Abstract
The roles of the ribosomal E site are not fully understood. Prior evidence suggests that deacyl-tRNA in the E site can prevent frameshifting. We hypothesized that if the E-site codon must dissociate from its tRNA to allow for frameshifting, then weak codon:anticodon duplexes should allow for greater frameshifting than stronger duplexes. Using the well-characterized Escherichia coli RF2 (prfB) programmed frameshift to study frameshifting, we mutagenized the E-site triplet to all Unn and Cnn codons. Those variants should represent a very wide range of duplex stability. Duplex stability was estimated using two different methods. Frameshifting is inversely correlated with stability, as estimated by either method. These findings indicate that pairing between the deacyl-tRNA and the E-site codon opposes frameshifting. We discuss the implications of these findings on frame maintenance and on the RF2 programmed frameshift mechanism.
Keywords: translation, codon:anticodon, duplex stability, frameshift, ribosome
INTRODUCTION
Translational roles for the E site have been among the most controversial topics in molecular biology. Ribosomes from representative organisms from all three living domains have an E site, and its role in accepting the deacylated tRNA during peptidyl transfer and translocation are well established (Noller et al. 2002). However, roles for the E site in aminoacyl-tRNA selection and reading frame maintenance have been more difficult to elucidate.
It is clear that codon-specific tRNA binding can occur in the E site (e.g., Triana-Alonso et al. 1995; for review, see Wilson and Nierhaus 2006), but a critical question is whether the E site is occupied long enough after translocation to have any significant translational role. Recent in vitro kinetic work suggests that deacylated tRNA remains in the E site until aminoacyl-tRNA bind in the A site but before GTP is hydrolyzed by EF–Tu (Dinos et al. 2005). However, other kinetic studies show that deacyl-tRNA readily dissociates from the E site in other standard buffer mixtures (Semenkov et al. 1996). Moreover, the kinetics of aminoacyl-tRNA selection can be explained without reference to an E-site tRNA (Wintermeyer et al. 2004). The differences among these in vitro studies suggest to us that the issue may only be resolved by in vivo studies.
Two in vivo studies suggest that the E site is occupied during the period after translocation and before the A site is filled. In one study, at least 75% of post-translocation ribosomes isolated from growing cells contain an E-site tRNA (Remme et al. 1989). However, since up to 25% of post-translocation ribosomes could lack an E-site tRNA, it is not clear that any residual tRNA has an important role. Another very interesting study shows that a slowly translated stop codon on a highly expressed message can cause intracellular depletion of a rare tRNA cognate to the E-site codon (i.e., the penultimate sense codon). The interpretation is that ribosomes stalled with the slowly translated terminator in the A site sequester the rare tRNA in the E site (Menez et al. 2000). This argues that the E site holds deacyl-tRNA for a significant period prior to termination at the A-site triplet, which is likely to be a slow reaction (Björnsson and Isaksson 1996; Freistroffer et al. 2000). On balance, therefore, it seems likely that deacyl-tRNA can reside in the E site following translocation. But what is it doing there?
Two studies show that the E-site codon can influence stop codon readthrough. Mottagui-Tabor et al. (1996) show that the next-to-last codon is related to readthrough, although that effect may be due to the nascent peptide rather than to the E-site tRNA. O'Connor et al. (1993) show that mutations near the 3′ end of the E-site tRNA can affect readthrough. That work specifically ties the E-site tRNA to translation, but the mechanism is not clear.
Others studies also provide evidence that the E site can have translational roles. In one study, Sergiev et al. (2005) show that mutation (C2394→G2394) of a 23S rRNA residue known to H bond with the 3′ end of E-site tRNA (Schmeing et al. 2003; Selmer et al. 2006) slightly increases frameshifting and nonsense codon readthrough. The mutation also inhibits binding of deacyl-tRNA in the E site and translocation (Sergiev et al. 2005), so it is possible that loss of deacyl-tRNA is the cause of the increased error rates. Of course, it is also possible that the translational defects are caused by some other, unknown effect of the mutation. In another system, tmRNA is subject to high-frequency frameshift when the resume codon is in the E site; that is, when there is no E-site codon:anticodon interaction, suggesting that an E-site duplex is important to hold the frame in at least this context (Trimble et al. 2004).
Finally, Marquez et al. (2004) provide evidence that a deacylated in the E site can contribute to reading frame maintenance, in vitro. In that report, tRNA cognate to the E-site message triplet can inhibit the Escherichia coli prfB (RF2) programmed frameshift. One complication with this in vitro experiment, however, is that the deacylated tRNA entered the E site from the solution rather than by translocation, which is presumably the only mechanism that occurs in vivo.
Altogether, these various studies suggest that the E site has translational roles. Here, we investigate the role of the E site in frame maintenance in vivo. We mutated the codon that resides at the E-site position of the RF2 programmed frameshift site at the time of the frameshift. Those mutations vary the codon:anticodon interactions that would occur in the E site if it were occupied during the frameshift. We find that frameshift frequencies vary depending on the codon and that the pattern of variation is neatly explained by differences in codon:anticodon stability. These results strongly suggest that the E site contains a paired codon:anticodon duplex and that the duplex must be disrupted to allow for frameshifting.
