Abstract
Gonadal function is critically dependant on regulated secretion of the gonadotropin hormones from anterior pituitary gonadotroph cells. Gonadotropin biosynthesis and release is triggered by the binding of hypothalamic GnRH to GnRH receptor expressed on the gonadotroph cell surface. The repertoire of regulatory molecules involved in this process are still being defined. We used the mouse LβT2 gonadotroph cell line, which expresses both gonadotropin hormones, as a model to investigate GnRH regulation of gene expression and differential display reverse transcription-polymerase chain reaction (RT-PCR) to identify and isolate hormonally induced changes. This approach identified Fanconi anemia a (Fanca), a gene implicated in DNA damage repair, as a differentially expressed transcript. Mutations in Fanca account for the majority of cases of Fanconi anemia (FA), a recessively inherited disease identified by congenital defects, bone marrow failure, infertility, and cancer susceptibility. We confirmed expression and hormonal regulation of Fanca mRNA by quantitative RT-PCR, which showed that GnRH induced a rapid, transient increase in Fanca mRNA. Fanca protein was also acutely upregulated after GnRH treatment of LβT2 cells. In addition, Fanca gene expression was confined to mature pituitary gonadotrophs and adult mouse pituitary and was not expressed in the immature αT3-1 gonadotroph cell line. Thus, this study extends the expression profile of Fanca into a highly specialized endocrine cell and demonstrates hormonal regulation of expression of the Fanca locus. We suggest that this regulatory mechanism may have a crucial role in the GnRH-response mechanism of mature gonadotrophs and perhaps the etiology of FA.
Keywords: anterior pituitary, gene regulation, gonadotropin-releasing hormone, mechanisms of hormone action, pituitary
INTRODUCTION
Reproduction requires regulated pulsatile release of the gonadotropin hormones, LH and FSH, from the gonadotroph cells of the anterior pituitary to stimulate gonadal function. The gonadotropin hormones are heterodimeric proteins, comprised of a common α subunit (αGSU) and a hormone-specific β subunit. Their biosynthesis and release is triggered by the binding of decapeptide GnRH to its cognate receptor (GnRHr), this stimulates second messenger signaling pathways, which, coupled with GnRHr number and turnover, combine to differentially regulate hormone biosynthesis [1-3]. Indeed, the pattern of GnRH administration is crucial for upregulating the mRNA levels of the constituent αGSU and β subunits that comprise LH and FSH, both in vivo [4, 5] and in vitro [6, 7].
Gonadotroph cell lines recapitulate the embryonic anterior pituitary temporal expression profile of gonadotropin genes [8-10]. Furthermore, because gonadotroph cells only comprise 10-15% of the cells in the anterior pituitary, cell lines afford greater manipulation of treatment conditions and provide a source of enriched cell-specific material. In this study, we used LβT2 cells, an immortalized gonadotroph cell line that is GnRH responsive, expresses GnRHr and all three gonadotropin subunits [9, 11, 12]. In addition, the second messenger signaling pathways required to transduce the GnRH signal are starting to be elucidated in these cells [13-16]. Thus, because LβT2 cells express all the features associated with mature pituitary gonadotrophs, they are a suitable cell model for investigating regulation of gene expression by GnRH.
To identify and isolate new and novel mRNAs that are differentially regulated by GnRH in gonadotrophs, which may be important for gonadotropin hormone biosynthesis, differential display (DD) reverse transcription-polymerase chain reaction (RT-PCR) [17] was performed on RNA extracted from untreated and GnRH-treated LβT2 cells. This approach identified that, among others, expression of Fanconi anemia a (Fanca) mRNA, was altered in response to GnRH treatment. Fanca is a member of a protein complex required for genome homeostasis [18, 19] and mutations in Fanca account for >60% of cases of Fanconi anemia (FA), an autosomal recessive inherited disorder. However, the pleiotropic phenotype of FA patients indicates that other cellular functions may also depend on the FA complex [20].
Therefore, we decided to investigate the expression profile of Fanca mRNA in detail and report that GnRH acutely upregulates the expression of both Fanca mRNA and protein. This suggests that Fanca may have a regulatory role in gonadotroph cells, and this is the first report of distinct hormonal regulation of this gene.
MATERIALS AND METHODS
Cell Culture
LβT2 and αT3-1 cells (obtained from P. Mellon, San Diego, CA) were cultured in DMEM (Sigma, Dorset, UK) supplemented with 10% FCS (Sigma) and 1% penicillin/streptomycin (Sigma). All gonadotroph cell culture plasticware was coated with a 1:30 dilution of Matrigel (Becton Dickinson Labware, Oxford, UK) in PBS (Sigma). Cells were treated with 1 μM of native GnRH (Peninsula, St. Helens, UK) for 15 min. This concentration is known to stimulate high levels of LH and FSH β subunit gene expression [21, 22], with an interpulse interval of 75 min, for either 3 or 6 pulses; fresh media was added and the cells harvested or cells were treated with one 15-min pulse and harvested 1, 2, or 4 h post-GnRH treatment into RNAzol B (AMS Biotechnology, Abingdon, UK) and stored at −70°C. Mouse L-cells and HeLa cells (ECACC, CAMR, Porton Down, UK) were passaged in growth media as described above, and all cells were grown in a humidified 5% CO2 atmosphere at 37°C.
Differential Display RT-PCR
RNA was extracted as per the manufacturer’s protocol and differential display was performed as described [17], with further modifications [23]. Briefly, cDNA was generated using a First Strand cDNA synthesis kit (Amersham Pharmacia Biotech, Little Chalfont, UK) and degenerate oligo(dT) primers (primers have 12 thymidine bases and a combination of two random bases TTTTTTTTTTTTVN where V and N = A, G, or C). The oligo(dT) primers were pooled into 3 × 24 μM mixes; T12VA, T12VC, and T12VG were used to prime first-strand cDNA subpopulations. DD-RT-PCR was performed using oligo(dT) and random 10-mer primers (Sigma-Genosys, Pampisford, UK), which are listed in Table 1. The 20-μl reaction mix contained 5 μl of cDNA (pipetted from a 1:131-μl dilution of the first-strand reaction), 0.5 μM random primer, 3 mM MgCl2, 2.4 μM T12VC, 2 μM dNTP’s (Amersham Pharmacia Biotech), 1 μl 35S-dATP (1000 Ci/mmol, Amersham Pharmacia Biotech), 0.3 μl AGS Gold Taq polymerase, 2 μl each reaction buffer and enhancer (Hybaid, Ashford, UK), and 1.2 μl H2O. The PCR reaction conditions were as described [23]. A 4-μl aliquot was loaded on a 6% acrylamide gel (HR-1000; Beckman Coulter UK Ltd, High Wycombe, UK) and electrophoresed on a GenomyxLR DNA analyzer (Beckman Coulter) at 2700 V for 2 h 15 min at 50°C. The gel was transferred to 3MM paper (Whatman, Fisher Scientific, Loughborough, UK) dried, and bands were visualized using BiomaxMR autoradiographic film (Amersham Pharmacia Biotech), excised and rehydrated in 150 μl low TE (10 mM Tris HCI, 0.1 mM EDTA, pH 7.4). The DNA was eluted at 100°C, ethanol precipitated, and resuspended in 10 μl of low TE. To facilitate subcloning, this was combined with a 40-μl reaction mix (3 mM MgCl2, 0.3 μl AGS Gold, 4 μl buffer, 4 μl Enhancer, 0.8 mM dNTP, 1.25 μM T12VN, 1.25 μM random primer, and 11 μl H2O) and amplified by PCR before splitting into four 10-μl aliquots and reamplified. The Fanca fragment was cloned into pT7-Blue (Novagen, CN Biosciences Ltd., Beeston, UK) using the Perfectly Blunt cloning kit (Novagen) and sequenced.
TABLE 1.
