Abstract
The 3′ poly(A) tail present on the majority of mature eukaryotic mRNAs is an important regulator of protein synthesis and mRNA stability. The poly(A) tail improves the efficiency of translation initiation through recruitment of PABP, enabling its interaction with eIF4F located at the mRNA 5′-end. Recent evidence has also implicated a possible role for PABP and the poly(A) tract in translation control at steps beyond the initiation phase. Similar to conventional mRNAs, plus-strand RNA virus genomes that utilize internal ribosome entry sites (IRESes) to promote cap-independent translation are influenced by PABP and poly(A) status. However, the relative contribution of these factors to translation initiation mediated by distinct IRESes is unclear. We have investigated cis- and trans-acting effects of poly(A) and PABP, respectively, on RNAs harboring IRESes from three diverse viruses: encephalomyocarditis virus (EMCV), hepatitis C virus (HCV), and coxsackievirus B3 (CBV3). A 3′ poly(A) tract enhanced translation of both capped and IRES-containing reporter RNAs. However, only CBV3 and capped transcripts were stabilized as a result of polyadenylation. Correspondingly, translation of polyadenylated CBV3 and capped RNAs displayed heightened sensitivity to the PABP inhibitor Paip2 compared with EMCV and HCV. Sucrose density gradient analyses suggested a stimulatory role for PABP and 3′ poly(A) in the CBV3 initiation phase, while assembly of HCV and EMCV RNAs into ribosomal complexes was little affected by either factor. Collectively, the observed differential effects of PABP and poly(A) on translation imply mechanistic differences between viral IRES elements and suggest modulating roles for PABP and the poly(A) tail at multiple phases of translation.
Keywords: poly(A)-binding protein, IRES, translation initiation, RNA decay
INTRODUCTION
Post-transcriptional control mechanisms involving regulated mRNA translation and decay are fundamental aspects of gene expression. A critical player in both processes is the 3′ poly(A) tail, a feature common to the vast majority of cellular mRNAs. Through association with its cytoplasmic binding partner, poly(A)-binding protein (PABP), the poly(A) tail participates in an intricate ribonucleoprotein (RNP) network that is believed to approximate mRNA termini, forming a “closed-loop” structure that is primed for recruitment of ribosomal complexes (Jacobson 1996). Essential to 5′–3′ communication is the widely conserved interaction between PABP and the eukaryotic initiation factor (eIF) 4G (Tarun and Sachs 1996; Imataka et al. 1998), a molecular scaffold that indirectly links the mRNA 5′ m7GTP cap to the 40S ribosomal subunit through interaction with eIF4E and eIF3 (for review, see Hershey and Merrick 2000). PABP–eIF4G interaction has been reported to functionally enhance affinity of eIF4E for the cap structure (Kahvejian et al. 2005), resulting in improved recruitment of the 43S preinitiation complex and subsequent formation of scanned 48S intermediates. In addition, PABP has been proposed to enhance initiation at the level of 60S subunit joining (Sachs and Davis 1989; Munroe and Jacobson 1990; Searfoss et al. 2001), apparently in a manner that also depends on its interaction with eIF4G (Kahvejian et al. 2005).
Beside its integral role in translation initiation of capped mRNAs, cytoplasmic PABP has also been implicated at other levels of post-transcriptional regulation, namely, mRNA decay and translation termination (Mangus et al. 2003). Eukaryotic release factor 3(eRF3) is a GTPase that facilitates termination through interaction with its partner eRF1, a protein that recognizes stop codons present within the ribosomal A-site (Kisselev et al. 2003). Interaction between PABP and eRF3 in yeast and mammalian cells (Hoshino et al. 1999; Cosson et al. 2002) has been suggested to enhance termination and promote hypothetical recycling of ribosomes into subsequent rounds of initiation (Uchida et al. 2002; Amrani et al. 2004). With respect to RNA decay, PABP protects mRNA from deadenylation (Bernstein et al. 1989; Ford et al. 1997; Wilusz et al. 2001a), the initial rate-limiting step for many mRNAs in the pathway to destruction (Wilusz et al. 2001b). Deadenylation precedes removal of the 5′-cap and subsequent exonucleolytic decay of the transcript body. For a typical cellular mRNA, it is conceivable that disruption of 5′–3′ interaction due to loss of the poly(A) tail results in a poorly translated and unstable template whose 5′-cap is susceptible to decapping enzymes.
Plus-strand RNA virus genomes serve directly as templates for synthesis of viral proteins and have evolved unconventional mechanisms of translation initiation. This is exemplified by internal ribosome entry sites (IRESes), RNA elements that promote initiation in a 5′-end- and cap-independent manner (for review, see Jang 2006). IRESes were first uncovered in viral genomes of picornaviruses (Jang et al. 1988; Pelletier and Sonenberg 1988) and have since been identified in hepatitis C virus (HCV) (Tsukiyama-Kohara et al. 1992) as well as a subpopulation of cellular mRNAs whose translation persists under conditions of generalized translational repression (Hellen and Sarnow 2001). For most IRESes, the precise molecular details of internal ribosome entry are unknown. Moreover, cis- and trans-acting factors that enable function differ substantially between IRES elements. Confirmed and putative IRESes display widely divergent sequence and structural characteristics. Consequently, assignment of IRES activity to a particular RNA requires appropriately controlled functional assays to ascertain cap- and 5′-end independence (Van Eden et al. 2004). With regard to utilization of canonical eIFs, IRESes by definition are eIF4E independent, but display remarkably variable requirements for other factors. For example, whereas the encephalomyocarditis virus (EMCV) IRES relies on the conventional set of eIFs (except eIF4E and intact eIF4G) (Pestova et al. 1996), that of the cricket paralysis virus appears to employ only ribosomal subunits in initiation (Wilson et al. 2000).
Enhancing effects of the poly(A) tail and PABP on IRES-dependent translation have been reported recently (Bergamini et al. 2000; Michel et al. 2001; Svitkin et al. 2001; Lopez de Quinto et al. 2002; Thoma et al. 2004a). However, the precise roles these factors play in translation initiation and stability of IRES-harboring RNAs are unclear. We previously reported that a poly(A) tail of sufficient length was able to potently enhance translation of HCV IRES-driven reporter RNAs in the absence of any pronounced effect on initiation (Bradrick et al. 2006). Here, we extend these findings by investigating the effects of PABP and the poly(A) tract on translation mediated by several distinct viral IRESes.