RESULTS
To study the role of the E site in reading frame maintenance, we mutated the codon corresponding to the E-site position of the RF2 programmed frameshift site that is cloned into a lacZ reporter (see Fig. 1 for an overview of the molecular mechanism of the programmed frameshift; Curran and Yarus 1988). Here, ribosomes stall at the slowly translated nonsense codon. Note that we substituted UAG for the WT UGA in all of our alleles (Curran and Yarus 1988; Curran 1993). This cuts the RF2 autoregulatory circuit, which could otherwise interfere with some experiments. The E-site codon (UAU in the WT) was varied to represent all 29 sense codons that begin with either U or C. We did not study the purine-starting codons because they might interfere with the adjacent, upstream Shine–Dalgarno-like element, which contains all purines.
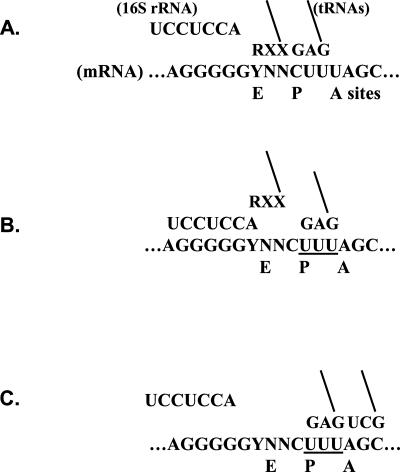
FIGURE 1.
Outline of the RF2 programmed frameshift mechanism. (A) Ribosome is paused during search to fill the A site. As is usual during elongation, the anti-Shine–Dalgarno of 16S rRNA is not engaged with the message. tRNAs occupy the E and P sites. The E site codon is varied to all YNN codons and is shown paired to an RXX anticodon. tRNALeu 2 is paired to CUU in the P site. (B) The Shine–Dalgarno engages, which dissociates the E and P site duplexes. The P site tRNA realigns onto the overlapping UUU triplet. (C) Translation resumes when the A site is filled and the Shine–Dalgarno disengages. See text for further details.
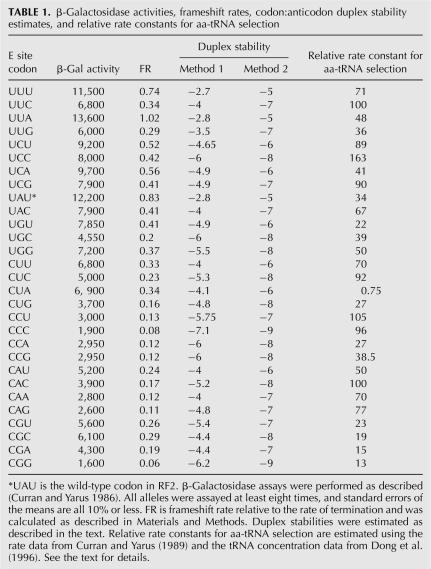
The β-galactosidase activities of the mutants were measured, and frameshifting rates (FR) were calculated as described in Materials and Methods. All alleles were assayed at least eight times, and standard errors were all 10% or less. β-Galactosidase activities and FR values are listed in Table 1. Technically, FR is the rate of the frameshift relative to the intrinsic rate of termination, which is assumed to be constant among alleles. This assumption is reasonable because, although termination may be sensitive to nearby nucleotides, the E-site codon is outside of the window known to have context effects in termination/suppression assays (for review, see Yarus and Curran 1992). The greatest FR value is for UUA, which at 1.02 frameshifts slightly faster than it terminates. The lowest FR value is for CGG (at 0.06), which frameshifts at a rate only 6% that of termination. The FR values vary over a 17-fold range, which shows that the E-site position is important to the RF2 frameshift mechanism.
TABLE 1.
β-Galactosidase activities, frameshift rates, codon:anticodon duplex stability estimates, and relative rate constants for aa-tRNA selection
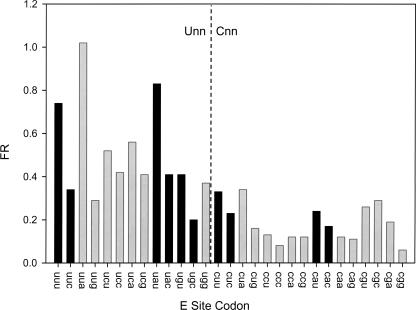
Two patterns are readily discernible in the data set. For one, codons that begin with U are generally more frameshift prone than codons that start with C (Fig. 2). Because U:A base pairs are generally weaker than G:C base pairs, this difference might be due to relatively facile dissociation of weaker duplexes at Unn codons, allowing more frameshifting. The other pattern is that for all five of the nnU/C codon pairs that are read by the same tRNA (Fig. 2, black bars), the U-ending codon frameshifts more than its C-ending partner. Because all of the U-enders are read with a wobble-type base pair while all C-enders are read with Watson–Crick pairs, this result is also consistent with the idea that weaker duplexes contribute to increased FR.
FIGURE 2.
