Abstract
In the course of systematic screening of the X-chromosome coding sequences in 250 families with nonsyndromic X-linked mental retardation (XLMR), two families were identified with truncating mutations in BRWD3, a gene encoding a bromodomain and WD-repeat domain–containing protein. In both families, the mutation segregates with the phenotype in affected males. Affected males have macrocephaly with a prominent forehead, large cupped ears, and mild-to-moderate intellectual disability. No truncating variants were found in 520 control X chromosomes. BRWD3 is therefore a new gene implicated in the etiology of XLMR associated with macrocephaly and may cause disease by altering intracellular signaling pathways affecting cellular proliferation.
For many years, it has been recognized that the presence of macrocephaly (a head circumference [HC] >2 SD above the mean) or microcephaly is more common in the mentally retarded population. Mentally retarded individuals are at least twice as likely to have macrocephaly than are their intellectually normal peers.1 Macrocephaly is also seen in ∼20% of autistic individuals.2,3 The neuropathological basis of macrocephaly is not well understood, but the presence of this feature can be helpful in restricting the differential diagnosis in a male with mental retardation. However, macrocephaly is common in a number of well-known X-linked mental retardation (XLMR) syndromes—such as fragile X syndrome, FG syndrome, and Lujan syndrome—and it is both clinically and genetically heterogeneous and, as an isolated feature, is a poor diagnostic discriminator. Families have been reported with apparently rare or unique XLMR macrocephaly syndromes. These include three families with macrocephaly, facial coarsening, obesity, and macro-orchidism.4–6 A further family with macrocephaly and macro-orchidism without facial coarsening has shown linkage to Xq12-q21.7 In this last family, the presence of macrocephaly was also observed in a number of intellectually normal males. Another family with XLMR and macrocephaly in both hemizygous males and heterozygous females has shown linkage to Xp11-Xq12.8 Relative macrocephaly and short stature are also features of MRX2, which has been mapped to Xq22.9,10
Here, we describe two families with mental retardation and macrocephaly and an X-linked pattern of inheritance. In each family, we have identified a different protein-truncating mutation in the BRWD3 gene (MIM 300553), located at Xq21.1. A third family with mild intellectual handicap but without macrocephaly had a missense sequence variant in the same gene. No sequence variants were detected in the XLMR-macrocephaly–affected families described by Johnson et al.7 and Turner et al.8
Our study cohort contained 250 families in which two or more males were affected with mental retardation. A sample from each family was selected and is being subjected to systematic sequence analysis of the coding exons and splice junctions of 676 Vega annotated genes (Vega Genome Browser) on the X chromosome by bidirectional, PCR-based direct sequencing.11,12 All samples had been prescreened and were excluded from further study if a karyotype abnormality was present or a mutation was identified in FMR1 or in one of the other 61 X-chromosome genes that are known to be associated with syndromic or nonsyndromic mental retardation.
Family 322 included a nephew and an uncle with mild-to-moderate mental retardation and an aunt with mild cognitive difficulties during early childhood (fig. 1). The pregnancy of proband III-6 was complicated by vaginal bleeding from 28 wk. He was delivered by lower-segment cesarean delivery because of placenta previa at 34 wk gestation, weighing 2.3 kg (50th–75th percentile) and with an HC of 33 cm (75th percentile). He had a left-sided unilateral cleft lip and disruption of the alveolar ridge. An unaffected paternal uncle was born with a cleft lip and palate. The proband's right testis was undescended and was brought down by orchiopexy at age 13 mo. He sat unsupported at age 10 mo, first said “Dad” at age 13 mo, walked alone at age 16 mo, and was toilet trained at age 3 years and 5 mo. Muscular hypotonia was noted in early life. He had limited functional language until age 8 years, despite having normal hearing for speech. He was affectionate but was shy and reserved with strangers. He attended a remedial class in mainstream primary and secondary school. At age 13 years, his combined IQ was 65 on the WISC (Wechsler Intelligence Scale for Children), with a verbal score in the 0.4th percentile, and his performance score was in the 37th percentile, with relative strengths in visual and spatial tests. He is now able to read and write simple material, but he has limited numeracy skills. He is independent for tasks of daily living. A normal echocardiogram was obtained after a murmur was heard at age 8 years. A CT scan of his head at age 17 years showed megalencephaly without hydrocephalus or other structural anomaly.