Differential display (DD) RT-PCR random 10-mer primers.
| DD-RT-PCR random 10-mer primer | DNA sequence |
|---|---|
| TK | 5′-CTTGATTGCC-3′ |
| A1 | 5′-ACAGAGCACA-3′ |
| A2 | 5′-ACGTATCCAG-3′ |
| MAX1 | 5′-GAGCATATCC-3′ |
| MAX2 | 5′-CACAGCTTGC-3′ |
| MAX3 | 5′-CCACAGAGTA-3′ |
| R21 | 5′-AGTCAGCCAC-3′ |
| R5 | 5′-AGGACCCTGG-3′ |
Northern blotting, Semiquantitative and Quantitative RT-PCR
Total RNA (40 μg) was fractionated, Northern blotted, and probed, then quantified as described [24]. Radiolabeled probes used corresponded to either the 5′ region (exons 1-11) or the 3′ region (exons 42-43) of Fanca cDNA.
RT-PCR was performed by reverse transcribing 1 μg RNA using First Strand cDNA synthesis kit (Amersham Pharmacia Biotech) and 1 μl was added to a 25-μl reaction containing 2 μM upstream and downstream primers and 10 μl Extensor High Fidelity Master Mix Buffer 2 (AB Gene, Epsom, UK). Standard PCR conditions were used and were identical for Fanca and penta zinc-finger protein 276 (Zfp276). For details of primers and PCR fragments, see Table 2. PCR products were verified by sequencing individual clones after ligation into TA-cloning vectors (Invitrogen, UK) using ABI big dye terminator reagents (ABI, Warrington, UK). DNA fragments were visualized on an ethidium bromide-stained agarose gel, which for Fanca was Southern blotted and probed with a radiolabeled probe (exons 7-17) before exposing to x-ray film.
TABLE 2.
Specific primers were used in PCR reactions to amplify first strand Fanca, Zfp276, and B-2-microglobulin cDNA.
| Specific primers* | DNA sequence | Location | PCR product (base pairs) |
|---|---|---|---|
| Fanca s | 5′-CTGTGTGAGCAGATAGGC-3′ | Exon 7 (640 bp) | 979 |
| Fanca as | 5′-TCACGCTCGGCAATGTCCC-3′ | Exon 17 (1619 bp) | |
| Fanca s | 5′-CAGCATGGTCACTGCGTTCC-3′ | Exon 14 (1263 bp) | 450 |
| Fanca as | 5′-CCTGAATATGCTGGCCTCCA-3′ | Exon 18 (1713 bp) | |
| Fanca s | 5′-GTGGTGGAGACCTGGAAGA-3′ | Exon 30 (2900 bp) | 211 |
| Fanca as | 5′-CGGCGTAGAACAGCCATG-3′ | Exon 32 (3111 bp) | |
| Fanca s | 5′-GCACTTTGCGTGGAGAGG-3′ | Exon 37 (3666 bp) | 129 |
| Fanca as | 5′-CAGGTAGGACGAGAGTAGAC-3′ | Exon 38 (3795 bp) | |
| Zfp276 s | 5′-CACTGTCCTCTGAGTACTGC-3′ | 5′ ATG 68 bp | 1238 |
| Zfp276 as | 5′-CGTCACCTGCTGAGTTCAAG-3′ | 3′ ATG 1170 bp | |
| B2m s | 5′-ATGGCTCGCTCGGTGACCCTGGT-3′ | Exon 1 (ATG) | 102 |
| B2m as | 5′-TGTTCGGCTTCCCATTCTCC-3′ | Exon 1 (102 bp) |
Primer annealing sites are expressed relative to the translational start site (ATG) in Fanca, Zfp276, and B-2-microglobulin (B2m). s, Sense; as, antisense; bp, base pairs.
Quantitative RT-PCR was performed for Fanca mRNA using a LightCycler Instrument (Roche Diagnostics, Lewes, East Sussex, UK), FastStart DNA Master SYBR Green I (Roche Diagnostics), and mouse beta-2-microglobulin (B2m) as an internal control. Specific primers are shown in Table 2. Each LightCycler reaction consisted of 5.8 μl dH2O, 1.2 μl MgCl2 (4 mM), 1 μl Primer Pair Mix (25 μM each), 1 μl LightCycler DNA Master SYBR Green I (Roche Diagnostics), and 1 μl cDNA. The LightCycler program used for Fanca and B2m real-time PCR was as follows: one cycle of 95°C for 10 min, 60 cycles of 95°C for 5 sec, 57°C for 5 sec, and 72°C for 15 sec, and a melting curve program (57-95°C). A standard curve of Fanca and B2m expression was determined using serially diluted cDNA made from mouse L-cell RNA. Results are expressed as arbitrary units of Fanca mRNA expression normalized against B2m mRNA expression levels and are a mean of three separate experiments performed in duplicate.
Western Blotting Analysis
LβT2 cells were left untreated or were treated with GnRH and harvested 2, 4, or 6 h later. Whole-cell extracts were prepared by washing the cell monolayer in ice-cold PBS with Complete (Roche Diagnostics) protease inhibitors, before scraping into a 1.5-ml centrifuge tube, which was frozen on dry ice. Total cellular protein was liberated by three rapid freeze-thaw cycles, and cellular debris was cleared by centrifugation at 4°C for 5 min at 20 000 × g. Protein concentration was determined using Bio-Rad protein assay reagent (Bio-Rad, Hemel Hempstead, UK), 50 μg of whole-cell extract was boiled in 1× loading buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.01% Bromophenol Blue, 1% β-mercaptoethanol), fractionated on a 6% SDS-PAGE gel, and electroblotted overnight at 25 V onto Immobilon-P (Millipore, Watford, UK) in 1× TGS/20% methanol (TGS; 25 mM Tris, pH 8.8, 250 mM glycine, 0.1% SDS). Blots were probed as described [5] with anti-mouse Fanca antisera raised to amino acids 1-271 (kind gift from Dr. Fre Arwert, VU University Medical Center, Amsterdam, The Netherlands) at a concentration of 1:1500, then stripped in 2% SDS, 50 mM Tris, pH 6.8, 0.7% β-mercaptoethanol at 55°C for 30 min before washing and reprobing with anti-mouse β-tubulin antisera at a concentration of 1:500 (Santa Cruz Biotechnology Inc., Santa Cruz, CA).
Bioinformatics and Statistics
All bioinformatic comparisons were made using Wisconsin Package Version 10.3 (Accelrys Inc., San Diego, CA) housed on the Medical Research Council’s Human Genome Mapping Project server (MRC, HGMP, Hinxton, UK). Statistical differences were determined using one-way analysis of variance (ANOVA), with P < 0.05 deemed significant.