RESULTS
A poly(A) tail enhances translation of capped and IRES-containing RNAs in HeLa S10 cytoplasmic extract
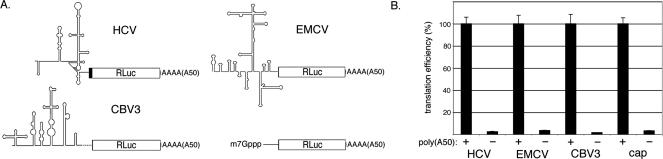
Three viral IRES elements were selected for analysis in an established in vitro translation system based on HeLa cytoplasmic extract that exhibits poly(A) responsiveness (Fig. 1A; Thoma et al. 2004b). The IRESes of EMCV, coxsackievirus B3 (CBV3), and HCV perform the same essential function in the infected cell: recruitment of cellular translation machinery for synthesis of viral proteins. This is illustrated by reports of viable virus chimeras that contain heterologous IRESes driving expression of viral proteins (Alexander et al. 1994; Gromeier et al. 1996; Lu and Wimmer 1996). Nevertheless, each IRES is unique in length, primary sequence, and structure (Fig. 1A). The CBV3 and EMCV IRESes have been classified into two distinct categories (types I and II) based upon functionality in rabbit reticulocyte lysate (Wimmer et al. 1993). Both IRES types, however, are thought to share mechanistic features, requiring the canonical set of initiation factors except for eIF4E and intact eIF4G. In contrast, the HCV IRES is capable of directly recruiting the 40S ribosomal subunit and does not depend on eIF4F components (eIFs 4G, 4A, and 4E) (Pestova et al. 1998). The positive-strand RNA genomes of these viruses also differ in the nature of their 3′-ends. Those of CBV3 and EMCV are polyadenylated (between 35 and 60 residues) (Ahlquist and Kaisberg 1979; data not shown), whereas the HCV genome terminates with a characteristic stable RNA structure known as the X region (Kolykhalov et al. 1996).
FIGURE 1.
A poly(A) tail enhances translation of capped and IRES-driven reporter RNAs in vitro. (A) Schematic representation of Rluc reporter RNAs containing the β-globin leader sequence or IRESes of HCV, EMCV, and CBV3. (B) Poly(A50)-plus and -minus versions of each RNA were translated in HeLa cytoplasmic extract for 30 min prior to measurements of RLuc activity. Translation of the polyadenylated RNA is set to 100% for each construct.
Previous experiments indicated that placement of a 50 residue poly(A) tract immediately downstream of the stop codon in subgenomic IRES-driven reporter RNAs significantly enhanced translation both in cells and in vitro, in part through relieving a translation termination defect exhibited by RNAs lacking any 3′-UTR (Bradrick et al. 2006; Dobrikova et al. 2006). The same was true for each reporter RNA assayed here in vitro; poly(A50) substantially enhanced synthesis of Renilla luciferase (RLuc) from capped- and IRES-containing RNAs by greater than 20-fold (Fig. 1B). Though a poly(A) tail uniformly increased translation, the overall efficiency of RLuc synthesis differed as a function of 5′-UTR identity. Under the conditions employed, HCV-A50 reporter RNA was translated approximately twofold higher than cap-A50 and approximately fivefold more that either CBV3-A50 or EMCV-A50 RNAs (see Materials and Methods; data not shown).
The poly(A) tail differentially influences RNA stability as a function of 5′-end identity
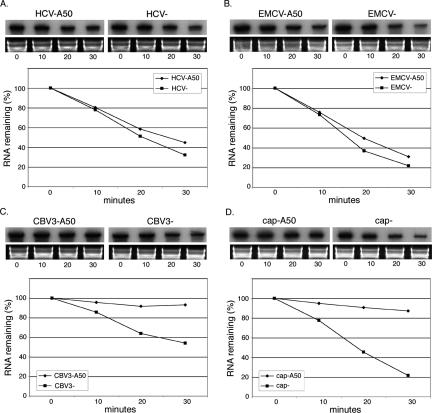
Intimate links exist between translation control and the regulated decay of mRNAs. It is therefore essential to evaluate RNA integrity in experiments concerning the role of cis-acting elements in translation efficiency. In the present case, discrepancies observed between expression of reporter RNAs with and without poly(A50) might reflect increased susceptibility of unpolyadenylated RNAs to cytoplasmic ribonucleases. The stability of each reporter RNA was evaluated as a function of poly(A) status by tracking the level of intact radiolabeled RNA over time in translation reactions (Fig. 2). Inherent rates of decay between poly(A)-minus reporter RNAs were similar except for that of CBV3, which exhibited two- to 2.5-fold more intact RNA after 30 min of incubation (Fig. 2C). Interestingly, the stabilizing effect of poly(A50) varied as a function of cis-acting elements present upstream of the RLuc open reading frame. A minor increase in stability was apparent for both EMCV and HCV RNAs with poly(A) tails (Fig. 2A,B). On the contrary, CBV3 and capped RNAs were exceptionally resistant to decay as a consequence of polyadenylation (Fig. 2C,D). After a 30-min incubation, ∼90% of each poly(A)-plus transcript remained intact, compared with ∼55% and ∼20% of unpolyadenyated CBV3 and capped RNAs, respectively. Consequently, enhancement of RLuc expression from these RNAs by poly(A) results, in part, from decreased kinetics of transcript degradation.
FIGURE 2.
A poly(A) tail differentially affects RNA decay depending on 5′-end identity. Levels of intact trace-radiolabeled RNA in translation reactions were monitored over the course of 30 min by denaturing PAGE and phosphorimaging: (A) HCV, (B) EMCV, (C) CBV3, and (D) cap. Ribosomal RNAs from corresponding samples are shown below panels displaying radioactive reporter RNAs. Graphs represent the average of two independent experiments.