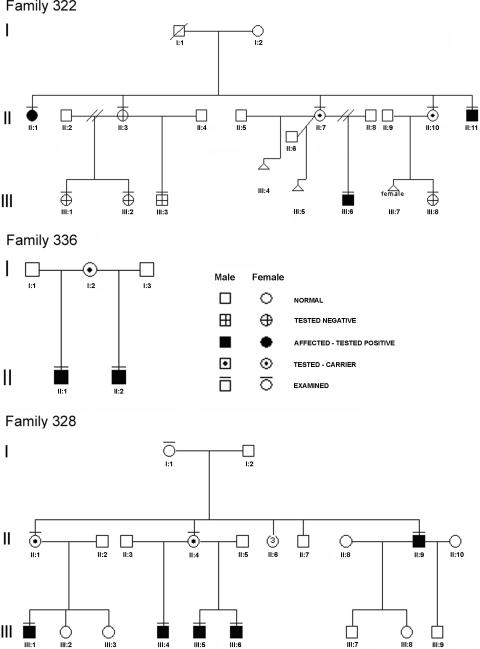
Figure 1. .
Family pedigrees. A blackened symbol indicates the presence of mental retardation; a horizontal bar above a symbol indicates that DNA was collected and analyzed.
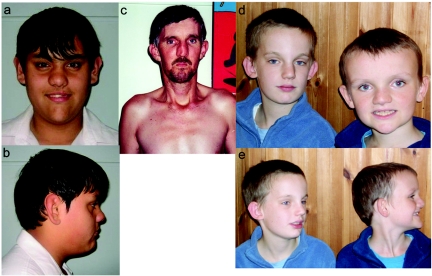
The height, weight, and HC of proband III-6 were consistently >97th percentile. When assessed at age 17 years, his height was 190 cm (≫97th percentile), weight was 88 kg (90th percentile), and HC was 62 cm (≫98th percentile). His facial features (fig. 2a and 2b) included a long face, pointed chin, and large, prominent ears. He had mild central obesity. His body proportions were normal, with an arm span of 192 cm. His hands and feet were large; hand length was 20.5 cm (97th percentile), middle-finger length was 8.6 cm (90th percentile), and the fingers were tapered. He had marked pes planus with shortened 4th and 5th toes. He had minimal facial or axillary hair. Genital pubertal development was at Tanner stage IV.
Figure 2. .
Clinical photography of families 322 and 336. a and b, Subject III-6 of family 322, aged 17 years. c, Subject II-11 of family 322. d and e, Brothers from family 336, aged 13 and 9 years. Note similar facial features, including prominent forehead; large, prominent ears; and pointed chin.
The affected uncle, II-11, of the proband (III-6) had a normal birth weight but grew rapidly in terms of height and weight during his teenage years. At age 38 years, he lives alone in a flat and receives a disability pension. His sister looks after his financial affairs. His height is 187 cm (90th percentile), weight is 73 kg (60th percentile), and HC is 59 cm (>98th percentile). He has frontal bossing; large, prominent ears; and a pointed chin (fig. 2c). His build is thin, with a midthoracic kyphosis and minimal pectus excavatum.
The proband’s mother (II-7) is 165 cm tall (50th percentile) and has an HC of 56.5 cm (60th percentile). She completed formal education until age 16 years. She has no distinguishing morphological features.
