Abstract
Fatal infantile lactic acidosis is a severe metabolic disorder characterized by the onset of lactic acidosis within the 1st d of life and early death. We found a combined respiratory-chain enzyme deficiency associated with mitochondrial DNA (mtDNA) depletion in a small consanguineous family with this disorder. To identify the disease-causing gene, we performed single-nucleotide polymorphism homozygosity mapping and found homozygous regions on four chromosomes. DNA sequencing revealed a homozygous 2-bp deletion in SUCLG1, a gene that encodes the α subunit of the Krebs-cycle enzyme succinate–coenzyme A ligase (SUCL). The mtDNA depletion is likely explained by decreased mitochondrial nucleoside diphosphate kinase (NDPK) activity resulting from the inability of NDPK to form a complex with SUCL.
The molecular basis of several disorders resulting from a decrease in mtDNA copy number—the mtDNA-depletion syndromes (MIM 251880)—has been elucidated over the past decade. Mutations in six genes have now been reported to cause mtDNA depletion in humans: TP, which is associated with mitochondrial neurogastrointestinal encephalopathy (MNGIE [MIM 603041])1; TK2, which causes mitochondrial myopathy2; DGUOK, associated with liver failure3; POLG, which can cause Alpers syndrome (MIM 203700)4; MPV17, which is responsible for isolated liver mtDNA depletion5; and SUCLA2, which causes encephalomyopathy.6
Fatal infantile lactic acidosis can be caused by various enzyme defects—such as pyruvate dehydrogenase (PDH) deficiency, pyruvate carboxylase deficiency, and deficiency of complex I of the respiratory chain—or by mtDNA mutations. The disorder has not, however, been reported in association with mtDNA depletion, and, in many cases, the underlying genetic defect remains unknown. Most cases are sporadic or occur in small families, which usually precludes the use of linkage analysis.
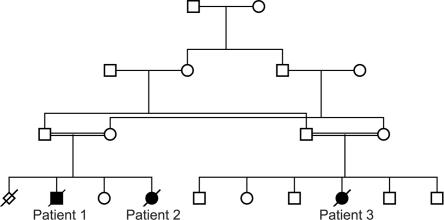
We identified a small consanguineous family of Pakistani origin with autosomal recessive fatal infantile lactic acidosis (fig. 1). The family gave informed consent for the study.
Figure 1. .
Pedigree of the family of Pakistani origin with fatal infantile lactic acidosis due to SUCLG1 mutations.
Patient 1 was the first child of first-cousin parents. A previous pregnancy had ended in a spontaneous abortion at 11 wk. Patient 1 was born at 40 wk gestation, with a birth weight of 2,150 g, a birth length of 48 cm, and a head circumference of 32 cm. His Apgar scores were normal. He was dysmature, with hypertrichosis and low hairline, and he had hepatomegaly and severe hypothermia. He was hypotonic, and, in the 1st d of life, he became severely acidotic, with a blood pH of 6.7. He had Cheyne-Stokes respiration and received respiratory support. Findings from an electroencephalogram were severely abnormal, with focal and paroxystic activity, sharp waves, and triphasic potentials bilaterally. Results of brain ultrasound were normal. A urine screening showed elevated levels of lactate and pyruvate and moderately elevated excretions of methylmalonate and methylcitrate. The patient died at age 4 d.
Patient 2 was the youngest sister of patient 1 (the other sister, older than patient 2, is healthy). Intrauterine growth retardation (IUGR) was suspected during the pregnancy. Because of her breech position, patient 2 was born by Cesarean delivery at 36 wk gestation, with a birth weight of 1,730 g, a length of 42.5 cm, and a head circumference of 30.5 cm. Cord blood pH and Apgar scores were normal. In the 1st d, she developed hypoglycemia, with blood glucose levels of 0.8–1.3 mmol/liter, and severe lactic acidosis. Results from a cerebral ultrasound were normal. A urine screening showed severely increased excretions of lactate and pyruvate, moderately elevated excretions of methylmalonate (67.7 μmol/mmol creatinine; reference <3.6 μmol/mmol creatinine) and methylcitrate (169.0 μmol/mmol creatinine; reference <7.6 μmol/mmol creatinine), and slightly elevated excretions of the Krebs-cycle intermediates fumarate, malate, citrate, and 2-oxoglutarate. Plasma and urine amino acids showed highly elevated taurine and glycine levels and moderately elevated alanine and lysine levels. From age 2 d, she received respiratory therapy. She died at age 3 d.
