Abstract
Deletions of chromosome 1q42-q44 have been reported in a variety of developmental abnormalities of the brain, including microcephaly (MIC) and agenesis of the corpus callosum (ACC). Here, we describe detailed mapping studies of patients with unbalanced structural rearrangements of distal 1q4. These define a 3.5-Mb critical region extending from RP11-80B9 to RP11-241M7 that we hypothesize contains one or more genes that lead to MIC and ACC when present in only one functional copy. Next, mapping of a balanced reciprocal t(1;13)(q44;q32) translocation in a patient with postnatal MIC and ACC demonstrated a breakpoint within this region that is situated 20 kb upstream of AKT3, a serine-threonine kinase. The murine orthologue Akt3 is required for the developmental regulation of normal brain size and callosal development. Whereas sequencing of AKT3 in a panel of 45 patients with ACC did not demonstrate any pathogenic variations, whole-mount in situ hybridization confirmed expression of Akt3 in the developing central nervous system during mouse embryogenesis. AKT3 represents an excellent candidate for developmental human MIC and ACC, and we suggest that haploinsufficiency causes both postnatal MIC and ACC.
Human geneticists have often taken advantage of structural chromosome rearrangements to map and clone human disease genes, including the causative genes for human brain and eye malformations such as holoprosencephaly (SHH, SIX3, TGIF, and ZIC2), Dandy-Walker malformation (ZIC1 and ZIC4), lissencephaly (DCX and LIS1), microphthalmia (SOX2), and anterior-chamber defects (FOXC1 and PAX6), among others.1–13 We have performed mapping studies of six patients with rearrangements of distal 1q42-q44 associated with several different brain malformations, including congenital and postnatal microcephaly (MIC), agenesis of the corpus callosum (ACC [MIM 217990]), cerebellar vermis hypoplasia (CVH), and polymicrogyria (PMG).
The abnormalities of three patients with MIC, ACC, and CVH consist of two large interstitial deletions of distal 1q42- or 1q43-q44 and a complex unbalanced rearrangement with two deleted and two inverted segments between 1q41 and 1q44. Another boy with postnatal MIC and ACC but not CVH has an apparently balanced reciprocal translocation between distal 1q44 and 13q32. The remaining patient had CVH and PMG but not MIC or ACC and had a terminal 1q44 deletion resulting from unbalanced segregation of a translocation between 1q44 and 11p15. We identified two genes—AKT3 and ZNF238—closest to the chromosome 1 translocation breakpoint, performed expression studies of mouse embryos, and sequenced both genes and an intervening highly conserved noncoding sequence in a cohort of 47 patients with ACC. Our results collectively implicate haploinsufficiency of AKT3 as a cause of human MIC and ACC.
Material and Methods
Study Subjects
We ascertained six children with structural rearrangements of distal chromosome 1q who presented at our local institutions or were referred to our research projects. Three were evaluated as newborns with complicated neonatal courses, whereas the remaining three were evaluated during early childhood because of developmental delay. All six had multiple congenital anomalies that prompted chromosome analysis.
Subjects with ACC
We (W.B.D. and E.H.S.) selected 55 patients with ACC and normal chromosome analysis from our large Brain Malformation Research Databases for sequencing of positional candidate genes identified in the patients with 1q4 deletion and translocation. This cohort included 11 patients with ACC only, 8 with ACC and congenital MIC (some with other brain anomalies), 10 with ACC and cerebellar hypoplasia, and 26 with ACC and other anomalies.