Viral IRES-mediated translation is differentially sensitive to inhibition of PABP
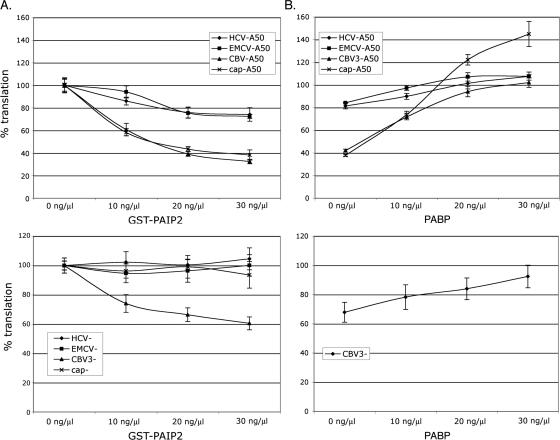
Since reporter RNAs responded differently to addition of a poly(A) tail with regard to decay, we chose to investigate whether cytoplasmic PABP, the trans-acting factor deemed responsible for effects of poly(A), might variably impact translation of distinct reporter RNAs. For this purpose we utilized recombinant GST-tagged Paip2, which binds PABP, displaces it from the poly(A) tail, and precludes its interaction with eIF4G (Khaleghpour et al. 2001; Karim et al. 2006). Addition of the fusion protein directly to translation reactions, instead of PABP depletion from extracts, was selected since the latter technique diminishes translation of capped-unpolyadenylated mRNA to a larger extent than the former (Svitkin and Sonenberg 2004; see below). Titrations of GST–Paip2 up to 30 ng/μL final concentration were performed in translation reactions programmed with poly(A)-plus and poly(A)-minus versions of each RNA (Fig. 3A). Two distinct patterns of sensitivity to Paip2 were observed depending on 5′-end identity. While translation of both EMCV-A50 and HCV-A50 was reduced ∼20% at 30 ng/μL GST–Paip2 compared with buffer-only control, cap-A50, and CBV3-A50 displayed threefold higher levels of inhibition (∼60% reduction) (Fig. 3A, top). Similar experiments performed with unpolyadenylated reporter RNAs produced different results (Fig. 3A, bottom). No Paip2-mediated interference was observed for cap-dependent, EMCV-, or HCV-mediated translation, indicating that the enhancing effects PABP on these RNAs depends upon presence of the poly(A) tail. Curiously, unpolyadenylated CBV3 RNA was uniquely inhibited upon titration of Paip2 (∼40% at 30 ng/μL GST–Paip2), though to a lesser extent than observed for CBV3-A50.
FIGURE 3.
Paip2-mediated inhibition of PABP differentially affects translation of reporter RNAs. (A) Sensitivity to PABP inhibition was determined for polyadenylated (top) and unpolyadenylated (bottom) versions of each RNA by titration of increasing amounts of GST–Paip2 into translation reactions incubated for 30 min. Reactions with “0 ng/μL” GST–Paip2 were set to 100% and contained only dialysis buffer. (B) Reversal of Paip2 activity by PABP add back. Recombinant PABP at the final concentrations indicated was used to supplement reactions containing 20 ng/μL GST–Paip2. Experiments with poly(A)-plus reporters are shown at top. Unpolyadenylated CBV3 RNA, the only one affected by Paip2, was also evaluated in PABP add-back experiments (bottom). Error bars represent calculated standard deviation values.
In order to confirm the specificity of PABP inhibition by Paip2, additional experiments were performed in which translation reactions containing 20 ng/μL GST–Paip2 (the concentration at which near maximum inhibition was observed) were supplemented with increasing amounts of recombinant PABP (Fig. 3B). As expected, PABP titration reversed the inhibitory effect of Paip2 on each polyadenylated RNA (Fig. 3B, top). Notably however, cap-A50 translation was elevated substantially (nearly 50%) over control reactions lacking either exogenous GST–Paip2 or PABP, suggesting that PABP may be limiting for cap-, poly(A)-dependent translation in cell-free reactions performed under the conditions utilized here. Due to its unique sensitivity to Paip2, unpolyadenylated CBV3 RNA was also assayed in PABP “addback” experiments (Fig. 3B, bottom). Similar to IRES-driven polyadenylated RNAs, PABP rescued CBV3- translation in the presence of Paip2, suggesting poly(A)-independent activity of PABP for this RNA. Collectively, these observations illuminate unexpected differences between distinct mRNA templates in their dependence on PABP for optimal translation.
Inhibition of PABP by Paip2 does not alter the rate of RNA decay
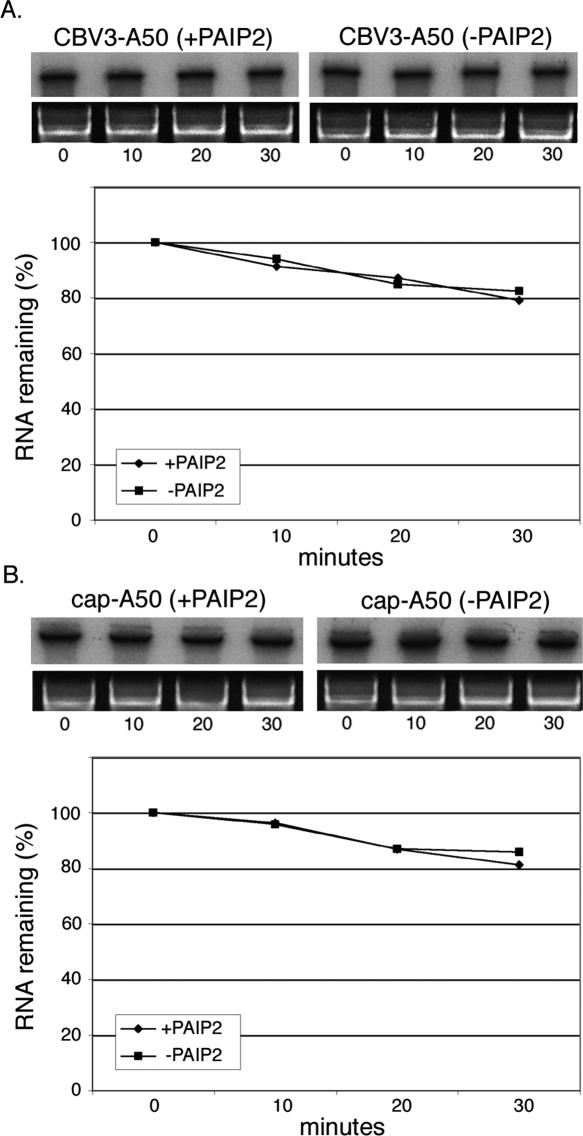
Since relatively enhanced sensitivity of cap-A50 and CBV3-A50 to Paip2 (Fig. 3A) coincided with stabilization of these same mRNAs by the poly(A) tail (Fig. 2C,D), we questioned whether Paip2 interference of PABP might increase rates of reporter RNA decay. Translation reactions with or without 30 ng/μL GST–Paip2 were programmed with 32P-labeled reporter transcripts (CBV3-A50 or cap-A50) and decay was monitored over a 30-min incubation (Fig. 4). Treatment with Paip2 did not induce degradation of either RNA compared with control reactions. Similar to experiments shown in Figure 2C, CBV3-A50 and cap-A50 RNAs remained predominantly intact after 30 min. Additionally, polyadenylated HCV and EMCV transcripts did not display altered decay rates in response to Paip2 treatment (data not shown). These results suggest direct participation of PABP in IRES- and cap-dependent translation processes in vitro without significant impact on RNA decay. Alternatively, it is possible that residual uninhibited PABP is present in sufficient quantities to confer RNA stabilization but not enhancement of translation.
FIGURE 4.