Patient 3 was the cousin of patients 1 and 2 and was also born to first-cousin parents. Five siblings are healthy. At 35 wk gestation, IUGR was suspected on ultrasound. She was born at 38 wk gestation, with a birth weight of 1,950 g and a birth length of 45 cm. Cord blood pH was 7.3, and Apgar scores were normal. She was dysmature, with polydactyly, and she was hypotonic and hypothermic. She developed lactic acidosis, with a pH of 6.6 and a base excess of −29.5. She received respiratory support. A urine screening showed massive excretions of lactic acid, moderate excretions of pyruvate and methylmalonate, and elevated excretions of glycine, taurine, and alanine. She died at age 2 d. No tissue was available for analysis.
Postmortem examination of muscle (from patient 2) by light microscopy showed normal fiber-type distribution with intracellular lipid accumulation (fig. 2C). No ragged-red fibers were seen. Postmortem histology of liver (from patient 1) showed microvesicular steatosis and sinusoidal dilatation.
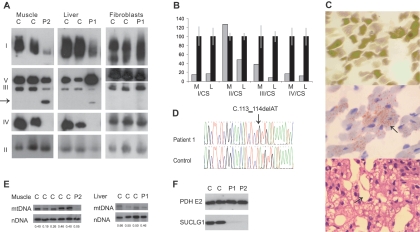
Figure 2. .
Histology and analysis of the respiratory chain and SUCLG1. A, BN-PAGE of muscle, liver, and fibroblasts, showing a decreased amount of fully assembled complexes I, III, and IV in muscle and liver in patients 1 and 2 (P1 and P2) with SUCLG1 mutations, compared with controls (C). In addition, a subcomplex from complex III is seen (arrow). B, Enzyme analysis of the respiratory chain related to citrate synthase (CS) shows a deficiency of complexes I, III, and IV in muscle (M) from patient 1 and in liver (L) from patient 2 (gray bars). Values±SDs for controls are shown by black bars. C, Histology of muscle and liver. ATPase staining (pH 9.4) (top) and oil red staining (middle) of muscle showing normal fiber-type distribution and intracellular lipid accumulation (arrow). The tissue fragmentation is a freeze artifact. Hematoxylin and eosin staining (bottom) of liver, showing sinusoidal dilatation (arrow) and microvesicular steatosis. D, Mutation analysis of SUCLG1 showing the homozygous c.113_114delAT mutation in patient 1. E, Analysis of the relative mtDNA copy number by Southern-blot analysis of muscle from patient 2 and liver from patient 1 compared with controls. The mtDNA/nuclear DNA (nDNA) ratio is shown below the bands. F, Immunoblot analysis of fibroblast mitochondria showing a complete absence of SUCLG1 protein in patients 1 and 2 compared with controls. An antibody against PDH E2 was used as a loading control.
Analysis of mitochondrial protein in muscle (from patient 2) and liver (from patient 1), by blue native PAGE (BN-PAGE),7 showed a decreased amount of fully assembled complexes I, III, and IV (fig. 2A). The amount of complex V was normal in muscle and increased in liver. The amount of complex II, which is encoded only by nuclear genes, was normal. In both muscle and liver, a subcomplex from complex III was seen at ∼320 kDa. BN-PAGE of fibroblast mitochondria from patient 1 showed normal amounts of fully assembled complexes I–V (fig. 2A).
In accordance with the findings of BN-PAGE, analysis of respiratory-chain enzyme activities8,9 showed a combined complex I, III, and IV deficiency in muscle and liver (fig. 2B). The enzyme analysis of fibroblasts was normal (results not shown).
The relative mtDNA copy number was determined by Southern-blot analysis, as reported elsewhere.10 Southern-blot analysis showed a decreased amount of mtDNA in muscle (15% of normal) and a slightly decreased amount of mtDNA in liver (63% of normal) (fig. 2E). Real-time quantitative PCR gave similar results (not shown).
