Sutphen, K., Moutafis, M., Sinha, S., Brewer, G. Structural re-modeling of an A+U-rich RNA element by cation or AUF1 binding. J. Biol. Chem. 2001b;276:38400–38409. doi: 10.1074/jbc.M106509200. [DOI] [PubMed] [Google Scholar]
- Wilson, G.M., Lu, J., Sutphen, K., Suarez, Y., Sinha, S., Brewer, B., Villanueva-Feliciano, E.C., Ysla, R.M., Charles, S., Brewer, G. Phosphorylation of p40AUF1 regulates binding to A+U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J. Biol. Chem. 2003a;278:33039–33048. doi: 10.1074/jbc.M305775200. [DOI] [PubMed] [Google Scholar]
- Wilson, G.M., Lu, J., Sutphen, K., Sun, Y., Huynh, Y., Brewer, G. Regulation of A+U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J. Biol. Chem. 2003b;278:33029–33038. doi: 10.1074/jbc.M305772200. [DOI] [PubMed] [Google Scholar]
- Winzen, R., Kracht, M., Ritter, B., Wilhelm, A., Chen, C.Y., Shyu, A.B., Muller, M., Gaestel, M., Resch, K., Holtmann, H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999;18:4969–4980. doi: 10.1093/emboj/18.18.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, E., van Nimwegen, E., Zavolan, M., Rajewsky, N., Schroeder, M., Magnasco, M., Darnell J.E., Jr Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]