Array Comparative Genomic Hybridization (aCGH)
aCGH was performed using a high-resolution BAC-tiling set from the University of California–San Francisco (UCSF) Comprehensive Cancer Center that contains ∼33,000 BAC PCR-based representations, each printed once on 18×36-mm2 chromium-coated slides.14 Before hybridization, 0.6 μg of fluorometry-quantified patient DNA and normal same-sex reference DNA were separately labeled with either Cy3 or Cy5 by random priming with separate fluor-conjugated 2-deoxycytidine 5-triphosphate and were purified from unincorporated nucleotides by spinning through a G-50 Sephadex column. Equal amounts of Cy3-labeled patient and Cy5-labeled reference DNA were mixed and precipitated with Cot 1 DNA, then resuspended in hybridization solution and applied to the array for 48 h. Arrays were washed in a 50% formamide solution and then were mounted in a 4',6-diamidino-2-phenylindole (DAPI) solution before imaging. Sixteen-bit 2,048×2,048 pixel DAPI, Cy3, and Cy5 images were collected using a custom CCD camera system,5 and the data were analyzed, using UCSF SPOT15 to automatically segment the array spots and to calculate the log2 ratios of the total integrated Cy3 and Cy5 intensities for each spot. A second custom program, SPROC, applied quality criteria to the measurements and mapped them to the genome, currently with use of the May 2004 freeze of the draft sequence (UCSC Genome Bioinformatics hg17). The detailed protocols for each of these methods are available on the UCSF Comprehensive Cancer Center Web site. All aCGH results were confirmed using FISH or loss-of-heterozygosity (LOH) studies. For LOH analysis, DNA from the probands (subjects LR05-202, LR06-076, and LR02-409), mothers (LR05-202-1 and LR06-076-1), and fathers (LR05-202-2 and LR06-076-2) was amplified with primers for markers spanning from 215.36 Mb to 246.19 Mb within 1q4. Primer sequences are available at the UCSC Genome Bioinformatics Web site. Products were resolved and sized by capillary electrophoresis with use of an ABI 3730XL (Applied Biosystems).
Deletion and Breakpoint Mapping by FISH
Deletion and breakpoint mapping by FISH was undertaken for patients LP94-079, LR04-249, LR02-409, and LR05-101. Chromosome preparations were produced from peripheral-blood lymphocytes by conventional techniques. BAC and fosmid clones from the regions of interest were identified from the Ensembl genome database. They were labeled with Spectrum Green or Spectrum Orange (Vysis), with use of nick translation. FISH was performed using standard methods, and images were captured using a cooled CCD camera and Smart Capture software (Applied Imaging). To provide confirmation for patients LP94-079, LR02-409, and LR05-101, each FISH experiment was undertaken using breakpoint-spanning clones for which 15 good-quality metaphase images were studied, as well as 5 or more good-quality metaphase images for the remaining clones. We had only a small archival sample for deceased patient LR04-249, which prevented the use of metaphase spreads. FISH signals were instead sought in interphase cells, and 5–10 cells were imaged.
Sequencing of AKT3, UC.43, and ZNF238
Primer sequences for the coding exons of AKT3 and ZNF238, as well as the ultraconserved element UC.43 located within intron 1 of AKT3,16 were obtained from the Primer3 database (primers and PCR conditions are available on request). Amplicons were engineered to span the entire transcribed sequence of the two genes, including the 5′ and 3′ UTRs, and the entire reported sequence of UC.43. However, primers designed to sequence the final 249 bp of the coding sequence of ZNF238 did not amplify on genomic DNA, so this region was not sequenced. The products were purified using Sephacryl media (GE Healthcare) with Durapore (Millipore) polyvinylidene difluoride columns, and amplicons were then sequenced by the DNA Sequencing and Genotyping Core at the University of Chicago. The reactions used a 1/4 volume BigDye 3.1 terminator kit protocol, with standard cycling conditions, for 35 cycles with use of 1.2 μl template and 1.2 μl primer. Runs were done on the 3730XL instrument from Applied Biosystems, with use of standard 2-h runs. Sequence data were analyzed using Sequencher (Gene Codes) sequence-analysis software and were compared with annotated sequence from the National Center for Bioinformatics.
