Abstract
Following peptide bond formation, transfer RNAs (tRNAs) and messenger RNA (mRNA) are translocated through the ribosome, a process catalyzed by elongation factor EF-G. Here, we have used a combination of chemical footprinting, peptidyl transferase activity assays, and mRNA toeprinting to monitor the effects of EF-G on the positions of tRNA and mRNA relative to the A, P, and E sites of the ribosome in the presence of GTP, GDP, GDPNP, and fusidic acid. Chemical footprinting experiments show that binding of EF-G in the presence of the non-hydrolyzable GTP analog GDPNP or GDP·fusidic acid induces movement of a deacylated tRNA from the classical P/P state to the hybrid P/E state. Furthermore, stabilization of the hybrid P/E state by EF-G compromises P-site codon–anticodon interaction, causing frame-shifting. A deacylated tRNA bound to the P site and a peptidyl-tRNA in the A site are completely translocated to the E and P sites, respectively, in the presence of EF-G with GTP or GDPNP but not with EF-G·GDP. Unexpectedly, translocation with EF-G·GTP leads to dissociation of deacylated tRNA from the E site, while tRNA remains bound in the presence of EF-G·GDPNP, suggesting that dissociation of tRNA from the E site is promoted by GTP hydrolysis and/or EF-G release. Our results show that binding of EF-G in the presence of GDPNP or GDP·fusidic acid stabilizes the ribosomal intermediate hybrid state, but that complete translocation is supported only by EF-G·GTP or EF-G·GDPNP.
Keywords: ribosome, chemical footprinting, translocation, elongation factor G, hybrid states
INTRODUCTION
The elongation phase of translation is an iterative process of the ribosome that decodes messenger RNA (mRNA) to synthesize a polypeptide chain. There are three principal steps that must occur during each cycle of elongation: (1) the mRNA codon is recognized by the correct aminoacyl transfer RNA (tRNA); (2) the peptidyl transferase reaction elongates the polypeptide chain by one amino acid; and (3) the remaining tRNAs bound to the ribosome are translocated to the next position, creating a vacant tRNA binding site for the next cycle. Ribosomal translocation is catalyzed in vivo by elongation factor G (EF-G), which requires GTP for its catalytic activity and hydrolyzes GTP in a ribosome-dependent manner, releasing EF-G from the ribosome (Spirin 1985).
The mechanism of translocation is not fully understood. The driving force behind the directionality of ribosomal translocation is at least partially determined by the acylation state of the 3′-CCA end of each tRNA. The 50S A, P, and E sites are specific for the aminoacyl, peptidyl, and deacylated tRNAs, respectively. As cognate aminoacyl-tRNA binds to the A site, the peptidyl transferase reaction transfers the peptidyl moiety from the P-site tRNA to the A-site tRNA. This reaction is followed by movement of the 3′-CCA ends of deacylated tRNA from the 50S P site to the E site and of peptidyl tRNA from the 50S A site to the P site, while their anticodon ends remain bound to the P and A sites of the 30S subunit, resulting in binding the P/E and A/P hybrid states, respectively (Moazed and Noller 1989b). The anticodon loop regions of the hybrid-state tRNAs, together with the associated mRNA, are then translocated on the 30S subunit, completing a single round of translocation. The first step of translocation has been observed to occur spontaneously in vitro, but the second step requires EF-G.
The ability of different guanine nucleotides to support EF-G–dependent translocation has developed into a matter of some controversy. Early studies from the Kaziro (Inoue-Yokosawa et al. 1974) and Spirin (Belitsina et al. 1975) groups established that EF-G binds to the ribosome and catalyzes translocation exclusively in its GTP form, in which the presence of either GTP or a nonhydrolyzable analog of GTP (GDPNP) supports translocation. Recent conflicting reports have challenged this view. Wintermeyer and colleages have reported that EF-G catalyzes translocation in the presence of GDPNP and GDP at similar rates (0.8 and 0.9 sec−1, respectively; Rodnina et al. 1997; Wilden et al. 2006). In contrast, Ehrenberg and co-workers have suggested that EF-G·GDPNP locks the ribosome in a distinct transition state in which the A-site codon–anticodon region is translocated relative to the 30S subunit, but translocation does not reach completion (Zavialov et al. 2005). The Ehrenberg group has also failed to observe translocation in the presence of GDP (Zavialov et al. 2005).
The role of EF-G in formation of the hybrid-state intermediate of translocation is not fully understood. Movement of ribosome-bound tRNAs into the hybrid state was initially observed by ribosomal RNA (rRNA)-directed chemical probing, which provided direct evidence that hybrid-state formation can occur spontaneously and prior to EF-G binding (Moazed and Noller 1989b). In contrast, no spontaneous hybrid-state formation was detected by cryogenic electron microscopic (cryoEM) reconstruction studies (Agrawal et al. 1999; Valle et al. 2003). Instead, movement of tRNAs into the hybrid state was observed when the ribosome was in complex with EF-G in the presence of GDPNP or GDP·fusidic acid (Valle et al. 2003), an antibiotic that prevents EF-G release after GTP hydrolysis (Bodley et al. 1970; Kuriki et al. 1970; Lin and Bodley 1976). Ehrenberg and co-workers have suggested that the hybrid state is initially induced by EF-G·GDP (Zavialov et al. 2005). In their model, hybrid-state formation precedes exchange of GDP for GTP, in which the ribosome serves as a guanine nucleotide exchange factor (GEF) for EF-G.
Thus, the precise sequence of events that occur during EF-G-catalyzed translocation bears further examination. In this study, we have used a combination of ribosomal RNA (rRNA)-directed chemical probing, peptidyl transferase activity measurements, and mRNA toeprinting to monitor the movements of tRNAs and mRNA induced by the interactions of EF-G with the ribosome in the presence of different nucleotides and antibiotics. Our results provide further supporting evidence for the hybrid-states model of translocation, suggest a connection between GTP hydrolysis and E-site tRNA release, and show that translocation does not occur in the presence of GDP.
RESULTS
Binding of EF-G to the ribosome stabilizes the hybrid-state conformation
In order to study the effect of EF-G on the movements of tRNA from the classical to the hybrid-state conformation, ribosome·tRNA·mRNA complexes were constructed with a single deacylated tRNA bound to the 30S P site. It was previously shown that a tRNA anticodon stem–loop bound to the 30S A site is minimally required for translocation of mRNA (Joseph and Noller 1998). Therefore, incubation of EF-G with ribosomes containing a vacant A site cannot result in translocation. Ribosomes containing a defined mRNA and fMet-tRNAfMet or N-Ac-Phe-tRNAPhe bound to the P site were reacted with puromycin to create complexes containing a single deacylated tRNA bound to the P site, which allows the deacylated tRNA to move into the 50S E site. Using chemical probing, we tested the effects of stable binding of EF-G to the ribosome on movement of deacylated tRNA under conditions disfavoring spontaneous hybrid-state formation (Moazed and Noller 1989b).
