Abstract
After the second transesterification step of pre-mRNA splicing, the Prp22 helicase catalyzes release of spliced mRNA by disrupting contacts in the spliceosome that likely involve Prp8. Mutations at Arg1753 in Prp8, which suppress helicase-defective prp22 mutants, elicit temperature-sensitive growth phenotypes, indicating that interactions in the spliceosome involving Prp8-R1753 might be broken prematurely at 37°C. Here we report that mutations in loop I of the U5 snRNA or in Prp18 can suppress the temperature-sensitive prp8-R1753 mutants. The same gain-of-function PRP18 alleles can also alleviate the growth phenotypes of multiple slu7-ts mutants, indicating a functional link between Prp8 and the second step splicing factors Prp18 and Slu7. These findings, together with the demonstration that changes at Arg1753 in Prp8 impair step 2 of pre-mRNA splicing in vitro, are consistent with a model in which (1) Arg1753 plays a role in stabilizing U5/exon interactions prior to exon joining and (2) these contacts persist until they are broken by the helicase Prp22.
Keywords: Prp8, Prp18, Prp22 helicase, mRNA release, mRNA splicing
INTRODUCTION
The spliceosome, an assembly of snRNAs (U1, U2, U4/U6, and U5) and proteins, catalyzes the excision of introns from pre-mRNAs in two successive transesterification reactions (Burge et al. 1999). The first step entails attack of the branch site 2′-OH on the 5′ splice site (5′ss) with formation of a branched lariat intermediate. The second step is the attack of the 3′-OH of the 5′ exon on the 3′ splice site (3′ss) to join the exons and expel the lariat-intron. For the second transesterification reaction to occur, the 3′ss phosphodiester must be identified and positioned for attack by the 3′ hydroxyl of exon 1.
Step 2 depends on integral spliceosome constituents such as U5 snRNA and Prp8 and nonspliceosomal proteins Prp16, Slu7, Prp18, and Prp22 (Umen and Guthrie 1995b; Grainger and Beggs 2005). ATP hydrolysis by the DEAH-box enzyme Prp16 promotes a conformational change in the spliceosome that leads to protection of the 3′ss from targeted RNase H cleavage (Schwer and Guthrie 1992). This change, which likely reflects binding of the 3′ss PyAG↓ in the catalytic center of the spliceosome, requires the ordered recruitment of Slu7, Prp18, and Prp22 to the spliceosome (James et al. 2002).
Interactions between the U5 snRNA and exon bases immediately adjacent to the splice sites assist in aligning the exons for step 2 catalysis (Newman 1997; Crotti et al. 2007). Whereas the nucleotides in the U5 snRNA “loop 1” that interact with exon sequences are phylogenetically invariant (Frank et al. 1994), the involved exon bases are not conserved in pre-mRNAs. Cross-linking data implicate the Prp8 protein, a component of the U5 snRNP, in stabilizing the inherently weak U5/exon interactions (Teigelkamp et al. 1995).
The Prp18 protein also plays a role in fortifying U5/exon contacts prior to exon joining (Bacíková and Horowitz 2005; Crotti et al. 2007). Structural analysis of a functional Prp18 fragment revealed five tightly packed α helices and an unstructured 36-amino acid loop between α helices 4 and 5 (Jiang et al. 2000). Included in the 36-amino acid segment are 19 amino acids, the so-called conserved region (CR), which are deleted in the temperature-sensitive prp18ΔCR mutant (Bacíková and Horowitz 2002). The idea that Prp18, via its CR, stabilizes U5/exon interactions was suggested by the finding that mutations in the U5 snRNA loop 1 suppress the growth phenotypes and the second step splicing defects elicited by prp18ΔCR mutants (Bacíková and Horowitz 2005). Furthermore, the second step of splicing with Prp18ΔCR, but not wild-type Prp18, is sensitive to mutations in exon bases adjacent to the splice sites that interact with loop 1 of U5 (Crotti et al. 2007).
The DEAH-box helicase Prp22 enters the spliceosome prior to the second transesterification step, after which it catalyzes the release of mRNA from the spliceosome (Schwer and Gross 1998). The ATPase and helicase activities of Prp22 are required for product release. ATPase-deficient Prp22 mutants are lethal, and Prp22 mutants that retain ATPase activity, yet fail to unwind RNA duplexes in vitro, are lethal or elicit severe cold-sensitive growth defects (Schwer and Meszaros 2000; Campodonico and Schwer 2002; Schneider et al. 2002, 2004). Mutations at Arg1753 in the 2413-amino acid Prp8 protein suppress the cold-sensitive growth phenotypes of helicase-defective Prp22 mutants, suggesting that wild-type Prp22 effects mRNA release by disrupting contacts in the spliceosome that involve Arg1753 of Prp8. Whether Prp22 breaks a direct connection between Prp8 and the mRNA or whether Prp22 breaks U5/exon interactions that might be stabilized by Prp8 is not known. In PRP22 wild-type cells, the prp8-R1753 mutant alleles elicit temperature-sensitive growth defects, suggesting that contacts involving Arg1753 are not formed properly or are broken prematurely at 37°C.