Analysis of Expression of Znf238 and Akt3 by In Situ Hybridization
Whole-mount in situ expression analysis was conducted on CD1 mouse embryos at embryonic day 10.5 (E10.5) and E12.5, as described elsewhere.17 For timed pregnancies, noon of the vaginal-plug date was E0.5. Antisense and sense probes were generated by in vitro transcription from Research Genetics mouse IMAGE clones 6417039 (Znf238) and 30089997 (Akt3), which were first sequenced to confirm identity.
Results
Clinical Details of Study Subjects
We studied a group of six patients with deletions of distal 1q42-q44 that are all associated with developmental brain malformations (table 1 and figs. 1 and 2).
Table 1. .
OFC and Brain Malformations in Patients with 1q4 Deletion[Note]
| OFC SDb |
||||
| Patienta | AKT3 | At Birth | Later (Age) | Brain Malformations |
| Gentile et al.18 | Deleted | −2 | −4 (8 mo) | MIC and partial ACC |
| Van Bever et al.19 | Deleted | −3 | NA | MIC, partial ACC, and CVH |
| LR04-249 | Deleted | −4 | NA | MIC, ACC, and CVH |
| LR05-202 | Deleted | −3 | −5 (2 years) | MIC, ACC, and CVH |
| LR02-409 | Deleted | −5c | −7 (5 years) | MIC, partial ACC, and CVH |
| LR06-076 | Deleted | −2d | −3.5 (4 years) | Mild MIC and partial ACC |
| LR05-101 | Disrupted | 50th | −2.5 (2 years) | MIC and ACC |
| de Vries et al.20 | Not tested | NA | −5 (2.5 years) | MIC and partial ACC |
| de Vries et al.21 | Not tested | −2 | −5 (1 year) | MIC and partial ACC |
| LP94-079 | Not deleted | 10th | NA | CVH and PMG |
| Daniel et al.22 | Not deleted | Normal | Normal (15 years) | None seen on CT scan |
Note.— NA=not available.
Patients other than those from the present study are listed by the authors who reported them.
Or percentile.
At age 15 wk.
At age 4 mo.
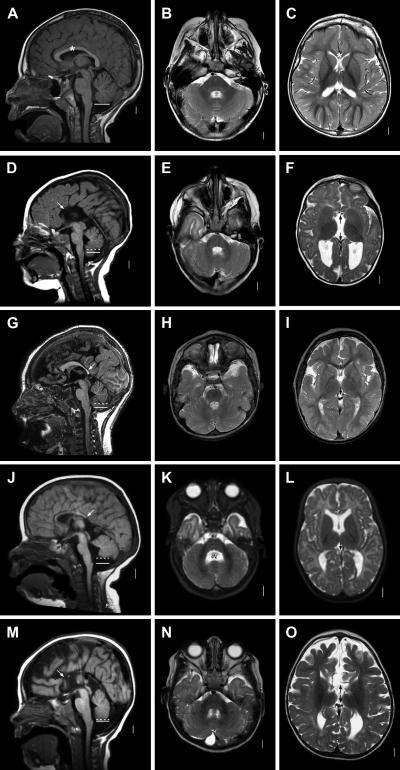
Figure 1. .
Brain-imaging abnormalities in patients with deletion 1q4, shown with T1-weighted midline sagittal (left column), T2-weighted low axial (middle column), and T2-weighted higher axial (right column) sequences in an unaffected 3-year-old child (LR06-130) (A–C) and in patients LR05-202 (D–F), LR06-076 (G–I), LR02-409 (J–L), and LR05-101 (M–O). In the normal scans, the corpus callosum is marked by an asterisk (*) (A and C), and the inferior margin of the vermis is marked by a solid white horizontal line at the level of the obex (A). In the images of the four patients, the corpus callosum is either absent (white arrows in panels D and M and two black arrows in panels F and O) or small with a narrow body and absent splenium (white arrow in panels G and J and single black arrow indicating absent posterior callosum in panels I and L). The cerebellar vermis is small in three patients who have the 1q4 deletion, with the inferior margin (dashed white lines in panels D, G, and J) located well above the level of the obex (solid white lines in panels D, G, and J). The vermis appears mildly small in the boy with the 1q;13q translocation, with a mildly reduced anterior-posterior diameter of the vermis resulting in an enlarged cisterna magna extending behind the cerebellum (M), even though the inferior vermis is located near the level of the obex (dashed and solid lines in panel M). The cerebellar hemispheres are normal (E, H, K, and N), and the fourth ventricle is normal (E, H, and N) or mildly enlarged (“4V” in panel K). Lower-resolution images of our other two patients (LP94-079 and LR04-249) are shown in figure 2.