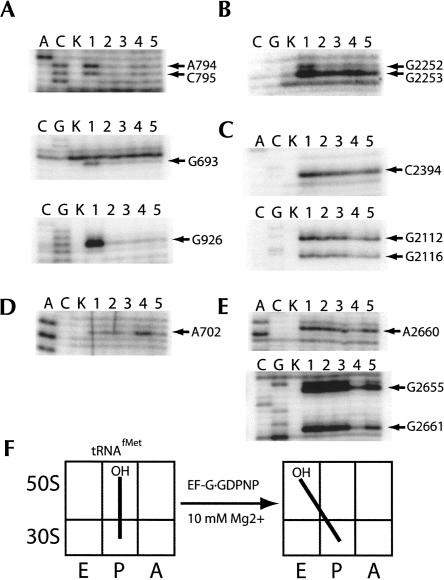
In the initial complex, fMet-tRNAfMet was used due to its propensity to remain in the classical P/P state at low (≤10 mM) Mg2+concentrations after deacylation by puromycin (Dorner et al. 2006; S. Joseph, M.V. Rodnina, H.F. Noller, unpubl.). Characteristic protections in 16S and 23S rRNA were observed for tRNA bound to the 30S and 50S P sites, respectively, upon addition of fMet-tRNAfMet (Fig. 1A–C, lanes 2). After deacylation of the P-site tRNA by puromycin, chemical protections indicative of classical P/P binding remained (Fig. 1A–C, lanes 3). In contrast, when EF-G·GDPNP was added, protections remained for the 30S P site (Fig. 1A, lane 4), but protections indicative of 50S P-site binding were diminished (Fig. 1B, lane 4), accompanied by the appearance of new protections that correspond to binding to the 50S E site (Fig. 1C, lane 4). These changes in chemical reactivity are indicative of tRNA movement from the classical P/P state to the hybrid P/E state (Moazed and Noller 1989b). This conclusion is further supported by the appearance of enhanced chemical reactivity for A702 of 16S rRNA, a characteristic signature of hybrid-state formation (Fig. 1D, lane 4; Moazed and Noller 1989b). Chemical protections in the sarcin–ricin loop (SRL) indicate stable binding of EF-G in the presence of GDPNP (Fig. 1E, lane 4). Finally, no significant changes in chemical reactivity appeared upon addition of EF-G·GTP, indicating that the apparent stabilization of the hybrid-state conformation is specific to the EF-G·GDPNP complex (Fig. 1, lane 5).
FIGURE 1.
Chemical footprinting of a ribosome·tRNAfMet·EF-G·GDPNP complex. Lanes A, C, and G, sequencing lanes; lane K, unmodified rRNA; lane 1, 70S ribosomes and mRNA32; lane 2, 70S ribosomes, mRNA32, and fMet-tRNAfMet; lane 3, 70S ribosomes, mRNA32, fMet-tRNAfMet, and puromycin; lane 4, 70S ribosomes, mRNA32, fMet-tRNAfMet, puromycin, and EF-G·GDPNP; lane 5, 70S ribosomes, mRNA32, fMet-tRNAfMet, puromycin, and EF-G·GTP. (A) 16S rRNA P site; (B) 23S rRNA P site; (C) 23S rRNA E site; (D) 16S rRNA inter-subunit bridge B7a; (E) 23S rRNA sarcin–ricin loop (SRL, EF-G binding site); (F) schematic indicating movement of tRNAfMet in the presence of EF-G·GDPNP.
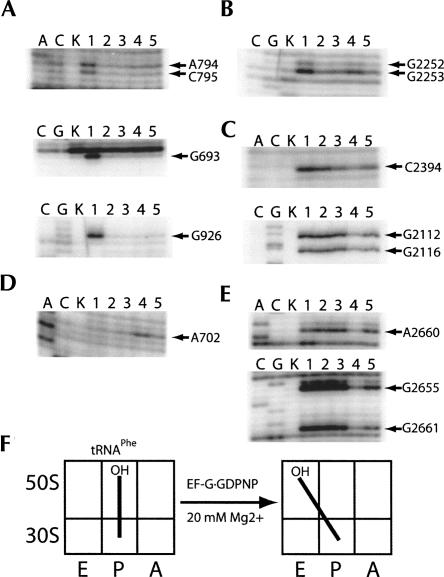
To further test the effects of EF-G·GDPNP, similar experiments were performed using N-Ac-Phe-tRNAPhe at high (20 mM) Mg2+concentrations. Formation of the P/E hybrid state with deacylated tRNAPhe was previously found to be favored by low Mg2+ concentrations (Moazed and Noller 1989b; S. Joseph, M.V. Rodnina, and H.F. Noller, unpubl.), and higher magnesium concentrations favor the classical P/P state. In the presence of N-Ac-Phe-tRNAPhe, protections indicative of both 30S and 50S P-site binding were observed (Figs. 2A,B, lanes 2). After reaction with puromycin, deacylated tRNAPhe remained in the classical P/P state, as expected (Fig. 2A–D, lanes 3). Upon addition of EF-G·GDPNP, protections in the 50S P site were diminished, accompanied by the appearance of 50S E-site protections (Fig. 2B,C, lanes 4), similar to what was observed for tRNAfMet. Protection of bases in the SRL (Fig. 2E, lane 4) confirmed that EF-G was stably bound to the complex. Enhancement in chemical reactivity was again observed for A702, providing further support for the stabilization of the hybrid state (Fig. 2D, lane 4).
FIGURE 2.
Chemical footprinting of a ribosome·tRNAPhe·EF-G·GDPNP complex. Lanes A, C, and G, sequencing lanes; lane K, unmodified rRNA; lane 1, 70S ribosomes and mRNA32; lane 2, 70S ribosomes, mRNA32, and N-Ac-Phe-tRNAPhe; lane 3, 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe, and puromycin; lane 4, 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe, puromycin, and EF-G·GDPNP; lane 5, 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe, puromycin, and EF-G·GTP. (A) 16S rRNA P site; (B) 23S rRNA P site; (C) 23S rRNA E site; (D) 16S rRNA inter-subunit bridge B7a; (E) 23S rRNA sarcin–ricin loop (SRL, EF-G binding site); (F) schematic indicating movement of tRNAPhe in the presence of EF-G·GDPNP.