RESULTS
Differential Display RT-PCR Analysis of GnRH-Treated LβT2 Cells
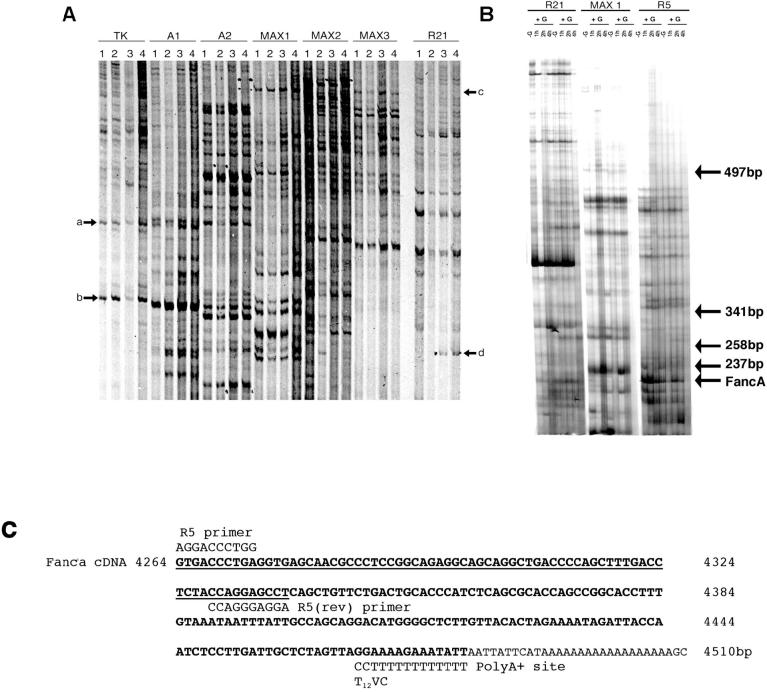
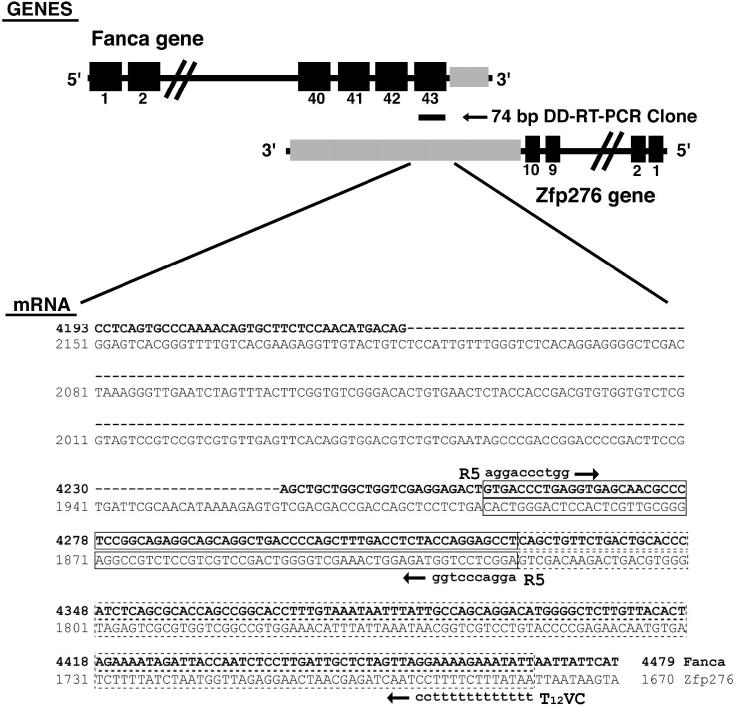
LβT2 cells were grown in vitro on Matrigel basement membrane, which contains collagen, other extracellular basement membrane proteins, growth factors, metalloproteinases, and factors that promote cell adherence. These factors may upregulate gene expression independent of GnRH, so to test this, two different treatment regimes were designed. In regime 1, cells were grown on uncoated or Matrigel-coated flasks, while for regime 2, cells were grown on Matrigel and left untreated or treated with GnRH. RNA was isolated and subjected to DD-RT-PCR analysis (Fig. 1A). There was no visible alteration in DD-RT-PCR generated transcripts between cells grown on uncoated or Matrigel-coated flasks, while addition of GnRH clearly affected transcript levels. This treatment regime has previously been used to regulate expression and trigger pulsatile release of LH from these cells [9]. However, we found that a short 15-min pulse of GnRH induced rapid changes in gene expression (Fig. 1B). The identified 216-base pair (bp) cDNA was isolated, subjected to a further two rounds of PCR amplification, cloned, and sequenced. Bioinformatic analysis of the 74-bp DNA sequence obtained produced an identical match to exon 43 of Fanca. The difference in size between the identified 216-bp DD-RT-PCR product and the amplified 74-bp cDNA clone was due to internal priming during the second round of PCR amplification, and the two sets of priming sites are shown in Figure 1C. This was a common feature (unpublished observation) due to the inherent redundancy of the PCR amplification and may reflect the poor processivity of some Taq polymerases and preferential cloning of small DNA fragments. The 216-bp DD-RT-PCR product visible on the gel was consistent with priming of Fanca mRNA by a T12VC-anchored primer (Fig. 1C). Bioinformatics also highlighted that the 74-bp cDNA clone obtained could possibly correspond to pentazinc finger protein 276 (Zfp276) because Zfp276 mRNA is encoded on the opposite DNA strand from Fanca and overlaps the Fanca gene locus at the 3′ end [25]. The orientation of these genes and region of DNA overlap between them is indicated in Figure 2. We concluded that Zfp276 was unlikely to be the original DD-RT-PCR clone because the mRNA polyA+ addition site and likely T12VC priming site maps a considerable distance downstream and there was no suitable T12VC annealing site within 200 bp of immediate flanking DNA sequence. However, because two transcripts, Fanca and Zfp276, matched the cloned DD-RT-PCR DNA fragment, additional expression analysis was needed to confirm that Fanca was differentially expressed.
FIG. 1.
Differential display RT-PCR analysis of GnRH-regulated transcripts isolated from LβT2 cells. A) Matrigel basement membrane was excluded as an inducer of gene expression in LβT2 gonadotroph cells while GnRH was shown to upregulate transcripts. Cells were grown without Matrigel (1) or were cultured on Matrigel (2, 3, and 4) and treated with 3 (3) or 6 (4) 15-min pulses of GnRH with an interpulse interval of 75 min. RNA was extracted and subjected to differential display (DD) RT-PCR using primers stated. Matrigel did not alter transcript expression (arrows a and b), while numerous transcripts altered after GnRH treatment (arrows c and d). B) In two separate experiments, LβT2 cells were left untreated (−G) or were treated with 1 × 15-min pulse of GnRH (+G), RNA was harvested (−G, 1, 2, and 4 h) and subjected to DD-RT-PCR. First-strand cDNA was generated using the T12VC downstream primer, then amplified by PCR using primers R21, MAX1, or R5. Arrow denotes location of Fanca. The location of DNA size markers, indicated as bp, are also shown. C) Bioinformatic line-up depicting the region of homology, shown underlined, between the cloned Fanca DD-RT-PCR product and mouse Fanca cDNA nucleotide sequence, and the likely internal R5 priming site. The location of DD-RT-PCR primers R5 and T12VC that generated the original 216-bp DD-RT-PCR product are also indicated.
FIG. 2.
Schematic representation of the Fanca locus showing the location of the differential display RT-PCR clone. Exons 40-43 of Fanca, shown as black boxes, overlap with the 3′ untranslated region (UTR) of Zfp276, shown in grey. The location of the cloned 74-bp differential display (DD) RT-PCR clone is indicated. Beneath the schematic, the regions of nucleotide identity between Fanca mRNA and Zfp276 mRNA have been aligned, Fanca mRNA sequence is shown in bold type, and spliced intronic sequence has been omitted and replaced with a dashed line, with Zfp276 mRNA in normal typeface. The amplified 74-bp DD-RT-PCR clone has been boxed with a solid line, and the original 216-bp DD-RT-PCR clone is indicated as an extension of the boxed region with dashed lines. The amplification primers R5 and T12VC are also shown on the corresponding Fanca sequence. No suitable priming sites were identified by bioinformatic analysis for the T12VC-anchored primer on the Zfp276 3′ UTR sequence.
Fanca mRNA Is Differentially Expressed in LβT2 Cells
Northern blotting of total RNA extracted from GnRH-treated LβT2 cells and probing with 5′ and 3′ fragments of mouse Fanca cDNA (Fig. 3A), confirmed that a full-length 4.5-kilobase (kb) transcript was expressed. The 5′ Fanca probe corresponded to 1-965 bp of the cDNA, which encodes exons 1-11. The 3′ Fanca probe corresponded to 4231-4500 bp of the mouse cDNA that encodes exons 42-43 and should cross-hybridize with the Zfp276 transcript that overlaps this coding region. Indeed, a large 4.5-kb Fanca transcript and a small 3.1-kb transcript corresponding to the reported size of Zfp276 mRNA [25] were detected. The Northern blot was stripped and reprobed with a ribosomal probe specific for 18s, which was used to control for loading (Fig. 3A).
FIG. 3.
