Abstract
Halogenation of bases is a widespread method used for solving crystal structures of nucleic acids. However, this modification may have important consequences on RNA folding and thus on the success of crystallization. We have used a combination of UV thermal melting, steady-state fluorescence, X-ray crystallography, and gel electrophoresis techniques to study the influence of uridine halogenation (bromination or iodination) on the RNA folding. The HIV-1 Dimerization Initiation Site is an RNA hairpin that can adopt an alternative duplex conformation and was used as a model. We have shown that, unexpectedly, the RNA hairpin/duplex ratio is strongly dependent not only on the presence but also on the position of halogenation.
Keywords: RNA, HIV, fluorescence, halogen, crystal
INTRODUCTION
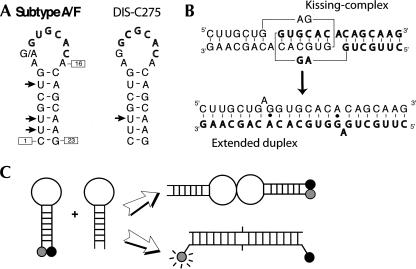
Similarly to the substitution of selenomethionines for methionines into proteins, the introduction of halogenated bases is a convenient way for solving the phase problem with crystals of nucleic acids (Pahler et al. 1990). We had used this strategy in order to solve crystal structures of a 23-nucleotide (nt) RNA corresponding to the HIV-1 genomic RNA Dimerization Initiation Site (DIS). Structures of the DIS were solved using the same DIS sequence in two different topologies (Fig. 1): as an extended duplex (Ennifar et al. 1999) and as a loop–loop complex involving two hairpins (Ennifar et al. 2001; Ennifar and Dumas 2006; Ennifar et al. 2006). To obtain the hairpin form, we found that the RNA concentration during the annealing procedure (heat denaturation in water followed by cooling to room temperature) was the crucial parameter. Indeed, with unmodified RNAs, a majority of hairpins could be obtained by that procedure only with an RNA concentration of less than 3 μM, whereas the effect of the cooling kinetics was insignificant (Bernacchi et al. 2005).
FIGURE 1.
(A) Sequence and secondary structure of HIV-1 subtype A and subtype F DIS stem–loops used in this study. Black arrows indicate uridines substituted into C5-bromouridines, C5-iodouridines, or C5-methyluridines. (B) Secondary structure of the kissing loop–loop complex and extended duplex formed upon dimerization. (C) Schematic drawing of the molecular beacon strategy used to distinguish between hairpins from duplex forms.
However, we recently observed that a DIS sequence containing a uridine 3 halogenated on C5 (either BrU3-DIS or IU3-DIS) allowed us to obtain kissing-loop complex crystals even after an annealing procedure performed at a relatively high RNA concentration (20 μM and higher), i.e., in conditions where only duplex crystals are obtained with the unmodified RNA. Strikingly, if the uridine 2, instead of the uridine 3, was brominated (BrU2) or iodinated (IU2) on C5, the resulting modified DIS sequence led mainly to duplex crystals, even after folding in water at an RNA concentration as low as 1 μM, i.e., in conditions where only kissing-loop complex crystals are normally obtained with the unmodified RNA. Therefore, far from being neutral, not only RNA halogenation but also the exact position of halogenation strongly affected the outcome of RNA folding. In the present study, we investigate the influence of the substitution of C5-bromouridines for uridines at three different positions on the DIS stem (U2, U3, and U6). A comparison with iodination at U2 and U3, as well as with methylation on the C5 of U3, is also performed.
RESULTS
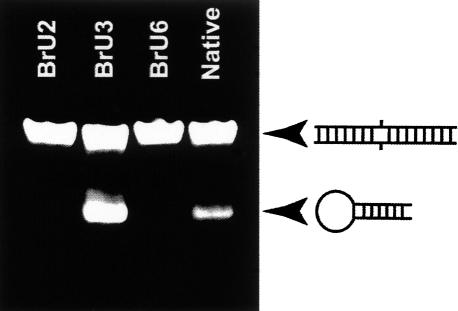
In order to assess the result of the annealing procedure on unmodified and halogenated RNA sequences, we performed polyacrylamide gel electrophoresis at room temperature in semidenaturing conditions. In such conditions, the loop–loop interaction is disrupted, which easily allows us to distinguish the hairpin from the duplex form (Theilleux-Delalande et al. 2000), and thus to estimate their respective amount. When unmodified RNA sequences were folded in water at 30 μM concentration, only a minor fraction of the hairpin form was obtained (Fig. 2), in agreement with our previous results (Bernacchi et al. 2005). On the contrary, when either BrU3-DIS or IU3-DIS was submitted to the same treatment, the resulting fraction of the hairpin form was as large as 50%. It was then thought that bromination or iodination of uridine was an important parameter to enhance the hairpin yield after such an annealing procedure. However, it appeared to be more complicated since changing the position of halogenation (X=Br or I) from XU3 to either XU2 or BrU6 (Fig. 2) reverted the outcome of the annealing procedure at 30 μM RNA to a majority of duplex. This surprisingly large difference between the XU3 DIS, on the one hand, and the unmodified or the XU2 and BrU6-DIS forms, on the other hand, was in line with the observation that the XU3-DIS had a strong propensity to crystallize as a kissing-loop complex.