Figure 2. .
Brain-imaging abnormalities in patients with deletion 1q4, shown with head CT scan (A–C) or with T1-weighted midline sagittal (D) and T2-weighted axial (E and F) MRI sequences in patients LP94-079 (A–C) and LR04-249 (D–F). In patient LP94-079, the cortex appears thickened in the posterior frontal and perisylvian regions (white arrow[s] in panels A–C), suggesting PMG, although the resolution is low. The lateral ventricles are located in the normal position close to the midline (A and B), suggesting that the corpus callosum is normal. The cerebellar vermis is small, since it is not seen at the level of the low midbrain (white arrowhead in panel A), and a small skull defect is seen beneath an occipital cephalocele (white arrowhead in panel C). In LR04-249, gyral pattern is poorly developed, even for 35 wk gestation, and the corpus callosum is absent (white arrow in panel D and two black arrows in panel E). The cerebellar vermis is small, with the inferior margin (dashed line in panel D) well above the level of the obex (solid line in panel D).
LP94-079
This girl was born at term, with a birth weight of 2.9 kg, length of 48 cm (both 25th percentile), and an occipitofrontal circumference (OFC) of 31.75 cm (10th percentile), and she had low blood glucose. Family history was significant for multiple miscarriages in close relatives and a paternal cousin who died with probable hydrocephalus; the pregnancy was complicated by polyhydramnios. Examination demonstrated a high forehead with vertical furrowing, hypertelorism, mildly large tongue, small jaw, low posterior hairline, widely spaced nipples, mild rhizomelic shortening of all limbs, short fingers with mildly broad distal phalanges, fifth-finger clinodactyly, abnormal palmar and plantar creases, and anteriorly displaced anus. She did not have organomegaly. Echocardiogram demonstrated patent foramen ovale, secundum atrial septal defect, small patent ductus, and moderate tricuspid valve insufficiency. Cranial CT scan showed thick and irregular cortex, consistent with PMG in the posterior frontal and perisylvian regions, best seen on the right, as well as severe CVH and a small occipital cephalocele (fig. 2A–2C). The corpus callosum appeared normal. She died at age 12 d from complications of supraventricular tachycardia.
LR04-249
This boy was born prematurely, at 35 wk gestation, because of preterm labor. His birth weight was 1.35 kg (−3 SD), length was 36.5 cm (−4 SDs), and OFC was 25 cm (−4 SD). His primary medical problems were intestinal obstruction, anemia secondary to ABO incompatability, and increasingly frequent episodes of desaturation, which led to his death at age 6 wk. He had obvious growth retardation, including severe MIC, low-set ears, long smooth philtrum, thin lips, small jaw, high-narrow palate, severe hypospadias, undescended testes, and sacral dimple. Further tests demonstrated patent foramen ovale, mild right-atrial enlargement, abnormal “domed” pulmonary valve, dilatation of the right pulmonary artery, microcolon with intestinal obstruction, small kidneys with renal insufficiency, 13 pairs of ribs, and butterfly vertebrae at T5 and T10. Brain magnetic resonance imaging (MRI) showed a very immature gyral pattern, even for 35 wk gestation, that suggests a possible cortical malformation (type uncertain); mildly enlarged and widely separated lateral ventricles; total ACC; and moderate CVH (fig. 2D–2F).