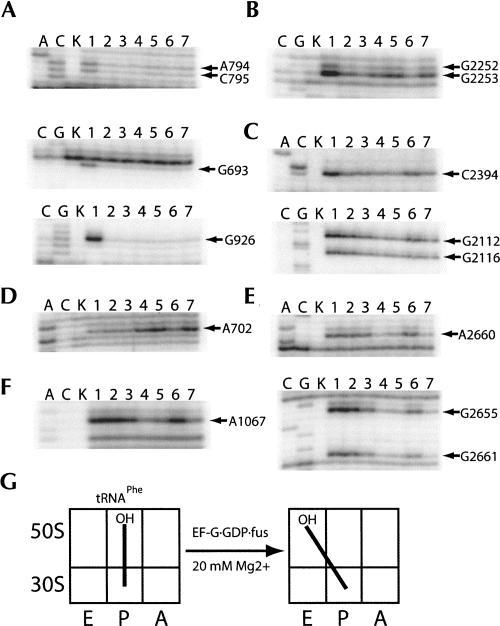
We then asked whether stabilization of the hybrid state is caused simply by binding of EF-G, or whether it requires binding of EF-G in its GDPNP form, by stabilizing the binding of EF-G to the ribosome with fusidic acid. N-Ac-Phe-tRNAPhe was bound to the 30S P site at 20 mM Mg2+ and reacted with puromycin as in the previous experiment. Protections were again observed for both the 30S and 50S P sites in the presence of tRNAPhe, following incubation with puromycin (Fig. 3A,B, lanes 2). After addition of EF-G and GTP (or GDP), ribosomal complexes showed no differences in tRNA-dependent protection (Fig. 3, lanes 3,6). In contrast, when either EF-G·GTP or EF-G·GDP was added in the presence of fusidic acid, the pattern of chemical protection changed, showing decreased protection of the 50S P site and appearance of protections in the 50S E site (Fig. 3B,C, lanes 4,7), similar to the changes observed for the EF-G·GDPNP complex. Enhanced DMS reactivity for A702 of 16S rRNA again confirms hybrid-state formation (Fig. 3D, lanes 4,7). Strong protections in the SRL and L11 rRNA regions show that, in the presence of fusidic acid, EF-G is stably bound, as expected (Fig. 3E,F, lanes 4,5,7; Moazed et al. 1988). The tRNA protection patterns and extent of EF-G binding show no difference between GDP and GTP in EF-G·fusidic acid complexes, and both are similar to the EF-G·GDPNP complexes.
FIGURE 3.
Chemical footprinting of a ribosome·tRNAPhe·EF-G·(GTP/GDP)·fusidic acid complex. Lanes A, C, and G, sequencing lanes; lane K, unmodified rRNA; lane 1, 70S ribosomes and mRNA32; lane 2, 70S ribosomes, mRNA32, and N-Ac-Phe-tRNAPhe, and puromycin; lane 3, 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe, puromycin, and EF-G·GTP; lane 4, 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe, puromycin, EF-G·GTP, and fusidic acid; lane 5, 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe, puromycin, and EF-G·GDPNP; lane 6, 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe, puromycin, and EF-G·GDP; lane 7, 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe, puromycin, EF-G·GDP, and fusidic acid. (A) 16S rRNA P site; (B) 23S rRNA P site; (C) 23S rRNA E site; (D) 16S rRNA inter-subunit bridge B7a; (E) 23S rRNA sarcin–ricin loop (SRL, EF-G binding site); (F) 23S rRNA GTPase-associated center (GAC, EF-G binding site); (G) schematic indicating movement of tRNAPhe in the presence of EF-G·(GTP or GDP)·fusidic acid.
Our data show that binding of EF-G, whether in its GDP or GDPNP form, stabilizes the hybrid P/E state. The pattern of chemical protection that is seen is remarkably similar to that previously observed for spontaneous hybrid-state formation (Moazed and Noller 1989b), indicating that the 70S ribosome has the inherent capability of adopting an authentic hybrid state in the absence of EF-G.
Binding of EF-G to the ribosome promotes mRNA back-slippage
Deacylated tRNA can be efficiently translocated by EF-G, but in some cases in which there is an available alternative cognate codon located upstream with a more optimal distance from the Shine–Dalgarno sequence, translocation of deacylated tRNA to the P site is accompanied by a re-pairing of the tRNA anticodon, an effect that has been called mRNA back-slippage (Fredrick and Noller 2002; McGarry et al. 2005). It has been proposed that movement of deacylated tRNA from the classical P/P state to the hybrid P/E state destabilizes codon–anticodon interaction, favoring mRNA back-slippage (McGarry et al. 2005). Based on our evidence that binding of EF-G stabilizes the hybrid state, two predictions can be made: first, binding of EF-G is predicted to be required for mRNA back-slippage at high magnesium ion concentrations; and second, EF-G·GDPNP, which binds stably to the ribosome, is predicted to be more efficient in promoting mRNA back-slippage than EF-G·GTP.
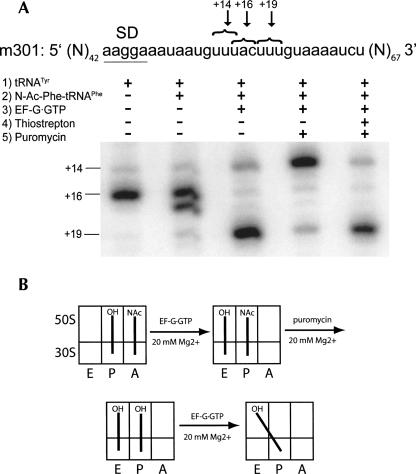
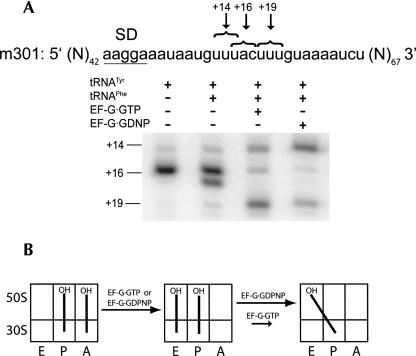
To test these predictions, mRNA toeprinting experiments were performed using mRNA 301, which has two alternative codons for tRNAPhe in close proximity to each other (+14 and +19, respectively; Fig. 4). Toeprinting experiments were done at 20 mM Mg2+ concentration, which suppresses spontaneous movement of deacylated tRNA from the P/P to the P/E state. Deacylated tRNATyr was first bound to the P site, resulting in the appearance of a prominent +16 band (+16 corresponds to the position on the mRNA where reverse transcriptase stops upon encountering the ribosome, relative to the first nucleotide of the P-site codon). N-Ac-Phe-tRNAPhe was then added to the A site to form a pre-translocation complex. The pre-translocation complex was then incubated with EF-G·GTP, and the sample was subsequently divided into three aliquots. The first aliquot was simply used for the primer extension reaction, which resulted primarily in a +19 band, corresponding to accurate translocation in ∼80% of ribosomes. The second aliquot was simply reacted with puromycin. The third aliquot was first incubated with thiostrepton, an antibiotic that inhibits EF-G binding to the ribosome (Cameron et al. 2002) but does not inhibit peptidyl-transferase activity (Pestka 1970), and subsequently reacted with puromycin. Deacylation of N-Ac-Phe-tRNAPhe bound to the ribosomal complex that was not treated with thiostrepton resulted in back-slippage of the mRNA, as visualized by the increase in intensity of a band corresponding to position +14 (85%). In contrast, deacylation of N-Ac-Phe-tRNAPhe in the presence of thiostrepton failed to promote back-slippage, since 80% of ribosomal complexes remained in the +19 position. Our results suggest that EF-G is required for mRNA back-slippage because it stabilizes the hybrid P/E state, specifically in the presence of deacylated tRNA, which is abolished by disruption of the interaction between EF-G and the ribosome with thiostrepton.