FIGURE 2.
Semidenaturing polyacrylamide gel showing the proportion of extended duplex and hairpin formed at 30 μM RNA concentration in water for unmodified and several brominated subtype F DIS.
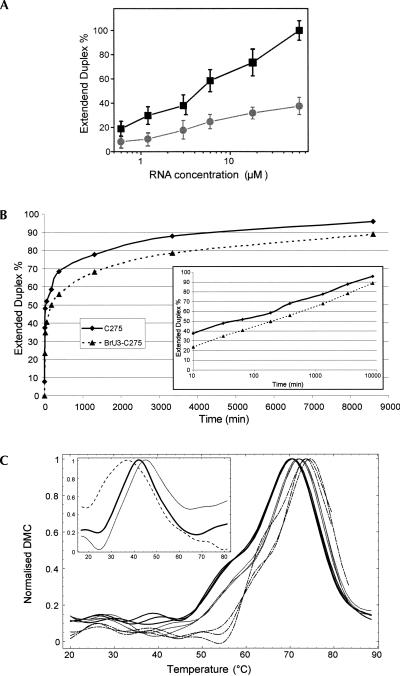
These first qualitative results on gels were then further investigated by using the molecular beacon strategy (Tyagi and Kramer 1996) and steady-state fluorescence as we already performed (Bernacchi et al. 2005). For that goal, DIS sequences were labeled at their 3′ end with a fluorescence dye and at their 5′ ends with a quencher. In the hairpin form, the two dyes are close from one another and the fluorescence is always quenched. However, with an excess of unlabeled strands, the two dyes are separated by 60 Å in the DIS duplex (apart for the negligible fraction of duplexes made of two labeled strands), which fully restores the fluorescence signal (Fig. 1C). It is then possible to distinguish the hairpin from the duplex form and to accurately quantify their relative amount immediately after annealing. Measurements were performed for RNA concentrations ranging from 300 nM to 60 μM of unmodified and BrU3-DIS RNA. In agreement with the previous gel experiment, we observed that BrU3-DIS requires much higher strand concentrations than the unmodified RNA to form duplex (Fig. 3A). At a 20 μM RNA concentration, the yield of duplex was only 25% for the BrU3-DIS, but 75% for the unmodified RNA. Together with the gel experiments, this clearly showed that the BrU3 modification strongly favors the hairpin over the duplex form after annealing in water, in comparison to the unmodified RNA.
FIGURE 3.
(A) Fluorescence quantification of the relative amount of extended duplex formed in water after folding. Values obtained for unmodified and BrU3 DIS sequences are represented as circles and squares, respectively. (B) Time-dependent hairpin–duplex conversion for the unmodified and BrU3-C275 DIS sequences upon incubation with salts at 37°C. The inset shows that the duplex yield varies linearly with the logarithm of the time over three orders of magnitude. The fractions of duplex at t=0 (i.e., before salt addition) are 8% for the unmodified sequence and undetectable for the BrU3 sequence. (C) Normalized differential melting curves of C275 DIS (black), BrU3-C275 DIS (gray), and BrU2-C275 DIS (dash-dot) in salts conditions. Three UV-melting curves are superimposed in each case to assess the reproducibility of measurements in saline condition. In absence of salt (insert), the maxima of the melting curves allowed us to define reliably the difference in “melting temperature” between C275 DIS (black), BrU3-C275 DIS (gray), and T3-C275 DIS (dashed), but overall, the melting curves were much more variable (data not shown), particularly at high temperature.
We then analyzed the hairpin–duplex conversion on a DIS sequence mutated in the self-complementary loop (DIS-C275). This mutant is unable to form a stable kissing-loop complex since the U275C mutation introduces two highly destabilizing A-C mismatches in the loop–loop helix. We have previously shown that this DIS-C275 sequence forms mainly hairpin, even after folding in water at high RNA concentration, and that the hairpin is spontaneously converted upon incubation in salts into duplex, very likely via a cruciform intermediate (Bernacchi et al. 2005). Here, after the folding step in water, we incubated in salts at 37°C either an unmodified C275-DIS sequence or a BrU3-C275-DIS sequence (20 μM RNA, 20 mM sodium cacodylate buffer at pH 7.0, 25 mM KCl, 2 mM MgCl2). The respective amounts of hairpin and duplex were then quantified from a native gel electrophoresis. As shown on Figure 3B, a clear difference was again observed between the two sequences immediately after folding in water (see Materials and Methods): 8% of duplex was obtained for the unmodified sequence, but no significant amount of it was detectable on the gel for the BrU3-C275 sequence. The effect of bromination of U3 is therefore comparable on the C275 mutant and on the wild-type DIS sequence. However, the bromination on U3 slows down, but does not prevent, the conversion to duplex. Additional comments on the kinetics of transition are made in the discussion.