FR for the YNN sense codons in the E site. Unn codons are on the left, and Cnn codons are on the right. The five pairs of codons ending in U or C and read by the same tRNA are darkly shaded. Standard errors of the mean are 10% or less.
Frameshift rate is inversely correlated with codon:anticodon duplex stability
To systematically investigate the role of duplex stability, we estimated the relative strengths of all of the codon:anticodon duplexes. There is a variety of sources of information about nucleic acid duplex stability in model systems; unfortunately, very little of these data may be directly applicable to codon:anticodon duplexes on the ribosome. Anticodons and neighboring residues contain modified nucleotides and are positioned within stacked helices within anticodon loops. All of these features are important for decoding. There are limited data on the effects of modified nucleotides on decoding (for examples and reviews, see Curran 1998; Agris et al. 2007); but it is not clear how those limited data can be applied in a systematic study of duplex stability. For all these reasons, we used two surrogate methods to estimate duplex stability. Neither method is likely to be fully representative, but they should give first-order estimates of relative kinetic stability for the various codon:anticodon pairs. And, critically, important features are likely to be common to both methods.
In the first method, we used the RNA duplex stability rules developed by Turner and colleagues (Freier et al. 1986). We treated modified nucleosides as unmodified. To calculate codon:anticodon duplex stability, we treated tRNA nucleotide 37, which is adjacent to the anticodon, as a 3′ dangling end. To that value, we added Turner's nearest-neighbor values for base pairs at the first and second codon positions. To that sum, we then added a value for terminal base pairs to account for the base pair at the third codon position. For codons thought to be actively read by two tRNAs (UCU, UCG, CCU, CCG) (Bjo¨rk 1995), we used the arithmetic average of the stabilities of the two duplexes. The values are listed in Table 1.
For our second method, we used a method similar to that used previously to study the stabilities of P-site duplexes after tRNA:message slippage (Curran 1993). In that work, we assigned relative stabilities to various base pairs and summed them for duplexes to estimate their stability. In the previous work, we tried two schemes, one in which A:U and G:C base pairs were equally stable, and another in which G:C pairs were more stable. In that previous work, both methods gave similar results. Here, because first-position Unn codons frameshift more than Cnn codons, we just used the scheme in which G:C pairs are more stable. Also in the previous work, we used Grosjean's study of the relative stabilities of a large number of anticodon:anticodon complexes using pairs of tRNAs with complementary (or nearly so) anticodons to assign values to various mispairs (Grosjean et al. 1978). But since the current work involves only cognate base pairs, a restricted set of the previous rules were used as follows: Watson–Crick base pairs with three H bonds (scored as “−3”) are more stable than Watson–Crick pairs with two H bonds (scored as “−2”), which are more stable than wobble pairs with two H bonds (scored as “−1”). To estimate relative duplex stability, we summed the values for the base pairs in each codon:anticodon duplex. And as above, we averaged the stabilities of duplexes for codons read by two tRNAs. The values are listed in Table 1.
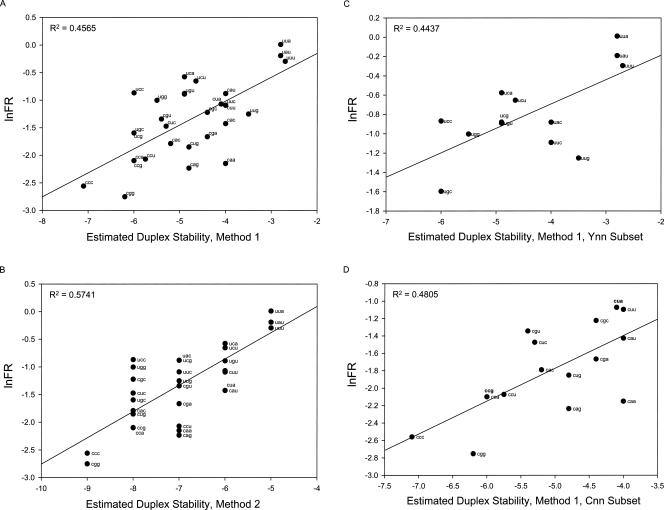
Each of these stability estimates was plotted versus an index of frameshifting. As described in Materials and Methods, FR is a linear scale. Because estimated duplex stability is logarithmic, we plotted stability versus the natural log of FR in Figure 3A,B. The plots are generally linear with the most stable duplexes having the least frameshifting and vice versa.
FIGURE 3.
Plots of ln(FR) versus estimates of codon:anticodon duplex stability; ln(FR) is plotted versus the first (A) and second (B) methods for estimating stability. (C, D) Plots for the Cnn and Unn subsets with FR plotted versus stabilities estimated with the first method.
Both plots show a strong relationship between frameshifting and duplex stability. Interestingly, perhaps, the ad hoc rules fit somewhat better than the Turner rules for this data set. But there is additional order in both of the plots. Unn codons tend to plot above the lines (more frameshifting), and Cnn codons tend to occur below them. Of course, this aspect of the pattern may occur simply because our methods misestimate the relative stabilities of first position U:A and C:G pairs. However, it is also possible that Unn codons are more frameshift prone for some factor other than base-pair stability. We will return to a plausible factor below. But because such a factor could contribute to the apparent correlation between duplex stability and frameshifting, we examined the Unn and Cnn codon sets separately to isolate them from any confounding factor associated with the first nucleotide. If duplex stability does indeed correlate with frameshifting, then this relationship should be observable for the isolated codon sets.