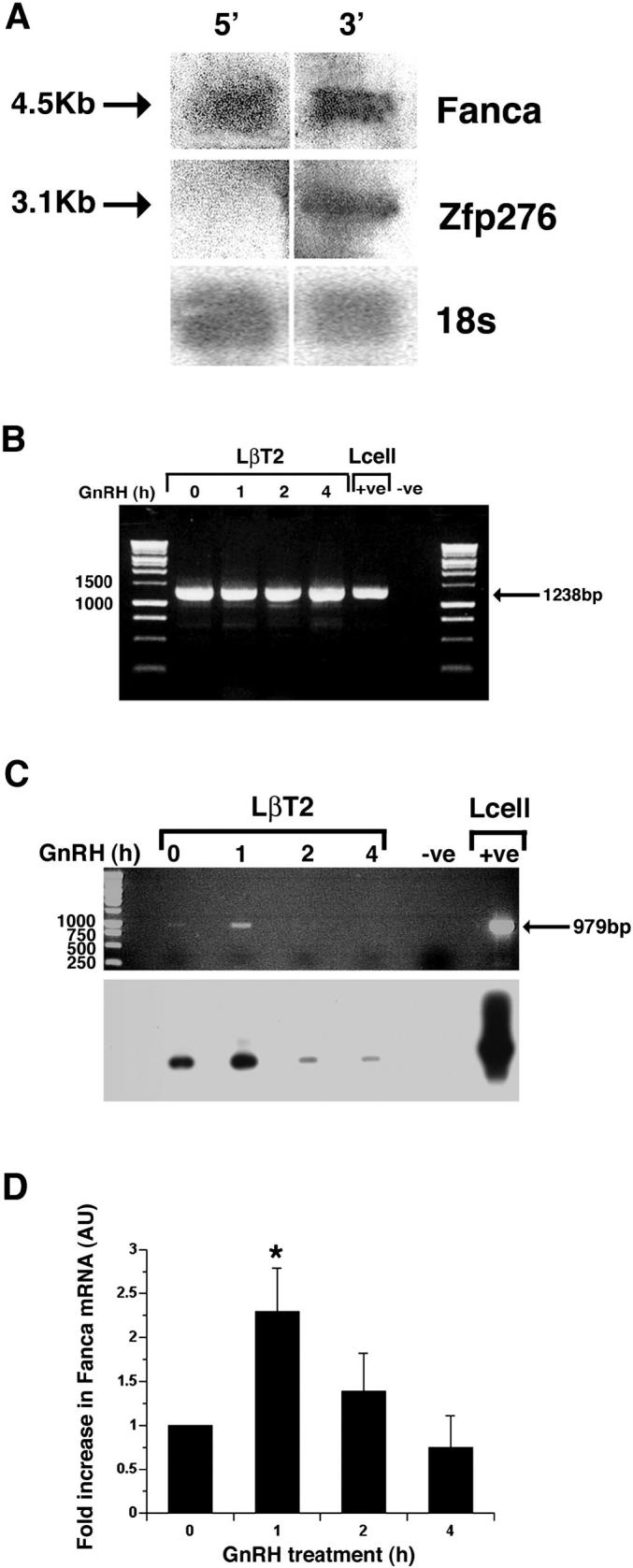
Characterization and quantification of Fanca and Zfp276 mRNA after GnRH treatment. A) Confirmation of expression of full-length 4.5-kb Fanca and 3.1-kb Zfp276 mRNA transcripts in LβT2 cells. Total RNA from LβT2 cells was fractionated on a formaldehyde gel, Northern blotted, and probed with radiolabeled probes corresponding to either the 5′ region (exons 1-11) or 3′ region (exons 42-43) of Fanca. The blot was then stripped and reprobed with an 18s probe. Specific Fanca, Zfp276, and 18s bands are indicated by arrows. B) Semiquantitative RT-PCR analysis of Zfp276 expression. LβT2 cells were left untreated (0) or treated with 1 × 15-min pulse of GnRH, then harvested 1, 2, and 4 h later, RNA was extracted, and first-strand cDNA made. PCR was performed to amplify full-length Zfp276 (1238 bp) and the products were visualized on an ethidium bromide-stained agarose gel. One specific 1238-bp PCR product was visible at all time points. L-cell cDNA was included as a positive control. An arrow denotes the 1238-bp PCR product, and DNA size markers are labeled. C) Semiquantitative RT-PCR analysis was performed for Fanca expression in LβT2 cells that were left untreated (0) or treated with 1 pulse of GnRH; harvested 1, 2, and 4 h later; RNA extracted, and cDNA made. Ethidium bromide staining identified one PCR product amplified from mRNA harvested from the 1-h time point. L-cell cDNA was included as a positive control. An arrow denotes the 979-bp PCR product, and size markers are labeled. Southern blotting analysis of the agarose gel with a 5′ Fanca probe confirmed that the Fanca PCR product was specific and present at all time points. D) LightCycler quantitative RT-PCR analysis of Fanca mRNA extracted from LβT2 cells either left untreated or treated with GnRH and harvested 1, 2, and 4 h later detected a consistent 2-fold increase in Fanca mRNA harvested 1 h after treatment. Results are shown as arbitrary units (AU) of Fanca mRNA normalized against the levels of internal control B-2-microglobulin (B2m) mRNA. This experiment was performed in duplicate and repeated three times. *P < 0.05 was determined as being significant by ANOVA one-way analysis of variance.
Although nonquantitative, there appeared to be no difference in expression levels of Zfp276 mRNA amplified by RT-PCR in untreated and treated cells (Fig. 3B). Interestingly, semiquantitative RT-PCR amplification of Fanca mRNA suggested that Fanca expression increased 1 h after GnRH treatment. Specific amplification of a 979-bp Fanca PCR product was confirmed by subsequent Southern blotting analysis, which also identified lower levels of expression in untreated, and a rapid reduction 2 and 4 h after hormone treatment (Fig. 3C).
Thus, quantitative RT-PCR was used to measure the rapid increase in Fanca mRNA after GnRH treatment. In this assay, Fanca mRNA clearly increased 2-fold 1 h after GnRH treatment (P < 0.05) and returned to unstimulated levels by 4 h (Fig. 3D). Taken together, these results indicate that Fanca, but not Zfp276, mRNA is acutely regulated by GnRH in LβT2 cells.
Analysis of Fanca Protein Expression
The rapid upregulation of Fanca mRNA may not be mirrored by an increase in protein, so Western blotting analysis was used to quantify Fanca protein levels in LβT2 cell protein extracts. First, protein extracts were made from untreated cells and from cells harvested 2 h after addition of GnRH, and were fractionated on a 6% SDS-PAGE gel, blotted onto polyvinylidene fluoride (PVDF) and incubated with Fanca antisera specific for amino acids 1-271 [26]. A specific 160-kDa band, corresponding to the expected size of Fanca protein, was detected in untreated and GnRH-treated extracts (Fig. 4A). Thus, the increase in levels of Fanca protein was measured over a longer time period following GnRH treatment and quantified (Fig. 4B). Again, Fanca protein levels increased 2-fold after addition of GnRH for 2 h (P < 0.001). This 2-fold increase was still evident 4 h after treatment with GnRH (P < 0.001), but by 6 h, Fanca protein levels had begun to fall (P < 0.05). This shows that a rapid increase in Fanca mRNA (Fig. 3D) is followed by a 2-4-h sustained increase in Fanca protein.
FIG. 4.