We then checked by UV melting the effect upon DIS hairpin stability of U3 and U2 bromination. According to our previous results, the hairpin was expected to be stabilized with the BrU3 modification and destabilized with the BrU2 modification. In order to overcome the difficulty introduced in the interpretation of the melting curves by the additional loop–loop interaction, we again used the DIS-C275 sequence, which is without effect on hairpin formation but prevents kissing-complex formation.
Melting experiments performed in water (Fig. 3C, inset) revealed, as anticipated, a stabilization for the BrU3-C275 hairpin (Tm=45 ± 0.8°C) in comparison to the unmodified C275 hairpin (Tm=41.5 ± 1°C). However, a stabilization was also observed for the BrU2-C275 DIS (Tm=43.5 ± 0.6°C), instead of the expected destabilization. In presence of salts (25 mM potassium acetate, 2 mM magnesium acetate, 20 mM sodium cacodylate at pH 7.0), the same global pattern was observed with BrU3-C275 (Tm=72.6 ± 0.6°C) and even BrU2-C275 (Tm= 74.0 ± 0.6°C), being slightly more stable than the unmodified sequence (Tm=70.8 ± 0.4°C).
We further analyzed the unmodified- versus brominated-RNA melting profile by investigating any possible effect of the pH on the brominated RNA. It is known that the pKa of the N3 of a uracil is affected by base substitutions and varies from 9.3 for uracil and 9.9 for thymine to 8.2 for 5-bromo-uracil (Smith 1957; Massoulie et al. 1966; Saenger 1984). Therefore, in our experimental conditions (pH 7.0), a small fraction of bromo-uridine is unprotonated, which might affect hairpin stability. To test this hypothesis, UV-melting experiments were conducted on C275 DIS and BrU3-C275 DIS in cacodylate buffer at pH 6.2 and 7.4 (in addition to previous experiments at pH 7.0). No difference due to pH was observed in melting profiles (data not shown), indicating that bromo-uridine deprotonation does not play a significant role in the BrU3-C275 DIS stabilization.
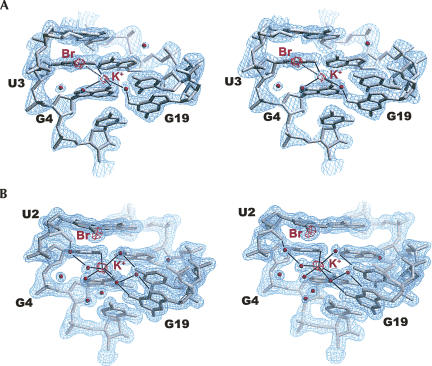
Since we previously solved the crystal structures of BrU2- and IU2-modified DIS duplex (Ennifar et al. 1999) and of BrU2- and BrU3-modified DIS kissing-loop complexes (Ennifar and Dumas 2006; Ennifar et al. 2006), these structures were scrutinized in order to understand the influence of halogenation. In addition, we also solved the structure of a IU3-modified DIS at 2.3 Å resolution in order to compare the effect of the iodination and of the bromination. A comparison of the hairpin stems of the unmodified BrU2-, IU3-, and BrU3-DIS kissing-loop complexes did not reveal any significant differences since the RMSD between their phosphate atoms ranged from 0.4 to 0.7 Å. These differences are of the order of, or just above, the expected errors on coordinates (0.3 Å from a Luzzati plot) (Luzzati 1952). We can thus conclude that the influence of bromination upon the DIS hairpin stability is not related to significant differences in RNA structures. However, a careful analysis of ion binding in the different structures revealed some possible hints about the influence of halogen position. Although some structures were obtained in different space groups and with unrelated crystal packing, all crystals were grown from very similar crystallization conditions containing KCl, MgCl2, spermine chloride, Na cacodylate (pH 6.5), and MPD as a precipitant. As a consequence, ion binding can be directly compared in all structures, provided these ions are not involved into crystal packing. In addition, to take into account that irrelevant differences in ion binding may exist between sites related by noncrystallographic symmetry (Ennifar et al. 2003), several comparable situations were analyzed. These comprised the BrU2-DIS kissing-loop complex (Protein Data Bank [PDB] ID 1ZCI) with four hairpins in the asymmetric unit, and four BrU3-DIS kissing-loop complex structures (PDB IDs 1Y3S, 2FCX, 2FCY, and 2FD0) with, in each case, two hairpins in the asymmetric unit. It appeared that in all kissing-loop complex structures (brominated, iodinated, or unmodified), a conserved potassium ion is tightly bound at the 3′U3pG45′ step, within the deep groove of the hairpin stem (Fig. 4). The cation was unambiguously identified thanks to its weak but measurable anomalous signal (Ennifar and Dumas 2006; Ennifar et al. 2006). It is located at 2.68 ± 0.15 Å from the O6 of G4 and at 2.75 ± 0.13 Å from the O4 of U3 (the estimated standard deviations [ESD] values 0.15 and 0.13 Å were obtained from the variability in the different structures). In BrU3- and IU3-DIS kissing-loop structures, this potassium is 3.78 ± 0.12 Å away from the halogen atom, but this distance increases to 4.66 ± 0.16 Å for BrU2 RNA. As a consequence, it may be thought that the Br-K+ (or I-K+) interaction in BrU3-DIS sequences contributes to the stem stability by linking the two strands since the bromine (or the iodine) is attached to U3 on one strand and to the potassium cation linked to G19 on the other strand through coordinated waters (Fig. 4A). This contrasts with the situation in the BrU2-DIS (and obviously also in the nonbrominated RNA), where such bromine-mediated cross-strand interaction does not exist (Fig. 4B). It is to be noticed that for the unmodified and BrU2-DIS, one can nevertheless perceive a water-mediated cross-strand interaction (Fig. 4B), albeit less direct than in the BrU3-DIS (Fig. 4A). Therefore, it may be thought that the interaction in the BrU3-DIS is more stabilizing than in the unmodified DIS and BrU2-DIS.
