Abstract
Piwi-interacting RNAs (piRNAs) are a novel class of small regulatory RNAs that are expressed specifically and abundantly in germ cells. Mammalian piRNAs are 26–31 nucleotides in length and bind to Piwi proteins, but their function and biogenesis remain elusive. We previously showed that mammalian piRNAs are 2′-O-methylated at their 3′ termini. The biosynthetic mechanism and function of this modification is unknown. Here, we report that the mouse homolog (mHEN1) of HEN1, a plant microRNA (miRNA) 2′-O-methyltransferase, is expressed specifically in testis and methylates 3′ termini of piRNAs in vitro. These findings provide insight into the biogenesis of piRNAs.
Keywords: piRNA, Piwi, Miwi, Mili, miRNA, microRNA, argonaute
INTRODUCTION
Small noncoding RNAs have emerged as universal regulators of gene expression. Three major classes of small regulatory RNAs have been identified so far: microRNAs (miRNAs), short-interfering RNAs (siRNAs), and Piwi-interacting RNAs (piRNAs). Although the size, which is ∼22 nucleotides (nt) for miRNAs and siRNAs and 26–31 nt for piRNAs, of these three classes differs, they all bind to proteins that belong to the argonaute family. Argonaute proteins are divided into Ago and Piwi subclades (Carmell et al. 2002). miRNAs and siRNAs bind to Ago proteins and silence gene expression (for review, see Bartel 2004; Pillai et al. 2007). In contrast, piRNAs are expressed only in the germline and bind to Piwi proteins (for review, see Kim 2006; O'Donnell and Boeke 2007). The biogenesis and function of mammalian piRNAs is unknown but like Drosophila piRNAs (also known as repeat-associated small interfering RNAs—rasiRNAs), they may be involved in silencing retrotransposons and repetitive elements in the germline (Aravin et al. 2003; Sarot et al. 2004; Vagin et al. 2006; Brennecke et al. 2007; Gunawardane et al. 2007; Pelisson et al. 2007; for review, see O'Donnell and Boeke 2007).
Though animal miRNAs contain 3′ hydroxyl termini (Elbashir et al. 2001; Hutvagner et al. 2001), plant miRNAs or other classes of small RNAs contain a modified 3′-terminal nucleotide. Arabidopsis miRNAs and siRNAs are 2′-O-methylated at their 3′ termini (Yu et al. 2005; Yang et al. 2006). We and the Suzuki laboratory have recently demonstrated that the 3′-teminal nucleotide of mouse piRNAs are also 2′-O-methylated (Kirino and Mourelatos 2007; Ohara et al. 2007) and the same is true for rat piRNAs and likely zebrafish piRNAs (Houwing et al. 2007). Moreover, Drosophila piRNAs and Caenorhabditis elegance endogenous siRNAs and 21U-RNAs contain a ribose modification at either the 2′ or 3′ position of the 3′-terminal nucleotide (Vagin et al. 2006; Ruby et al. 2006).
The Chen laboratory has shown that in Arabidopsis, 2′-O-methylation of miRNAs and siRNAs is mediated by the methyltransferase protein HEN1. HEN1 is a 942 amino acid protein with a N-terminal double-strand RNA-binding domain (dsRBD) and a C-terminal methyltransferase domain (Park et al. 2002). HEN1 methylates 21–24 nt miRNA/miRNA* or siRNA/siRNA* duplexes with 2 nt overhangs in vitro, but does not methylate single-stranded RNAs (Yu et al. 2005; Yang et al. 2006). In hen1 mutants, small RNAs lack methylation and have additional nucleotides, primarily uridines, on their 3′ ends, suggesting that one function of small RNA methylation is to protect the 3′ ends of the small RNAs from an as yet unidentified uridine-addition activity in vivo (Li et al. 2005; Yu et al. 2005).
To address the biogenesis and functional role of terminal methylation of piRNAs, it is necessary to identify the methyltransferase involved. A stretch of ∼230 amino acids at the C terminus of HEN1 protein (containing the methyltransferase domain but lacking dsRBD) shows 40%–50% similarity to protein sequences predicted from cDNAs of metazoan, yeast, and cyanobacteria species (Park et al. 2002). These observations prompted us to test whether the mouse homolog of HEN1 (mHEN1) might be a 2′-O-methyltransferase of piRNAs. In this study we characterized the expression and the methyltransferase activity of mHEN1, demonstrating that mHEN1 is specifically expressed in testis and is capable of methylating 3′ termini of piRNA in vitro.