Here we tested whether the contacts with spliced mRNA that are broken by the Prp22 helicase (1) involve the U5 snRNA in addition to the U5 snRNP component Prp8, and (2) are established prior to exon joining. We show that U5 snRNAs carrying specific mutations in loop 1 suppress the temperature-sensitive growth defects of prp8-R1753 mutants and exacerbate the growth defects of prp22-cs cells. The temperature sensitivity of prp8-R1753 mutants and of multiple slu7-ts alleles was also relieved by gain-of-function PRP18 alleles V191A and S162P, indicating a close functional relationship between Prp8, Prp18, and Slu7. In vitro studies show that Prp8-R1753 mutants slow the second step of splicing certain pre-mRNAs containing mutations in the exons adjacent to the splice sites. Our findings are consistent with a model whereby Arg1753 of Prp8 collaborates with Prp18 to stabilize U5/exon contacts that are important for the second transesterification step, and that the Prp22 helicase disrupts interactions between the U5 snRNP and mRNA to elicit mRNA release.
RESULTS
Mutations in U5 snRNA loop 1 suppress the growth phenotype of Prp8-R1753 mutants
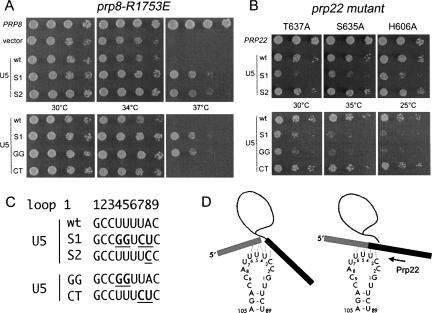
If Prp8-R1753 mutants are temperature sensitive because they fail to stabilize contacts of the U5 snRNP with exon sequences, then mutations in the U5 snRNA that increase pairing might rescue the growth defects. Because exon sequences are not conserved, Prp8-R1753 mutants likely influence splicing of a subset of precursor RNAs. To identify U5 alleles that rescue the splicing defects of affected RNAs, we screened for dominant suppressors of the temperature-sensitive phenotype of prp8-R1753E cells. We introduced into prp8-R1753E cells that contained a wild-type U5 gene (SNR7) a plasmid library of U5 mutants in which the nucleotides of loop 1 were randomized (Newman and Norman 1991). Of approximately 15,000 transformants analyzed, seven grew at 37°C, the nonpermissive temperature for prp8-R1753E cells. Plasmids were recovered from these strains and the U5 genes analyzed (Fig. 1C). U5-S1, which was found in six isolates, carried four mutations in loop 1 (U4G, U5G, U7C, A8U). The U5-S2 allele, which was isolated once, contained a single A8C change. Plasmids bearing PRP8, wild-type U5, U5-S1, and U5-S2 were reintroduced into prp8-R1753E SNR7 cells and growth of the resulting strains was compared (Fig. 1A). Whereas PRP8 restored growth to prp8-R1753E cells at 37°C, an extra copy of the gene for wild-type U5 snRNA did not. However, the U5-S alleles supported growth at nonpermissive temperature. U5-S1 and U5-S2 also suppressed the temperature sensitivity of prp8-R1753K and prp8-R1753A cells (not shown).
FIGURE 1.
Genetic interactions between Prp8, Prp22, and the U5 snRNA. (A) Fivefold serial dilutions of prp8-R1753E cells bearing plasmids (URA3 CEN) for expression of wild-type Prp8, wild-type U5 snRNA, and the mutants U5-S1, U5-S2, U5-GG, and U5-CT were spotted to minimal agar medium lacking uracil. The plates were photographed after 3 d of incubation at 30°C, 34°C, and 37°C. (B) prp22-T637A, prp22-S635A, and prp22-H606A were transformed with plasmids expressing wild-type Prp22 and U5 snRNAs as indicated at the left. Serial dilutions (10-fold) were spotted to agar medium and photographed after 3 d of incubation at 30°C and 35°C, and 4 d at 25°C. (C) Alignment of the wild-type U5 loop 1 sequence with those of the suppressors U5-S1 and U5-S2. Mutated nucleotides in the suppressor U5′s are in bold and underlined. The loop 1 sequences in U5-GG and U5-CT are U4G, U5G and U7C, A8U, respectively. (D) Model for interactions between the U5 loop and exon sequences prior to step 2 (left) and after exon joining (right). The dashed lines indicate putative base-pairing interactions between the U5 loop 1 and exon sequences (Newman 1997). Exons 1 and 2 are drawn as gray and black rectangles, respectively; the intron is indicated by a line. Loop 1 nucleotides, which are numbered 1–9 according to (Newman and Norman 1991), correspond to nucleotides 93–101 in the U5 snRNA (Frank et al. 1994).
To determine which of the four base changes in U5-S1 were responsible for suppression, we generated two U5 double mutants, U5-GG (U4G U5G) and U5-CT (U7C A8U), and analyzed their effects on growth of prp8-R1753E cells. Whereas U5-CT had no beneficial effect, prp8-R1753E cells harboring U5-GG grew as well as cells carrying the U5-S1 allele (Fig. 1A).
Although the sequence or length of loop 1 does not appear to affect the assembly of the U5 snRNP particle in vitro (O'Keefe et al. 1996; O'Keefe and Newman 1998; Ségault et al. 1999; McGrail et al. 2006), the function of the U5 snRNP might be disrupted by mutations in the loop sequence (Frank et al. 1992; O'Keefe 2002). We therefore tested whether U5-S1, U5-S2, U5-GG, and U5-CT supported viability of a U5Δ strain. U5-S2 and U5-CT supported normal growth of U5Δ cells at all temperatures (not shown; Bacíková and Horowitz 2005). U5-GG cells grew poorly, forming only pinpoint colonies after more than 6 d (not shown). U5-S1 failed to complement a U5Δ strain at 18°C, 30°C, or 37°C (not shown). The finding that U5-S1 was lethal yet suppressed the growth defect of prp8-R1753 mutants argues that (1) the altered U5 snRNA functions specifically with the mutated Prp8 to splice certain precursor RNAs, and (2) wild-type U5 snRNA is required for splicing of other essential RNAs.