FIGURE 4.
Detailed stereo views showing the potassium cation in the DIS RNA for the BrU3 at 2.0 Å resolution (A, PDB ID 2FCZ) and for the BrU2 at 1.6 Å resolution (B, PDB ID 1ZCI). The 2Fo-Fc electron density map contoured at 1.4 σ above mean level is represented in blue, and the anomalous difference map contoured at 5.0 σ above mean level is represented in red. Ligands of the potassium cation and some H-bonds are depicted with black lines.
In order to check this assumption, we have performed two new melting experiments. The first one was performed with BrU3 and unmodified DIS-C275 after extensive dialysis to exchange potassium by either lithium or ammonium cations. Since a small, but significant, Tm increase by 1.7°C was observed in presence of potassium for the BrU3 versus the unmodified DIS-C275, and since we hypothesized that the additional stabilization was provided by a cross-strand interaction mediated by a potassium cation, we expected that removing potassium would suppress the increase in Tm. However, this was not the case, and the same stabilization was observed for the BrU3 DIS-C275 whether or not potassium was replaced by lithium or ammonium (data not shown). The second experiment was performed after substitution of BrU3 for a ribothymine (T3-C275 DIS sequence), thus substituting a bromine atom for a methyl group, which should disfavor a stabilizing interaction with a cation and increases base hydrophobicity. A significant destabilization was observed in water (Tm=38.5 ± 1°C) (Fig. 3B), and in presence of salts, the T3-C275 was indeed less stable than the BrU3 DIS sequence since it behaved as the unmodified C275 (data not shown). Therefore, these experimental results do not allow to derive a clear-cut conclusion. We can nevertheless suggest that if monovalent ions are involved into a stabilization of the BrU3-DIS hairpin, this would not result from of a specific potassium binding site but rather from a more diffuse ion binding (Draper 2004).
DISCUSSION
Several previous studies have reported a strong influence of base modifications on RNA folding. The role of nucleobase methylation on a short hairpin–duplex equilibrium (a situation similar to the DIS sequence) was systematically investigated, which showed that several slightly different methylations can seriously affect RNA secondary structure and stability in a very different way (Micura et al. 2001). Another RNA conformational switch was reported for the spliced leader RNA from Leptomonas collosoma: a competition between two alternative secondary structures was seen to be influenced by the presence of a bromodeoxycytidine in lieu of a cytidine. In presence of bromodeoxycytidine, one of the two conformations had its melting temperature increased by 5°C (LeCuyer and Crothers 1994). An even more dramatic structural effect was observed for the bromination of the DNA sequence d(CCAGTACTGG). This sequence folds as a duplex, but it folds as a four-way junction when a bromo-uridine is substituted for the second thymine in the sequence (Hays et al. 2003). In this situation, analyzed in detail by Auffinger et al. (2004), the bromine atom, being involved in a crucial halogen bond (∼3.0 Å distance) with an adjacent phosphate oxygen, appears to be responsible for the conformational change. A similar situation was reported for the DNA sequence d(GCGAAGCT). Here again, two alternate conformations were observed by X-ray crystallography depending on whether or not the second cytosine was halogenated as d(GXGAAGCT), where X is a 5-bromo- or 5-iodo-cytidine. A “base-intercalated duplex” structure was observed for halogenated sequences (Sunami et al. 2004), whereas a parallel-stranded duplex containing a C:C+ base pair was observed for the nonhalogenated sequence (Sunami et al. 2002). It was suggested that the cytosine protonation required for the formation of the C:C+ base-pairing is affected by a decrease of pKa of the halogenated cytosine (Sunami et al. 2004). In addition, the base-intercalated duplex (i.e., without C:C+ base-pairing) is likely stabilized by halogen bonds between the nonprotonated cytosine and a phosphate group (Auffinger et al. 2004).