LR05-202
This girl was born at term after a pregnancy complicated by abnormal triple-screen results. However, level II ultrasound was normal, so the family did not proceed with amniocentesis. At birth, she had MIC with a birth OFC of 30.5 cm (−3 SD), two ventricular septal defects, and a single palmar crease. OFC was 38.5 cm at age 7 mo (−4 SD) and 41 cm at age 2 years (−5 SD). She was noted to have delays in cognitive, fine and gross motor, and social skills. Seizures began at age 8.5 mo and led to an evaluation that included brain MRI. At age 2.5 years, her seizures were well controlled with phenobarbitol treatment. A review of systems revealed corrected myopia; three small ventricular septal defects, two of which had closed; mild eczema; history of urinary-tract infections; mildly distended right renal pelvis on kidney ultrasound; and allergy to milk. She had hypotonia, a broad nasal bridge, low-set ears, short webbed neck, epicanthal folds, and small jaw. She was able to crawl, pull herself to a standing position, and vocalize, although she does not use intelligible words. Brain MRI at age 8 mo revealed a normal gyral pattern, total ACC, mildly enlarged third and lateral ventricles, markedly reduced white-matter volume, small bilateral Probst bundles, and mild CVH (fig. 1D–1F).
LR06-076
This boy was born at term; birth OFC is not available, although it was described as borderline small. His OFC was 39.5 cm at age 4 mo (−2 SD), 41 cm at age 9 mo, 45 cm at age 30 mo, and 46 cm at age 4 years (last three values approximately −3.5 SD). Generalized seizures began at age 2.5 years and have been partially controlled with phenobarbital and valproic acid treatment. On examination at age 4 years, he had bilateral ear pits, small jaw, bilateral supernumerary nipples, abnormal foreskin of the penis, undescended left testicle, intermittent esotropia, and generalized spasticity with brisk reflexes. He did not understand speech and was not able to walk or talk. Brain MRI demonstrated a normal gyral pattern, diffusely decreased white-matter volume, partial ACC with thin posterior body and absent splenium, and normal cerebellum (fig. 1G–1I).
LR02-409
This girl was born at 36 wk gestation by emergency cesarean section because of placenta previa, with a birth weight 1.9 kg and length of 42 cm (both 3rd percentile). Her OFC was 33.3 cm at age 15 wk (−4 SD for corrected age of 11 wk) and fell to −5 SD by age 1 year. She began having seizures at age 8 mo. At age 5 years, she was small for her age, with weight of 10.45 kg (−4 SD), length of 93 cm (−3 SD), and OFC of 41.5 cm (−7 SD). She was unable to walk or use any words, suggesting severe mental retardation. On examination, she was strikingly dysmorphic, with MIC, bitemporal narrowing, full cheeks, prominent epicanthal folds, anteverted nares, and tented upper lip with a long philtrum. She also had a mixed sensorineural and conductive hearing loss, left esotropia, and atrial septal defect. Brain MRI demonstrated a normal gyral pattern, partial ACC with thin body and absent splenium, and mild CVH (fig. 1J–1L).
LR05-101
This boy was born at term after an uncomplicated pregnancy, except that a prenatal ultrasound at 22 wk gestation did not detect the septum pellucidum. His birth weight was 3.88 kg (75th percentile), length was 54 cm (90th percentile), and OFC was 35 cm (50th percentile). Developmental delay and generalized hypotonia were noted at age 3 mo because of lack of head control and visual tracking, and seizures began at age ∼1 year. By age 2 years, he had global developmental delay, axial hypotonia, limb spasticity, and poor feeding that required placement of a nasogastric tube. He was not dysmorphic. Examination revealed a small penis. Serial exams demonstrated postnatal MIC, and his OFC was at the 25th percentile by age 3 mo and at the 2nd percentile by age 12 mo. His OFC at age 28 mo was 46.3 cm (−2.5 SD). Brain MRI demonstrated a normal gyral pattern, reduced white matter, mildly enlarged and widely separated lateral ventricles, complete ACC with Probst bundles along the medial hemispheric walls, and mild CVH (fig. 1M–1O).