RESULTS AND DISCUSSION
mHEN1 is a 395 amino acid, 44.9 kDa protein. Using mHEN1 as a query, we have retrieved homologs from human, Drosophila, C. elegance, Tetrahymena, and Schizosaccharomyces pombe. The sequence alignment (Supplemental Fig. 1) shows that these proteins share conserved regions, including the methyltransferase domain, as previously detected (Park et al. 2002). Expression of mHEN1 mRNA in various mouse tissues was analyzed by RT-PCR. As shown in Figure 1, mHEN1 is expressed specifically in testis, like mouse piRNA and Piwi proteins (Kim 2006).
FIGURE 1.
The mRNA of mouse HEN1 (mHEN1) is expressed specifically in testis. Analysis of mHEN1 expression in mouse tissues. EF-1β was used as a control of a ubiquitously expressed mRNA.
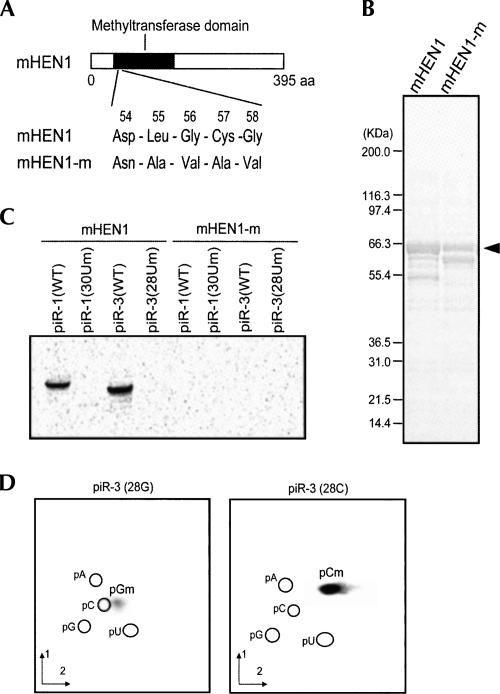
To test whether mHEN1 is a piRNA methyltransferase, we expressed in Escherichia coli recombinant, full-length GST-tagged mHEN1 (GST-mHEN1) and a catalytically inactive mHEN1 mutant (GST-mHEN1-m), where the potential active methyltransferase motif of mHEN1 was mutated (Fig. 2A,B). To test for methyltransferase activity, purified GST-mHEN1 or GST-mHEN1-m was incubated with synthetic piRNAs, piR-1 (30 nt), or piR-3 (28 nt), in the presence of S-adenosyl-L-[methyl-14C] methionine ([14C] SAM), which served as a methyl group donor. As shown in Figure 2C, GST-mHEN1, but not GST-mHEN1-m, methylated both piR-1 and piR-3. Since mHEN1 was unable to methylate piR-1 and piR-3 with 2′-O-methyluridine at their 3′ termini (Fig. 2C; piR-1 [30Um] and piR-3 [28Um]), the methylation was predicted to occur at the hydroxy group on ribose 2′ position. To identify the methylated nucleotides produced by the mHEN1 methylation, we performed the methylation reaction using piR-3 with 3′-terminal G (piR-3 [28G]) or C (piR-3 [28C]), followed by two-dimensional thin-layer chromatography (2D-TLC) analysis of the resultant methylated, [14C]-labeled nucleotides. The spots obtained from the methylated piR-3 (28G) or piR-3 (28C) corresponded to 5′-monophospho-2′-O-methylguanosine (pGm) or 2′-O-methylcytidine (pCm), respectively (Fig. 2D). These results indicate that mHEN1 methylates a 3′-terminal nucleotide of piRNA and forms a 2′-O-methyl modification. We also analyzed the time course of the in vitro methylation reaction by mHEN1 (Supplemental Fig. 2). The methylation reaction containing 67 μM (3 μg) of mHEN1 and 17 μM of RNA substrate, piR-1, or piR-3, gave a specific activity of 0.17 or 0.25 pmol/min/μg mHEN1, respectively.