FIGURE 4.
Interaction of EF-G with the ribosome is required for mRNA back-slippage. (A) A pre-translocation complex was made by binding deacylated tRNATyr to the P site and N-Ac-Phe-tRNAPhe to the A site in the presence of mRNA 301. Some of the pre-translocation complexes were incubated with EF-G and GTP to allow translocation to proceed. Newly formed post-translocation complexes were separated into three aliquots. The first aliquot was left unperturbed, the third was initially incubated with thiostrepton to disrupt the interaction of EF-G with the ribosome, and the second and third aliquots were each incubated with puromycin to deacylate the P-site–bound N-Ac-Phe-tRNAPhe as indicated. The position of the ribosome along the mRNA was mapped by toeprinting. A toeprint band at +16 corresponds to pre-translocation complex, a band at +19 is the product of accurate translocation, and a band at +14 is the product of mRNA back-slippage. (B) Schematic illustrating mechanism of back-slippage. Incubation of the ribosome with puromycin after translocation results in a deacylated tRNAPhe bound in the P site. Binding of EF-G to the ribosome induces a movement of tRNAPhe from the classical P/P state to the hybrid P/E state, which destabilizes tRNA·mRNA interactions and results in mRNA back-slippage. Thiostrepton disrupts interaction of EF-G with ribosome and prevents mRNA back-slippage.
If this interpretation is correct, then we would predict that EF-G·GDPNP should be more efficient at inducing mRNA back-slippage than EF-G·GTP. We formed a pre-translocation complex by binding deacylated tRNATyr to the 30S P site followed by binding deacylated tRNAPhe to the 30S A site, resulting in the appearance of the characteristic +16/+17 doublet (Fig. 5). In the presence of EF-G·GTP at 25 mM Mg2+, ∼70% of the mRNA was moved to position +19, corresponding to accurate translocation, and 30% of the message back-slipped to position +14. In contrast, after incubation of the pretranslocation complex with EF-G·GDPNP, only 20% of the mRNA was accurately translocated to position +19, with ∼80% back-slippage. These results further support the interpretation that EF-G·GDPNP stabilizes the hybrid P/E state.
FIGURE 5.
EF-G·GDPNP is efficient in promoting mRNA back-slippage. (A) A pre-translocation complex was made by binding deacylated tRNATyr to the P site and deacylated tRNAPhe to the A site in the presence of mRNA 301. Some of the pre-translocation complexes were incubated with EF-G·GTP or EF-G·GDPNP as indicated. The position of the ribosome along the mRNA was mapped by toeprinting. A toeprint band at +16 corresponds to the pre-translocation complex, a band at +19 is the product of accurate translocation, and a band at +14 is the product of mRNA back-slippage. (B) Schematic illustrating the mechanism of mRNA back-slippage. When deacylated tRNAPhe is translocated from the A site to the P site, it moves from the classical P/P state to the hybrid P/E state as EF-G binds to the ribosome. Hybrid-state formation results in a destabilization of the codon–anticodon interactions and results in mRNA back slippage. EF-G·GDPNP binds to the ribosome more stably and promotes mRNA back-slippage more efficiently than EF-G·GTP.
EF-G catalyzes translocation in its GTP form
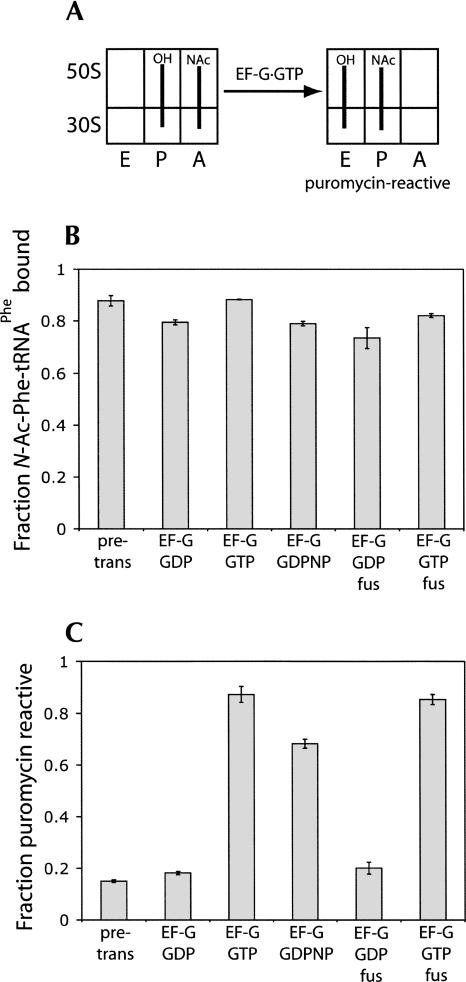
We then tested whether EF-G can catalyze translocation in the presence of GDPNP (Inoue-Yokosawa et al. 1974; Belitsina et al. 1975) and GDP (Rodnina et al. 1997) using three independent assays to monitor movement of tRNA and mRNA. Ribosomes were first incubated with mRNA and deacylated tRNAfMet to fill the P site, followed by binding of N-Ac-Phe-tRNAPhe to the A site. The pre-translocation complex was then subjected to different combinations of EF-G, guanine nucleotides, and antibiotics. In the first assay, translocation of N-Ac-Phe-tRNAPhe from the A to the P site was tested by appearance of puromycin reactivity (Fig. 6). N-Ac-Phe-tRNAPhe is unreactive with puromycin in the A site but becomes reactive when bound to the P site (Spirin 1985). Initially, the pre-translocation complex had low levels of puromycin reactivity (Fig. 6C). Incubation with EF-G·GTP, EF-G·GTP·fusidic acid, or EF-G·GDPNP led to increased puromycin reactivity, corresponding to movement of N-Ac-Phe-tRNAPhe into the P site. In contrast, ribosomal complexes incubated with EF-G·GDP and EF-G·GDP·fusidic acid showed negligible increases in reactivity. All complexes bound N-Ac-Phe-tRNAPhe to a comparable level, showing that these differences are not due to loss of tRNA binding but to the inability of EF-G·GDP to support translocation (Fig. 6B).
FIGURE 6.
Puromycin reactivity of pre-translocation state ribosome complexes incubated with EF-G. (A) Diagram indicating the puromycin reactivity of the pre- and post-translocation state ribosome complexes. Deacylated tRNAfMet was bound to the 30S P site, and N-Ac-[14C]Phe-tRNAPhe was bound to the 30S A site. (B) Histograms indicate the relative amount of N-Ac-[14C]Phe-tRNAPhe that is bound to the ribosome. (C) Histograms indicate the fraction of ribosome-bound N-Ac-[14C]Phe-tRNAPhe that is puromycin reactive: (pre-trans) the pre-translocation complex as described in A; (GDPNP) non-hydrolyzable GTP analog; (fus) fusidic acid.