Precise Mapping of Chromosome 1q Abnormalities by FISH and aCGH
We mapped the 1q4 breakpoints of all six patients by either BAC and fosmid FISH or by aCGH (fig. 3).
Figure 3. .
Precise delineation of chromosome rearrangements in distal 1q42-q44 in study patients. Deletion mapping of patients LR02-409 and LR05-202 defines a minimal deleted region of 1.25 Mb between BACs RP11-657L23 and RP1-241M7. This region is deleted in four patients, all of whom had ACC and MIC. Patient LP94-079 has a subtelomeric deletion that is distal to RP1-241M7 and had neither ACC or MIC. In patient LR05-101, the breakpoint of the t(1;13) balanced de novo translocation lies within this region and is situated with BAC clone RP11-351N5 and fosmid clone G248P89661B9. This is upstream of the first exon of AKT3.
LP94-079
Chromosome analysis and FISH detected an unbalanced translocation inherited from the proband's father: 46,XX,der(1)t(1;11)(q44;p15.3)pat. The extent of the deletion was determined by mapping the chromosome 1 breakpoint of the balanced translocation in the father. The clone RP11-518I10 (242.57–242.65 Mb) at 1q44 was split between the translocation derivatives, indicating that all DNA telomeric to this probe was present in one copy in this child (fig. 3).
LR04-249
Chromosome analysis showed a large interstitial deletion: 46,XY,del(1)(q42.1q44) de novo. FISH analysis demonstrated that the deletion extended from BAC clone RP11-87P4 (230.78–230.9 Mb) to RP11-399B15 (243.22–243.42 Mb), a region of >13 Mb.
LR05-202
Chromosome analysis was interpreted as normal, but telomeric probes detected a de novo deletion of 1qter. aCGH demonstrated a large subtelomeric de novo deletion beginning after BAC RP11-147E13 (ending at 235.38 Mb) and extending to the telomere, a distance of 11.9 Mb (fig. 4). This deletion was also confirmed using LOH analysis with STRP (microsatellite) markers beginning with marker D1S227 (215.36 Mb) and extending to the subtelomeric marker D1S2682 (246.19 Mb). Some of these interspersed markers were uninformative, since both parents carried the same allele(s), but, for all of these, the patient was homozygous for an allele carried by the mother, consistent with the pattern established by the unambiguously deleted markers (table 2).
Figure 4. .
aCGH demonstrating deletions (arrows) in patients with the 1q4 deletion. Data are represented as the corrected log2 ratios between control and patient DNA. A, LR05-202. B, LR06-076. C, LR02-409. The distal region that is not deleted in LR06-076 is marked with an arrowhead.
Table 2. .
LOH Analysis of Patients with the 1q4 Deletion[Note]
| Position |
|||||||||
| Marker | Marshfield Map (cM) |
Starting Chromosome | LR05-202 | LR05-202-1 | LR05-202-2 | LR06-076 | LR06-076-1 | LR06-076-2 | LR02-409 |
| D1S227 | 238.52 | 1:215,361,062 | 70/70 (Heterozygous) | 68/70 | 70/70 | 68/68 (Uninformative) | 68/68 | 64/68 | 68/72 (Heterozygous) |