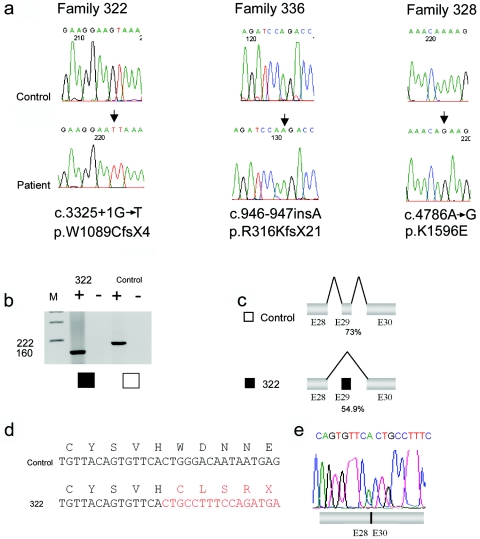
In family 322, a mutation of the highly conserved +1 position of the 5′ donor splice site of intron 29 of the BRWD3 gene, c.3325+1G→T (GenBank accession number NM_153252), was identified. The affected individuals II-11 and III-6 carry this mutation, and individual II-7 was identified as being heterozygous for the mutation. The mutation was not identified in an intellectually normal male cousin, III-3. Analysis of cDNA isolated from a lymphoblastoid cell line derived from III-6 demonstrates that this mutation leads to complete skipping of exon 29 and the introduction of a premature stop codon, p.W1089CfsX4 (GenBank accession number NP_694984) (fig. 3). The c.3325+1G→T mutation changes the 5′ donor splice-site strength from 73.2% to 54.9% (fig. 3). X-inactivation studies of three carrier females—II-1, II-7, and II-10—showed high-to-moderate skewing, with values of 96:4, 100:0, and 86:14, respectively. In all three carriers, it is the chromosome carrying the BRWD3 c.3325+1G→T mutation that is preferentially inactivated (data not shown).
Figure 3. .
Mutation analysis of BRDW3. a, DNA-sequence chromatograms showing the mutations in BRWD3 in genomic DNA from families 322, 336, and 328. The position of the mutation is indicated with an arrow. Control and affected-male sequence analyses are shown. b, RT-PCR analysis of the BRWD3 gene on lymphocyte RNA of proband III-6 from family 322 (blackened square) and a control individual (unblackened square). Primer sequences and conditions are available on request. M indicates pUC19/HpaII size marker, and plus (+) and minus (−) signs indicate the presence or absence of reverse transcriptase, respectively. c, Schematic diagrams showing BRWD3 splicing in a control individual (unblackened square) and patient III-6 from family 322 (blackened square) with the c.3325+1G→T mutation. The mutation results in aberrant splicing—that is, exclusion of exon 29 (dark gray) from the mature BRWD3 mRNA. The percentages given show exon 29 5′ splice-site consensus strength with and without the c.3325+1G→T mutation. In panel d, the sequence resulting from the splice-site mutation is illustrated in red, and the control cDNA sequence is in black. e, Sequence chromatogram of the BRWD3 RT-PCR product of patient III-6, showing the absence of exon 29 from the mRNA.
Family 336 was referred to clinical geneticists for investigation of their intellectual disability. The family includes two affected half-brothers, who share the same mother (fig. 1). Both boys were removed from the care of the mother because of neglect and have been adopted. There has been no contact with the extended biological family. The boys’ mother is reported to have an intellectual disability. Her HC was recorded as 57 cm (97th percentile).