To identify the disease-causing gene, DNA from patients 1 and 2 was used in a genomewide search for homozygosity with the Affymetrix GeneChip 10K array, version 2.0 (Affymetrix).10 Because of the small family size, the SNP analysis showed several homozygous regions of variable sizes. Because of the close relationship between the parents (first cousins), the homozygous regions identical by descent in each patient were expected to be large, ∼28 cM.11 We searched for candidate genes in homozygous regions, arbitrarily chosen to be at least 10 Mb, that were common to the two patients. We found four such regions, on chromosomes 2, 3, 4, and 5 (table 1). The Mitop2 database was used to search for genes encoding mitochondrial proteins in these regions. Of the 15 RefSeq genes found, we considered the SUCLG1 gene (GenBank accession number NM_003849) on chromosome 2, which encodes the α subunit of the Krebs-cycle enzyme succinate–coenzyme A (CoA) ligase (SUCL) (also termed “succinyl-CoA synthetase” and “succinate thiokinase”), to be a candidate gene because of the involvement of the ATP-forming β subunit of SUCL, encoded by SUCLA2, in patients with a combined respiratory-chain deficiency and mtDNA depletion in muscle,6 elevated levels of urine methylmalonate and methylcitrate—which was also found in patients with SUCLA2 mutations10,12—and the excretion of Krebs-cycle metabolites in patient 2.
Table 1. .
Regions >10 Mb That Were Homozygous in Both Affected Siblings
| Chromosome | SNPs | Interval Size (Mb) |
No. of RefSeq Genes in Mitop2 |
| 2p12-q13 | rs934310–rs1002016 (57 SNPs) | 29.0 | 11 |
| p26.3-p25.1 | rs3856832–rs3846122 (80 SNPs) | 13.0 | 3 |
| q25-q28.3 | rs1008326–rs925084 (79 SNPs) | 24.0 | 1 |
| 5q21.3-q23.1 | rs961426–rs1918159 (52 SNPs) | 10.8 | 0 |
Sequencing of SUCLG1 (table 2) revealed a homozygous 2-bp deletion, c.113_114delAT, in exon 2 in both patients (fig. 2D). The parents and a healthy sister were heterozygous for the mutation. The mutation leads to a change of the reading frame and a premature stop codon and is expected to lead to the synthesis of a prematurely truncated protein.
Table 2. .
Primers for Amplification of SUCLG1[Note]
| Primer(5′→3′) |
||
| Exon | Forward | Reverse |
| 1 | AATTTGTTCAGGCGACTGCT | GGCGCCAGGAAGACAGTA |
| 2 | GCGCGTGCATTAAAGAATTT | TTCTGCAACAATCATGTGTTATTT |
| 3 | GCTTTTGCTTCTCTTGGGCT | CAAAGAATGCTCGCTCTTCC |
| 4–5 | TGTCCTTTTCTTACCCCAAGA | GAGTTTTGAGGGTTTAAGGCA |
| 6 | TCACTCGAAGTGTTTGGTAATTT | CTCATCCAATGAAGACACCAC |
| 7 | AAATTCCATGGTTCACCCTT | ACTTCTGAAACAAGCCTCTGAT |
| 8 | CATGAATTTGAGGTCCCGTT | CCACACACAGAGAAAGCCTG |
| 9 | TCAAACACCCTCATCCTGGT | CAAACTGCTGCTGGGTTACA |
Note.— The PCR products were sequenced with the Big Dye Terminator v1.1 and were analyzed on an ABI 3130 (Applied Biosystems).
A rabbit polyclonal antibody against SUCLG1 was generated using a synthetic peptide specific to human SUCLG1 (CGTTIYKEFEKRKML) (Rockland Immunochemicals). Immunoblot analysis of fibroblast mitochondria showed the complete absence of SUCLG1 protein in the affected patients (fig. 2F).