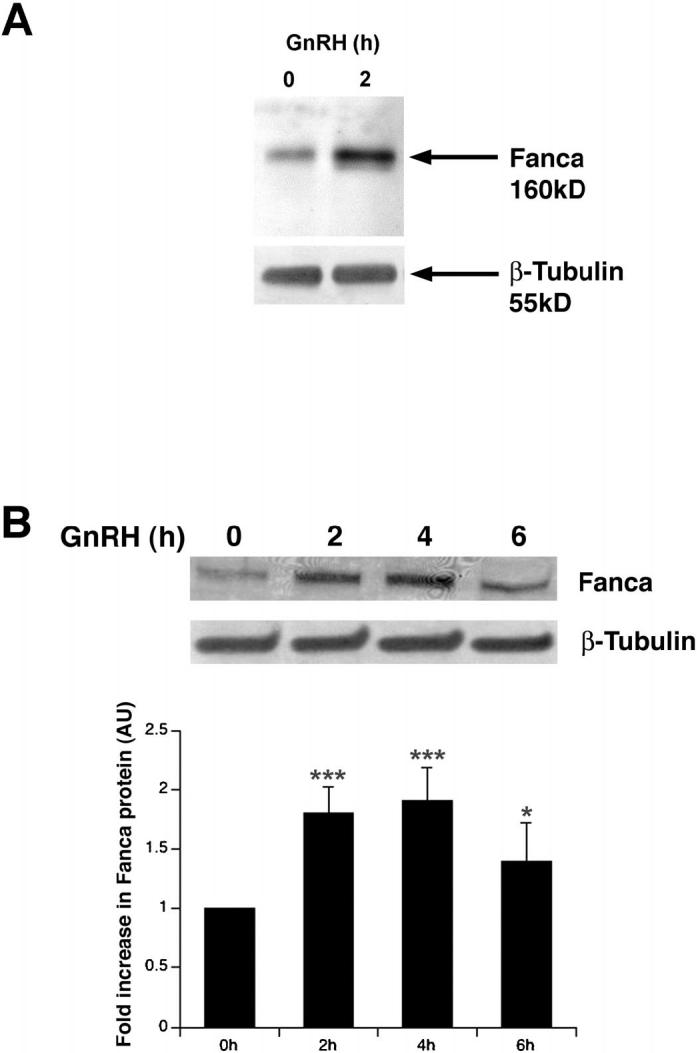
Western blotting analysis of Fanca protein. A) Western blotting analysis of protein extracts from untreated, 0, and extracts harvested 2 h after GnRH treatment identified Fanca protein. Cellular protein was fractionated, transferred to PVDF, and probed with anti-mouse Fanca antisera before being stripped and reprobed with anti-mouse β-tubulin. Arrows indicate the 160-kDa Fanca and 55-kDa β-tubulin proteins. B) Western blotting analysis of whole-cell protein extracts from untreated (0) and hormone-treated cells harvested 2, 4, and 6 h later. Blots were probed as above in A. ANOVA one-way analysis of variance determined that the increases in Fanca protein levels after hormone treatment were significant: ***P < 0.001; ***P < 0.001; *P < 0.05.
In Vivo and Temporal Expression Profile of Fanca mRNA
We next determined if Fanca was expressed in the pituitary in vivo. Mouse pituitary RNA was extracted, reverse transcribed, and cDNA amplified using specific primers to Fanca. DNA bands corresponding to exons 7-18, 14-18, and 30-32 were identified (Fig. 5A). This analysis confirmed pituitary expression of Fanca in adult mice. Because LβT2 cells are derived from embryonic Day 16.5 mouse pituitaries, we also investigated if Fanca mRNA was expressed in a different immature gonadotroph cell line (Fig. 5B). As expected, Fanca was expressed in LβT2 cells, HeLa, and L cells, but no corresponding PCR product was detected in immature precursor αT3-1 gonadotroph cells.
FIG. 5.
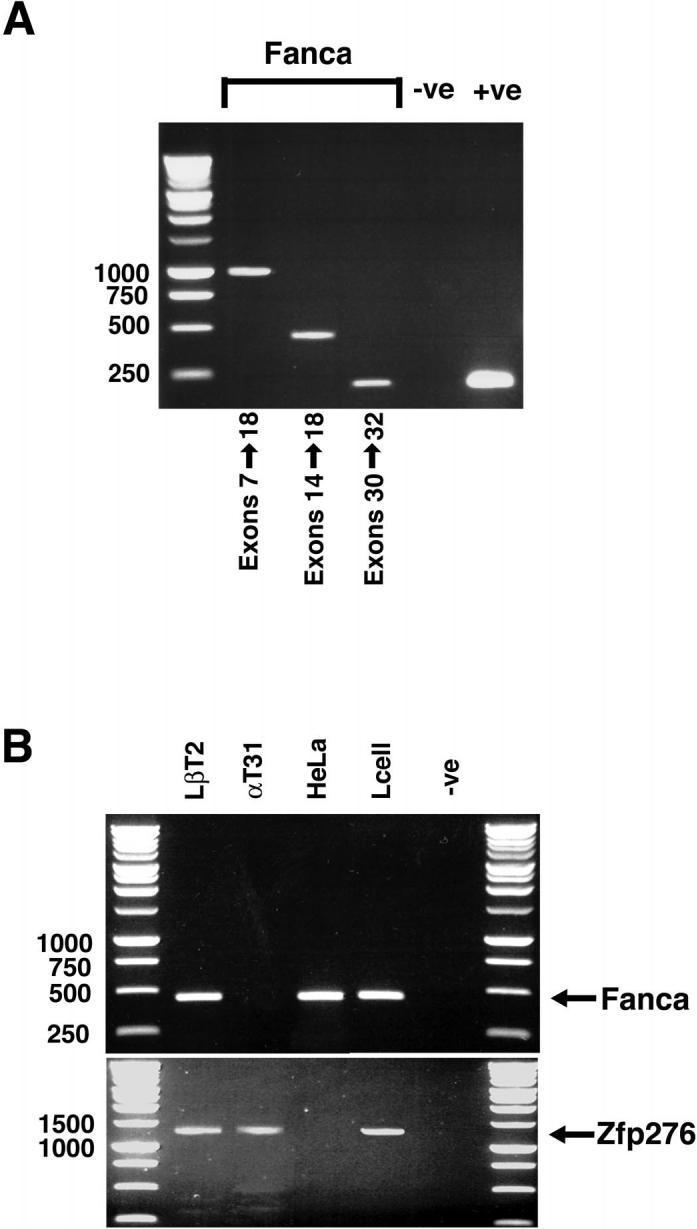
Fanca is expressed in adult mouse pituitary. A) RT-PCR analysis of RNA extracted from adult mouse pituitary was performed using primers that amplified exons 7-18, exons 14-18, and exons 30-32 of mouse Fanca. Ethidium bromide staining identified PCR products for all regions amplified. A control GAPDH PCR product was also amplified, confirming the integrity of the mouse pituitary cDNA. B) RNA was extracted from LβT2, αT3-1, HeLa, and L cells; reverse transcribed; and first-strand cDNA was analyzed by PCR for expression of Fanca. The PCR primers corresponded to exons 7-18 of mouse Fanca cDNA and ethidium bromide staining identified a 450-bp product in LβT2, HeLa, and L cells. No Fanca expression was detectable when amplifying αT3-1 cDNA. In this experiment, expression of Zfp276 was used as an internal control to check PCR conditions and RNA integrity. A specific 1238-bp PCR product, corresponding to Zfp276, was amplified from LβT2, αT3-1, and L cell first-strand cDNA.
DISCUSSION
Murine Fanca mRNA has a predicted size of 4503 bp, which encodes a 160-kDa protein [27, 28]. Fanca mRNA is relatively highly expressed in lymphoid tissues, testes, and ovary in adult mice and is activated as early as Embryonic Day 7, largely in cells of epithelial origin [27, 28]. This is the first report of hormonal regulation of Fanca and of Fanca expression in a specialized endocrine cell. Although there have been reports of smaller Fanca mRNA molecules being expressed in brain [27], none of these were localized to the pituitary, and they may correspond to the recently identified Zfp276 gene [25], because in our hands, neither Northern nor RT-PCR analysis indicated any variation in size of Fanca mRNA in LβT2 cells.