U5-S1 (U4G, U5G, U7C, A8U) exacerbates the growth defects of helicase-defective Prp22 mutants
Bases C3 to U6 of the U5 loop are thought to interact with bases in the exons adjacent to the splice sites (Newman 1997; Crotti et al. 2007). The multiple mutations in U5-S1 (specifically U4G, U5G) might enhance the base-pairing of U5 with exon sequences in one or more pre-mRNAs. If Prp22 normally disrupts those contacts to release the mRNA, then U5-S1 and U5-GG might exacerbate the phenotype of helicase-defective prp22-cs mutants. To test this, prp22-T637A, prp22-S635A, and prp22-H606A cells (containing wild-type alleles for Prp8 and U5) were transformed with centromeric plasmids carrying either PRP22, wild-type U5, or the various U5 mutants and then plated at various temperatures (not shown; Fig. 1B). PRP22 restored growth to each of the prp22-cs mutants. Whereas an extra copy of wild-type U5, U5-S2, or U5-CT did not affect growth of the prp22-cs mutants at any temperature, T637A, S635A, and H606A cells bearing U5-S1 or U5-GG grew more slowly at 30°C, 35°C and 25°C, their respective semipermissive temperatures (Fig. 1B).
The findings that the snr7 alleles U5-S1 and U5-GG suppressed the temperature-sensitive growth phenotype of prp8-R1753 mutants and enhanced cold sensitivity of helicase-defective prp22 mutants are consistent with a model wherein the Prp22 helicase disrupts interactions between the U5 snRNP and mRNA (Fig. 1D).
Prp8-R1753 mutants are defective for the second step of splicing in vitro
To determine whether Prp8-R1753 mutants were defective for pre-mRNA splicing in vitro, we prepared extracts from isogenic PRP8, prp8-R1753K, and prp8-R1753E cells and assayed their splicing activities in vitro. ACT1 pre-mRNA was spliced with comparable efficiency in all three extracts at 23°C and 32°C, and preincubation at 37°C did not result in selective inhibition of mRNA formation in Prp8-R1753K and Prp8-R1753E extracts (not shown). Thus, splicing of ACT1 pre-mRNA in vitro appears to be insensitive to mutations at Arg1753 of Prp8 that cause temperature-sensitive growth defects in vivo. Although we cannot rule out the possibility that Prp8-R1753 mutant proteins cannot be heat inactivated in vitro, it seems more likely that the function of Arg1753 in Prp8 might not be rate limiting for splicing of ACT1.
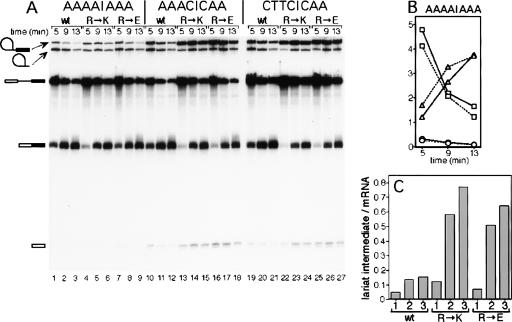
If Prp8-R1753 stabilizes the interaction of U5 with the exons, then altering the exon sequences so that their interaction with U5 is weakened might affect splicing in prp8-R1753 mutant extracts. Modified ACT1 precursors with mutations at exon sequences that affect the second step of prp18ΔCR splicing have been identified (Crotti et al. 2007). For example, AAAA|AAA (the last four bases of exon 1 and the first three bases of exon 2 are separated by “|”) was spliced better in prp18ΔCR extract than ACT1 (TCTG|AGG), while the mutants AAAC|CAA and CTTC|CAA were spliced worse (Crotti et al. 2007). We assayed splicing of these three mutant pre-mRNAs in PRP8, prp8-R1753K, and prp8-R1753E extracts at 32°C (Fig. 2). The AAAA|AAA pre-mRNA was spliced with similar rates and efficiencies in extracts from wild-type or Prp8 mutant cells (Fig. 2B). However, splicing of CTTC|CAA and AAAC|CAA pre-mRNAs was impaired, insofar as splicing intermediates accumulated to higher levels, especially in mutant extracts. The relative amounts of intermediates (expressed as the ratio of intermediates/mRNA) from the CTTC|CAA and AAAC|CAA pre-mRNAs were increased four- to sixfold compared to AAAA|AAA pre-mRNA at 5, 9, and 13 min (Fig. 2A,C). Splicing in wild-type extracts was less sensitive to mutations in the exons. Because different extracts were used, we cannot assess whether mutations at Arg1753 have an effect on the first step of splicing. The observed second step defects likely reflect the failure of Prp8-R1753 mutants to stabilize U5/exon contacts required to configure the active site for step 2 catalysis.
FIGURE 2.