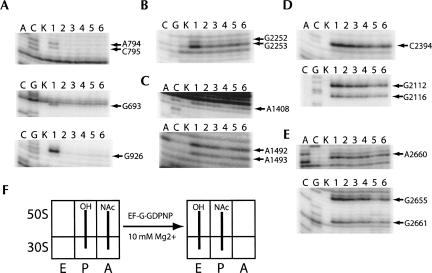
In the second assay, we monitored the positions of tRNA during translocation, using chemical footprinting (Moazed and Noller 1989b) to record changes in the interactions between tRNA and 16S and 23S rRNA at the A, P, and E sites. Binding of fMet-tRNAfMet caused protection of bases A794, C795, G693, and G926 of 16S rRNA and G2252 and G2253 of 23S rRNA, indicating occupation of the P site on both the 30S and 50S subunits (Fig. 7A,B, lanes 2); incubation with puromycin had no effect on the chemical protection pattern (Fig. 7A,B, lanes 3). After addition of N-Ac-Phe-tRNAPhe and formation of the pre-translocation complex, chemical protections characteristic of A-site tRNA binding were observed (A1408, A1492, and A1493; Fig. 7C, lane 4). Addition of EF-G·GTP resulted in disappearance of the A-site protections (Fig. 7C, lane 6) consistent with translocation of N-Ac-Phe-tRNAPhe from the A site to the P site. Incubation of the pre-translocation complex with EF-G·GDPNP also resulted in disappearance of the A-site protections (Fig. 7C, lane 5), indicative of translocation and in agreement with the puromycin reactivity assay. Interestingly, chemical protections in the sarcin–ricin loop of 23S rRNA around positions 2660–2665, indicative of EF-G binding (Moazed et al. 1988), were observed in the EF-G·GDPNP complex (Fig. 7E, lane 5), indicating that EF-G was stably bound to the 70S ribosome in the post-translocation state. Finally, an unexpected difference between the EF-G·GTP and EF-G·GDPNP complexes was observed for the appearance of chemical protections in the 50S E site. In the presence of EF-G·GDPNP, protection of C2394, G2112, and G2116 was observed (Fig. 7D, lane 5), corresponding to movement of tRNAfMet from the P site to the 50S E site. In contrast, protection of E-site bases is only barely detectable for EF-G·GTP (Fig. 7D, lane 6). This difference suggests that dissociation of deacylated tRNA from the E site is promoted by GTP hydrolysis and/or EF-G release.
FIGURE 7.
Chemical footprinting of the pre-translocation complex in the presence of EF-G·GDPNP. Lanes A, C, and G, sequencing lanes; lane K, unmodified rRNA; lane 1, 70S ribosomes and mRNA32; lane 2, 70S ribosomes, mRNA32, and fMet-tRNAfMet; lane 3, 70S ribosomes, mRNA32, fMet-tRNAfMet, and puromycin; lane 4, 70S ribosomes, mRNA32, fMet-tRNAfMet, puromycin, and N-Ac-Phe-tRNAPhe; lane 5, 70S ribosomes, mRNA32, fMet-tRNAfMet, puromycin, N-Ac-Phe-tRNAPhe, and EF-G·GDPNP; lane 6, 70S ribosomes, mRNA32, fMet-tRNAfMet, puromycin, N-Ac-Phe-tRNAPhe, and EF-G·GTP. (A) 16S rRNA P site; (B) 23S rRNA P site; (C) 16S rRNA A site; (D) 23S rRNA E site; (E) 23S rRNA sarcin–ricin loop (SRL, EF-G binding site); (F) schematic indicating movement of deacylated tRNAfMet (P site) and N-Ac-Phe-tRNAPhe (A site) in the presence of EF-G·GDPNP.
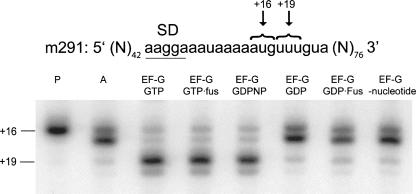
In the third assay, we followed the movement of mRNA on the ribosome using a toeprinting assay (Fig. 8; Joseph and Noller 1998). Binding of elongator tRNAMet resulted in the appearance of a prominent band at position +16. Binding of N-Ac-Phe-tRNAPhe results in appearance of the characteristic doublet (positions +16 and +17), corresponding to binding of tRNA to the A site (Jerinic and Joseph 2000). After addition of EF-G·GTP, EF-G·GTP·fusidic acid, or EF-G·GDPNP, a single, prominent band appeared at position +19, indicating translocation of the mRNA by one codon. In contrast, EF-G·GDP or EF-G·GDP·fusidic acid failed to support translocation of mRNA. Thus, results from three different assays support the conclusion that EF-G stabilizes the intermediate hybrid state when bound with GDPNP or GDP·fusidic acid but supports complete translocation exclusively in its GTP (GTP, GDPNP, or GTP·fusidic acid) form.
FIGURE 8.
Toeprinting analysis of mRNA translocation catalyzed by EF-G in the presence of different nucleotides and antibiotics. The pre-translocation complex was assembled by binding deacylated tRNAMet (P site), N-Ac-Phe-tRNAPhe (A site), and m291 mRNA to the ribosome. The pre-translocation complex was incubated with EF-G in the presence of GTP, GDPNP, GDP, and fusidic acid as indicated. The toeprint bands at +16 and +19 correspond to the pre- and post-translocation states of the ribosome, respectively.
DISCUSSION
Cryo-EM reconstructions (Frank and Agrawal 2000; Valle et al. 2003) have shown that binding of EF-G to the ribosome induces a counter-clockwise rotation of the small subunit accompanied by movement of tRNA into a binding state proposed to correspond to the P/E hybrid state (Moazed and Noller 1989b). Complete EF-G–dependent translocation requires a full-length deacylated tRNA in the P site and at least a tRNA anticodon stem–loop bound to the A site (Joseph and Noller 1998). Using complexes containing only a single deacylated tRNA bound to the P site, which cannot undergo a complete round of translocation, we observed that stable binding of EF-G (in the presence of either GDPNP or GDP·fusidic acid) induces movement of tRNA from the classical P/P state to the P/E hybrid state at Mg2+ concentrations that otherwise suppress spontaneous hybrid-state formation (Figs. 1–3). EF-G-induced back-slippage (McGarry et al. 2005) provides further evidence for hybrid-state stabilization by EF-G (Fig. 5). In complexes containing two tRNAs, which allow completion of translocation, EF-G·GDPNP remained bound to the classical-state ribosome (Fig. 7E), in agreement with earlier findings (Belitsina et al. 1975, 1979). An alternative interpretation of these results could be that N-Ac-Phe-tRNAPhe has the ability to bind in the hybrid P/E state, as there are findings to suggest that aminoacylated tRNA can bind to the 50S E site (Wang et al. 2006). We conclude that this is not the case in our complexes for two reasons: First, chemical protection of C2394 in 23S rRNA is observed, which is specific for the deacylated 3′-CCA region of tRNA; and second, there is no enhancement of chemical reactivity for A702 in 16S rRNA, which is indicative of the hybrid P/E state (data not shown). These observations suggest that EF-G is able to bind to both the hybrid and classical states of the ribosome. However, our results (Figs. 1–3), demonstrating stabilization of the hybrid state by EF-G, together with recent reports (Valle et al. 2003; Zavialov and Ehrenberg 2003), suggest that EF-G may have a higher affinity for the hybrid state.