Because DD-RT-PCR amplifies the transcriptome, low-abundance messages are represented with no preselection bias from the user [17, 29]. This approach was particularly suited toward identification and isolation of Fanca cDNA. Differential GnRH regulation of Fanca mRNA was confirmed by a combination of semiquantitative and quantitative RT-PCR. However, the DD-RT-PCR technique does generate high numbers of false positives, and a number of strategies have been suggested to eliminate this [23, 30, 31]. These were taken into account during the design of our experiments and briefly include use of a time course, so differences were easily recognized, RNA extraction and cDNA synthesis was done simultaneously, and samples from different experiments were electrophoresed in duplicate to ensure repeatability. Thus, we established that, although Fanca mRNA was expressed at very low levels in the LβT2 cell line, GnRH regulation was still rapid and transient, with mRNA returning to unstimulated levels after 2 h. In contrast, when using microarray analysis, the design of the cDNA microarray determines which transcripts are identified. Although Fanca was omitted from the microarray, rapid transient increases in transcripts were measured in response to GnRH treatment, most corresponded to immediate early gene products that had returned to baseline levels by 3 h post-GnRH treatment, and early growth factor-1 (Egr-1) and c-Jun were induced over a wide range of GnRH concentrations (1 nM up to 1 μM) [32, 33]. Because we and others all measure significant increases in gene expression in LβT2 cells after one treatment with a pharmacological dose of GnRH, this indicates that, although the concentration of GnRH is important, the timing between GnRH pulses is also a critical factor. It is not clear if the same changes in gene expression would be induced with a pulsatile GnRH treatment regime as in the normal physiological state, although our original experiment did address this by using 15-min pulses separated by an interpulse interval of 75 min (Fig. 1A), we found it was not ideally suited to isolation of differentially expressed transcripts and that a shorter time course of induction of gene expression was preferable.
The rapid, transient increase in Fanca mRNA levels measured in response to GnRH was followed by a similar, but longer, increase in Fanca protein levels. This suggests that levels of Fanca mRNA are tightly controlled, and we hypothesize that hormone treatment either increases Fanca gene transcription and/or stabilizes Fanca mRNA. Measuring steady-state mRNA levels by quantitative RT-PCR does not distinguish between these two possibilities. Our observation that Fanca mRNA is highly regulated in a mature GnRH-responsive gonadotroph cell line, an endocrine tissue, may explain why researchers had difficulties in detecting Fanca gene expression by in situ hybridization in embryonic mouse testes, but did detect expression in adult testes [27, 28, 34]. In keeping with the rapid, short-lived peak in Fanca mRNA, we consistently found that hormonal stimulation also increased levels of Fanca protein. Fanca protein increased 2 h after treatment with GnRH, but persisted for longer, only starting to decline 6 h after treatment. Because we used whole-cell extracts in this study, we have yet to analyze if this increase in protein also results in a recompartmentalization of Fanca within the cell in response to hormone, but prior treatment of cells with cycloheximide, an inhibitor of translation, blocks the GnRH-induced increase in Fanca protein levels (unpublished results), suggesting that the increase in Fanca protein is indeed due to de novo translation. The hormonally induced increase in Fanca protein suggests it may be a component of a rapid response mechanism in these cells. Indeed, the action of Fanca in immune cells is upstream of the immediate early response genes [35], suggesting that Fanca may act as part of a signal-transduction cascade. We and others have noted that expression of Fanca protein is low because detection requires either reasonable amounts of starting material [27] or an enriched population of cells, indicating that relatively small changes in protein expression could have a large impact. Interestingly, expression levels and posttranslational modification are critically important for many proteins involved in signal transduction [36].
The hormonal regulation of expression and temporal pattern of Fanca gene activation indicates that this molecule may be important for mature gonadotroph cell function. While the existence of a complementary Zfp276 transcript complicated our analysis, especially because it was coexpressed in LβT2 cells, there was no clear evidence to indicate that Zfp276 mRNA was the original DD-RT-PCR fragment or that it was hormonally regulated. Furthermore, although Zfp276 mRNA was expressed, neither Fanca mRNA or protein (data not shown) was detected in αT3-1 cells, which are a GnRH-responsive gonadotroph cell line derived from embryonic Day 13.5 pituitaries, that exclusively express αGSU, but not LH or FSH β subunit [10, 37]. This indicates that Fanca gene expression is activated late in pituitary development because LβT2 cells are derived from embryonic Day 16.5 pituitaries, and indeed, we confirmed that Fanca was expressed in adult pituitary.
The pleiotropic nature of the FA syndrome, which in humans is an autosomal recessive disorder characterized by bone marrow failure, aplastic anemia, and variable predisposition to cancers of the gynecologic system among other clinical features, including infertility [18, 38, 39], has made it difficult to ascribe particular phenotypic features to mutations in any particular region of Fanca or any of the other Fanconi complementation groups [40, 41]. Targeted disruption of Fanca in mice has reproduced some of the associated FA phenotypes described above, but the most consistent of these appears to be a severe reduction in fertility [34, 42, 43]. Furthermore, although the pituitary was not examined, the testes were identified as a major site of Fanca gene expression, and these mice developed ovarian granulosa cell tumors [34], a phenotype also known to be consistent with elevated plasma levels of pituitary LH [44]. Elevated gonadotropin levels have also been reported in FA patients [45], and there is evidence that FA impacts on reproduction [46, 47]. Taken together, these observations suggest a role for Fanca in reproduction, especially gonadal function, but how this impacts on the pituitary is under further investigation.
There may be a link between FA and development of pituitary neoplasms because pituitary tumors develop through various mechanisms [48]. FA is a relatively rare disease, while the incidence of pituitary adenoma within the general population is high (∼20%), and gonadotroph cell neoplasms account for ∼35% of these [49]. However, a specific subset of pituitary tumors (<2%) are caused by mutations in the MEN1 gene, which encodes the transcriptional repressor MENIN [50] and interestingly, MENIN has recently been shown to interact with FANCD2, the downstream target of the FA complex [51].
In addition, a number of FA genes have been implicated in ovarian tumorigenesis in man and mouse [52, 53] and Fanca is required for gonadal function [34]. GnRH, acting through its receptor, is a key autocrine/paracrine regulator of ovarian and testicular function [54] and has a role in development of GnRH-responsive ovarian, breast, and prostate cancer [55]. This evidence and the prevalence of endocrinopathies in patients support a role for the FA genes in endocrine signaling [20]. Thus, we hypothesize that GnRH regulation of Fanca gene expression may be important for the normal endocrine function of the pituitary and possibly reproductive organs.
In conclusion, Fanca was identified in and isolated from mouse anterior pituitary gonadotrophs in a DD-RT-PCR screen for transcripts regulated by GnRH. This broadens the expression profile of Fanca into highly specialized endocrine tissues and establishes hormonal regulation of the Fanca locus. The acute hormonal regulation of the molecule indicates that Fanca may have a role in mediating GnRH responsiveness in mature gonadotrophs.
ACKNOWLEDGMENTS
We would like to thank Professor Pamela Mellon (San Diego, CA) for supplying LβT2 cells, Dr. Fre Arwert (Amsterdam, The Netherlands) for the generous gift of a mouse Fanca cDNA and anti-mouse Fanca-specific antisera, Julie Bell for guidance in using the LightCycler, and Dimitra Karali for technical support.
Footnotes
R.L. and L.C. were supported by M.R.C. Ph.D. research studentships during the course of this work, and these two authors made equal contributions to this work.