Inhibition of PABP does not affect stability of reporter RNAs. Decay of polyadenylated CBV3 (A) and capped (B) RNAs were examined in the presence and absence of GST–Paip2 (30 ng/μL) as shown in Figure 2. Graphs represent the average of two independent experiments.
Effects of PABP and the poly(A) tail on ribosome recruitment mediated by viral IRESes
The interaction of PABP, bound to the mRNA poly(A) tail, with eIF4G at the mRNA 5′-end, enhances ribosome recruitment by capped mRNAs. With regard to viral IRES-driven translation, a direct role for PABP and the poly(A) tail in the initiation phase has not been demonstrated. To examine the role of these factors in viral IRES function, sucrose density gradient sedimentation assays were conducted to evaluate the first step in initiation, recruitment of the 40S ribosomal subunit and associated eIFs. Translation reactions programmed with radiolabeled reporter mRNAs were supplemented with the nonhydrolysable GTP analog GMPPNP to prevent 60S subunit joining (Hershey and Merrick 2000), allowing analysis of 48S complex formation. Linear-absorbance profiles obtained during gradient fractionation indicated the presence of ribosomal subunits and intact monosomes at distinct gradient depths (Fig. 5). Analysis of radioactivity levels in collected fractions reproducibly allowed discrimination of two prominent peaks for each reporter RNA, corresponding to a ribosome-free RNP and 48S complex (Fig. 6). However, as evidenced by the presence of radioactive RNA in heavier fractions (14–18), 60S subunit joining was incompletely inhibited by GMPPNP.
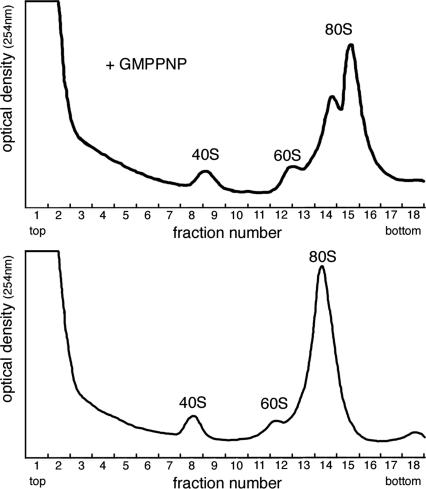
FIGURE 5.
Linear absorbance profiles of sucrose gradients. Optical density traces are plotted versus fraction number for reactions containing GMPPNP (above) and cycloheximide (below). Peaks of ribosomal subunits (40S and 60S) and monosomes (80S) are indicated. Gradients for reactions containing GMPPNP were centrifuged under conditions that increased separation of RNP and 48S complexes (see Materials and Methods). GMPPNP treatment resulted in two separate closely migrating peaks around 80S.
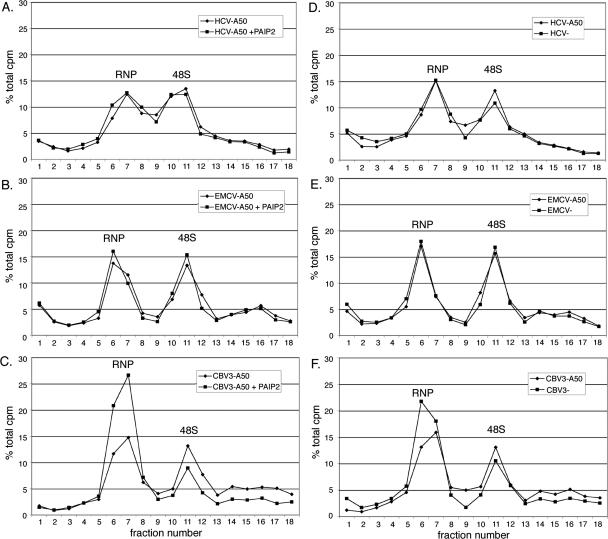
FIGURE 6.
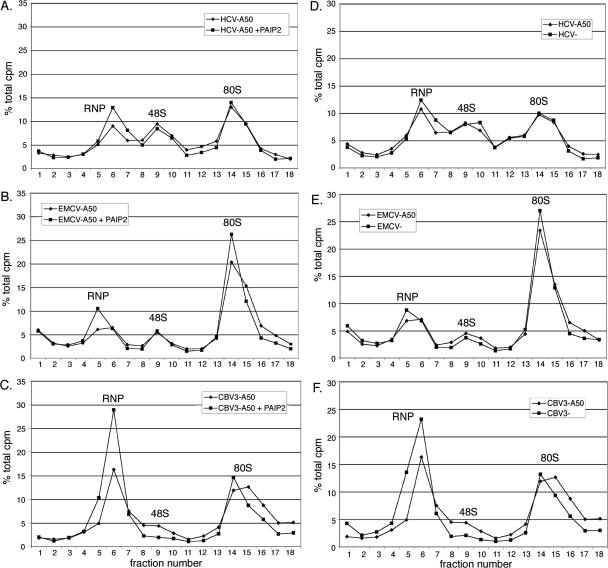
PABP and poly(A) differentially stimulate formation of 48S ribosomal complexes as a function of viral IRES identity. In vitro translation reactions were assembled with radiolabeled reporter RNA and then separated on 5%–20% linear sucrose gradients before fractionation (see Materials and Methods for details). Peaks of radioactive counts formed by ribonucleoprotein complexes (RNP) and 48S complexes are indicated. The effect of GST–Paip2 (30 ng/μL final concentration) on initiation was evaluated using intact, polyadenylated HCV (A), EMCV (B), and CBV3 (C) mRNAs. In corresponding experiments, the effect of a poly(A) tail on 48S formation was examined for each IRES: HCV (D), EMCV (E), and CBV3 (F).