FIGURE 2.
mHEN1 has piRNA methyltransferase activity in vitro. (A) A diagram of mHEN1 protein showing the putative methyltransferase domain. The sequences of the domain (54–58 amino acids) are mutated in mHEN1-m as indicated. (B) Coomassie-blue stained SDS-PAGE of the affinity purified GST-mHEN1 and GST-mHEN1-m proteins. The full-length mHEN1 proteins are indicated by the arrowhead. (C) The RNA substrates (shown in Table 1) were incubated with mHEN1 protein and [14C] SAM. [14C]-labeled RNA was resolved by UREA-PAGE and detected by autoradiography. (D) 2D-TLC analysis of in vitro methylated nucleotides derived from piR-3 (28G) and piR-3 (28C). The positions of 5′monophosphonucleotides, pA, pG, pC, and pU, detected by UV shadowing are indicated. Solvent dimensions 1 and 2 are detailed in Materials and Methods.
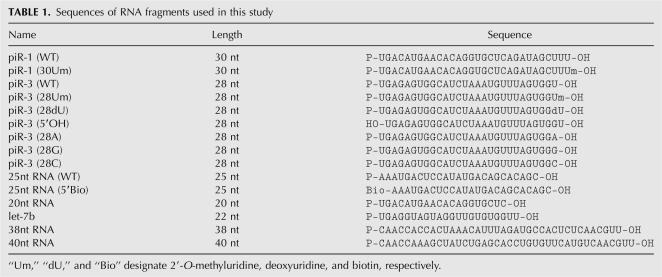
We next analyzed the substrate specificity of mHEN1 by testing various molecules as RNA substrates in the in vitro methylation reaction (Fig. 3A,B; Table 1). piR-3 having 3′-terminal 2′-O-methyluridine (piR-3 [28Um]) or deoxyuridine (piR-3 [28dU]) was not methylated by mHEN1, confirming that mHEN1 methylates the hydroxy group at the ribose 2′ position of the 3′-terminal nucleotide (Fig. 3A). mHEN1 showed no reduction of the methylation activity to piR-3 lacking 5′ phosphate (piR-3 [5′OH]) or 25 nt RNA bearing biotin at the 5′ terminus (25nt RNA [5′Bio]) compared to wild-type piRNA (Fig. 3A). These results suggest that mHEN1 does not recognize the 5′ termini of substrate RNAs. Interestingly, the methylation reaction using piR-3 bearing U (piR-3 [WT]), A (piR-3 [28A]), G (piR-3 [28G]), or C (piR-3 [28C]) at the 3′ terminus revealed different methylation efficiency of mHEN1 to each base, in vitro (Fig. 3A,B). The methylation efficiencies detected by liquid scintillation counting were A (259%)>C(137%)>U(100%)>G(44%). We also carried out methylation reaction with 20 nt (20nt RNA), 22 nt (let-7b), 38 nt (38nt RNA), and 40 nt (40nt RNA) RNA substrates, which have cytidine or uridine at their 3′ termini (Fig. 3A,B). Though the methylation efficiency is less than that of ∼28 nt RNA, mHEN1 is able to methylate 20–40 nt single-stranded RNAs, suggesting that mHEN1 does not strictly discriminate the length of RNA substrate. In vivo, mHEN1 likely requires other factors, possibly Piwi proteins, to distinguish piRNAs from other small RNAs.
FIGURE 3.
In vitro mHEN1 methylation of various RNA substrates (A) Methylation of indicated RNA substrates was detected by liquid scintillation counting. The amount of input RNA was defined as 100. The averages of five independent experiments with SD values are shown. (B) Methylation of RNA substrates was analyzed by UREA-PAGE and autoradiography.
TABLE 1.