REFERENCES
- 1.Stanislaus D, Pinter JH, Janovick JA, Conn PM. Mechanisms mediating multiple physiological responses to gonadotropin-releasing hormone. Mol Cell Endocrinol. 1998;144:1–10. doi: 10.1016/s0303-7207(98)00126-9. [DOI] [PubMed] [Google Scholar]
- 2.Brown P, McNeilly AS. Transcriptional regulation of pituitary gonadotrophin subunit genes. Rev Reprod. 1999;4:117–124. doi: 10.1530/ror.0.0040117. [DOI] [PubMed] [Google Scholar]
- 3.Shacham S, Harris D, Ben-Shlomo H, Cohen I, Bonfil D, Przedecki F, Lewy H, Ashkenazi IE, Seger R, Naor Z. Mechanism of GnRH receptor signaling on gonadotropin release and gene expression in pituitary gonadotrophs. Vitam Horm. 2001;63:63–90. doi: 10.1016/s0083-6729(01)63003-6. [DOI] [PubMed] [Google Scholar]
- 4.Charlton HM, Halpin DM, Iddon C, Rosie R, Levy G, McDowell I, Megson A, Morris J, Bramwell A, Speight A, Ward B, Broadhead J, Davey-Smith G, Fink G. The effects of daily administration of single and multiple injections of gonadotropin releasing hormone on pituitary and gonadal function in the hypogonadal (HPG) mouse. Endocrinology. 1983;113:535–544. doi: 10.1210/endo-113-2-535. [DOI] [PubMed] [Google Scholar]
- 5.Brown P, McNeilly J, Evans J, Crawford G, Walker M, Christian H, McNeilly A. Manipulating the in vivo mRNA expression profile of FSHβ to resemble that of LHβ does not promote a concomitant increase in intracellular storage of follicle-stimulating hormone. J Neuroendocrinol. 2001;13:50–62. doi: 10.1046/j.1365-2826.2001.00594.x. [DOI] [PubMed] [Google Scholar]
- 6.Mercer JE, Clements JA, Funder JW, Clarke IJ. Regulation of follicle stimulation hormone beta and common alpha subunit mRNA by gonadotropin releasing hormone and estrogen in the sheep pituitary. Neuroendocrinology. 1989;50:321–326. doi: 10.1159/000125240. [DOI] [PubMed] [Google Scholar]
- 7.Bedecarrats GY, Kaiser UB. Differential regulation of gonadotrophin subunit gene promoter activity by pulsatile gonadotropin-releasing hormone (GnRH) in perfused LβT2 cells: role of GnRH receptor concentration. Endocrinology. 2003;144:1802–1811. doi: 10.1210/en.2002-221140. [DOI] [PubMed] [Google Scholar]
- 8.Japon MA, Rubinstein M, Low J. In situ hybridization analysis of anterior pituitary hormone gene expression during fetal mouse development. J Histochem Cytochem. 1994;42:1117–1125. doi: 10.1177/42.8.8027530. [DOI] [PubMed] [Google Scholar]
- 9.Turgeon JL, Kimura Y, Waring DW, Mellon PL. Steroid and pulsatile gonadotropin-releasing hormone (GnRH) regulation of luteinizing hormone and GnRH receptor in a novel gonadotrope cell line. Mol Endocrinol. 1996;10:439–450. doi: 10.1210/mend.10.4.8721988. [DOI] [PubMed] [Google Scholar]
- 10.Windle JJ, Weiner RI, Mellon PL. Cell lines of the pituitary gonadotrope lineage derived by targeted oncogenesis in transgenic mice. Mol Endocrinol. 1990;4:597–603. doi: 10.1210/mend-4-4-597. [DOI] [PubMed] [Google Scholar]
- 11.Graham KE, Nusser KD, Low MJ. L beta T2 gonadotroph cells secrete follicle stimulating hormone (FSH) in response to activin A. J Endocrinol. 1999;162:R1–R5. doi: 10.1677/joe.0.162r001. [DOI] [PubMed] [Google Scholar]
- 12.Pernasetti F, Vasilyev VV, Rosenberg SB, Bailey JS, Huang HJ, Miller WL, Mellon PL. Cell-specific transcriptional regulation of follicle-stimulating hormone-β by activin and gonadotropin releasing hormone in the LβT2 pituitary gonadotrope cell model. Endocrinology. 2001;142:2284–2295. doi: 10.1210/endo.142.6.8185. [DOI] [PubMed] [Google Scholar]
- 13.Liu F, Austin DA, Mellon PL, Olefsky JM, Webster NJG. GnRH activates ERK1/2 leading to the induction of c-fos and LHβ protein expression in LβT2 cells. Mol Endocrinol. 2002;16:419–434. doi: 10.1210/mend.16.3.0791. [DOI] [PubMed] [Google Scholar]
- 14.Yokoi T, Ohmichi K, Tasaka K, Kimura A, Kanda Y, Hayakawa J, Tahara M, Hisamoto K, Kurachi H, Murata Y. Activation of the luteinizing hormone beta promoter by gonadotropin-releasing hormone requires c-JUN NH2-terminal protein kinase. J Biol Chem. 2000;275:21639–21647. doi: 10.1074/jbc.M910252199. [DOI] [PubMed] [Google Scholar]
- 15.Liu F, Usui I, Guojing Evans L, Austin DA, Mellon PL, Olefsky JM, Webster NJG. Involvement of both Gq/11 and Gs proteins in gonadotropin-releasing hormone receptor-mediated signaling in LβT2 cells. J Biol Chem. 2002;277:32099–32108. doi: 10.1074/jbc.M203639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris D, Bonfil D, Chuderland D, Kraus S, Seger R, Naor Z. Activation of MAPK cascades by GnRH: ERK and Jun N-terminal kinase are involved in basal and GnRH-stimulated activity of the glycoprotein hormone LHβ subunit promoter. Endocrinology. 2002;143:1018–1025. doi: 10.1210/endo.143.3.8675. [DOI] [PubMed] [Google Scholar]
- 17.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1991;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 18.Auerbach AD, Buchwald M, Joenje H. Genetic Basis of Human Cancer. New York: McGraw-Hill; 1998. Fanconi anemia; pp. 317–332. [Google Scholar]
- 19.Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001;2:446–457. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- 20.Wajnrajch MP, Gertner JM, Huma Z, Popovic J, Lin K, Verlander PC, Batish SD, Giampietro PF, Davis JG, New MI, Auerbach AD. Evaluation of growth and hormonal status in patients referred to the international Fanconi anemia registry. Pediatrics. 2001;107:744–754. doi: 10.1542/peds.107.4.744. [DOI] [PubMed] [Google Scholar]
- 21.Abbas MM, Evans JJ. Regulation of c-fos protein in gonadotrope cells by oxytocin and gonadotropin-releasing hormone. Neuroendocrinology. 2000;71:292–300. doi: 10.1159/000054549. [DOI] [PubMed] [Google Scholar]
- 22.Coss D, Jacobs SB, Bender CE, Mellon PL. A novel AP-1 site is critical for maximal induction of the follicle-stimulating hormone beta gene by gonadotropin-releasing hormone. J Biol Chem. 2004;279:152–162. doi: 10.1074/jbc.M304697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miele G, MacRae L, McBride D, Manson J, Clinton M. Elimination of false positives generated through PCR reamplification of differential display cDNA. Biotechniques. 1998;25:138–144. doi: 10.2144/98251rr02. [DOI] [PubMed] [Google Scholar]
- 24.Brown P, McNeilly AS. Steroidogenic factor-1 (SF-1) and the regulation of expression of luteinising hormone and follicle stimulating hormone b-subunits in the sheep anterior pituitary in vivo. Int J Biochem Cell Biol. 1997;29:1513–1524. doi: 10.1016/s1357-2725(97)00082-4. [DOI] [PubMed] [Google Scholar]
- 25.Wong JCY, Alon N, Norga K, Kruyt FAE, Youssoufian H, Buchwald M. Cloning and analysis of the mouse Fanconi anemia group A cDNA and an overlapping penta zinc finger cDNA. Genomics. 2000;67:273–283. doi: 10.1006/geno.2000.6252. [DOI] [PubMed] [Google Scholar]