Both the Unn and Cnn codon sets show correlations between duplex stability and frameshifting with both stability estimation methods. The plots for the first method are shown in Figure 3, C and D. The plots with Method 2 are not shown, but the R 2 values exceed 0.45. Altogether, the correlations strongly support the conclusions that, prior to the frameshift, the E-site tRNA is base paired to its codon and that this pairing resists frameshifting.
There may be an additional factor, however. As noted above, the Unn codons may be relatively frameshift prone for another reason. A first position U could potentially strengthen the Shine–Dalgarno interaction that stimulates frameshifting. Crystal structures of ribosomes in complex with mRNA show that a U adjacent to the purine run can be included within the Shine–Dalgarno interaction by base pairing to A in 16S rRNA (Yusupova et al. 2001). In that work, the message has a spacing of 5 nucleotides (nt) between the purine run and the P-site codon. The RF2 mRNA has only a 3 nt spacer, and it is not clear that the first position U could participate in the Shine–Dalgarno interaction; but if it does, that could partially explain why alleles with Unn E-site codons are generally more frameshift prone that those with Cnn. It is important to note that a Unn effect on the Shine–Dalgarno interaction must be independent of the duplex stability effect. That is because the duplex must dissociate before the U could become available for participation in the Shine–Dalgarno interaction.
Finally, because the first nucleotide may have an effect independent of duplex stability, we recalculate the fold effect on FR separately for each codon set. The fold effect among the Unn codons is 5.1, with the highest FR at UUA and the least at UGC, and the range among the Cnn codons is 5.7-fold, with CUA having the greatest and CGG the least FR. These extreme values exemplify the base composition pattern, with U:A base pairs facilitating and G:C pairs inhibiting frameshifting.
Frameshifting does not correlate with the stability of the rephased complex
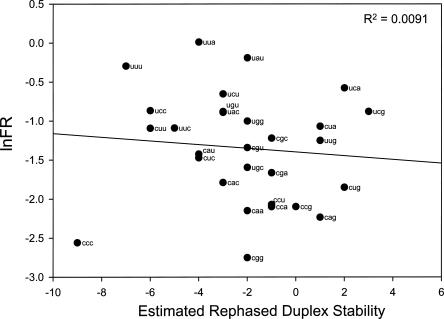
To determine whether frameshifting correlates with the stability of potential duplexes that might form in the rightward shifted reading frame, we plotted ln(FR) against the stabilities of the duplexes that could result after mRNA slippage. Stabilities were estimated using the same method used to estimate rephased duplex stabilities for the P site in a previous study assuming that G:C are more stable than A:U (Curran 1993). There is essentially no correlation (R 2 = 0.009; Fig. 4), thus our data set does not provide evidence that the E-site tRNA base pairs to the message in the rightward frame following message realignment.
FIGURE 4.
Plot of ln(FR) versus estimated stability of the potential rephased duplexes.
FR attributed to a tRNA in the E site does not correlate with its selection rate in the A site
To determine whether the E and A sites respond to the same molecular properties, we compared FR with the estimated rate constant for aa-tRNA selection at the A site. We previously estimated the rates of aminoacyl-tRNA (aa-tRNA) selection at the A site (Curran and Yarus 1989). In that work, we changed the A-site codon of the RF2 programmed frameshift site to all of the Ynn sense codons and measured β-galactosidase activities of the various alleles. If frameshifting is assumed to compete with normal translation, then frameshifting should be inversely correlated with the rate of aa-tRNA selection. In that work, our estimated rates of aa-tRNA selection correlated with codon usage in highly expressed E. coli genes. The data also provide evidence that tRNAs can have different rates of aa-tRNA selection at different cognate codons. tRNAs that read nnU/C codon pairs compete more effectively with frameshifting at the nnC codon.
With that background, if we can account for differences in the concentrations of the various tRNAs, we can estimate the relative rate constants for aa-tRNA selection at these codons. We did that in the previous work, but at the time we had only crude estimates of relative concentrations of some tRNAs. Since then, Dong et al. (1996) have performed an extensive analysis of tRNA concentrations at various growth rates. We used their concentrations for cells growing at 1.6 doublings per hour, which is the approximate growth rate of cultures in our β-galactosidase assays. The estimated rates of aa-tRNA selection from Curran and Yarus (1989) were divided by the tRNA concentrations from Dong et al. (1996) to estimate the relative rate constants for selection. We made one modification in the Curran and Yarus data set. The data from Curran (1995) indicate that the clones representing the CGA and CGC codons were probably switched; therefore, we switched their values for the current study.