(A) Splicing of three exon-mutant substrates in vitro using extracts from wild-type PRP8 or mutant prp8 cells. ACT1 substrate with the exon mutations shown at the top of each panel was spliced in vitro in wild-type PRP8 extract (wt), in prp8-R1753K extract (R→K), or in prp8-R1753E extract (R→E), as indicated above each panel. Aliquots were withdrawn at the times indicated along the top of each gel and analyzed by denaturing PAGE. Autoradiographs of the gels are shown. The positions of the pre-mRNA, splicing intermediates, and mRNA are depicted at the left. (B) Relative molar amounts of RNA species derived from the wild-type (lanes 1–3) and prp8-R1753E (lanes 7–9) time courses are graphed for the AAAA|AAA pre-mRNA. The pre-mRNA is represented by squares, lariat intermediate by circles, and mRNA by triangles; the dashed lines represent wild-type extracts and the solid lines prp8-R1753E extracts. (C) Graphical representation of the relative amounts of the intermediates/mRNA for the 9 min time points (wt extract: lanes 2,11,20; for R→K: lanes 5,14,23; for R→E: lanes 8,17,26). The number beneath each bar indicates the pre-mRNA: 1: AAAA|AAA; 2: AAAC|CAA; and 3: CTTC|CAA.
Mutations in PRP18 suppress the temperature sensitivity of prp8-R1753 mutants
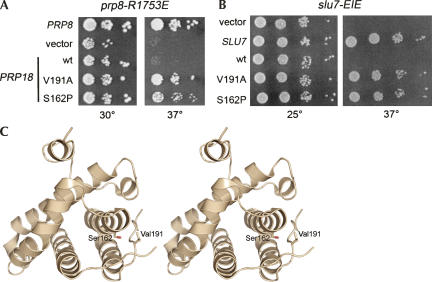
Two lines of evidence suggested that Prp8 and Prp18 have closely related functions: (1) prp8-R1753E and prp18ΔCR mutations were suppressed by mutations in the gene for U5 (Fig. 1; Bacíková and Horowitz 2005) and (2) the second step of splicing of pre-mRNAs with exon mutations was impaired in extracts with mutant Prp8-R1753 proteins or with Prp18ΔCR protein (Fig. 2; Crotti et al. 2007). If Prp18 and Prp8 cooperate in stabilizing U5/exon interactions, then mutations in Prp18 might suppress the phenotype of prp8-R1753E mutants at 37°C. To identify gain-of-function mutations in PRP18, a Prp18-mutant library was generated by error-prone PCR and the plasmid library (URA3 CEN) was transformed into prp8-R1753E PRP18 cells. Of approximately 3000 Ura+ cells that were plated, 4 grew at 37°C. The plasmids were recovered from the suppressor strains and the PRP18 genes were sequenced. One of the dominant PRP18 suppressor alleles carried a single mutation, resulting in a Val-191-Ala change. The other three suppressors contained a Ser-162-Pro change and were judged to be independent isolates insofar as they bore other unique changes (Materials and Methods). The single V191A or S162P changes in Prp18 were sufficient to restore growth to prp8-R1753E PRP18 cells at 37°C (Fig. 3A). V191A and S162P also suppressed the temperature sensitivity of prp8-R1753K and prp8-R1753A cells (not shown). The PRP18-V191A and PRP18-S162P alleles were fully functional in lieu of wild-type PRP18, insofar as they restored wild-type growth to prp18Δ cells (not shown).
FIGURE 3.
Gain-of-function mutations in Prp18. (A) Serial 10-fold dilutions of prp8-R1753E cells bearing plasmids that carry the PRP8 gene or various PRP18 alleles as indicated at the left were plated to agar medium lacking uracil. Cells containing the empty plasmid (vector) were analyzed in parallel. The plates were photographed after 3 d of incubation at 30°C and 37°C. (B) A photograph of slu7-EIE cells containing the indicated expression plasmids after 4 and 2 d of incubation at 25°C and 37°C, respectively, is shown. (C) Stereo representation of Prp18-(80–251) shown as a ribbon diagram (Jiang et al. 2000). Ser162 and Val191are shown as sticks.
The close functional relationship between Prp8 and Prp18 was underscored by the finding that prp18 alleles with mutations in three functionally important regions (Bacíková and Horowitz 2002), including prp18ΔCR, were synthetic lethal with prp8-R1753 mutants (not shown). Taken together, the genetic data support the idea that Prp8 (through Arg1753) and Prp18 (via its CR) cooperate to stabilize contacts between the U5 snRNA and exon sequences during the second step of splicing (Fig. 1D).
Prp18-V191A and Prp18-S162P suppress the temperature sensitivity of slu7 mutants
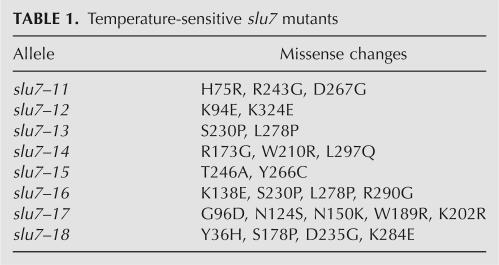
Prp18 interacts with Slu7, and together they recruit Prp22 to the spliceosome (Zhang and Schwer 1997; James et al. 2002). To explore the functional connection between Slu7 and Prp18 further, we screened the Prp18 mutant library for suppressors of the temperature-sensitive growth defect of slu7-EIE, an alanine cluster (E215A–I216A–E217A) mutant of Slu7. The EIE mutations abolished Slu7 binding to Prp18 in vitro and in a two-hybrid assay in vivo (James et al. 2002). We reasoned that Prp18 mutants selected by the screen might restore interaction with Slu7-EIE. The Prp18-mutant library was introduced into slu7-EIE PRP18 cells, and candidate suppressors were selected at 37°C. Plasmids carrying the dominant PRP18 alleles were isolated (seven from a total of approximately 5000 transformants) and sequenced. Five isolates contained a Ser-162-Pro change, and two carried Val-191-Ala mutations, the same prp18 mutations that were isolated as suppressors of prp8-R1753E. The single amino acid changes S162P or V191A sufficed for suppression (Fig. 3B).