- 26.Waisfisz Q, De Winter JP, Kruyt FAE, de Groot J, van der Weel L, Dijkmans LM, Arwert F, Scheper RJ, Youssoufian H, Hoatlin ME, Joenje H. A physical complex of the Fanconi anemia proteins FANCG/XRCC9 and FANCA. Proc Natl Acad Sci U S A. 1999;96:10320–10325. doi: 10.1073/pnas.96.18.10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van de Vrugt HJ, Cheng NC, de Vries Y, Rooimans MA, de Groot J, Scheper RJ, Zhi Y, Hoatlin ME, Joenje H, Arwert F. Cloning and characterization of murine Fanconi anemia group A gene: Fanca protein is expressed in lymphoid tissues, testes and ovary. Mamm Genome. 2000;11:326–331. doi: 10.1007/s003350010060. [DOI] [PubMed] [Google Scholar]
- 28.Abu-Issa R, Eichele G, Youssoufian H. Expression of the Fanconi anemia group A gene (Fanca) during mouse embryogenesis. Blood. 1999;94:818–824. [PubMed] [Google Scholar]
- 29.Liang P. SAGE genie: a suite with panoramic view of gene expression. Proc Natl Acad Sci U S A. 2002;99:11547–11548. doi: 10.1073/pnas.192436299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang P. Factors ensuring successful use of differential display. Methods. 1998;16:361–364. doi: 10.1006/meth.1998.0690. [DOI] [PubMed] [Google Scholar]
- 31.Luce MJ, Burrows PD. Minimizing false positives in differential display. Biotechniques. 1998;24:766–768. doi: 10.2144/98245bm16. [DOI] [PubMed] [Google Scholar]
- 32.Wurmbach E, Yuen T, Ebersole BJ, Sealfon SC. Gonadotropin-releasing hormone receptor-coupled gene network. J Biol Chem. 2001;276:47195–47201. doi: 10.1074/jbc.M108716200. [DOI] [PubMed] [Google Scholar]
- 33.Yuen T, Wurmbach E, Ebersole BJ, Ruf F, Pfeffer RL, Sealfon SC. Coupling of GnRH concentration and the GnRH receptor-activated gene program. Mol Endocrinol. 2002;16:1145–1153. doi: 10.1210/mend.16.6.0853. [DOI] [PubMed] [Google Scholar]
- 34.Wong JCY, Alon N, Mckerlie C, Huang JR, Meyn MS, Buchwald M. Targeted disruption of exons 1 to 6 of the Fanconi anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum Mol Genet. 2003;12:2063–2076. doi: 10.1093/hmg/ddg219. [DOI] [PubMed] [Google Scholar]
- 35.Pipaon C, Casado JA, Bueren JA, Fernandez-Luna JL. Jun N-terminal kinase activity and early growth-response factor-1 gene expression are down-regulated in Fanconi anemia group A lymphoblasts. Blood. 2004;103:128–132. doi: 10.1182/blood-2003-06-2091. [DOI] [PubMed] [Google Scholar]
- 36.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 37.Alarid ET, Holley S, Hayakawa M, Mellon PL. Discrete stages of anterior pituitary differentiation recapitulated in immortalized cell lines. Mol Cell Endocrinol. 1998;140:25–30. doi: 10.1016/s0303-7207(98)00025-2. [DOI] [PubMed] [Google Scholar]
- 38.Alter BP. Cancer in Fanconi anemia, 1927-2001. Cancer. 2003;97:425–440. doi: 10.1002/cncr.11046. [DOI] [PubMed] [Google Scholar]
- 39.Bagby GC. Genetic basis of Fanconi anemia. Curr Opin Hematol. 2003;10:68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 40.Faivre L, Guardiola P, Lewis C, Dokal I, Edell W, Zatterale A, Altay C, Poole J, Stones D, Kwee ML, van Weel-Sipman M, Havenga C, Morgan N, de Winter J, Digweed M, Savoia A, Pronk J, de Ravel T, Jansen S, Joenje H, Gluckman E, Mathew CG. Association of complementation group and mutation type with clinical outcome in Fanconi anemia. Blood. 2000;96:4064–4070. [PubMed] [Google Scholar]
- 41.Adachi D, Oda T, Yagasaki H, Nakasato K, Taniguchi T, D’Andrea AD, Asano S, Yamashita T. Heterogeneous activation of the Fanconi anemia pathway by patient-derived FANCA mutants. Hum Mol Genet. 2002;25:3125–3134. doi: 10.1093/hmg/11.25.3125. [DOI] [PubMed] [Google Scholar]
- 42.Cheng NC, van de Vrugt HJ, van der Valk MA, Oostra AB, Krimpenfort P, de Vries Y, Joenje H, Berns A, Arwert F. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum Mol Genet. 2000;9:1805–1811. doi: 10.1093/hmg/9.12.1805. [DOI] [PubMed] [Google Scholar]
- 43.Rio P, Segovia JC, Hanenberg H, Casado JA, Martinez J, Gottsche K, Cheng NC, van de Vrugt HJ, Arwert F, Joenje H, Bueren JA. In vitro phenotypic correction of hematopoietic progenitors from Fanconi anemia group A knockout mice. Blood. 2002;100:2032–2039. [PubMed] [Google Scholar]
- 44.Risma KA, Clay CM, Nett TM, Wagner T, Yun J, Nilson JH. Targeted overexpression of luteinizing hormone in transgenic mice leads to infertility, polycystic ovaries, and ovarian tumours. Proc Natl Acad Sci U S A. 1995;92:1322–1326. doi: 10.1073/pnas.92.5.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berkovitz GD, Zinkham WH, Migeon CJ. Gonadal function in two siblings with Fanconi’s anemia. Horm Res. 1984;19:137–141. doi: 10.1159/000179880. [DOI] [PubMed] [Google Scholar]
- 46.Alter BP, Frissora CL, Halperin DS, Freedman MH, Chitkara AD, Alvarez E, Lynch L, Adler-Brecher B, Auerbach AD. Fanconi’s anaemia and pregnancy. Br J Haematol. 1991;77:410–418. doi: 10.1111/j.1365-2141.1991.tb08593.x. [DOI] [PubMed] [Google Scholar]
- 47.Bargman GJ, Shahidi NT, Gilbert EF, Opitz JM. Studies of malformation syndromes of man XLVII: disappearance of spermatogonia in the Fanconi anemia syndrome. Eur J Pediatr. 1977;125:169–174. doi: 10.1007/BF00480592. [DOI] [PubMed] [Google Scholar]
- 48.Melmed S. Mechanisms for pituitary tumorigenesis: the plastic pituitary. J Clin Invest. 2003;112:1603–1618. doi: 10.1172/JCI20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asa SL, Ezzat S. The pathogenesis of pituitary tumours. Nat Rev Cancer. 2002;2:836–849. doi: 10.1038/nrc926. [DOI] [PubMed] [Google Scholar]
- 50.Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, Emmert-Buck MR, Debelenko LV, Zhuang Z, Lubensky IA, Liotta LA, Crabtree JS, Wang Y, Roe BA, Weisemann J, Boguski MS, Agarwal SK, Kester MB, Kim YS, Heppner C, Dong Q, Spiegel AM, Burns AL, Marx SJ. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276:404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 51.Jin S, Mao H, Schnepp RW, Sykes SM, Silva AC, D’Andrea AD, Hua X. Menin associates with FANCD2, a protein involved in repair of DNA damage. Cancer Res. 2003;63:4204–4210. [PubMed] [Google Scholar]
- 52.Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, Grompe M. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, Mok SC, D’Andrea AD. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 54.Kang SK, Choi KC, Yang HS, Leung PC. Potential role of gonadotrophin-releasing hormone (GnRH)-I and GnRH-II in the ovary and ovarian cancer. Endocr Relat Cancer. 2003;10:169–177. doi: 10.1677/erc.0.0100169. [DOI] [PubMed] [Google Scholar]
- 55.Schally AV. LH-RH analogues: I. Their impact on reproductive medicine. Gynecol Endocrinol. 1999;13:401–409. doi: 10.3109/09513599909167587. [DOI] [PubMed] [Google Scholar]