Sequences of RNA fragments used in this study
Here we have demonstrated that mouse HEN1 is expressed specifically in testis and possesses in vitro methyltransferase activity to form ribose 2′-O-methyl modification at the 3′-terminal nucleotide of piRNA. HEN1 homologs in Drosophila or C. elegance might modify piRNAs (rasiRNAs) or 21U-RNAs, respectively. In contrast to plant HEN1, which contains dsRBD and methylates double-stranded RNAs (Yang et al. 2006), mHEN1 lacks a dsRBD and methylates single-stranded RNAs. These results suggest a model in which single-stranded piRNA precursors are methylated after their 3′ ends are generated by an as yet unknown mechanism. In plants, 2′-O-methylation protects miRNAs from addition of uridine-rich tails to the 3′ terminus and from degradation (Li et al. 2005; Yu et al. 2005). 2′-O-methylation of piRNAs might have a similar function to protect the terminal nucleotide from undesirable enzymatic activities. This modification may also facilitate the recognition of piRNAs by Piwi proteins rather than Ago proteins, since the 2′-O-methyl on the 3′-terminal nucleotide decreases the binding affinity by the PAZ domain in animal Ago proteins (Ma et al. 2004). Further studies on the putative cofactors and the functional significance of the piRNA methylation will enrich our understanding of piRNA biogenesis and function.
MATERIALS AND METHODS
RT-PCR analysis
Total RNA samples from various mouse tissues (Ambion) were first treated with RQ1 DNase (Promega). Single-strand cDNAs were prepared by reverse transcription from 0.2 μg of the total RNA sample in 20 μL reaction mixture using each gene-specific reverse primer (described below) and SuperScript II reverse transcriptase (Invitrogen). Each PCR reaction was performed using a 1/25 dilution of the RT products and Pfu turbo DNA polymerase (Stratagene). The PCR was carried out for 30 cycles of 95°C for 30 sec, 57°C for 30 sec, and 72°C for 1.5 min. The following primer pairs were used for the RT-PCR; mHEN1, forward (5′-AGTGGATCGCCATGAACCCAAGAAGG-3′) and reverse (5′-GGCCAGATCATCTGAATCCAAGTGTTC-3′); mouse EF-1β, forward (5′-TTACCTGGCGGACAAGAGCT-3′) and reverse (5′-CCAATTTAGAGGAGCCCCACA-3′).
Expression and purification of mHEN1 protein from E. coli
The ORF of mouse HEN1 (NM_025723) was confirmed by RACE using FirstChoice RACE-Ready cDNA (Ambion) and amplified by RT-PCR from the total RNA of mouse testicles using the following primers:
mHEN1-F-BamHI (5′-AGCTGAGGATCCATGGAAATGGCAGAAAGCATACCGTGCAAT-3′), and
mHEN1-R-XhoI (5′-TGACCTCTCGAGTCAGGAGTTCCCCAACCAGAACAACTTGAG-3′).
The RT-PCR product was cloned into pGEX-6P-2 (GE Healthcare) for producing GST-mHEN1 fusion protein expression in E. coli and verified by sequencing. The cloned pGEX-6P-2 was mutated by site-directed mutagenesis to change amino acids in the putative methyltransferase domain (“54D-L-G-C-G58” to “54N-A-V-A-V58” as described by Yu et al. 2005) by using the following primers:
mHEN1-QC-F (5′-CGCCATGAACCCAAGAAGGTTGCCAACGCTGTAGCCGTAGATGCCAAGCTCCTAAAGCTGCTA-3′), and
mHEN1-QC-R (5′-TAGCAGCTTTAGGAGCTTGGCATCTACGGCTACAGCGTTGGCAACCTTCTTGGGTTCATGGCG-3′).
These constructs were expressed in E. coli BL21 Codon Plus (RIL) (Stratagene) at 16°C overnight in the presence of 50 μM IPTG to produce mHEN1 fusion proteins with amino-terminal GST regions, which were subsequently purified using Glutathione Sepharose 4B resin (GE Healthcare–Amersham Biosciences).