Mutations at Ser162 and Val191 in Prp18 did not restore interaction with Slu7-EIE in a 2-hybrid assay (not shown), arguing that suppression occurs independent of the physical interaction between Slu7 and Prp18. Suppression was not allele specific insofar as S162P and V191A suppressed the temperature sensitivity of eight slu7 mutants containing lesions throughout the polypeptide (Table 1), and of two previously described slu7(125–382) and slu7(163–382) alleles (not shown; Zhang and Schwer 1997). Yet, Prp18-V191A and Prp18-S162P did not bypass the requirement for Slu7 in vivo or in vitro (not shown).
TABLE 1.
Temperature-sensitive slu7 mutants
We surmise that V191A and S162P suppress slu7 mutants by improving the functionality of Prp18. Consistent with this idea, we found that introducing a Val-191-Ala change into mutant Prp18s (R151E-R152E, R151A-R152A, H118E-R151A) that were defective for Slu7 interaction partially suppressed the temperature sensitivity of prp18-R151E-R152E, prp18-R151A-R152A, and prp18-H118E-R151A cells (Bacíková and Horowitz 2002), without restoring Slu7 interaction (not shown).
DISCUSSION
The temperature-sensitive PRP8 allele R1753K was isolated as a suppressor of the cold sensitivity of prp22-S635A, an allele encoding a helicase-defective Prp22 protein that is unable to release mature mRNA from the spliceosome (Schwer and Meszaros 2000; Schneider et al. 2004). The present study of prp8-R1753 mutants provides insights to the mechanism of the second step of splicing and the release of mature mRNA from the spliceosome. The major findings are (1) mutations in loop 1 of U5 snRNA suppress the temperature sensitivity of prp8-R1753 mutants; (2) the second step of splicing certain pre-mRNAs is impaired in cell-free extracts from prp8-R1753 mutant cells; (3) gain-of-function mutations in PRP18 suppress prp8-R1753 and multiple slu7 mutants; and (4) mutations in loop 1 of U5 snRNA exacerbate the growth defects of prp22-cs mutants.
Multiple mutations at Arg1753 (R1753K, R1753A, R1753Q, R1753E) suppressed helicase-defective prp22 mutants and elicited temperature-sensitive growth defects in PRP22 cells, leading to the proposal that Arg1753 makes contacts in the spliceosome that are loosened in the prp8-R1753 mutants (Schneider et al. 2004). Our findings that (1) mutations in loop 1 of U5 snRNA alleviate the temperature sensitivity of prp8-R1753 mutants, and (2) splicing of transcripts with mutations in exon sequences is impaired in prp8-R1753 mutant extracts implicate the U5 snRNA and exon bases in contacts that are strengthened by Arg1753. A role for Prp8 in stabilizing U5/exon interactions has been suggested based on cross-linking studies, which placed Prp8 close to bases in loop 1 of U5 and to bases at the ends of both exons (Teigelkamp et al. 1995; Dix et al. 1998; Turner et al. 2006). Using site-specific photocross-linking and proteolytic analyses, Turner et al. (2006) have established a physical map of Prp8 contacts with the U5 and U6 snRNAs and the 5′ss and branch point in the pre-mRNA. None of the analyzed RNA–protein cross-links mapped in the vicinity of Arg1753, which could indicate that (1) Arg1753 makes contact with the U5/3′exon RNA, (2) Arg1753 does not interact directly with the RNA, or (3) the interaction cannot be captured by photocross-linking. In any event, our results provide functional evidence for an involvement of Prp8 in stabilizing U5/exon interactions.
The gain-of-function PRP18-V191A and PRP18-S162P alleles suppress prp8-R1753E and also a variety of slu7 mutants (Fig. 3). In the crystal structure of Prp18Δ79 (Jiang et al. 2000), Ser162 and Val191 are near each other (Fig. 3C). Val191 is located within Prp18′s conserved region. Prp18 proteins from human, plant, fly, and worm contain an alanine at the equivalent position; thus, the V191A mutation increases the identity of the Saccharomyces cerevisiae protein with other Prp18s. Ser162 is situated in helix3, which contacts the conserved loop in the crystal structure. The spatial proximity of the two suppressor mutations suggests that they are mechanistically similar, perhaps altering the conformation and/or the flexibility of Prp18′s conserved loop. The isolation of S162P and V191A in two independent screens for dominant suppressors and the finding that prp18 mutations suppressed multiple slu7 alleles support the idea that the mutations improve the functionality of Prp18, likely by enhancing the ability of Prp18 to stabilize contacts between U5 and the exons. The notion that Prp18′s conserved region plays an important role in facilitating U5/exon interactions comes from genetic and biochemical studies of prp18ΔCR, an allele that encodes a Prp18 protein lacking the conserved loop (Bacíková and Horowitz 2002, 2005; Crotti et al. 2007).