This is the first report, to our knowledge, of mutations in SUCLG1, which we show here to cause fatal infantile lactic acidosis associated with mtDNA depletion. SUCL is a dimeric enzyme that catalyzes a reversible reaction in which succinyl-CoA and either adenosine diphosphate (ADP) or guanosine diphosphate (GDP) are converted to succinate and ATP or guanosine triphosphate (GTP), depending on which β subunit is present (fig. 3).13 The α subunit of SUCL forms a heterodimer with either of its β subunits encoded by SUCLA2 and SUCLG2, resulting in an ATP/ADP–specific SUCL (A-SUCL) and a GTP/GDP–specific SUCL (G-SUCL). A-SUCL and G-SUCL are located in the mitochondrial matrix, and it is likely that they both catalyze substrate-level phosphorylation in the Krebs cycle, although this has not been conclusively established for G-SUCL.14 Both enzymes may also catalyze the reverse reaction to supply succinyl-CoA for heme synthesis and ketone-body activation.
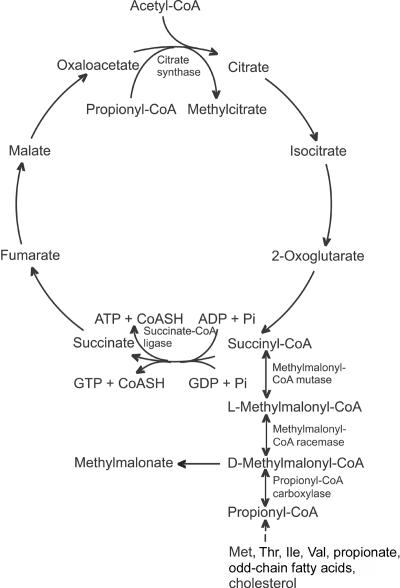
Figure 3. .
The Krebs cycle and methylmalonate metabolism. CoASH = coenzyme A. Pi = inorganic phosphate.
Enzymatic assays and western- and northern-blot analyses have shown that the two enzymes are widely expressed in human tissues, with G-SUCL predominantly expressed in anabolic tissues, such as liver and kidney, and A-SUCL in catabolic tissues, such as brain, heart, and skeletal muscle.13,15 The pattern of expression and activity of the two SUCL enzymes may reflect the high ATP energy demand in catabolic tissues, such as brain and muscle. Substrate-level generation of GTP may, on the other hand, be more important in anabolic tissues, such as liver and kidney, where it is used by the enzymes phosphoenolpyruvate carboxykinase and GTP-AMP phosphotransferase, and it is involved in various processes, such as protein synthesis and possibly mitochondrial membrane fusion.14
What links the deficiency in SUCL with mtDNA depletion? One strong possibility is a decrease in the activity of nucleoside diphosphate kinase (NDPK). Immunoprecipitation experiments that use mitochondria from rat liver β cells show that antiserum against SUCL coimmunoprecipitates NDPK.16 Further, NDPK forms a complex with SUCL in rabbit heart mitochondria,17 and NDPK and SUCL were shown to copurify in Pseudomonas aeruginosas.18 NDPKs are protein kinases that use ATP to synthesize nonadenylic nucleoside triphosphates for nucleic acid synthesis. A decreased activity of NDPK due to the lack of interaction with SUCL could explain the mtDNA depletion and, hence, the combined respiratory-chain enzyme deficiency in muscle and liver.
The analysis of the respiratory chain in fibroblasts gave normal results, despite the absence of SUCLG1 protein. A likely explanation is that, in replicating tissues such as fibroblasts, deoxyribonucleotide triphosphates (dNTPs) are transported from the cytoplasm into the mitochondria via the deoxynucleotide carrier, and these tissues are therefore less dependent on mitochondrial NDPK activity for the de novo synthesis of dNTPs.
Mutations were recently reported in the SUCLA2 gene, which encodes the ATP-forming β subunit of SUCL.6 The disorder is associated with a combined respiratory-chain enzyme deficiency and mtDNA depletion in muscle, similar to what is found in the family reported here. In patients from the Faroe Islands, we found a founder mutation in SUCLA2 that causes skipping of exon 4, resulting in the absence of functional protein.10 Despite the similar findings—that is, a combined oxidative phosphorylation deficiency and mtDNA depletion—patients with SUCLA2 and SUCLG1 mutations differ markedly in their clinical presentations. The patients with SUCLG1 mutations had an extremely severe phenotype with antenatal manifestations of the disorder—severe lactic acidosis in the 1st d of life and death within 2–4 d. The phenotypes of patients with SUCLA2 mutations were milder; these patients were generally healthy at birth but presented with severe hypotonia and muscle weakness at age 3–6 mo. Subsequently, they developed a Leigh-like disorder with hearing impairment and dystonia, and they had a life span of up to 21 years. The much more severe disorder in patients with SUCLG1 mutations is likely caused by the complete absence of both A-SUCL and G-SUCL, whereas the milder phenotype of patients with SUCLA2 mutations may be explained by the presence of functional G-SUCL. In addition, in patients with SUCLA2 mutations, no liver symptoms have been reported, which may be explained by the relatively low expression of SUCLA2 and the high expression of SUCLG2 in liver tissue.