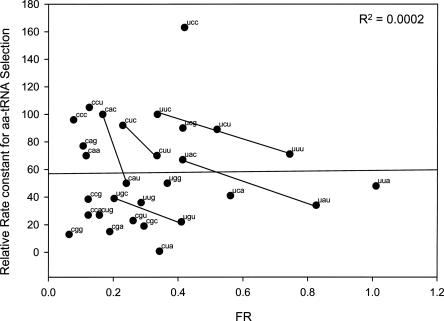
The relative rate constant estimates are listed in Table 1, and the plot of FR versus estimated relative rate constant is shown in Figure 5. There is essentially no correlation between these values, which shows that the assays measure different aspects of tRNA function on the ribosome. We suggest that FR is a measure of kinetic stability to duplex dissociation, while aa-tRNA selection is a more complicated process involving duplex association and various other interactions between the ribosome and the EF–Tu–tRNA complex.
FIGURE 5.
Plot of FR versus estimated relative rate of aa-tRNA selection at the A site. Data are from Table 1.
One feature that is shared by both assays is that, among the five codon pairs ending with pyrimidines and read by the same tRNA (connected by lines in Fig. 5), the C-ending codon is superior. That is, the duplex at the C-ender is more stable, as indicated by a lower FR, and is read more quickly, as indicated by a higher relative rate constant for aa-tRNA selection.
DISCUSSION
We studied the E-site codon during prfB (RF2) frameshifting. The codon was varied by mutagenesis, and all of the Unn and Cnn triplets were examined. Frameshifting activity, as estimated from β-galactosidase activities of lacZ fusions, varies about 17-fold among these alleles. Baranov et al. (2002) have presented an analysis on a smaller set of E-site codons and also observed significant effects on frameshifting. Of the codons that the two studies have in common, there is some but not complete concordance. We do not have a full explanation for differences, but our analysis was made on a much larger codon set using the standard β-galactosidase assay, which in our hands is linear over several orders of magnitude (Curran and Yarus 1987). In addition, all codons were assayed at least eight times, and all standard errors of the mean are 10% or less. Thus, we are confident that our data are sound, and certainly the broad patterns observed for our large data set are reliable.
Our analysis strongly suggests that codon:anticodon duplex stability is inversely related to frameshifting. One complication of that analysis is that duplex stabilities estimated from model systems may not be fully representative of duplexes on the ribosome. However, we used two methods to estimate duplex stability: one using stabilities determined for RNA duplexes in solution (Freier et al. 1986), and the other based on general principles of codon:anticodon stability (Curran 1993; Lim and Curran 2001). Both methods gave similar results. Thus, we are confident in the conclusion that the E-site duplex must dissociate to allow for RF2 frameshifting. This conclusion is consistent with observations that tRNA cognate to the E-site triplet can impede RF2 frameshifting in vitro (Marquez et al. 2004).
Mottagui-Tabor et al. (1996) have shown that the E-site codon can affect readthrough of UGA nonsense codons. However, in that case, the effect was related to the isoelectric point of the next-to-last amino acid. Our data do not show any such correlation (not shown). Moreover, the six leucine codons in our data set span nearly the entire range of frameshift rates (Table 1, 1.02 for UUA to 0.16 for CUG). These differences are explicable by differences in codon:anticodon stability but not by any difference in the amino acid.
Our data are consistent with the broader issue that the E site is occupied and can inhibit frameshifting prior to filling the A site. That idea was suggested by Nierhaus as a possible justification for the existence of the E site (e.g., Blaha and Nierhaus 2001; Wilson and Nierhaus 2006). Theoretical work argues that the E-site duplex is necessary to hold the frame until the A site is occupied (Lim et al. 2005). It is argued that reading frame maintenance during translocation requires that the codon:anticodon duplex for the peptidyl-tRNA be continuously shielded by a ribosomal component (a steric restriction element [SRE]) as it moves from the A to the P site. This shielding provides a kinetic barrier to duplex dissociation, which would otherwise occur on a timescale faster than translocation (Lim and Curran 2001; Lim et al. 2005). Then, to check the qualities of duplexes as they form in the A site, the SRE should oscillate between the P and A sites. This means that the frame is vulnerable to loss while the SRE is between sites. One solution is to use a kinetically stabilized E-site duplex to hold the frame (Lim et al. 2005). Our current data suggest that the E site can indeed help prevent frame loss as predicted by those studies.
In all likelihood, the Shine–Dalgarno interaction is important for dislodging both the E- and P-site tRNAs from their codons. There is direct evidence that the P-site duplex opposes frameshifting. Tsuchihashi and Brown (1992) showed that a relatively unstable duplex between tRNALys and AAG facilitated slipping in the E. coli dnaX programmed frameshift. In addition, among RF2 alleles that have various P-site codons, those that are read by third-position wobble base pairs frameshift more than codons read by third-position Watson–Crick base pairs (Curran 1993), which suggests that strong P-site duplexes inhibit frameshifting. Thus, both tRNAs must be dislodged from their in-phase triplets, and it is likely that the Shine–Dalgarno interaction stimulates these dissociation events (see Fig. 1).