Prp8-R1753 mutants resemble the prp18ΔCR allele in three aspects. First, the U5 allele U5-S2 (A8C) suppresses the growth defect of prp8-R1753E and of prp18ΔCR, and it improves the step 2 splicing defect in prp18ΔCR cells (Bacíková and Horowitz 2005). U5-S2 (A8C) alleviates temperature sensitivity in the absence or presence of wild-type U5, indicating that A8C is a gain-of-function mutation. Second, Prp8-R1753E and prp18ΔCR are also suppressed by mutations in the bases of loop 1 of U5 that interact directly with the exons (Newman 1997; Crotti et al. 2007); U5-GG (U4G U5G) and U5-U4A suppress prp8-R1753E and prp18ΔCR, respectively. These U5 alleles suppress only in the presence of wild-type U5, suggesting that they are necessary for splicing a subset of pre-mRNAs that rely on Prp8 or Prp18 to strengthen U5/exon interactions, whereas wild-type U5 is required for splicing other essential transcripts. Third, the Prp8-R1753 mutants and Prp18ΔCR impair the second step of splicing pre-mRNAs with exon mutations that, based on genetic and biochemical analyses, weaken interaction with U5 (Newman 1997; Crotti et al. 2007). Taken together, the findings are consistent with the idea that Prp8 and Prp18 cooperate to stabilize U5/exon interactions that are important for the second transesterification reaction.
The PRP18 alleles V191A and S162P suppress the growth defects of several slu7 mutants (Fig. 3). SLU7 was originally isolated in a synthetic lethal screen with the U5-U98A mutant that carries a U to A change at position 6 in loop 1 (Frank et al. 1992). Slu7 affects 3′ss choice in yeast and in human cell extracts; when presented with competing 3′ss, yeast Slu7 is necessary for selection of the distal PyAG↓ (Frank and Guthrie 1992) and human Slu7 is reported to discriminate against aberrant AGs (Chua and Reed 1999). In vitro studies suggested that human Slu7 plays a role in properly holding exon 1 within the spliceosome (Chua and Reed 1999). The gain-of-function mutations in Prp18 identified here are not allele specific, but suppress multiple slu7 mutants. We surmise that suppression occurs at the stage when the U5/exon contacts are formed during the second step of splicing. Whether Slu7 plays a direct role in stabilizing contacts between U5 and exon bases is not known.
Prp8-R1753 mutants affect the second step of splicing certain pre-mRNAs in vitro. Prp8 is essential early in splicing (Jackson et al. 1988; Brown and Beggs 1992), and a role for Prp8 during the second step of splicing is well documented. For example, prp8-101 is defective for the second step of splicing precursor RNAs that have suboptimal 3′ splice sites (Umen and Guthrie 1995a), and several prp8 alleles suppress intronic mutations including changes at the 3′ss PyAG↓ (Umen and Guthrie 1995a, 1996; Collins and Guthrie 1999; Siatecka et al. 1999; Konarska et al. 2006). Our findings indicate that Prp8 also plays a role in modulating the second step of splicing based on exonic sequences, extending earlier experiments that showed that Prp8 contacts the exons during the second step (Teigelkamp et al. 1995; Turner et al. 2006; McPheeters et al. 2000; McPheeters and Muhlenkamp 2003).
Unlike Prp8 and U5/exon pairing, other interactions that facilitate the formation of the active site for step 2 catalysis might not be maintained after exon ligation when Prp22 helicase acts to release mRNA. This is indicated by the findings that the gain-of-function mutants U5-S2 (A8C) and the PRP18 alleles V191A and S162P do not exacerbate the phenotypes of prp22-cs mutants (data not shown; Fig. 1B). Our results are consistent with the idea that the Prp22 disrupts contacts between the U5 snRNP and mRNA. The biochemical characteristics of the Prp22 helicase, the 3→5′ directionality, and the requirement for a single-stranded RNA segment of ≥20 nt (Tanaka and Schwer 2005) have implications for the positioning of Prp22 on the RNA target (Fig. 1D).
MATERIALS AND METHODS
Plasmids
The plasmid library for the U5-loop I mutants was generously provided by Andy Newman (MRC Laboratory of Molecular Biology, Cambridge, United Kingdom). Mutations in the SNR7 gene to generate U5-GG and U5-CT were introduced using the two-stage overlap PCR method. PCR products were restricted and the 590 base pair (bp) ClaI–HindIII fragments were inserted into pSE360 (URA3 CEN) and pRS413 (HIS3 CEN) vectors. To generate a Prp18 mutant library, a genomic HincII/SphI fragment spanning the PRP18 coding sequence plus 1417 and 211 bp upstream and downstream of the ATG and stop codon, respectively, was inserted into pUC18. The plasmid pUC18-PRP18 was used as the template for PCR amplification of the PRP18 ORF in vitro with Taq DNA polymerase. The standard PCR reaction mixture was modified to contain 0.2 mM MnCl2, and the concentration of dATP was reduced to 1/5 compared to the other three dNTPs (Leung et al. 1989). The PCR products were digested with BglII (which cleaves immediately downstream of the ATG) and EcoRV (which cleaves 40 bp downstream of the stop codon), and the mutagenized Prp18 DNA fragment was ligated into pUC18-PRP18 to replace the wild-type segment. The ligation mixes were transformed into Escherichia coli dg98 and a pooled plasmid library was prepared from approximately 6000 ampicillin-resistant colonies harvested directly from agar plates. From the pUC18-prp18 mutant library DNA, a 2.3-kb DNA fragment was excised, ligated into pSE360 (URA3 CEN), and transformed. The p360-prp18 mutant library DNA was prepared from approximately 50,000 transformants. To gauge the mutation frequency, an aliquot of the plasmid pool was transformed into E. coli. Plasmid DNA was recovered from six individual ampicillin-resistant colonies and the PRP18 gene was sequenced. Each of the plasmids contained one or more lesions, but none carried mutations at Ser162 or Val191, indicating that these changes were not overrepresented in the library.