The situation of the BrU3-DIS RNA is likely different from these previous cases and is not of immediate interpretation since the bromine atom is not engaged in a direct interaction with the RNA itself as in the four-way junction mentioned before. It is, however, clear that the BrU3 modification yields a significant increase of the amount of the hairpin monomer over the duplex dimer. For the time being, we can only push forward two nonexclusive hypotheses. The first one is that the stabilization is indeed of ionic character, but without need of a specific cation binding site and with only diffuse binding (Draper 2004). The second one is that there exists some favorable interaction between BrU3 (or IU3) and the neighboring base U2 it is stacked on. However, in the latter case, the stabilization would not be the result of a cross-strand interaction.
Finally, the observation of the transition to duplex upon addition of salts (Fig. 3B) deserves additional comments. It is striking that the kinetics of transition, be it for the unmodified or the brominated sequence, shows nonclassical behavior. Indeed, the fraction of duplex varies linearly with the logarithm of the incubation time at least over three orders of magnitude (whereas “classical kinetics” would involve an exponential-like time dependence). The observed kinetics implies that one cannot consider, even formally, an extrapolation to infinite time (as for classical kinetics) since this would lead to an absurd infinite amount of duplex. However, one may extrapolate the straight lines in Figure 3B to 100% of duplex, which then provides us with an estimate of a finite time for the full transition to duplex. As a result, it appears that the brominated sequence reached the full transition to duplex after 7.8 d and the unmodified sequence after 7.1 d. Although such an extrapolation is certainly to be taken with caution, it nevertheless yields a reasonable estimate of the lag of the brominated sequence behind the unmodified sequence (17 h) for full transition to duplex. We will only suggest a possible explanation for these observations. According to our previous work, a simple scheme involving single-strand intermediates from hairpin to duplex cannot explain the observed kinetics; very likely, the transition occurs via hairpins engaged in cruciform intermediates (Bernacchi et al. 2005). However, according to this scheme, when the duplex species has sufficiently accumulated, there is no reason why duplexes would not be involved themselves in cruciform intermediates with hairpins (and with other duplexes as well). With this line of view, since a duplex has two identical ends, it is conceivable that complex supramolecular assemblies, trapping several strands, would need to be “undone” for allowing duplex formation. We think such a cascade of transient complexity is a plausible explanation of such unusual kinetics. This remains to be proven.
Conclusions
The DIS RNA stem–loop represents another example of nucleic acid structure that can undergo important structural changes upon apparently neutral halogenation of bases. We have shown that in comparison with the unmodified RNA, a BrU3 or IU3 modification strongly shifts the hairpin–duplex ratio toward the hairpin monomer, whereas BrU2, IU2, or BrU6 modifications tend to favor the duplex form. It should be emphasized that the consequences of halogenation are not limited to RNA annealing in water but also affect the kinetics of subsequent hairpin–duplex conversion in presence of salts. The analysis of several brominated DIS crystal structures suggested that halogenation of uridine 3 does not perturb the RNA structure. In spite of our efforts, however, the exact cause of the modification of the hairpin–duplex ratio remains unclear. In particular, the results of UV-melting experiments were only partially consistent with the observations. It would certainly be of interest to extend the present study to halogenation of C5 of uridines with fluorine and chlorine.
It is occasionally observed that halogenated nucleic acids do not crystallize in conditions successfully used for native sequences. These failures are commonly attributed to changes in crystallization conditions, as frequently observed for sequence variations. If this holds true in some cases, the increasing examples of halogen-driven nucleic acid conformational switches makes us suggest that several unsuccessful crystallizations of halogenated sequences were due to unnoticed change in RNA conformation, and not in crystallization conditions.
Our results, as well as those previously mentioned (LeCuyer and Crothers 1994), also lead one to question the basis of studies aimed at determining the influence of loop sequences on the outcome of competing hairpins in two alternative two-dimensional (2D) structures (Nagel et al. 2006). Indeed, in such kind of studies, the design of a sequence leading to two competing 2D structures implies differences not only in the two alternative loops but also in the two alternative hairpin-stem sequences. Therefore, if a change as limited as the displacement of a single halogenation in a hairpin stem (without any modification of the loop sequence) yields such important differences on hairpin closing, one cannot rule out that even a little change in the hairpin–stem sequences might interfere significantly with the influence of the alternative loops.