Zavialov et al. have recently proposed that EF-G catalyzes movement of tRNAs into the hybrid state in its GDP form (Zavialov et al. 2005). In our experiments, neither stable EF-G binding nor movement of tRNA into the hybrid state was detected using GDP in the absence of fusidic acid. Recent studies by Wilden et al. have concluded that EF-G·GDP translocates a pre-translocation complex at rates similar to that of EF-G·GDPNP (Rodnina et al. 1997; Wilden et al. 2006). However, it has been suggested that translocation in the presence of GDP can be explained by contamination of GDP with trace amounts of GTP (Zavialov et al. 2005). Indeed, we found that commercially available GDP preparations contain significant levels of GTP. Using both mRNA toeprinting and puromycin reactivity assays, we observed translocation in the presence of crude GDP (data not shown). However, when using GDP that was purified by ion-exchange chromatography (Zavialov et al. 2005), no translocation was detected.
Although our results as well as published cryo-EM studies provide evidence that EF-G binds to the hybrid-state conformation of the ribosome, stabilization of the hybrid state alone is not sufficient to induce completion of translocation. Indeed, spontaneous formation of the hybrid state does not lead to translocation at any appreciable rate (Moazed and Noller 1989b). Moreover, our peptidyl transferase activity measurements, rRNA-directed chemical probing, and mRNA toeprinting show that EF-G catalyzes translocation of a pre-translocation complex exclusively in its GTP form, while EF-G·GDP·fusidic acid, which is able to stabilize the hybrid state, does not support completion of translocation. Our findings are in good agreement with most previous reports (Inoue-Yokosawa et al. 1974; Belitsina et al. 1975, 1979; Modolell et al. 1975; Girbes et al. 1976).
How does EF-G catalyze the second step of translocation? It has been proposed that domain IV of EF-G occupies the 30S A site, displacing the anticodon stem–loop of tRNA as the 30S subunit rotates clockwise from the hybrid state back to the classical state (Abel and Jurnak 1996; Czworkowski and Moore 1997). Alternatively, EF-G·GTP (or EF-G·GDPNP) could induce the formation of a second, yet undescribed, intermediate state of translocation, similar to the “unlocked” state proposed by Spirin (Spirin 1969, 1985). Such an intermediate might be short-lived, and its transient stabilization by EF-G could be important for driving translocation to completion. Sparsomycin, an antibiotic that binds to the 50S subunit near the peptidyl-transferase center and triggers efficient translocation (Fredrick and Noller 2003), might also stabilize this second hypothetical intermediate state. Recent kinetic (Pan et al. 2007) and single-molecule FRET (Munro et al. 2007) studies have provided evidence for the existence of an additional intermediate state of translocation. Thus, EF-G most likely decreases the energy barrier for translocation through several contributions (Wintermeyer et al. 2001), one of which is stabilization of the hybrid state. Although translocation occurs efficiently with GDPNP, it has been shown that GTP hydrolysis accelerates translocation by about 50-fold in comparison to translocation in the presence of EF-G·GDPNP (Rodnina et al. 1997). Thus, GTP hydrolysis and inorganic phosphate release may trigger another structural rearrangement that further increases the rate of translocation (Peske et al. 2000; Savelsbergh et al. 2003).
In agreement with previous reports (Semenkov et al. 1996, and references therein), we observe release of deacylated tRNA from the E site following translocation with EF-G·GTP. In contrast, we find that deacylated tRNA is retained in the E site following translocation in the presence of EF-G·GDPNP. This observation is in agreement with the findings of Zavialov et al. (2005), who showed that, in the presence of EF-G·GDPNP, deacylated tRNA can be chased from the ribosomal complex only with an excess of cognate tRNA. Thus, GTP hydrolysis and/or EF-G release appear to promote release of tRNA from the E site. Cryo-EM reconstructions have revealed that binding of EF-G to the ribosome induces an inward movement of the L1 stalk of the 50S subunit, possibly stabilizing the binding of deacylated tRNA to the E site (Valle et al. 2003). We suggest that GTP hydrolysis and EF-G release induce an outward movement of the L1 stalk to facilitate the escape of tRNA from the E site.
MATERIALS AND METHODS
Preparation of ribosomes and ribosomal ligands
tRNAfMet was purchased from MP Biomedicals; tRNAPhe and tRNAMet were purchased from Sigma; defined mRNA32 (5′-GGCAAGGAGGUAAAAAUGUUUAAACGUAAAUCUACU-3′) was synthesized by Integrated DNA Technologies. fMet-tRNAfMet, N-Ac-Phe-tRNAPhe, and EF-G with 6-histidine tag were prepared and purified as previously described (Moazed and Noller 1989b; Wilson and Noller 1998; Dorner et al. 2006). GTP, GDP, GDPNP, puromycin, and fusidic acid were purchased from Sigma. GDP was further purified from contaminating GMP and GTP using ion-exchange chromatography essentially as described (Zavialov et al. 2005) but with some modifications. Crude GDP was loaded onto a Resource Q column (6 mL, Pharmacia) in 20 mM Tris·HCl (pH 7.5). Pure GDP was eluted with a gradient of NH4Cl (0–400 mM, 60 mL).
Tight-couple 70S ribosomes and ribosomal subunits were prepared from Escherichia coli MRE600 as previously described (Lancaster et al. 2002; Hickerson et al. 2005). For toeprinting, wild-type 30S subunits were activated by incubation for 10 min at 42°C in buffer A (50 mM Hepes·KOH [pH 7.5], 20 mM MgCl2, 100 mM NH4Cl, 6 mM β-mercaptoethanol) and associated with a 1.5-molar excess of wild-type 50S subunits for 10 min at 37°C in buffer A.