Strains
The prp8-R1753E strain [Mata ura3–1 trp1–1 his3–11,15 leu2–3,112 ade2–1 can1–100 Δprp8∷kanMX] carries the prp8-R1753E allele on pRS413 (HIS3 CEN) (Schneider et al. 2004). prp22-T637A, prp22-S635A, and prp22-H606A strains were made by replacing p360-PRP22 (URA3 CEN) with the respective p358-prp22 mutant (TRP1 CEN) in a prp22Δ strain using the plasmid shuffle method (Schwer and Meszaros 2000). Strain slu7-EIE [Mata leu2 his7 trp1 ura3 slu7∷hisG] harbors the slu7-E215A-I216A-E217A allele on a TRP1 CEN plasmid (James et al. 2002). To generate strains for extract preparation, the PRP8 gene was disrupted by insertion of the kanMX marker in BJ2168 cells harboring p360-PRP8 (URA3 CEN) (Schneider et al. 2004). Using the plasmid shuffle procedure, we then replaced the URA3-marked PRP8 by the wild-type or the R1753K and R1753E mutant alleles on TRP1 CEN plasmids.
Screen for suppressors of prp8-R1753E
The prp8-R1753E cells were transformed with the plasmid libraries of U5 loop I or of prp18 mutants. Ura+ colonies were selected at 37°C. Plasmids were recovered from individual colonies, amplified in bacteria, and the genes for the U5 snRNA or for Prp18 were sequenced. The four independently isolated prp18 suppressors carried the following mutations: (1) S162P and a silent change at Thr31, (2) S162P, (3) V191A, and (4) K141E and S162P.
Screen for prp18 suppressors of the temperature-sensitive growth defect of slu7-EIE cells
The prp18 mutant library DNA was introduced into slu7-EIE cells, and Ura+ colonies were selected at 37°C. The plasmid DNAs were recovered and analyzed after amplification in E. coli. Three individual isolates carried a single S162P change, and one isolate contained a V191A mutation. Three other suppressors contained the following mutations: (1) K125E, I176V, V191A; (2) K141E, S162P; (3) S162P, and silent mutations at Leu109 and His203.
Isolation of temperature-sensitive slu7 mutants
A DNA fragment spanning the Slu7 coding region was amplified by Taq DNA polymerase under mutagenic conditions (Leung et al. 1989). The PCR product was restricted and inserted into p358-Slu7-(B), thereby replacing the wild-type DNA fragment. Ligation mixes were transformed into E. coli dg98 and a pooled plasmid library was prepared from approximately 30,000 ampicillin-resistant colonies harvested directly from agar plates. The mutant library was transformed into a slu7Δ strain harboring SLU7 on a URA3-marked plasmid. Trp+ transformants were selected at 25°C and replica plated to 5-FOA twice. One thousand 5-FOA survivors were patched to rich medium (YPD) and replica-plated to 25°C and 37°C. The TRP1 slu7 plasmids were isolated from cells that grew at 25°C, but failed to grow at nonpermissive temperature, amplified in E. coli, and the SLU7 genes were sequenced to determine the lesions.
Extract preparation and in vitro splicing reactions
Yeast whole cell extract from BJ-Prp8, BJ-Prp8-R1753K, or BJ-Prp8-R1753E cells was prepared by grinding in liquid nitrogen as described (Ansari and Schwer 1995). Labeled precursor RNAs for in vitro splicing were synthesized using SP6 RNA polymerase and α-32P-UTP from plasmids pDBN1a (AAAA|AAA), pDBN6a (AAAC|CAA) and pDBN7a (CTTC|CAA) that had been restricted with NdeI (Crotti et al. 2007). Splicing reactions were carried out at 32°C and analyzed as described (Lin et al. 1985). The RNA species were visualized by autoradiography and quantified using a phosphorimager. To determine the relative molar amounts, the values were normalized for the length of the respective RNA species (Crotti et al. 2007).
ACKNOWLEDGMENTS
We are grateful to Andy Newman for providing the U5 mutant library. This work was supported by NIH grant GM50288 to B.S. and by NIH grant GM57267 and USUHS intramural grant C017HO to D.S.H.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.572807.