The elder boy was born at 38 wk gestation after a labor with prolonged fetal bradycardia, resulting in a cesarean delivery. He weighed 2.72 kg (9th–25th percentile), and his HC was 37 cm (98th percentile). During infancy, his height was in the 2nd percentile, and his HC was in the 97th percentile. He crawled at age 19 mo and walked at age 2 years. He went into foster care at age 4 years. At age 6 years, he could walk but not run and was not toilet trained. He had few words and limited speech. He had an undescended testis. He is being educated in a support class and has a moderate learning disability. He is able to read and write and has improved language skills. He has no sense of danger. He is clumsy, falls frequently, has difficulties with fine-motor control, and is prone to temper tantrums. At age 13 years, his height was 137 cm (0.4th–2nd percentile), his weight was 30 kg (0.4th–2nd percentile), and his HC was 57 cm (97th percentile). His face is long with a broad, prominent forehead and a small pointed chin (fig. 2d and 2e). His ears are cupped and posteriorly rotated.
His half-brother was born at 38 wk gestation by elective cesarean delivery and weighed 3.680 kg (>75th percentile), with an HC of 37 cm (98th percentile). He was placed in emergency foster care at age 5 mo and was subsequently adopted with his brother. At age 7 mo, developmental delay was noted. He crawled at age 14 mo, and he walked at age 18 mo. He was noted to have mild muscular hypotonia. At age 22 mo, his speech was assessed at a 9-mo level, and global development was assessed at a 12-mo level. A Griffiths assessment at age 39 mo showed that locomotor skills were age appropriate, but language and hand-eye coordination were at the 29-mo level. Results of a hearing assessment during early childhood were normal. At age 9 years, he attended a regular school with additional assistance and has a mild learning disability. He has no sense of danger and has behavioral problems, with frequent temper tantrums and head banging. At age 9 years, his height was 134 cm (50th percentile), weight was 30 kg (50th percentile), and HC was 57 cm (>97th percentile). He has macrocephaly, frontal bossing, a pointed chin, and prominent ears with cupped helices.
A BRWD3 frameshift mutation, c.946-947insA, which leads to a predicted truncated protein p.R316KfsX21, was identified in both affected males from this family (see fig. 3). The biological mother was not available for assessment, and no further follow-up has been possible, because of the children’s adoption proceedings.
Family 328 was reported elsewhere by Gedeon et al. as family E.13 The family includes three brothers, a cousin, and an uncle, all with borderline-to-mild mental retardation (fig. 1). A developmental assessment of one of the males during early childhood identified speech delay as more marked than delay in other domains. None of the affected males had macrocephaly or tall stature in childhood. They were noted to have large, prominent ears (ear length >2 SD) and, in retrospect, a triangular face and pointed chin. The family has been contacted but, at present, has declined clinical follow-up.
A missense BRWD3 variant, c.4786A→G (p.K1596E), was identified in all affected males of family 328 (see fig. 3). This variant segregates with the XLMR phenotype in the individuals assessed. However, the location of BRWD3 disagrees with the suggestive (LOD=1.8) linkage interval (DXS207–DXS426) obtained for this family and reported elsewhere.13 This apparent discrepancy and likely incorrect localization in the work of Gedeon et al.13 was due to a wrong initial diagnosis of the condition of one of the boys (III-6) (fig. 1). At the time of the linkage mapping of this family, the boy was aged only 1 year and was classified as “unaffected.” More than 10 years later, this individual was reassessed and received the diagnosis of mild mental retardation. The two-point LOD score (5.2) for pedigree 328 was recalculated with correct clinical assignments, and the BRWD3 c.4786A→G+/− genotypes were identified. A maximum LOD score of 2.11 at BRWD3 (θ=0) was obtained. X-linked recessive inheritance and a disease frequency of 1:10,000 was assumed, with penetrance set at 1.00.
SIFT and Polyphen analysis of the sequence variant K1596E predicts this change to be deleterious to the function of the protein. Analysis of conservation of the protein sequence across species shows that the residue K1596 is conserved across mammalian species but is not conserved in Xenopus, Drosophila, or Caenorhabditis elegans (see fig. 4) (BLAST; ClustalW). Analysis of X inactivation showed random or slightly skewed inactivation of the chromosome carrying the c.4786A→G (p.K1596E) mutation. Whereas subject I-1 showed almost random (65:35) inactivation on the basis of the evaluation of the androgen receptor gene (AR), subjects II-1 and II-4 showed modestly skewed X-chromosome inactivation, at 80:20 and 84:16, respectively.