Finally, in addition to halogenation of uridines and cytosines for X-ray crystallography, other modifications are commonly used in structural studies of nucleic acids: ribose modification for EPR spectroscopy (Macosko et al. 1999), 2′-fluoro-nucleotides (Kreutz et al. 2005, 2006) or 5-fluoropyrimidines (Marshall and Smith 1977; Rastinejad et al. 1995) for NMR spectroscopy, introduction of various fluorescent bases for fluorescence studies (Qin and Pyle 1999; Yamana et al. 1999; Ben Gaied et al. 2005), or, more recently, incorporation of selenium for crystal structure determination (Wilds et al. 2002; Hobartner and Micura 2004). Our results highlight the importance of checking the possible influence on RNA folding of even subtle nucleic acids modifications.
MATERIAL AND METHODS
Sample preparation
Native and modified DIS RNA sequences (wild-type and C275 mutant) were purchased from Dharmacon and purified as already described (Ennifar et al. 2003). Native or brominated molecular beacon sequences were labeled on the 5′ terminus using 5 (and 6)-carboxyfluorescein (FAM) as a fluorescent dye, and on the 3′ terminus with 4-(4′-dimethylethyl-aminophenylazo) benzoic acid (DABCYL), as a fluorescence quencher. Unlike unlabeled sequences, all molecular beacon RNA were purified on denaturant polyacrylamide gel. All RNA sequences were extensively washed in water and concentrated using a Centricon 10K (Millipore).
Gel electrophoresis
RNA was diluted to a final concentration of 30 μM in water, heated for 3 min at 90°C for denaturation, and then cooled in water at 0°C. One volume of a 10× dimerization buffer (250 mM KCl, 20 mM MgCl2, 250 mM Na cacodylate at pH 7.0) was then added. For time-course experiments, a 20 μL aliquot of RNA sample (representative of no incubation) was immediately flash-frozen in ethanol maintained at −70°C in dry ice and kept frozen until the end of the experiment. RNA was incubated at 37°C, and 20 μL aliquots were frozen at variable time intervals. At the end of the experiment, 5 μL of gel loading solution was added, and samples were then loaded on a 15% polyacrylamide gel, Tris borate 1×, 1 mM MgCl2. Gels were run at 5 W at room temperature (semi-denaturing conditions) or at 4°C (native conditions).
Fluorescence measurements
Fluorescence emission spectra were recorded at 20°C on a Fluoromax-2 spectrofluorometer (Horiba–Jobin Yvon). Steady-state fluorescence measurements were performed by mixing 60 nM of molecular beacon RNA with an excess of unlabeled RNA (ranging from 300 nM–60 μM). Measurements were achieved in water to directly probe the result of the folding without addition of any salts. The fluorescence signal was found to be stable with time and reproducible in such conditions. Fluorescence signal of solutions containing either 60 nM of molecular beacon only or 60 nM of a singly labeled 5′-FAM DIS were used as references for 100% hairpin and 100% duplex, respectively. A 5′-FAM BrU3 DIS sequence was used to take into account the effect of the bromination on the fluorescein signal. Experiments using various concentrations of unlabeled RNA were repeated using singly labeled 5′-FAM DIS to evaluate the influence of the excess of RNA on the fluorescent dye.
UV thermal melting
Absorption spectra were recorded on a Uvicon XL spectrophotometer. Melting curves were recorded at a 260-nm wavelength between 15°C and 90°C at 0.8°C min−1. The RNA concentration was 3 μM. Measurements were achieved in 2 mM MgCl2, 25 mM KCl, 20 mM Na cacodylate (pH 7.0). Alternatively, KCl was replaced by 25 mM LiCl or 25 mM (NH4)2SO4 for testing the effect of potassium. We also performed measurements in water at 0.3°C min−1, or in salts with cacodylate buffer at pH 6.2 and 7.4.
X-ray crystallography
The BrU2-DIS kissing complex was crystallized as described by Ennifar and Dumas (2006). The BrU3 or IU3 DIS kissing complex was obtained by mixing 7 μL of a solution containing 500 μM RNA in 150 mM KCl, 25 mM Na cacodylate (pH 6.5), 2 mM MgCl2 with 2 μL of a solution made with 30% 2, 4-methylpentanediol (MPD), 50 mM spermine chloride (or alternatively 1.0 μL of a solution made with 30% MPD, and 5 mM lividomycin or neomycin sulfate). Sitting drops were equilibrated at 37°C over a reservoir made with 40% MPD, 300 mM KCl, 50 mM Na cacodylate (pH 6.5), 20 mM MgCl2. Crystals were transferred at 20°C prior flash-freezing in liquid ethane and data collection at SLS or ESRF synchrotrons. Structures were solved and refined up to 1.65 and 1.85 Å resolution for BrU2- and BrU3-DIS, respectively, as described by Ennifar and Dumas (2006) and Ennifar et al. (2006). IU3-DIS crystals were collected at 1.55 Å wavelength (to maximize anomalous signal of the iodine) on the ID29 beamline at the ESRF. Data were processed with Denzo and Scalepack (Otwinowski and Minor 1996) and refined at 2.3 Å resolution using CNS (Brunger et al. 1998). BrU2 or IU2-DIS duplex crystals were obtained in almost identical conditions than BrU2-DIS kissing-loop complex crystals as described by Ennifar et al. (1999). We were unable to grow crystals of BrU3- or IU3-DIS duplex. Potassium and bromide positions were revealed accurately using anomalous signal of these atoms collected at the absorption K-edge for bromide and at low-energy wavelengths for potassium, in a way to maximize f″ values.