Filter binding and puromycin reactivity
Nitrocellulose filter binding of ribosome-bound tRNA and puromycin reactivity to measure the peptidyltransferase activity were performed as previously described (Moazed and Noller 1989b). 70S tight-couple ribosomes (0.8 μM) were incubated in reaction buffer (80 mM HEPES·KOH, pH 7.8, 100 mM NH4Cl, 10 mM MgCl2, 1 mM dithiothreitol [DTT]) with mRNA 32 (1.5 μM), deacylated tRNAfMet (1.5 μM), and N-Ac-[14C]Phe-tRNAPhe (1.0 μM), for 20 min at 37°C. Subsequent addition of EF-G with different nucleotides and/or antibiotics was performed at 37°C for 10 min. Binding of tRNA was measured by addition of each reaction mixture (6 μL) onto a nitrocellulose membrane, which was subsequently washed with 5 mL of ice-cold reaction buffer three times. An aliquot of each reaction mixture (5 μL) was then incubated with puromycin (1 mM) at room temperature for 30 min. The N-Ac-Phe·puromycin products were separated from the reaction mixture by addition of 1 eq of extraction buffer (MgSO4-saturated 0.3 M NaOAc, pH 5.3) and 750 μL of ethyl acetate. The ethyl acetate fraction was then separated form the aqueous phase and incubated with scintillation fluid, and the amount of radioactivity was then measured.
Chemical footprinting
Pre-translocation complexes were formed by addition of 70S tight-couple ribosomes (0.5 μM), mRNA 32 (1.0 μM), and fMet-tRNAfMet (1.0 μM) in reaction buffer M10 (80 mM HEPES·KOH, pH 7.8, 100 mM NH4Cl, 10 mM MgCl2, 1 mM dithiothreitol) and was incubated for 20 min at 37°C. P-site–bound tRNA was then deacylated by addition of puromycin (1 mM), which was subsequently removed by centrifugation through a Sephadex G-50 column. Addition of N-Ac-Phe-tRNAPhe (0.8 μM) to the 30S A site was performed at 37°C for 30 min. The resulting complex was then incubated with EF-G and different nucleotides and/or antibiotics for 10 min at 37°C. Single tRNA·ribosome complexes were formed by addition of 70S tight-couple ribosomes (0.5 μM), mRNA 32 (1.0 μM), and either fMet-tRNAfMet or N-Ac-Phe-tRNAPhe (1.0 μM) in reaction buffer M10 or M20 (80 mM HEPES·KOH, pH 7.8, 100 mM NH4Cl, 20 mM MgCl2, 1 mM DTT), respectively. Subsequent addition of puromycin was performed at 37°C for 20 min, and addition of EF-G with different nucleotides and/or antibiotics was performed at 37°C for 10 min.
Chemical footprinting analysis was performed as previously described (Moazed and Noller 1989a, 1990). Briefly, ribosome function complexes were incubated with kethoxal or dimethyl sulfate (DMS) for 8 min at 37°C. Ribosomal RNA from the modification reactions was then purified by phenol/chloroform extraction and used for primer extension analysis.
Toeprinting analysis
Toeprinting experiments were performed as previously described (Fredrick and Noller 2002). For the experiment in Figure 8, 70S ribosomes (1 μM) were incubated with tRNAMet (2 μM) and mRNA 291 (2 μM) pre-annealed to 32P-labeled primer (Fredrick and Noller 2002) in buffer A (50 mM HEPES·KOH [pH 7.5], 20 mM MgCl2, 100 mM NH4Cl, 6 mM β-mercaptoethanol) for 20 min at 37°C. N-Ac-Phe-tRNAPhe (2 μM) was added to P-site tRNA-bound complexes followed by incubation for 30 min at 37°C. Ribosomal complexes were diluted to a final concentration of 0.5 μM with buffer A. Translocation was carried out by incubation of EF-G (1.5 μM), different nucleotides (GTP, GDP, GDPNP—all 0.5 mM) and, in some cases, fusidic acid (0.1 mM) with pre-translocation complexes, containing tRNAMet in the P site and N-Ac-Phe-tRNAPhe in the A site, for 10 min at 37°C. The position of the ribosome along the mRNA was monitored using a primer-extension reaction as described previously (Fredrick and Noller 2002). Viomycin (1 mM) was added to the primer-extension reaction to prevent translocation in the presence of deoxy-GTP.
For the mRNA back-slippage experiment in Figure 4, pretranslocation complexes were assembled by binding deacylated tRNATyr to the P site in the presence mRNA 301 and binding N-Ac-Phe-tRNAPhe to the A site essentially as described above. Pre-translocation complexes (0.5 μM) were incubated with EF-G (1.5 μM) and GTP (0.5 mM) for 10 min at 37°C. The sample, now in the post-translocation state, was split into three aliquots. One aliquot was used for the primer-extension reaction. The second aliquot of the pre-translocation complex was incubated with puromycin (1 mM) alone for 15 min at 37°C. The third aliquot was incubated with thiostrepton (12.5 μM) for 5 min at 37°C and then with puromycin (1 mM) for 15 min at 37°C.
For the experiment in Figure 5, the pre-translocation complex was assembled by binding deacylated tRNATyr to the P site in the presence mRNA 301 and binding deacylated tRNAPhe to the A site in a reaction buffer containing 50 mM HEPES·KOH (pH 7.5), 25 mM MgCl2, 100 mM NH4Cl, and 6 mM β-mercaptoethanol. Then, the pre-translocation complex was incubated with EF-G (1.5 μM) and either GTP or GDPNP (both 0.5 mM). No viomycin was added to the extension reaction for the experiments of Figures 4 and 5.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.601507.