Figure 4. .
Protein alignments of vertebrate BRWD3 orthologues. Only partial sequences, corresponding to the human BRWD3 protein p.1551–1611, are shown. The lysine (K) residue at position 1596 is conserved, as indicated by an arrow. The amino acid residues that do not match the consensus sequence are highlighted with black boxes.
In this report, we have identified 2 of 250 probands from XLMR-affected families with truncating mutations in the BRWD3 gene. Mutations in this gene therefore account for ∼1% of XLMR. Neither of the identified truncating mutations was present in 520 control X chromosomes. A single synonymous sequence variant (c.5100T→C [pG1700G]) was identified in the remaining patient samples. This variant is unlikely to be pathogenic, particularly since the carrier has a disease-causing mutation in CUL4B.11 Interestingly, no other variants were identified in 520 control X chromosomes after sequencing all coding exons of BRWD3. The role of the c.4786A→G missense variant in family 328 is unclear. Although it may be responsible for XLMR in this family, the absence of shared phenotypic features with families 322 and 336 may equally indicate that it is a rare, innocuous sequence polymorphism. However, the milder clinical phenotype and absence of macrocephaly in family 328, compared with the two families with truncating mutations, may reflect the milder pathogenicity of this mutation or may reflect the common variations in phenotype typically described in XLMR-affected families published elsewhere.11
Both of the families with truncating mutations had macrocephaly noted in infancy, prominent ears, and mild-to-moderate developmental delay. A disproportionate delay in language skills has been documented in both families. The one congenital malformation seen in these families was a cleft lip in the proband of family 322. We think this is likely to relate to the paternal history of oral clefting rather than to the mutation in BRWD3.
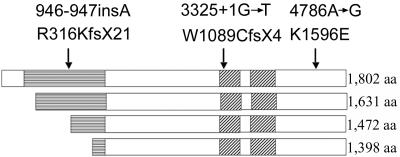
The BRWD3 gene was first identified when the breakpoints of an X-autosome translocation t(X;11) in a B-cell chronic lymphocytic leukemia (B-CLL) cell line were cloned.14 In the leukemic cells, the translocation disrupted the production of the BRWD3 transcript. BRWD3 maps to Xq21.1 (79.81–79.95 Mb). The gene is ubiquitously expressed, including in the brain and during embryonic development. BRWD3 encodes a protein containing eight WD40 domains and two bromodomains. BRWD3 shares homology with at least two other human genes: ABCG2 (MIM 603756) and WDR11 (MIM 606417) (also called “BRWD1” and “BRWD2,” respectively). Thirteen splice variants have been identified.14 From these splice variants, four possible translated products that contain the bromodomains are possible, calculated to produce polypeptides of 1,802, 1,631, 1,472, and 1,398 aa (fig. 5). It is interesting to note that the c.946-947insA mutation occurs before the translation start site of the two shorter protein isoforms. In the two longer protein isoforms, the c.946-947insA leads to a frameshift and an early truncation of the protein within the WD40-repeat region. The c.3325+1G→T mutation also causes a frameshift that results in truncation of the protein before the first bromodomain in all of the protein isoforms. Given the similarities of the phenotype between families 322 and 336, it seems likely that the two shorter protein isoforms do not play a significant role in the pathology of this disorder.
Figure 5. .
Schematic representation of the four predicted major isoforms of BRWD3. The amino terminus is to the left of the image. The WD40-repeat region is indicated by boxes marked with horizontal lines, and the bromodomains are indicated by the boxes marked with diagonal lines. The positions of the mutations relative to the translated products are indicated by arrows.