| D1S549_G | 239.66 | 1:217,710,236 | 188/196 (Heterozygous) | 188/196 | 188/188 | 184/188 (Heterozygous) | 184/192 | 180/188 | 188/188 |
| D1S2833 | 245.05 | 1:228,814,635 | 95/107 (Heterozygous) | 95/95 | 105/107 | 97/97 (Hemizygous, maternal) | 95/97 | 101/101 | 105/105 |
| D1S2709 | 247.23 | 1:230,096,973 | 199/201 (Heterozygous) | 195/201 | 197/199 | 195/199 (Heterozygous) | 199/199 | 195/199 | 197/201 (Heterozygous) |
| D1S3462_G | 247.23 | 1:230,017,109 | 231/237 (Heterozygous) | 231/237 | 234/237 | 234/243 (Heterozygous) | 237/243 | 231/234 | 231/243 (Heterozygous) |
| D1S2800 | 252.12 | 1:232,524,978 | 213/213 (Uninformative) | 209/213 | 209/213 | 209/215 (Heterozygous) | 202/209 | 215/215 | 215/217 (Heterozygous) |
| D1S235_G | 254.64 | 1:233,960,378 | 202/213 (Heterozygous) | 213/213 | 202/202 | 213/215 (Heterozygous) | 213/213 | 215/219 | 202/202 |
| D1S2850 | 256.26 | 1:234,697,246 | 152/152 (Uninformative) | 152/156 | 152/152 | 154/154 (Uninformative) | 154/154 | 150/154 | 150/154 (Heterozygous) |
| D1S2670 | 262.96 | 1:237,615,801 | 165/165 (Uninformative) | 165/175 | 165/175 | 173/177 (Heterozygous) | 177/177 | 165/173 | 165/175 (Heterozygous) |
| D1S2785 | 266.27 | 1:238,943,306 | 183/183 (Hemizygous, maternal) | 183/183 | 179/179 | 177/177 (Hemizygous, maternal) | 177/179 | 175/183 | 173/183 (Heterozygous) |
| D1S304 | 267.51 | 1:239,355,104 | 175/175 (Uninformative) | 175/175 | 171/175 | 173/173 (Uninformative) | 173/175 | 173/173 | 171/171 |
| D1S2842 | 273.46 | 1:240,939,182 | 345/345 (Hemizygous, maternal) | 345/351 | 341/353 | 349/349 (Hemizygous, maternal) | 349/349 | 345/345 | 345/345 |
| D1S1609_G | 274.53 | 1:242,132,480 | 243/243 (Uninformative) | 239/243 | 239/243 | 239/239 (Hemizygous, maternal) | 239/239 | 243/248 | 248/248 |
| D1S423 | 277.8 | 1:243,612,280 | 92/92 (Hemizygous, maternal) | 92/94 | 94/94 | 92/92 (Uninformative) | 92/92 | 92/92 | 92/92 |
| D1S2836 | 285.75 | 1:244,936,701 | 256/256 (Hemizygous, maternal) | 243/256 | 243/246 | 243/243 (Uninformative) | 243/243 | 243/246 | 248/250 (Heterozygous) |
| D1S2682_G | 288.29 | 1:246,196,829 | 149/149 (Hemizygous, maternal) | 149/149 | 130/151 | 130/149 (Heterozygous) | 130/130 | 130/149 | 130/145 (Heterozygous) |
Note.— Allele sizes from the probands (LR05-202, LR06-076, and LR02-409), mothers (LR05-202-1 and LR06-076-1), and fathers (LR05-202-2 and LR06-076-2) are listed for each marker (with associated interpretation). No DNA was available at the time of the study for the parents of proband LR02-409, so only heterozygosity is noted. Chromosome numbering is derived from the hg18 genome assembly (March 2006). Maternal=maternally inherited region.
LR06-076
Chromosome analysis was normal, but aCGH detected a small deletion of 1q44 between BACs RP11-158M3 (237.51–237.68 Mb) and RP11-606D18 (245.33–245.50 Mb), demonstrating a deletion of 7.6A Mb between 237.68 Mb and 245.33 Mb. This interstitial deletion was confirmed by a second aCGH analysis and by LOH analysis (fig. 4 and table 2).