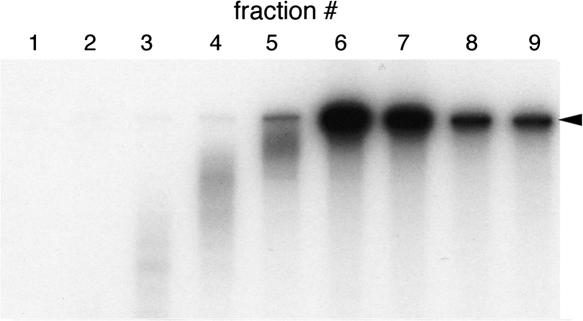
We first examined whether PABP influenced the rate of initiation on polyadenylated IRES-containing reporter RNAs. The presence of GST–Paip2 minimally affected 48S formation mediated by the HCV and EMCV IRESes (Fig. 6A,B). In comparison, PABP inhibition substantially abrogated 48S formation on the CBV3 IRES and concomitantly increased apparent RNP peak size by ∼80% (Fig. 6C, note that the area under the peak is the critical variable). These data directly implicate PABP participation in 40S subunit recruitment by the type I CBV3 IRES. Additional analyses were conducted to assay effects of a 3′ poly(A) tail on initiation mediated by each viral IRES. Similar to results obtained with Paip2, a poly(A) tract failed to reproducibly affect the efficiency of 48S formation on the HCV or EMCV IRESes (Fig. 6D,E). In contrast, loss of the poly(A) tract resulted in several distinct alterations in the gradient distribution of CBV3 reporter RNA (Fig. 6F). Overall, unpolyadenylated CBV3 RNA was less abundant in the 48S peak and heavier fractions (14–18) with a correspondingly higher portion in the RNP, indicating differential rates of initiation. However, the magnitude of these effects was less than those observed with Paip2 treatment. Interestingly, we also consistently observed higher levels of radioactive counts in lighter fractions (1–5) as a function of poly(A) tail loss (Fig. 6F; see below). Since poly(A) stabilized CBV3 reporter RNA (Fig. 2C) to a higher degree than other IRES-containing reporter RNAs, we reasoned that this radioactivity might correspond to degraded RNA fragments and/or free nucleotides. To confirm this, RNA was extracted from the top half of the CBV3-A50 gradient (fractions 1–9) and subjected to denaturing PAGE analysis. As shown in Figure 7, intact reporter RNA was found predominately in fractions 6–9, while radioactive counts found in fractions 1–5 were produced by degraded RNA fragments and/or nucleotides. Thus, cpm levels in lighter gradient fractions are indicative of the extent of RNA degradation.
FIGURE 7.
Migration of intact reporter RNA in sucrose gradients. CBV3-A50 RNA was extracted from equal volumes of fractions 1–9 and analyzed by denaturing PAGE. The arrowhead indicates the position of full-length reporter RNA. No RNA was resolved in fractions 1 and 2.
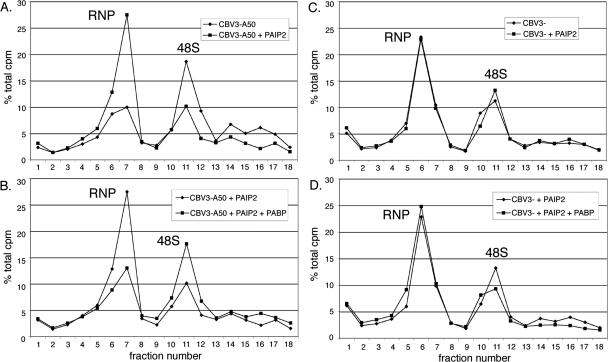
In order to confirm and extend data implicating PABP/poly(A) in type I IRES function, additional experiments were conducted to investigate (1) whether recombinant PABP could reverse the inhibitory effect of Paip2 on CBV3 IRES-mediated 48S formation, and (2) whether Paip2 is able to abrogate initiation on unpolyadenylated CBV3 reporter RNA (Fig. 8). We chose to perform these experiments using a separate preparation of HeLa S10 extract to control for batch-to-batch variation. Utilizing the same conditions as assays presented in Figure 6, polyadenylated CBV3 mRNA was somewhat more efficiently recruited into 48S complexes (Figs. 6C, 8A, cf. “CBV3-A50” patterns). Paip2 again significantly inhibited ribosome recruitment by the CBV3 IRES as evidenced by changes in RNP and 48S peak sizes, as well as RNA in heavier gradient fractions (Fig. 8A). As expected, supplementation of reactions containing Paip2 with recombinant PABP (30 ng/μL) restored 48S assembly to nearly identical levels observed for mock treatment (Fig. 8B). This indicates that the inhibitory effect of Paip2 on CBV3 IRES function is specific with respect to both PABP (Fig. 8A,B) and the CBV3 IRES (Fig. 6). Gradient centrifugation experiments performed simultaneously with unpolyadenlyated CBV3 mRNA yielded different results. Neither Paip2 (Fig. 8C) alone nor PABP plus Paip2 (Fig. 8D) significantly affected 48S formation on the CBV3 IRES, suggesting that the enhancing effect of PABP on ribosome recruitment by CBV3 requires the presence of a 3′ poly(A) tail.
FIGURE 8.
PABP specifically enhances 48S complex formation on the CBV3 IRES in presence of a poly(A) tail. Analyses similar to those shown in Figure 6 were conducted exclusively with CBV3 mRNAs. Reactions were mock-treated, supplemented with Paip2 (30 ng/mL), or supplemented with both Paip2 and PABP (30 ng/mL). Effects of Paip2 (A,B) and PABP “addback” (B,D) on 48S assembly are shown for reporter RNAs containing (A,B) or lacking (C,D) a poly(A) tail.
Finally, we evaluated the effects of Paip2 and the poly(A) tract on incorporation of radiolabeled reporter RNAs into 80S ribosomal complexes by performing reactions assembled in the presence of cycloheximide (an inhibitor of elongation) instead of GMPPNP. Each RNA formed a characteristic distribution across the gradient depending on the viral IRES present (Fig. 9). As with experiments analyzing 48S formation, PABP interference or poly(A) tail deletion appeared to produce more significant effects on CBV3-mediated initiation (Fig. 9C,F) than other IRESes (Fig. 9A,B,D,E). Paip2 treatment increased the size of the CBV3-A50 RNP peak by ∼70% and correspondingly reduced the amount of intact RNA in ribosome-containing fractions (Fig. 9C) (see Fig. 5 for absorbance profile). However, rather than resulting in a discrete reduction in 80S peak height (Fig. 9C, fraction 14), as might be expected, both PABP inhibition and poly(A) tail removal consistently induced several unexpected changes in the shape of the RNA distribution in fractions 8–18. First, in the absence of Paip2, the 80S complex formed on CBV3-A50 RNA sediments somewhat faster, forming a relatively broad peak (Fig. 9C,F; cf. cpm levels in fractions 15–18). Secondly, both poly(A) deletion and Paip2 treatment reduced the amount of RNA migrating in fractions 8–13, which contain 48S complexes and perhaps other undefined intermediates. Finally, removal of the poly(A) tract was associated with increased radioactivity in lighter gradient fractions (1–4), indicating elevated decay (Fig. 9F). The basis for altered sedimentation patterns in fractions 8–18 as a function of PABP/poly(A) status are not entirely clear. We speculate that 80S particles formed on unpolyadenylated CBV3 RNA, or in the presence of Paip2, lack PABP and perhaps other factors that affect sedimentation velocity. Nevertheless, taken together with sucrose density gradient analyses presented in Figures 6 and 8, these findings directly implicate roles for the poly(A) tail and PABP in first-round initiation mediated by the type I CBV3 IRES.
FIGURE 9.