Data about Drosophila (Berkeley Drosophila Genome Project) indicate that the putative BRWD3 orthologue acts as a positive regulator of the JAK/STAT–signaling pathway.15 Up-regulation of the JAK/STAT pathway is associated with hemocyte proliferation and melanoma progression in the fly. Down-regulation of BRWD3 gene expression leads to reduced JAK/STAT–signaling activity and reduces tumor progression in the presence of a second gain-of-function mutation within the pathway.15 In Drosophila, the JAK/STAT pathway is involved in intracellular signaling, growth, and embryogenic development. The pathway is conserved in vertebrates.16 Inherited mutations in genes in the JAK/STAT pathway have not yet been implicated in human disease, although dysregulation of the pathway through somatic-activating mutations has been reported in human myeloproliferative and other hematological disorders.17
A putative mode of action of BRWD3 may be surmised from its structure. Bromodomains are typically present in chromatin-associated proteins, many of which have a chromatin-modifying function.18 The bromodomain region in the P/CAF protein has been shown to be crucial for chromatin binding via lysine-acetylated peptides associated with major acetylation sites on histones H3 and H4.19 Removal of the bromodomain in chromatin-regulatory proteins such as Swi/Snf2 and the SAGA complex removes their ability to remain anchored to chromatin.20,21 RNA-interference studies in Drosophila indicate that the BRWD3 gene functions downstream in the JAK/STAT pathway, which would be compatible with the protein having a chromatin-binding function and hence having an effect on DNA transcription.15 The presence of WD40 domains may facilitate the formation of protein complexes.14
In summary, we have identified truncating mutations in the BRWD3 gene in two families with a constellation of features including macrocephaly; mild-to-moderate delay, with specific weaknesses in early language development associated with frontal bossing; and prominent ears. The identified mutations would produce a loss-of-function effect on the pathway. Loss of BRWD3 function seems likely to lead to loss of normal developmental signaling and implicates the JAK/STAT pathway in the etiology of XLMR. Screening for mutations in this gene may be appropriate for individuals with macrocephaly and a family history compatible with XLMR.
Acknowledgments
We thank the families for their long-term cooperation. The work was supported by Australian National Health and Medical Research Council program grant 400121; the State of New South Wales (NSW) Health Department, through their support of the NSW GOLD Service; National Institute of Child Health and Human Development grant HD26202, a grant from the South Carolina Department of Disabilities and Special Needs; and the Wellcome Trust.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Berkeley Drosophila Genome Project, http://fruitfly.org/ (for splice-site prediction by neural network)
- BLAST, http://www.ncbi.nlm.nih.gov/blast/
- ClustalW, http://www.ebi.ac.uk/clustalw/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for BRWD3 [accession number NM_153252] and p.W1089CfsX4 [accession number NP_694984])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BRWD3, ABCG2, and WDR11)
- Vega Genome Browser, http://vega.sanger.ac.uk/index.html
References
- 1.Smith RD, Ashley J, Hardesty RA, Tulley R, Hewitt J (1984) Macrocephaly and minor congenital anomalies in children with learning problems. J Dev Behav Pediatr 5:231–236 10.1097/00004703-198410000-00001 [DOI] [PubMed] [Google Scholar]
- 2.Fombonne E, Roge B, Claverie J, Courty S, Fremolle J (1999) Microcephaly and macrocephaly in autism. J Autism Dev Disord 29:113–119 10.1023/A:1023036509476 [DOI] [PubMed] [Google Scholar]