One of the reasons we considered SUCLG1 a candidate gene was the excretion of methylmalonate and methylcitrate in our patients, similar to that found in patients with SUCLA2 mutations. Both methylmalonyl-CoA mutase and methylmalonyl-CoA racemase are reversible enzymes, and, therefore, accumulation of succinyl-CoA due to a deficiency of succinate-CoA ligase can lead to an increased concentration of d-methylmalonyl-CoA (fig. 3). An enzyme capable of hydrolyzing d-methylmalonyl-CoA to methymalonate and CoA—the methylmalonyl-CoA hydrolase—has been described elsewhere,19 which may explain the elevated free methylmalonate found in body fluids of SUCL-deficient patients.
This study demonstrates the power of using homozygosity mapping to identify disease-causing genes. The method may be applied even in small consanguineous families, provided that one has some knowledge of the function or localization of the gene product. In this case, although the candidate regions altogether comprised ∼77 Mb, the number of candidate genes could be limited to 15 because of the assumed mitochondrial localization.
Both the α subunit and the ATP-forming β subunit of SUCL have now been shown to cause respiratory-chain disorders with mtDNA depletion, and therefore SUCLG2, which encodes the GTP-forming β subunit, should be considered a candidate gene for other mtDNA-depletion disorders, especially of the liver, because of the high expression of this subunit in liver.
Acknowledgments
We thank the family for their participation and the Rigshospitalet Microarray Centre for the SNP analyses. This work was supported by The Danish National Health Research Council, The Novo Nordic Foundation, The P. A. Messerschmidt and Wife Foundation, The A. P. Moller Foundation for the Advancement of Medical Science, Jacob Madsen and Wife Olga Madsen’s Foundation, The Rosalie Petersen Foundation, and The Vanfoere Foundation. E.A.S. is a senior scientist of the Canadian Institutes of Health Research and an international scholar of the Howard Hughes Medical Institute.
Web Resources
The accession number and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SUCLG1 [accession number NM_003849])
- Mitop2, http://www.mitop2.de
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for hepatocerebral mtDNA depletion syndromes, MNGIE, and Alpers syndrome)
References
- 1.Nishino I, Spinazzola A, Hirano M (1999) Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science 283:689–692 10.1126/science.283.5402.689 [DOI] [PubMed] [Google Scholar]
- 2.Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O (2001) Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat Genet 29:342–344 10.1038/ng751 [DOI] [PubMed] [Google Scholar]
- 3.Mandel H, Szargel R, Labay V, Elpeleg O, Saada A, Shalata A, Anbinder Y, Berkowitz D, Hartman C, Barak M, et al (2001) The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nat Genet 29:337–341 10.1038/ng746 [DOI] [PubMed] [Google Scholar]
- 4.Naviaux RK, Nguyen KV (2004) POLG mutations associated with Alpers’ syndrome and mitochondrial DNA depletion. Ann Neurol 55:706–712 10.1002/ana.20079 [DOI] [PubMed] [Google Scholar]
- 5.Spinazzola A, Viscomi C, Fernandez-Vizarra E, Carrara F, D’Adamo P, Calvo S, Marsano RM, Donnini C, Weiher H, Strisciuglio P, et al (2006) MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat Genet 38:570–575 10.1038/ng1765 [DOI] [PubMed] [Google Scholar]