PABP and poly(A) exert differential effects on 80S assembly depending on IRES identity. In vitro translation reactions containing 0.5mM cycloheximide were performed as in Figure 6 and then separated on 5%–20% linear sucrose gradients (see Fig. 5 for absorbance trace, indicating positions of ribosomal subunits and monosomes). Effects of Paip2 (A–C) and a poly(A) tail (D–F) on initiation are shown for HCV (A,D), EMCV (B,E), and CBV3 (C,F) reporter RNAs. Peaks formed by ribonucleoprotein complexes (RNP), 48S complexes, and 80S ribosome-RNA complexes are indicated.
DISCUSSION
The accumulated data presented here lend additional support to the significant role that PABP plays in translation control, but illuminate intriguing differences between RNA templates in their functional requirement for poly(A) and PABP. Each RNA examined displayed substantial reduction in overall translation efficiency due to poly(A) tail loss (Fig. 1B). For HCV, this result contrasts with a previous study (Svitkin et al. 2001) in which a 3′ poly(A) tract minimally enhanced translation of a monocistonic reporter. The source of this discrepancy might be traced to differences in reporter construct design: unpolyadenylated reporter RNAs characterized here possess 3′-UTR truncations that, at least in HeLa cytoplasmic extract, result in delayed polypeptide release from the ribosome and overall translation repression, a defect that is relieved by the poly(A) tail (Bradrick et al. 2006). Similar phenomena have been recently described in yeast and mammalian in vivo systems where mRNAs with truncated or short 3′-UTRs are poorly translated and exhibit association with heavy polysomes (Inada and Aiba 2005; Akimitsu et al. 2007), findings that suggest termination defect(s) for these RNAs.
Interestingly, contrasting effects of polyadenylation on RNA decay rates were observed as a function of 5′ identity (Fig. 2). The fact that capped and CBV3 transcripts were potently stabilized by the poly(A) tail, while EMCV and HCV reporters were not, suggests the presence of 5′–3′ interactions for the former RNAs that protect against cytoplasmic nuclease activity. Evidence for the existence of an RNP bridge that brings together ends of the poliovirus (a relative of CBV3) genome has been reported (Herold and Andino 2001). This RNP is based upon interaction of PABP located at the poly(A) tail with poly(rC)-binding protein 2 (PCBP2) bound to a structured element at the genomic 5′-end known as the cloverleaf (Andino et al. 1990). PCBP2–cloverleaf interaction itself has been previously shown to impede RNA degradation (Murray et al. 2001). Thus, while proposed to function in the initiation of viral RNA synthesis, putative 5′–3′ interactions involving PABP and PCBP2 may also enhance the integrity and translation of CBV3 and related viral genomes in the hostile cytoplasmic environment early during infection. Previous findings implicating PCBP2 in type I, but not type II, IRES function support this intriguing notion (Walter et al. 1999).
Though only minor effects on decay were observed for EMCV and HCV as a function of poly(A) status, it will be interesting to explore whether viral genomic regions outside of those present in subgenomic reporter constructs confer enhanced resistance to cellular nucleases for these viruses. For example, the EMCV IRES RNA predominantly used in translation studies lacks a significant portion of upstream 5′-UTR sequence that is not required for IRES activity (Jang and Wimmer 1990; Duke et al. 1992); the 5′-UTR fragment in this study corresponds to nucleotides 282–836 of the EMCV genome. Although upstream EMCV 5′-UTR sequence lacks an obvious cloverleaf domain at the 5′-end, alternative RNA elements might participate in long-range interactions that could impact RNA decay susceptibility.
Two distinct patterns of inhibition by Paip2 were observed in translation reactions (Fig. 3). The HCV IRES functions independently of eIF4G and was expectedly insensitive to PABP interference, in agreement with previous findings (Svitkin et al. 2001). Formation of 48S particles on the EMCV IRES using purified factors requires eIF4G (Pestova et al. 1996) and, in contrast to HCV, the EMCV genome terminates with a poly(A) tail that presumably binds PABP. The nearly identical effects of Paip2 on HCV and EMCV IRES-dependent translation were therefore unexpected. Given these findings, we suggest that, at least in vitro, EMCV IRES function does not require, or is minimally affected by, the canonical PABP–eIF4G interaction. CBV3 IRES- and cap-dependent translation were more Paip2 sensitive than other RNAs and displayed similar trends of inhibition. Additionally, translation of unpolyadenylated CBV3 RNA, though exceedingly weak to begin with, was moderately inhibited by Paip2, suggesting a possible trans-acting effect (i.e., poly(A)-independent) of PABP for this IRES. However, density gradient assays failed to detect an effect of Paip2 on unpolyadenylated CBV3 mRNA. While it is difficult to explain these findings, future investigation into the mechanisms by which PABP participates in translation may help to reconcile these observations.
Comprehensive analyses of ribosomal complexes by sucrose density gradient fractionation revealed differential effects of PABP and the poly(A) tail on viral IRES-mediated translation initiation (Figs. 6, 8, 9). Treatment with Paip2 or poly(A) tract deletion decreased CBV3 RNA association with ribosomal components, but affected other IRES-containing RNAs to lesser extents. This suggests that at least first-round initiation by HCV and EMCV IRESes does not require PABP or 5′–3′ communication involving a poly(A) tail in vitro. Indeed, even though CBV3 initiation was reproducibly depressed by Paip2 or poly(A) tail loss, the observed effects may be considered subtle compared with IRES manipulations that almost completely block assembly of 48 complexes and 80S monosomes (data not shown; Ji et al. 2004; Otto and Puglisi 2004). We hypothesize that viral IRESes possess intrinsic capacity to recruit ribosomes to the start codon in the absence of other sequences and, in the case of CBV3, PABP/poly(A) tail simply enhances rather than enables IRES function. Nevertheless, multiple observations including enhanced stability conferred by poly(A), increased sensitivity to Paip2 and abrogated initiation suggest that type I CBV3 IRES function relies more heavily on PABP than other IRESes studied here. We propose that CBV3 and related type I picornavirus IRESes are able to directly bind eIF4G early in infection to facilitate 43S initiation complex recruitment in a manner that is productively enhanced through PABP–eIF4G interaction. Alternatively, PABP may exert its trans-activity exclusively through interaction with IRES-bound PCBP2.
Does the poly(A) tail or PABP participate in translation at levels other than initiation? Despite apparently minimal impact on initiation by HCV and EMCV IRESes, Paip2 still repressed translation of these RNAs ∼20%. The magnitude of this effect is modest but, though speculative at present, could represent action of PABP at the level of termination. Poly(A) tail deletion also produced minor effects on efficiency of initiation for HCV and EMCV while substantially reducing overall RLuc synthesis, suggesting defects at the level of termination. Further investigation will be required to elucidate possible mechanisms for PABP and the poly(A) tail in translation processes that occur downstream of initiation. The HCV and EMCV IRESes may prove useful tools in such analyses, given their capacity to efficiently initiate independent of 3′-untranslated RNA elements.