ACKNOWLEDGMENTS
This work was supported by the “Agence Nationale de Recherche sur le SIDA” (ANRS). S.B. is a recipient of a Sidaction fellowship. We thank G. Bec for his constant assistance, C. Schulze-Briese and E. Pohl (SLS, Viligen, Switzerland) for support on the PXI beamline, and B. Pertuiset (Perbio France) for her help in obtaining brominated molecular beacons. We also thank J. Kondo and P. Auffinger for fruitful discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.408507.
REFERENCES
- Auffinger, P., Hays, F.A., Westhof, E., Ho, P.S. Halogen bonds in biological molecules. Proc. Natl. Acad. Sci. 2004;101:16789–16794. doi: 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Gaied, N., Glasser, N., Ramalanjaona, N., Beltz, H., Wolff, P., Marquet, R., Burger, A., Mely, Y. 8-Vinyl-deoxyadenosine, an alternative fluorescent nucleoside analog to 2′-deoxyribosyl-2-aminopurine with improved properties. Nucleic Acids Res. 2005;33:1031–1039. doi: 10.1093/nar/gki253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi, S., Ennifar, E., Toth, K., Walter, P., Langowski, J., Dumas, P. Mechanism of hairpin–duplex conversion for the HIV-1 dimerization initiation site. J. Biol. Chem. 2005;280:40112–40121. doi: 10.1074/jbc.M503230200. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Draper, D.E. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennifar, E., Dumas, P. Polymorphism of bulged-out residues in HIV-1 RNA DIS kissing complex and structure comparison with solution studies. J. Mol. Biol. 2006;356:771–782. doi: 10.1016/j.jmb.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Ennifar, E., Walter, P., Dumas, P. An efficient method for solving RNA structures: MAD phasing by replacing magnesium with zinc. Acta Crystallogr. 2001;D57:330–332. doi: 10.1107/s0907444900017558. [DOI] [PubMed] [Google Scholar]
- Ennifar, E., Walter, P., Dumas, P. A crystallographic study of the binding of 13 metal ions to two related RNA duplexes. Nucleic Acids Res. 2003;31:2671–2682. doi: 10.1093/nar/gkg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennifar, E., Yusupov, M., Walter, P., Marquet, R., Ehresmann, B., Ehresmann, C., Dumas, P. The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges. Structure. 1999;7:1439–1449. doi: 10.1016/s0969-2126(00)80033-7. [DOI] [PubMed] [Google Scholar]
- Ennifar, E., Paillart, J.C., Bodlenner, A., Walter, P., Weibel, J.M., Aubertin, A.M., Pale, P., Dumas, P., Marquet, R. Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: From crystal to cell. Nucleic Acids Res. 2006;34:2328–2339. doi: 10.1093/nar/gkl317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays, F.A., Vargason, J.M., Ho, P.S. Effect of sequence on the conformation of DNA Holliday junctions. Biochemistry. 2003;42:9586–9597. doi: 10.1021/bi0346603. [DOI] [PubMed] [Google Scholar]
- Hobartner, C., Micura, R. Chemical synthesis of selenium-modified oligoribonucleotides and their enzymatic ligation leading to an U6 SnRNA stem-loop segment. J. Am. Chem. Soc. 2004;126:1141–1149. doi: 10.1021/ja038481k. [DOI] [PubMed] [Google Scholar]
- Kreutz, C., Kahlig, H., Konrat, R., Micura, R. Ribose 2′-F labeling: A simple tool for the characterization of RNA secondary structure equilibria by 19F NMR spectroscopy. J. Am. Chem. Soc. 2005;127:11558–11559. doi: 10.1021/ja052844u. [DOI] [PubMed] [Google Scholar]
- Kreutz, C., Kahlig, H., Konrat, R., Micura, R. A general approach for the identification of site-specific RNA binders by 19F NMR spectroscopy: Proof of concept. Angew. Chem. Int. Ed. Engl. 2006;45:3450–3453. doi: 10.1002/anie.200504174. [DOI] [PubMed] [Google Scholar]
- LeCuyer, K.A., Crothers, D.M. Kinetics of an RNA conformational switch. Proc. Natl. Acad. Sci. 1994;91:3373–3377. doi: 10.1073/pnas.91.8.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzati, V.P. Traitement statistique des erreurs dans la determination des structures cristallines. Acta Crystallogr. 1952;5:802–810. [Google Scholar]