After dissociation, however, the two sites have different roles in the frameshift mechanism. It is clear that the P-site tRNA is required to establish the new frame as suggested (Jacks et al. 1988; Weiss et al. 1990; Tsuchihashi and Brown 1992; for discussion, see Baranov et al. 2004). Our earlier work shows that frameshifting is strongly dependent on the stability of the rephased P-site duplex (Curran 1993). In contrast, our current data do not show any correlation between frameshifting and the potential rephased duplexes in the E site. Therefore, any such pairing in the E site is unimportant to the frameshift. In all likelihood, the E-site tRNA does not reassociate with the message in the new frame. Our data do not allow us to determine whether the tRNA actually leaves the ribosome—it could occupy a subsite within the E site in which it is not paired with the message (Yusupov et al. 2001; Selmer et al. 2006).
Our analysis supports the suggestion (Baranov et al. 2002) that the first nucleotide of the E-site codon participates in the Shine–Dalgarno interaction. Crystal structural data show that A of 16S rRNA can hybridize to a synthetic message having a U 3′ to the purine run of a Shine–Dalgarno (Yusupova et al. 2001). However, in that case, the U was the fifth nucleotide upstream of the P-site codon, and it is not clear that such an interaction would occur when the message U is only 3 nt upstream of the P site. But in our plots of estimated stability versus ln(FR) (Fig. 3A,B), it is clear that Cnn codons frameshift less and Unn codons more than expected based on duplex stability alone. One possible explanation is that we have over- and underestimated the relative stabilities of Unn and Cnn codons, respectively. We cannot rule that out, but it seems likely that if anything our estimates using Turner data would have erred in the other direction. In those estimates, we treated nucleotide 37 (3′ to the anticodon) as the 3′ dangling end. Because we could not quantitatively account for them, we ignored base modifications, but there is abundant evidence that the hypermodifications at position A37 of tRNAs that read Unn codons (ms2i6A) increase the stability of codon:anticodon duplexes (Nishimura 1972; Houssier and Grosjean 1985; Bouadloun et al. 1986; Li et al. 1997). Thus, by not accounting for the modifications we may have systematically underestimated not overestimated the stabilities at Unn codons. Therefore, it is probable that the Unn versus Cnn effect is due to some factor other than duplex stability. The only obvious factor is that the U of Unn codons might participate in the Shine–Dalgarno and thereby facilitate frameshifting. Baranov et al. (2002) have made the same suggestion based on their analysis of a smaller set of E-site codons.
It is also worth noting that the magnitude of effects on RF2 frameshifting differs greatly among the codons in the P (>1000-fold) (Curran 1993) and E (∼17-fold) sites. One obvious reason is that the major E-site effect is holding the frame against loss, while the major P-site effect is for establishing the new frame. All of the E-site duplexes are presumably cognate and therefore span a relatively narrow range of stability. In contrast, rephased complexes range from those that are fully cognate to those with three mispairs. Thus, the range of rephased stabilities must vary greatly in the P site among alleles, largely (if not totally) accounting for the greater range of effects on frameshifting.
The RECODE database (Baranov et al. 2003) catalogs 47 putative RF2 frameshift sites from various bacteria. Only a small number of these sites have been experimentally tested for frameshifting. Of these sites, almost half (23 of 47) contain UAU, which is the same E-site codon as in the highly frameshift-prone E. coli RF2. The next most common E-site codon is UCU (14 of 47), which frameshifts at ∼35% in our study (Table 1). Generally, the various other sites have at least two Us or a U and an A. Only five sites do not begin with a U, and only one of those begins with a purine (AGU for Treponena pallidum). All of the codons used (possibly except for AGU, which we did not study) frameshift at frequencies of 20% or greater in our study (Table 1). Thus, these sites are consistent with a frameshift-dependent mechanism for RF2 expression.
Finally, although an E-site duplex may be important for holding the frame, in general codon identity may not be critical. In their statistical analysis designed to detect codon context effects in E. coli genes, Yarus and Folley (1985) found that nucleotides surrounding sense codons are highly biased, which suggests that message context affects translation at sense codons. In highly expressed genes, strong biases occur primarily for neighboring nucleotides and, to a lesser extent, triplets. Strong biases do not extend further upstream, which suggests that the identities of E-site codons do not appreciably influence translation at the A site. It is important to note that this does not mean that the E-site duplex is not important; instead, it may mean that all cognate E-site interactions are adequate for any translational role, including effects on reading frame maintenance.
MATERIALS AND METHODS
E. coli strain and plasmids
All cloning methods and assays were performed using strain MY600 (Curran and Yarus 1986), which has the genotype E. coli K12 Δlac-pro, ara, thi. All plasmids were constructed starting with pJC27 (Curran and Yarus 1986), which encodes a pseudo-wild-type lacZ as well as chloramphenicol resistance. It has a P15A origin of replication. Based on numerous assays, it produces 27,000 units of β-galactosidase activity, which is used at the 100% value for FR calculations (below). All E-site derivatives were made by replacing a 20-nt section (between HindIII and BamHI sites) of the polylinker just inside the 5′ end of lacZ with 30-nt synthetic oligonucleotides encoding the RF2 frameshift site and having the desired E-site codons. These constructs require frameshifting to produce the β-galactosidase enzyme.