REFERENCES
- Abel, K., Jurnak, F. A complex profile of protein elongation: Translating chemical energy into molecular movement. Structure. 1996;4:229–238. doi: 10.1016/S0969-2126(96)00027-5. [DOI] [PubMed] [Google Scholar]
- Agrawal, R.K., Penczek, P., Grassucci, R.A., Burkhardt, N., Nierhaus, K.H., Frank, J. Effect of buffer conditions on the position of tRNA on the 70 S ribosome as visualized by cryoelectron microscopy. J. Biol. Chem. 1999;274:8723–8729. doi: 10.1074/jbc.274.13.8723. [DOI] [PubMed] [Google Scholar]
- Belitsina, N.V., Glukhova, M.A., Spirin, A.S. Translocation in ribosomes by attachment–detachment of elongation factor G without GTP cleavage: Evidence from a column-bound ribosome system. FEBS Lett. 1975;54:35–38. doi: 10.1016/0014-5793(75)81062-3. [DOI] [PubMed] [Google Scholar]
- Belitsina, N.V., Glukhova, M.A., Spirin, A.S. Elongation factor G-promoted translocation and polypeptide elongation in ribosomes without GTP cleavage: Use of columns with matrix-bound polyuridylic acid. Methods Enzymol. 1979;60:761–779. doi: 10.1016/s0076-6879(79)60070-8. [DOI] [PubMed] [Google Scholar]
- Bodley, J.W., Zieve, F.J., Lin, L. Studies on translocation. IV. The hydrolysis of a single round of guanosine triphosphate in the presence of fusidic acid. J. Biol. Chem. 1970;245:5662–5667. [PubMed] [Google Scholar]
- Cameron, D.M., Thompson, J., March, P.E., Dahlberg, A.E. Initiation factor IF2, thiostrepton and micrococcin prevent the binding of elongation factor G to the Escherichia coli ribosome. J. Mol. Biol. 2002;319:27–35. doi: 10.1016/S0022-2836(02)00235-8. [DOI] [PubMed] [Google Scholar]
- Czworkowski, J., Moore, P.B. The conformational properties of elongation factor G and the mechanism of translocation. Biochemistry. 1997;36:10327–10334. doi: 10.1021/bi970610k. [DOI] [PubMed] [Google Scholar]
- Dorner, S., Brunelle, J.L., Sharma, D., Green, R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat. Struct. Mol. Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, J., Agrawal, R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Fredrick, K., Noller, H.F. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol. Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- Fredrick, K., Noller, H.F. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- Girbes, T., Vazquez, D., Modolell, J. Polypeptide-chain elongation promoted by guanyl-5′-yl imidodiphosphate. Eur. J. Biochem. 1976;67:257–265. doi: 10.1111/j.1432-1033.1976.tb10657.x. [DOI] [PubMed] [Google Scholar]
- Hickerson, R., Majumdar, Z.K., Baucom, A., Clegg, R.M., Noller, H.F. Measurement of internal movements within the 30 S ribosomal subunit using Forster resonance energy transfer. J. Mol. Biol. 2005;354:459–472. doi: 10.1016/j.jmb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Inoue-Yokosawa, N., Ishikawa, C., Kaziro, Y. The role of guanosine triphosphate in translocation reaction catalyzed by elongation factor G. J. Biol. Chem. 1974;249:4321–4323. [PubMed] [Google Scholar]
- Jerinic, O., Joseph, S. Conformational changes in the ribosome induced by translational miscoding agents. J. Mol. Biol. 2000;304:707–713. doi: 10.1006/jmbi.2000.4269. [DOI] [PubMed] [Google Scholar]
- Joseph, S., Noller, H.F. EF-G–catalyzed translocation of anticodon stem–loop analogs of transfer RNA in the ribosome. EMBO J. 1998;17:3478–3483. doi: 10.1093/emboj/17.12.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriki, Y., Inoue, N., Kaziro, Y. Formation of a complex between GTP, G factor, and ribosomes as an intermediate of ribosome-dependent GTPase reaction. Biochim. Biophys. Acta. 1970;224:487–497. doi: 10.1016/0005-2787(70)90581-2. [DOI] [PubMed] [Google Scholar]
- Lancaster, L., Kiel, M.C., Kaji, A., Noller, H.F. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell. 2002;111:129–140. doi: 10.1016/s0092-8674(02)00938-8. [DOI] [PubMed] [Google Scholar]
- Lin, L., Bodley, J.W. Binding interactions between radiolabeled Escherichia coli elongation factor G and the ribosome. J. Biol. Chem. 1976;251:1795–1798. [PubMed] [Google Scholar]
- McGarry, K.G., Walker, S.E., Wang, H., Fredrick, K. Destabilization of the P site codon–anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Mol. Cell. 2005;20:613–622. doi: 10.1016/j.molcel.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed, D., Noller, H.F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989a;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Noller, H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989b;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Noller, H.F. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16 S rRNA. J. Mol. Biol. 1990;211:135–145. doi: 10.1016/0022-2836(90)90016-F. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Robertson, J.M., Noller, H.F. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- Modolell, J., Girbes, T., Vazquez, D. Ribosomal translocation promoted by guanylylimido diphosphate and guanylyl-methylene diphosphonate. FEBS Lett. 1975;60:109–113. doi: 10.1016/0014-5793(75)80429-7. [DOI] [PubMed] [Google Scholar]
- Munro, J.B., Altman, R.B., O'Connor, N., Blanchard, S.C. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, D., Kirillov, S.V., Cooperman, B.S. Kinetically competent intermediates in the translocation step of protein synthesis. Mol. Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske, F., Matassova, N.B., Savelsbergh, A., Rodnina, M.V., Wintermeyer, W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Mol. Cell. 2000;6:501–505. doi: 10.1016/s1097-2765(00)00049-6. [DOI] [PubMed] [Google Scholar]
- Pestka, S. Thiostrepton: A ribosomal inhibitor of translocation. Biochem. Biophys. Res. Commun. 1970;40:667–674. doi: 10.1016/0006-291x(70)90956-3. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V., Savelsbergh, A., Katunin, V.I., Wintermeyer, W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Savelsbergh, A., Katunin, V.I., Mohr, D., Peske, F., Rodnina, M.V., Wintermeyer, W. An elongation factor G-induced ribosome rearrangement precedes tRNA–mRNA translocation. Mol. Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- Semenkov, Y.P., Rodnina, M.V., Wintermeyer, W. The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli . Proc. Natl. Acad. Sci. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin, A.S. A model of the functioning ribosome: Locking and unlocking of the ribosome subparticles. Cold Spring Harb. Symp. Quant. Biol. 1969;34:197–207. doi: 10.1101/sqb.1969.034.01.026. [DOI] [PubMed] [Google Scholar]
- Spirin, A.S. Ribosomal translocation: Facts and models. Prog. Nucleic Acid Res. Mol. Biol. 1985;32:75–114. doi: 10.1016/s0079-6603(08)60346-3. [DOI] [PubMed] [Google Scholar]
- Valle, M., Zavialov, A., Sengupta, J., Rawat, U., Ehrenberg, M., Frank, J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Wang, B., Zhou, J., Lodder, M., Anderson R.D., 3rd, Hecht, S.M. Tandemly activated tRNAs as participants in protein synthesis. J. Biol. Chem. 2006;281:13865–13868. doi: 10.1074/jbc.C600018200. [DOI] [PubMed] [Google Scholar]
- Wilden, B., Savelsbergh, A., Rodnina, M.V., Wintermeyer, W. Role and timing of GTP binding and hydrolysis during EF-G–dependent tRNA translocation on the ribosome. Proc. Natl. Acad. Sci. 2006;103:13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K.S., Noller, H.F. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell. 1998;92:131–139. doi: 10.1016/s0092-8674(00)80905-8. [DOI] [PubMed] [Google Scholar]
- Wintermeyer, W., Savelsbergh, A., Semenkov, Y.P., Katunin, V.I., Rodnina, M.V. Mechanism of elongation factor G function in tRNA translocation on the ribosome. Cold Spring Harb. Symp. Quant. Biol. 2001;66:449–458. doi: 10.1101/sqb.2001.66.449. [DOI] [PubMed] [Google Scholar]
- Zavialov, A.V., Ehrenberg, M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- Zavialov, A.V., Hauryliuk, V.V., Ehrenberg, M. Guanine-nucleotide exchange on ribosome-bound elongation factor G initiates the translocation of tRNAs. J. Biol. 2005;4:9. doi: 10.1186/jbiol24. [DOI] [PMC free article] [PubMed] [Google Scholar]