- 3.Miles JH, Hadden LL, Takahashi TN, Hillman RE (2000) Head circumference is an independent clinical finding associated with autism. Am J Med Genet 95:339–350 [DOI] [PubMed] [Google Scholar]
- 4.Atkin JF, Flaitz K, Patil S, Smith W (1985) A new X-linked mental retardation syndrome. Am J Med Genet 21:697–705 10.1002/ajmg.1320210411 [DOI] [PubMed] [Google Scholar]
- 5.Baraitser M, Reardon W, Vijeratnam S (1995) Nonspecific X-linked mental retardation with macrocephaly and obesity: a further family. Am J Med Genet 57:380–384 10.1002/ajmg.1320570303 [DOI] [PubMed] [Google Scholar]
- 6.Clarke RD, Baraister M (1987) A new X-linked mental retardation syndrome. Am J Med Genet 26:13–15 10.1002/ajmg.1320260104 [DOI] [PubMed] [Google Scholar]
- 7.Johnson JP, Nelson R, Schwartz CE (1998) A family with mental retardation, variable macrocephaly and macro-orchidism, and linkage to Xq12-q21. J Med Genet 35:1026–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner G, Gideon A, Mulley J (1994) X-linked mental retardation with heterozygous expression and macrocephaly: pericentromeric gene localization. Am J Med Genet 51:575–580 10.1002/ajmg.1320510456 [DOI] [PubMed] [Google Scholar]
- 9.Proops R, Mayer M, Jacobs PA (1983) A study of mental retardation in children in the Island of Hawaii. Clin Genet 23:81–96 [DOI] [PubMed] [Google Scholar]
- 10.Hu LJ, Blumenfeld-Heyberger S, Hanauer A, Weissenbach J, Mandel JL (1994) Non-specific X-linked mental retardation: linkage analysis in MRX2 and MRX4 families revisited. Am J Med Genet 51:569–574 10.1002/ajmg.1320510455 [DOI] [PubMed] [Google Scholar]
- 11.Tarpey PS, Raymond FL, O’Meara S, Edkins S, Teague J, Butler A, Dicks E, Stevens C, Tofts C, Avis T, et al (2007) Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am J Hum Genet 80:345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarpey PS, Stevens C, Teague J, Edkins S, O’Meara S, Avis T, Barthorpe S, Buck G, Butler A, Cole J, et al (2006) Mutations in the gene encoding the sigma 2 subunit of the adaptor protein 1 complex, AP1S2, cause X-linked mental retardation. Am J Hum Genet 79:1119–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gedeon A, Kerr B, Mulley J, Turner G (1994) Pericentromeric genes for non specific X-linked mental retardation. Am J Med Genet 51:553–564 10.1002/ajmg.1320510453 [DOI] [PubMed] [Google Scholar]
- 14.Kalla C, Nentwich H, Schlotter M, Mertens D, Wildenberger K, Dohner H, Stilgenbauer S, Lichter P (2005) Translocation t(X;11)(q13;q23) in B-cell chronic lymphocytic leukemia disrupts two novel genes. Genes Chromosomes Cancer 42:128–143 10.1002/gcc.20131 [DOI] [PubMed] [Google Scholar]
- 15.Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M (2005) Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature 436:871–875 10.1038/nature03869 [DOI] [PubMed] [Google Scholar]
- 16.Hombria JC, Brown S (2002) The fertile field of Drosophila Jak/STAT signalling. Curr Biol 12:R569–R575 10.1016/S0960-9822(02)01057-6 [DOI] [PubMed] [Google Scholar]
- 17.Campbell PJ, Green AR (2006) The myeloproliferative disorders. N Engl J Med 355:2452–2466 10.1056/NEJMra063728 [DOI] [PubMed] [Google Scholar]
- 18.de la Cruz X, Lois S, Sanchez-Molina S, Martinez-Balbas MA (2005) Do protein motifs read the histone code? Bioessays 27:164–175 10.1002/bies.20176 [DOI] [PubMed] [Google Scholar]
- 19.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491–496 10.1038/20974 [DOI] [PubMed] [Google Scholar]
- 20.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL (2002) Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369–379 10.1016/S0092-8674(02)01005-X [DOI] [PubMed] [Google Scholar]
- 21.Syntichaki P, Topalidou I, Thireos G (2000) The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature 404:414–417 10.1038/35006136 [DOI] [PubMed] [Google Scholar]