REFERENCES
- Ansari, A., Schwer, B. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 1995;14:4001–4009. doi: 10.1002/j.1460-2075.1995.tb00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacíková, D., Horowitz, D.S. Mutational analysis identifies two separable roles of the Saccharomyces cerevisiae splicing factor Prp18. RNA. 2002;8:1280–1293. doi: 10.1017/s1355838202023099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacíková, D., Horowitz, D.S. Genetic and functional interaction of evolutionarily conserved regions of the Prp18 protein and the U5 snRNA. Mol. Cell. Biol. 2005;25:2107–2116. doi: 10.1128/MCB.25.6.2107-2116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.D., Beggs, J.D. Roles of PRP8 protein in the assembly of splicing complexes. EMBO J. 1992;11:3721–3729. doi: 10.1002/j.1460-2075.1992.tb05457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T.H., Sharp, P.A. In: The RNA world. 2nd ed. Gesteland R.F., et al., editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1999. pp. 525–560. [Google Scholar]
- Campodonico, E., Schwer, B. ATP-dependent remodeling of the spliceosome: Intragenic suppressors of release-defective mutants of Saccharomyces cerevisiae Prp22. Genetics. 2002;160:407–415. doi: 10.1093/genetics/160.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, K., Reed, R. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature. 1999;402:207–210. doi: 10.1038/46086. [DOI] [PubMed] [Google Scholar]
- Collins, C.A., Guthrie, C. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes & Dev. 1999;13:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti, L.B., Bacíková, D., Horowitz, D.S. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes & Dev. 2007;21:1204–1216. doi: 10.1101/gad.1538207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix, I., Russell, C.S., O'Keefe, R.T., Newman, A.J., Beggs, J.D. Protein–RNA interactions in the U5 snRNP of Saccharomyces cerevisiae . RNA. 1998;4:1675–1686. doi: 10.1017/s1355838298412998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, D., Guthrie, C. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes & Dev. 1992;6:2112–2124. doi: 10.1101/gad.6.11.2112. [DOI] [PubMed] [Google Scholar]
- Frank, D.N., Patterson, B., Guthrie, C. Synthetic lethal mutations suggest interactions between U5 small nuclear RNA and four proteins required for the second step of splicing. Mol. Cell. Biol. 1992;12:5197–5205. doi: 10.1128/mcb.12.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, D.N., Roiha, H., Guthrie, C. Architecture of the U5 small nuclear RNA. Mol. Cell. Biol. 1994;14:2180–2190. doi: 10.1128/mcb.14.3.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger, R.J., Beggs, J.D. Prp8 protein: At the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.P., Lossky, M., Beggs, J.D. Cloning of the RNA8 gene of Saccharomyces cerevisiae, detection of the RNA8 protein, and demonstration that it is essential for nuclear pre-mRNA splicing. Mol. Cell. Biol. 1988;8:1067–1075. doi: 10.1128/mcb.8.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, S.A., Turner, W., Schwer, B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA. 2002;8:1068–1077. doi: 10.1017/s1355838202022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., Horowitz, D.S., Xu, R.M. Crystal structure of the functional domain of the splicing factor Prp18. Proc. Natl. Acad. Sci. 2000;97:3022–3027. doi: 10.1073/pnas.97.7.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska, M.M., Vilardell, J., Query, C.C. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol. Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Leung, D.W., Chen, E., Goeddel, D.V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- Lin, R.J., Newman, A.J., Cheng, S.C., Abelson, J. Yeast mRNA splicing in vitro. J. Biol. Chem. 1985;260:14780–14792. [PubMed] [Google Scholar]
- McGrail, J.C., Tatum, E.M., O'Keefe, R.T. Mutation in the U2 snRNA influences exon interactions of U5 snRNA loop 1 during pre-mRNA splicing. EMBO J. 2006;25:3813–3822. doi: 10.1038/sj.emboj.7601258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters, D.S., Muhlenkamp, P. Spatial organization of protein–RNA interactions in the branch site-3′ splice-site region during pre-mRNA splicing in yeast. Mol. Cell. Biol. 2003;23:4174–4186. doi: 10.1128/MCB.23.12.4174-4186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters, D.S., Schwer, B., Muhlenkamp, P. Interaction of the yeast DExH-box RNA helicase Prp22p with the 3′ splice site during the second step of nuclear pre-mRNA splicing. Nucleic Acids Res. 2000;28:1313–1321. doi: 10.1093/nar/28.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, A.J. The role of U5 snRNP in pre-mRNA splicing. EMBO J. 1997;16:5797–5800. doi: 10.1093/emboj/16.19.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, A., Norman, C. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell. 1991;65:115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- O'Keefe, R.T. Mutations in U5 snRNA loop 1 influence the splicing of different genes in vivo. Nucleic Acids Res. 2002;30:5476–5484. doi: 10.1093/nar/gkf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe, R.T., Newman, A.J. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J. 1998;17:565–574. doi: 10.1093/emboj/17.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe, R.T., Norman, C., Newman, A.J. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Schneider, S., Hotz, H.R., Schwer, B. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J. Biol. Chem. 2002;277:15452–15458. doi: 10.1074/jbc.M112473200. [DOI] [PubMed] [Google Scholar]
- Schneider, S., Campodonico, E., Schwer, B. Motifs IV and V in the DEAH box splicing factor Prp22 are important for RNA unwinding, and helicase-defective Prp22 mutants are suppressed by Prp8. J. Biol. Chem. 2004;279:8617–8626. doi: 10.1074/jbc.M312715200. [DOI] [PubMed] [Google Scholar]
- Schwer, B., Gross, C.H. Prp22, an RNA-dependent ATPase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, B., Guthrie, C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, B., Meszaros, T. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 2000;19:6582–6591. doi: 10.1093/emboj/19.23.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségault, V., Will, C.L., Polycarpou-Schwarz, M., Mattaj, I.W., Branlant, C., Lührmann, R. Conserved loop I of U5 snRNA is dispensible for both catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol. Cell. Biol. 1999;19:2782–2790. doi: 10.1128/mcb.19.4.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siatecka, M., Reyes, J.L., Konarska, M.M. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes & Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, N., Schwer, B. Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry. 2005;44:9795–9803. doi: 10.1021/bi050407m. [DOI] [PubMed] [Google Scholar]
- Teigelkamp, S., Newman, A.J., Beggs, J.D. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, I.A., Norman, C.M., Churcher, M.J., Newman, A.J. Dissection of Prp8 protein defines multiple interactions with crucial RNA sequences in the catalytic core of the spliceosome. RNA. 2006;12:375–386. doi: 10.1261/rna.2229706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen, J.G., Guthrie, C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes & Dev. 1995a;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- Umen, J.G., Guthrie, C. The second catalytic step of pre-mRNA splicing. RNA. 1995b;1:869–885. [PMC free article] [PubMed] [Google Scholar]
- Umen, J.G., Guthrie, G. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Schwer, B. Functional and physical interaction between the yeast splicing factors Slu7 and Prp18. Nucleic Acids Res. 1997;25:2146–2152. doi: 10.1093/nar/25.11.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]