Calculations of frameshift rate (FR)
β-Galactosidase assays were performed as described (Curran and Yarus 1986). All alleles were assayed at least eight times, and standard errors of the mean were all 10% or less. To calculate frameshift frequencies, β-galactosidase activities of mutants were divided by that of the WT control (pJC27). Because frameshift frequencies are not linear (they saturate at 100%), to facilitate analysis we transformed the data to a linear scale. This allowed for direct comparisons among alleles. The RF2 autoregulatory mechanism requires that frameshifting and termination are in kinetic competition (Curran and Yarus 1988). Assuming that the rate of termination is constant, then frameshift frequencies of the various alleles can be used to estimate their rates of frameshifting, relative to the termination rate. We used this property to calculate the FR, relative to the constant termination rate. Equations 1–4 are essentially from earlier work (Curran and Yarus 1988). Equation 5 is simply the reciprocal of Equation 4.
When frameshifting is assumed to compete with termination, then
where R f is the rate of the frameshift and R t is the rate of termination. Then, inverting and factoring gives
Then by rearrangement,
Finally, the inverse is taken so that R f is reported relative to R t:
ACKNOWLEDGMENTS
We are grateful for the technical assistance of several student assistants, Matthew Byrd, Hugh Bender, Caitlin DeBoey, Elissa Gorey, Sarah Kuhn, and James Phipps. We also thank Dr. Valery Lim for comments. This work was supported by NIH grant GM077194 to J.F.C.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.638707.
REFERENCES
- Agris, P.F., Vendeix, F.A.P., Graham, W.D. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Baranov, P.V., Gesteland, R.F., Atkins, J.F. Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 2002;3:373–377. doi: 10.1093/embo-reports/kvf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov, P.V., Gurvich, O.L., Hammer, A.W., Gesteland, R.F., Atkins, J.F. RECODE 2003. Nucleic Acids Res. 2003;31:87–89. doi: 10.1093/nar/gkg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov, P.V., Gesteland, R.F., Atkins, J.F. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA. 2004;10:221–230. doi: 10.1261/rna.5122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk, G.R. Biosynthesis and function of modified nucleosides. In: Söll D., RajBhandary U., editors. tRNA: Structure, biosynthesis and function. ASM Press; Washington, DC: 1995. pp. 165–205. [Google Scholar]
- Björnsson, A., Isaksson, L.A. Accumulation of a mRNA decay intermediate by ribosomal pausing at a stop codon. Nucleic Acids Res. 1996;24:1753–1757. doi: 10.1093/nar/24.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaha, G., Nierhaus, K.H. Features and functions of the ribosomal E site. Cold Spring Harb. Symp. Quant. Biol. 2001;66:135–146. doi: 10.1101/sqb.2001.66.135. [DOI] [PubMed] [Google Scholar]
- Bouadloun, F., Srichaiyo, T., Isaksson, L.A., Björk, G.R. Influence of modification next to the anticodon in tRNA on codon context sensitivity of translational suppression and accuracy. J. Bacteriol. 1986;166:1022–1027. doi: 10.1128/jb.166.3.1022-1027.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran, J.F. Analysis of effects of tRNA:message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site. Nucleic Acids Res. 1993;21:1837–1843. doi: 10.1093/nar/21.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran, J.F. Decoding with the A:I wobble pair is inefficient. Nucleic Acids Res. 1995;23:683–688. doi: 10.1093/nar/23.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran, J.F. Modified nucleosides in translation. In: Grosjean H., Benne R., editors. Modification and editing of RNA: The alteration of RNA structure and function. ASM Press; Washington, DC: 1998. pp. 493–516. [Google Scholar]
- Curran, J.F., Yarus, M. Base substitutions in the tRNA anticodon arm do not degrade the accuracy of reading frame maintenance. Proc. Natl. Acad. Sci. 1986;83:1638–1642. doi: 10.1073/pnas.83.17.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran, J.F., Yarus, M. Reading frame selection and transfer RNA anticodon loop stacking. Science. 1987;238:1545–1550. doi: 10.1126/science.3685992. [DOI] [PubMed] [Google Scholar]
- Curran, J.F., Yarus, M. Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J. Mol. Biol. 1988;203:75–83. doi: 10.1016/0022-2836(88)90092-7. [DOI] [PubMed] [Google Scholar]
- Curran, J.F., Yarus, M. Rates of tRNA selection by 29 specific codons in vivo. J. Mol. Biol. 1989;209:65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- Dinos, G., Kalpaxix, D.L., Wilson, D.N., Nierhaus, K.H. Deacylated tRNA is released from the E site upon occupation but before GTP is hydrolyzed by EF–Tu. Nucleic Acids Res. 2005;33:5291–5296. doi: 10.1093/nar/gki833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H., Nilsson, L., Kurland, C.G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- Freier, S.M., Kierzek, R., Jaeger, J.A., Sugimoto, N., Caruthers, M.H., Nielson, T., Turner, D.H. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer, D.V., Kwiatkowski, M., Buckingham, R.H., Ehrenberg, M. The accuracy of codon recognition by polypeptide release factors. Proc. Natl. Acad. Sci. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean, H.J., De Henau, S., Crothers, D.M. On the physical basis for ambiguity in genetic coding interactions. Proc. Natl. Acad. Sci. 1978;75:610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houssier, C., Grosjean, H. Temperature jump relaxation studies on the interactions between transfer RNAs with complementary anticodons. The effect of modified bases adjacent to the anticodon triplet. J. Biomol. Struct. Dyn. 1985;3:387–408. doi: 10.1080/07391102.1985.10508425. [DOI] [PubMed] [Google Scholar]