- 6.Elpeleg O, Miller C, Hershkovitz E, Bitner-Glindzicz M, Bondi-Rubinstein G, Rahman S, Pagnamenta A, Eshhar S, Saada A (2005) Deficiency of the ADP-forming succinyl-CoA synthase activity is associated with encephalomyopathy and mitochondrial DNA depletion. Am J Hum Genet 76:1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schagger H, von Jagow G (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199:223–231 10.1016/0003-2697(91)90094-A [DOI] [PubMed] [Google Scholar]
- 8.Birch-Machin MA, Briggs HL, Saborido AA, Bindoff LA, Turnbull DM (1994) An evaluation of the measurement of the activities of complexes I-IV in the respiratory chain of human skeletal muscle mitochondria. Biochem Med Metab Biol 51:35–42 10.1006/bmmb.1994.1004 [DOI] [PubMed] [Google Scholar]
- 9.Krahenbuhl S, Talos C, Wiesmann U, Hoppel CL (1994) Development and evaluation of a spectrophotometric assay for complex III in isolated mitochondria, tissues and fibroblasts from rats and humans. Clin Chim Acta 230:177–187 10.1016/0009-8981(94)90270-4 [DOI] [PubMed] [Google Scholar]
- 10.Ostergaard E, Hansen FJ, Sorensen N, Duno M, Vissing J, Larsen PL, Faeroe O, Thorgrimsson S, Wibrand F, Christensen E, et al (2007) Mitochondrial encephalomyopathy with elevated methylmalonic acid is caused by SUCLA2 mutations. Brain 130:853–861 10.1093/brain/awl383 [DOI] [PubMed] [Google Scholar]
- 11.Genin E, Todorov AA, Clerget-Darpoux F (1998) Optimization of genome search strategies for homozygosity mapping: influence of marker spacing on power and threshold criteria for identification of candidate regions. Ann Hum Genet 62:419–429 10.1017/S000348009800712X [DOI] [PubMed] [Google Scholar]
- 12.Carrozzo R, Dionisi-Vici C, Steuerwald U, Lucioli S, Deodato F, Di GS, Bertini E, Franke B, Kluijtmans LA, Meschini MC, et al (2007) SUCLA2 mutations are associated with mild methylmalonic aciduria, Leigh-like encephalomyopathy, dystonia and deafness. Brain 130:862–874 10.1093/brain/awl389 [DOI] [PubMed] [Google Scholar]
- 13.Johnson JD, Mehus JG, Tews K, Milavetz BI, Lambeth DO (1998) Genetic evidence for the expression of ATP- and GTP-specific succinyl-CoA synthetases in multicellular eucaryotes. J Biol Chem 273:27580–27586 10.1074/jbc.273.42.27580 [DOI] [PubMed] [Google Scholar]
- 14.Lambeth DO (2006) Reconsideration of the significance of substrate-level phosphorylation in the citric acid cycle. Biochem Mol Biol Educ 34:21–29 10.1002/bmb.2006.49403401021 [DOI] [PubMed] [Google Scholar]
- 15.Lambeth DO, Tews KN, Adkins S, Frohlich D, Milavetz BI (2004) Expression of two succinyl-CoA synthetases with different nucleotide specificities in mammalian tissues. J Biol Chem 279:36621–36624 10.1074/jbc.M406884200 [DOI] [PubMed] [Google Scholar]
- 16.Kowluru A, Tannous M, Chen HQ (2002) Localization and characterization of the mitochondrial isoform of the nucleoside diphosphate kinase in the pancreatic beta cell: evidence for its complexation with mitochondrial succinyl-CoA synthetase. Arch Biochem Biophys 398:160–169 10.1006/abbi.2001.2710 [DOI] [PubMed] [Google Scholar]
- 17.Kadrmas EF, Ray PD, Lambeth DO (1991) Apparent ATP-linked succinate thiokinase activity and its relation to nucleoside diphosphate kinase in mitochondrial matrix preparations from rabbit. Biochim Biophys Acta 1074:339–346 [DOI] [PubMed] [Google Scholar]
- 18.Kavanaugh-Black A, Connolly DM, Chugani SA, Chakrabarty AM (1994) Characterization of nucleoside-diphosphate kinase from Pseudomonas aeruginosa: complex formation with succinyl-CoA synthetase. Proc Natl Acad Sci USA 91:5883–5887 10.1073/pnas.91.13.5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovachy RJ, Copley SD, Allen RH (1983) Recognition, isolation, and characterization of rat liver D-methylmalonyl coenzyme A hydrolase. J Biol Chem 258:11415–11421 [PubMed] [Google Scholar]