MATERIALS AND METHODS
Plasmids and in vitro transcription
Construction of RLuc reporter plasmids containing the HCV and CBV3 IRESes has been reported previously (Bradrick et al. 2006; Dobrikova et al. 2006). The EMCV reporter was generated by insertion of nucleotides 282–836 (corresponding to NC_001479) (Duke et al. 1992) into vector prepared from the CBV3 plasmid using NotI and SacI. The β-globin construct encoding poly(A50) was assembled by insertion of the BsrGI–ClaI fragment from CBV3 reporter plasmid into pTNT-RLuc (Bradrick et al. 2006). For generation of plasmids encoding Paip2 and PABP, standard RT–PCR from HeLa cytoplasmic RNA was used to amplify entire ORFs for subsequent cloning into prokaryotic expression vectors. The sequences of forward and reverse primers, respectively, for Paip2 amplification were: 5′-cgtggatccaaagatccaagtcgcagcagtactagcccaagc-3′ and 5′-atgcggccgctcaaatatttccgtacttcaccccaggaac-3′. The sequences of forward and reverse primers, respectively, for PABP amplification were: 5′-atggctagcaaccccagtgcccccagctaccc-3′ and 5′-atccccgggaacagttggaacaccggtggcactgttaactgc-3′. The Paip2 ORF was cloned into the BamHI and NotI sites of pGEX4T-1 (Amersham) and the PABP ORF was inserted into the NheI and SmaI sites of pTYB2 (New England Biolabs). All clones were verified by sequence analysis. Reporter plasmids were linearized with XbaI or ClaI and used for in vitro transcription (Megascript, Ambion) to produce unpolyadenylated and polyadenlyated RNAs, respectively. β-globin leader-containing reporter RNAs were capped using guanylyltransferase (Ambion) according to the manufacturer's protocol, and IRES RNAs were left uncapped. RNAs were diluted to working concentration (30 ng/μL) and inspected for quality by agarose gel electrophoresis.
Recombinant proteins
Escherichia coli BL21 cells transformed with PABP or GST–Paip2 expression plasmids were induced with IPTG (0.5 mM) and cultured for 6 h at 30°C before harvesting and lysis by sonication. Untagged PABP protein was purified using the IMPACT-CN system (New England Biolabs) according to the manufacturer's protocol. GST–Paip2 protein was purified using a GSTrap FF column (Amersham). Both recombinant proteins were dialyzed against hypotonic buffer (10 mM HEPES at pH 7.5, 0.5 mM MgOAc2, 10 mM KOAc, 2 mM DTT). Recombinant GST–Paip2 impacted translation reactions identically to a previously described, nearly identical GST–Paip2 fusion protein (Svitkin et al. 2001).
In vitro translation and RNA stability assays
Extracts were prepared similarly to the method described by Bergamini et al. (2000). Briefly, HeLa S3 cells were grown in suspension to a density of 1 × 106 cell/mL, washed multiple times with ice-cold PBS, and resuspended in 1 vol of hypotonic buffer before dounce-mediated lysis. Cellular debris was removed by centrifugation, and supernatant (S10 extract) frozen in aliquots at –80°C. Before use, extract was adjusted to 1 mM CaOAc2 and treated with micrococcal nuclease (10 U/mL) for 10 min at room temperature. EGTA was then added to 2 mM final concentration and the extract was clarified by centrifugation. In vitro translation reactions (20 μL) were performed at 37°C for the indicated time intervals and contained the following: 40% extract, 3 ng/μL reporter RNA, 2.5 mM MgOAc2, 120 mM KOAc (40 mM for reactions with capped RNAs), 20 mM creatine phosphate, 0.1 mg/mL creatine kinase, 0.1 mM spermidine, 60 μM amino acids, 16 mM HEPES (pH 7.4), 0.8 mM ATP, and 0.1 mM GTP. Reactions were stopped with addition of EDTA (10 mM final concentration) and RLuc levels were measured by enzymatic assay (Promega). Reactions (50 μL) performed to assay RNA decay were assembled using 32P-body-labeled RNA transcripts. At the time points indicated, 10 μL aliquots were removed into trizol reagent (Invitrogen) for RNA extraction and subsequent analysis by denaturing 4% PAGE. Band intensities were determined using a PhosphorImager (Molecular Dynamics).
Sucrose density gradient analysis
Reactions (50 μL) programmed with radiolabeled RNA and containing 0.5 mM cycloheximide were incubated at 37°C for 5 (HCV and EMCV) or 10 (CBV3) min before addition of 0.4 mL ice-cold gradient buffer (Otto and Puglisi 2004). In 48S formation experiments GMPPNP (Sigma) was added to 1.8 mM final concentration. Diluted reactions were layered onto 5%–20% linear sucrose gradients and spun in an SW-41 rotor for 3.5 h at 35,000 rpm (3.7 h at 37,000 rpm for 48S experiments) before fractionation (Isco). Each fraction was subjected to liquid scintillation counting. Figures display representative experimental data from at least two independent replicates for each assay performed. Peak size comparisons were estimated by weighing paper under peaks using an analytical balance (Kahvejian et al. 2005).
ACKNOWLEDGMENTS
We thank Y. Svitkin for providing Paip2 expression plasmid and J. Keene for antibody to PABP. We also thank C. Nicchitta, M. Garcia-Blanco, and R. Wheeler Walters for helpful discussions. This work was supported by Public Health Service grants to S.S.B. (DK067781) and M.G. (CA124756).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.556107.