Methyltransferase assay
The in vitro methyltransferase assay was performed as described (Yu et al. 2005; Yang et al. 2006) with slight modification. A reaction mixture (30 μL) containing 50 mM Tris-HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 5% glycerol, 40 units RNase inhibitor (Promega), 0.15 μCi S-adenosyl-L-[methyl-14C] methionine (58.7 mCi/mmol, GE Healthcare–Amersham Biosciences), 3 μg purified protein, and 500 pmol RNA substrate was incubated at 37°C for 90 min (or 10–90 min in the time course experiment). For the detection of methylated RNA by PAGE, the reaction was treated by phenol extraction and ethanol precipitation and analyzed on 15% UREA-PAGE. The [14C]-methylated RNAs were visualized by storage phosphor autoradiography. For the detection of methylation by [14C] count, the sample was spotted onto a Whatman 3MM filter disk. The disk was washed two times with 10% TCA for 15 min and then washed with ethanol for 5 min, followed by complete drying. The [14C] radioactivity was measured by liquid scintillation counting.
Two-dimensional thin-layer chromatography (2D-TLC)
For detection of 2′-O-methylated nucleotide, 2D-TLC analysis was performed as described by Kirino and Mourelatos (2007). Briefly, in vitro methylated piRNA was first digested completely by Nuclease P1 (Sigma). The resultant 5′-monophosphonucleotides were analyzed by 2D-TLC (cellulose on glass plate; Sigma) with a solvent system consisting of isobutyric acid/concentrated ammonia/H2O (66:1:33 by volume) in the first dimension, and 2-propanol/HCl/H2O (70:15:15 by volume) in the second dimension. [14C]-methylated nucleotide spots were visualized by storage phosphor autoradiography. The spots were assigned by comparing them to spots obtained from a mixture of synthetic piR-1 RNA (5′-UGACAUGAACACAGGUGCUCAGAUAGCUUU-3′) and 27 nt RNA containing 2′-O-methyl modification of all bases (5′-UmCmUmUmCmGmCmCmAmAmUmAmUmUmUmAmCmGmUmGmCmUmGmCmUmAmAm-3′). These synthetic RNAs were digested by Nuclease P1, analyzed by 2D-TLC, and visualized by UV shadowing.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.med.upenn.edu/camb/faculty/ggr/mourelatos.html.
ACKNOWLEDGMENTS
We are grateful to Drs. M.C. Siomi, H. Siomi, K. Saito (University of Tokushima) and members of our laboratory for stimulating discussions. This research was supported by a Human Frontier Science Program Long Term Fellowship to Y.K. and by NIH grants GM0720777, NS053839, and P30-HD026979, and by the Philadelphia Foundation to Z.M.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.659307.
NOTE ADDED IN PROOF
During the proof stages of this study, two groups reported that the Drosophila homolog of HEN1 is the Drosophila piRNA methylase (Horwich et al. 2007; Saito et al. 2007).
REFERENCES
- Aravin, A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., Tuschl, T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Aravin, A.A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila . Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Xuan, Z., Zhang, M.Q., Hannon, G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane, L.S., Saito, K., Nishida, K.M., Miyoshi, K., Kawamura, Y., Nagami, T., Siomi, H., Siomi, M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′-end formation in Drosophila . Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Horwich, M.D., Li, C., Matranga, C., Vagin, V., Farley, G., Wang, P., Zamore, P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007 doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Houwing, S., Kamminga, L.M., Berezikov, E., Cronembold, D., Girard, A., van den Elst, H., Filippov, D.V., Blaser, H., Raz, E., Moens, C.B., et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Kim, V.N. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes & Dev. 2006;20:1993–1997. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- Kirino, Y., Mourelatos, Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- Li, J., Yang, Z., Yu, B., Liu, J., Chen, X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis . Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.B., Ye, K., Patel, D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, K.A., Boeke, J.D. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara, T., Sakaguchi, Y., Suzuki, T., Ueda, H., Miyauchi, K., Suzuki, T. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., Chen, X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana . Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson, A., Sarot, E., Payen-Groschene, G., Bucheton, A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J. Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R.S., Bhattacharyya, S.N., Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Ruby, J.G., Jan, C., Player, C., Axtell, M.J., Lee, W., Nusbaum, C., Ge, H., Bartel, D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans . Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Saito, K., Sakaguchi, Y., Suzuki, T., Suzuki, T., Siomi, H., Siomi, M.C. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes & Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot, E., Payen-Groschene, G., Bucheton, A., Pelisson, A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, V.V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Ebright, Y.W., Yu, B., Chen, X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R.W., Steward, R., Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]