- Jacks, T., Madhani, H.D., Masiarz, F.R., Varmus, H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.-N., Esberg, B., Curran, J.F., Björk, G.R. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J. Mol. Biol. 1997;271:209–221. doi: 10.1006/jmbi.1997.1176. [DOI] [PubMed] [Google Scholar]
- Lim, V.I., Curran, J.F. Analysis of codon:anticodon interactions within the ribosome provides new insights into codon reading and the genetic code structure. RNA. 2001;7:942–957. doi: 10.1017/s135583820100214x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, V.I., Curran, J.F., Garber, M. Ribosomal elongation cycle: Energetic, kinetic, and stereochemical aspects. J. Mol. Biol. 2005;351:470–480. doi: 10.1016/j.jmb.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Marquez, V., Wilson, D.N., Tate, W.P., Triana-Alonso, F., Nierhaus, K.H. Maintaining the translational reading frame: The influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Menez, J., Huergue-Hamard, V., Buckingham, R.H. Sequestration of specific tRNA species cognate to the last sense codon of an overproduced gratuitous protein. Nucleic Acids Res. 2000;28:4725–4732. doi: 10.1093/nar/28.23.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottagui-Tabor, S., Björnsson, A., Isaksson, L.A. The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J. 1996;13:249–257. doi: 10.1002/j.1460-2075.1994.tb06255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, S. Minor components in transfer RNA: Their characterization, location and function. Prog. Nucleic Acid Res. Mol. Biol. 1972;12:49–85. [PubMed] [Google Scholar]
- Noller, H.F., Yusupov, M.M., Yusupova, G.Z., Baucom, A., Cate, J.D. Translocation of tRNA during protein synthesis. FEBS Lett. 2002;514:11–16. doi: 10.1016/s0014-5793(02)02327-x. [DOI] [PubMed] [Google Scholar]
- O'Connor, M., Willis, N.M., Bossi, L., Gesteland, R.F., Atkins, J.F. Functional tRNAs with altered 3′ ends. EMBO J. 1993;12:2559–2566. doi: 10.1002/j.1460-2075.1993.tb05911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remme, J., Margus, T., Villems, R., Nierhaus, K.H. The third ribosomal tRNA-binding site, the E site, is occupied in native polysomes. Eur. J. Biochem. 1989;183:281–284. doi: 10.1111/j.1432-1033.1989.tb14925.x. [DOI] [PubMed] [Google Scholar]
- Schmeing, T.M., Moore, P.B., Steitz, T.A. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;7:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer, M., Dunham, C.M., Murphy, F.V., Weixlbaumer, A., Petry, S., Kelley, A.C., Weir, J.R., Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Semenkov, Y.P., Rodnina, M.V., Wintermeyer, W. The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli . Proc. Natl. Acad. Sci. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergiev, P.V., Lesnyalk, D.V., Kiparisov, S.V., Burakovsky, D., Leonov, L., Bogdanov, A.A., Brimacombe, R., Dontsova, O. Function of ribosomal E-site: A mutagenesis study. Nucleic Acids Res. 2005;33:6048–6056. doi: 10.1093/nar/gki910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triana-Alonso, F.J., Chakraburtty, K., Nierhaus, K.H. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E-site factor. J. Biol. Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- Trimble, M.J., Minnicus, A., Williams, K.P. tRNA slippage at the tmRNA resume codon. RNA. 2004;10:805–812. doi: 10.1261/rna.7010904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi, Z., Brown, P.O. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNALys and AAG lysine codon. Genes & Dev. 1992;6:511–519. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- Weiss, R.B., Dunn, D.M., Atkins, J.F., Gesteland, R.F. Ribosomal frameshifting from −2 to +50 nucleotides. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:159–183. doi: 10.1016/s0079-6603(08)60626-1. [DOI] [PubMed] [Google Scholar]
- Wilson, D.N., Nierhaus, K.H. The E-site story: The importance of maintaining two tRNAs on the ribosome during protein synthesis. Cell. Mol. Life Sci. 2006;63:2715–2737. doi: 10.1007/s00018-006-6125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermeyer, W., Peske, F., Beringer, M., Gromadski, K.B., Savalsbergh, A., Rodnina, M.V. Mechanisms of elongation on the ribosome: Dynamics of a macromolecular machine. Biochem. Soc. Trans. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- Yarus, M., Curran, J.F. The translational context effect. In: Hatfield D., Lee B., Pirtle R., editors. Transfer RNA in protein synthesis. CRC Press; Boca Raton, FL: 1992. pp. 319–365. [Google Scholar]
- Yarus, M., Folley, L.S. Sense codons are found in specific contexts. J. Mol. Biol. 1985;182:529–540. doi: 10.1016/0022-2836(85)90239-6. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Zh., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H.D., Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Yusupova, G.Zh., Yusupov, M.M., Cate, J.H.D., Noller, H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]