- Macosko, J.C., Pio, M.S., Tinoco I., Jr, Shin, Y.K. A novel 5 displacement spin-labeling technique for electron paramagnetic resonance spectroscopy of RNA. RNA. 1999;5:1158–1166. doi: 10.1017/s1355838299990830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, A.G., Smith, J.L. Nuclear-spin-labeled nucleic acids. 1 19F nuclear magnetic resonance of Escherchia coli 5-fluorouracil-5S-RNA. J. Am. Chem. Soc. 1977;99:635–636. doi: 10.1021/ja00444a066. [DOI] [PubMed] [Google Scholar]
- Massoulie, J., Michelson, A.M., Pochon, F. Polynucleotide analogues. VI. Physical studies on 5-substituted pyrimidine polynucleotides. Biochim. Biophys. Acta. 1966;114:16–26. [PubMed] [Google Scholar]
- Micura, R., Pils, W., Hobartner, C., Grubmayr, K., Ebert, M.O., Jaun, B. Methylation of the nucleobases in RNA oligonucleotides mediates duplex-hairpin conversion. Nucleic Acids Res. 2001;29:3997–4005. doi: 10.1093/nar/29.19.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel, J.H., Flamm, C., Hofacker, I.L., Franke, K., de Smit, M.H., Schuster, P., Pleij, C.W. Structural parameters affecting the kinetics of RNA hairpin formation. Nucleic Acids Res. 2006;34:3568–3576. doi: 10.1093/nar/gkl445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter C.W., Sweet R.M., editors. Methods in enzymology. Academic Press; New York: 1996. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pahler, A., Smith, J.L., Hendrickson, W.A. A probability representation for phase information from multiwavelength anomalous dispersion. Acta Crystallogr. A. 1990;46:537–540. doi: 10.1107/s0108767390002379. [DOI] [PubMed] [Google Scholar]
- Qin, P.Z., Pyle, A.M. Site-specific labeling of RNA with fluorophores and other structural probes. Methods. 1999;18:60–70. doi: 10.1006/meth.1999.0757. [DOI] [PubMed] [Google Scholar]
- Rastinejad, F., Evilia, C., Lu, P. Studies of nucleic acids and their protein interactions by 19F NMR. Methods Enzymol. 1995;261:560–575. doi: 10.1016/s0076-6879(95)61025-1. [DOI] [PubMed] [Google Scholar]
- Saenger, W. Principles of nucleic acid structure. Springer; New York: 1984. [Google Scholar]
- Smith, J.D. Methods in enzymology. Vol. 12. Academic Press; London: 1975. Paper electrophoresis of nucleic acids components; pp. 350–361. [Google Scholar]
- Sunami, T., Kondo, J., Kobuna, T., Hirao, I., Watanabe, K., Miura, K., Takenaka, A. Crystal structure of d(GCGAAAGCT) containing a parallel-stranded duplex with homo base pairs and an anti-parallel duplex with Watson–Crick base pairs. Nucleic Acids Res. 2002;30:5253–5260. doi: 10.1093/nar/gkf639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunami, T., Kondo, J., Hirao, I., Watanabe, K., Miura, K., Takenaka, A. Structure of d(GCGAAGC) (hexagonal form): A base-intercalated duplex as a stable structure. Acta Crystallogr. D Biol. Crystallogr. 2004;60:90–96. doi: 10.1107/s0907444903024703. [DOI] [PubMed] [Google Scholar]
- Theilleux-Delalande, V., Girard, F., Huynh-Dinh, T., Lancelot, G., Paoletti, J. The HIV-1(Lai) RNA dimerization. Thermodynamic parameters associated with the transition from the kissing complex to the extended dimer. Eur. J. Biochem. 2000;267:2711–2719. doi: 10.1046/j.1432-1327.2000.01292.x. [DOI] [PubMed] [Google Scholar]
- Tyagi, S., Kramer, F.R. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Wilds, C.J., Pattanayek, R., Pan, C., Wawrzak, Z., Egli, M. Selenium-assisted nucleic acid crystallography: Use of phosphoroselenoates for MAD phasing of a DNA structure. J. Am. Chem. Soc. 2002;124:14910–14916. doi: 10.1021/ja021058b. [DOI] [PubMed] [Google Scholar]
- Yamana, K., Iwase, R., Furutani, S., Tsuchida, H., Zako, H., Yamaoka, T., Murakami, A. 2′-Pyrene modified oligonucleotide provides a highly sensitive fluorescent probe of RNA. Nucleic Acids Res. 1999;27:2387–2392. doi: 10.1093/nar/27.11.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]