REFERENCES
- Ahlquist, P., Kaisberg, P. Determination of the length distribution of poly(A) at the 3′ terminus of the virion RNAs of EMC virus, poliovirus, rhinovirus, RAV-61 and CPMV and of mouse globin mRNA. Nucleic Acids Res. 1979;7:1195–1204. doi: 10.1093/nar/7.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimitsu, N., Tanaka, J., Pelletier, J. Translation of nonSTOP mRNA is repressed post-initiation in mammalian cells. EMBO J. 2007;26:2327–2328. doi: 10.1038/sj.emboj.7601679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, L., Lu, H.H., Wimmer, E. Polioviruses containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: Genetic hybrids and the expression of a foreign gene. Proc. Natl. Acad. Sci. 1994;91:1406–1410. doi: 10.1073/pnas.91.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani, N., Ganesan, R., Kervestin, S., Mangus, D.A., Ghosh, S., Jacobson, A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- Andino, R., Rieckhof, G.E., Baltimore, D. A functional ribonucleoprotein complex forms around the 5′-end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Bergamini, G., Preiss, T., Hentze, M.W. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA. 2000;6:1781–1790. doi: 10.1017/s1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, P., Peltz, S.W., Ross, J. The poly(A)–poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol. Cell. Biol. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradrick, S.S., Walters, R.W., Gromeier, M. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson, B., Couturier, A., Chabelskaya, S., Kiktev, D., Inge-Vechtomov, S., Philippe, M., Zhouravleva, G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI +] propagation. Mol. Cell. Biol. 2002;22:3301–3315. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikova, E.Y., Grisham, R.N., Kaiser, C., Lin, J., Gromeier, M. Competitive translation efficiency at the picornavirus type 1 internal ribosome entry site facilitated by viral cis and trans factors. J. Virol. 2006;80:3310–3321. doi: 10.1128/JVI.80.7.3310-3321.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke, G.M., Hoffman, M.A., Palmenberg, A.C. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J. Virol. 1992;66:1602–1609. doi: 10.1128/jvi.66.3.1602-1609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, L.P., Bagga, P.S., Wilusz, J. The poly(A) tail inhibits the assembly of a 3′ to 5′ exonuclease in an in vitro RNA stability system. Mol. Cell. Biol. 1997;19:4552–4560. doi: 10.1128/mcb.17.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromeier, M., Alexander, L., Wimmer, E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. 1996;93:2370–2375. doi: 10.1073/pnas.93.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen, C.U., Sarnow, P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Herold, J., Andino, R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey, J.W.B., Merrick, W.C. The pathway and mechanism of initiation of protein synthesis. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 33–38. [Google Scholar]
- Hoshino, S., Imai, M., Kobayashi, T., Uchida, N., Katada, T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- Imataka, H., Gradi, A., Sonenberg, N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada, T., Aiba, H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 2005;24:1584–1595. doi: 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, A. Poly(A) metabolism and translation: The closed loop model. In: Hershey J.W.B., et al., editors. Translational control. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1996. pp. 451–480. [Google Scholar]
- Jang, S.K. Internal initiation: IRES elements of picornaviruses and hepatitis C virus. Virus Res. 2006;119:2–15. doi: 10.1016/j.virusres.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Jang, S.K., Wimmer, E. Cap-independent translation of encephalomyocarditis virus RNA: Structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes & Dev. 1990;4:1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- Jang, S.K., Krausslich, H.G., Nicklin, M.J., Duke, G.M., Palmenberg, A.C., Wimmer, E. A segment of the 5′-nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, H., Fraser, C.S., Yu, Y., Leary, J., Doudna, J.A. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl. Acad. Sci. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian, A., Svitkin, Y.V., Sukarieh, R., M'Boutchou, M.N., Sonenberg, N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes & Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim, M.M., Svitkin, Y.V., Kahvejian, A., De Crescenzo, G., Costa-Mattioli, M., Sonenberg, N. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc. Natl. Acad. Sci. 2006;103:9494–9499. doi: 10.1073/pnas.0603701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour, K., Svitkin, Y.V., Craig, A.W., DeMaria, C.T., Deo, R.C., Burley, S.K., Sonenberg, N. Translational repression by a novel partner of human poly(A)-binding protein, Paip2. Mol. Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- Kisselev, L., Ehrenberg, M., Frolova, L. Termination of translation: Interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolykhalov, A.A., Feinstone, S.M., Rice, C.M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto, S., Saiz, M., de la Morena, D., Sobrino, F., Martinez-Salas, E. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Res. 2002;30:4398–4405. doi: 10.1093/nar/gkf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H.H., Wimmer, E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc. Natl. Acad. Sci. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus, D.A., Evans, M.C., Jacobson, A. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, Y.M., Borman, A.M., Paulous, S., Kean, K.M. Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation. Mol. Cell. Biol. 2001;21:4097–4109. doi: 10.1128/MCB.21.13.4097-4109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe, D., Jacobson, A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, K.E., Roberts, A.W., Barton, D.J. Poly(rC)-binding proteins mediate poliovirus mRNA stability. RNA. 2001;7:1126–1141. doi: 10.1017/s1355838201010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, G.A., Puglisi, J.D. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- Pelletier, J., Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Shatsky, I.N., Hellen, C.U. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Shatsky, I.N., Fletcher, S.P., Jackson, R.J., Hellen, C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes & Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, A.B., Davis, R.W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Searfoss, A., Dever, T.E., Wickner, R. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: Relations of Pab1p, eukaryotic translation initiation factor 5B (Fun12p), and Ski2p-Slh1p. Mol. Cell. Biol. 2001;21:4900–4908. doi: 10.1128/MCB.21.15.4900-4908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin, Y.V., Sonenberg, N. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol. Biol. 2004;257:155–170. doi: 10.1385/1-59259-750-5:155. [DOI] [PubMed] [Google Scholar]
- Svitkin, Y.V., Imataka, H., Khaleghpour, K., Kahvejian, A., Liebig, H.D., Sonenberg, N. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA. 2001;7:1743–1752. [PMC free article] [PubMed] [Google Scholar]
- Tarun, S.Z., Sachs, A.B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Thoma, C., Bergamini, G., Galy, B., Hundsdoerfer, P., Hentze, M.W. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol. Cell. 2004a;15:925–935. doi: 10.1016/j.molcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Thoma, C., Ostareck-Lederer, A., Hentze, M.W. A poly(A) tail-responsive in vitro system for cap- or IRES-driven translation from HeLa cells. Methods Mol. Biol. 2004b;257:171–180. doi: 10.1385/1-59259-750-5:171. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara, K., Iizuka, N., Kohara, M., Nomoto, A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, N., Hoshino, S., Imataka, H., Sonenberg, N., Katada, T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in cap/poly(A)-dependent translation. J. Biol. Chem. 2002;277:50286–50292. doi: 10.1074/jbc.M203029200. [DOI] [PubMed] [Google Scholar]
- Van Eden, M.E., Byrd, M.P., Sherrill, K.W., Lloyd, R.E. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, B.L., Nguyen, J.H., Ehrenfeld, E., Semler, B.L. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999;5:1570–1585. doi: 10.1017/s1355838299991483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E., Pestova, T.V., Hellen, C.U., Sarnow, P. Initiation of protein synthesis from the A-site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J., Gao, M., Jones, C.L., Wilusz, J., Peltz, S.W. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA. 2001a;7:1416–1424. [PMC free article] [PubMed] [Google Scholar]
- Wilusz, C.J., Wormington, M., Peltz, S.W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001b;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- Wimmer, E., Hellen, C.U., Cao, X. Genetics of poliovirus. Annu. Rev. Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]