LR02-409
The initial chromosome analysis detected an apparently balanced paracentric inversion of distal 1q. In view of the girl's phenotype, chromosome analysis was repeated and demonstrated a complex rearrangement of distal 1q, with two paracentric inversions 17.36 Mb apart—inv(1)(q32.2q42.2) and inv(1)(q43q44)—each containing a small interstitial deletion. The proximal inversion extends from RP11-434B7 (211.48–211.60 Mb) in 1q32.2 to RP11-99J16 (228.96–229.01 Mb) in 1q42.2, whereas the distal inversion extends from RP11-433N10 (236.57–236.76 Mb) in 1q43 to CTB-160H23 at 1q44. In addition, we found two deletions in the region. The proximal 4-Mb deletion was located in 1q41-q42.12 and included the clones RP11-332J14 (218.70–218.87 Mb) and RP11-192M1 (224.12–224.28 Mb). The distal deletion was located in 1q43-q44. It included the clones RP11-553N16 (240.00–240.15 Mb) and RP1-241M7 (242.49–242.57 Mb) and was flanked by the nondeleted clones RP11-80B9 (238.96–239.13 Mb) and RP11-518L10 (242.57–242.65 Mb). The deletion is therefore <3.5 Mb (fig. 5). These deletions were confirmed with aCGH (fig. 4). LOH analysis was performed without parental samples, but results were consistent with the deletion boundaries defined by FISH and aCGH (table 2).
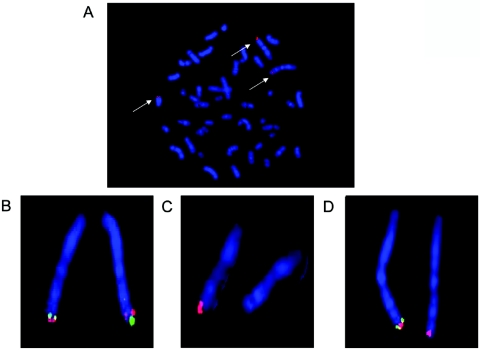
Figure 5. .
FISH mapping of patients LR05-101 (A) and LR02-409 (B–D). Analysis of LR05-101, who has a de novo 46,XY,t(1;13)(q44;q32) translocation, shows signals for fosmid G248P89661B9 (red) located on the normal and derived chromosomes 1, as well as on the derived chromosome 13. This confirms that it spans the breakpoint on chromosome 1q44. Analysis of LR02-409 with probes RP11-90B9 (green) and RP11-399B15 (red [B]) shows the distal inversion, since the position of the two probes is reversed on the two chromosome 1 homologs. The associated deletion inside the distal inversion is demonstrated by loss of RP11-553N16 (red [C]) and by loss of RP11-411G13 (green [D]) compared with RP11-438H8 (red [D]).
LR05-101
Chromosome analysis demonstrated an apparently balanced reciprocal translocation: 46,XY,t(1;13)(q44;q32) de novo. The breakpoint on 1q44 was mapped to clone RP11-351N5 (241.98–242.13 Mb), since this clone gave signals on the normal chromosome 1, as well as both derivative chromosomes in FISH experiments. This clone contains the AKT3 gene (Ensembl). Fine mapping of the chromosome 1 breakpoint with use of fosmid clones was undertaken. The breakpoint was found to lie in clone G248P89661B9 (242.08–242.12 Mb), a 38-kb clone whose proximal end lies 4.5 kb upstream of the first exon of AKT3 (fig. 5). The ZNF238 gene is located on the distal side of the breakpoint, 210 kb away from AKT3 (fig. 3). The chromosome 13 breakpoint was mapped between fosmids G248P81419A12 and G248P86855F5, a region that contains no known genes. aCGH analysis demonstrated that no deletions larger than two BACs were found in this patient (data not shown), consistent with the balanced translocation identified by FISH.
Sequencing of AKT3, UC.43, and ZNF238
We sequenced all 13 coding exons of AKT3, as well as an ultraconserved element (UC.43) that lies within the first intron of AKT3. Ultraconserved elements such as UC.43 are defined as sequences of >200 bp with 100% conservation among human, rat, and mouse.16 All regions were sequenced in at least 47 patients. No pathogenic mutations were defined. The sequence of UC.43 was normal in 43 patients. The remaining 4 of 47 patients, as well as 1 of 8 control subjects, had a heterozygous SNP (C→T at position 111153 of AL662889). In addition, we sequenced the first 1,315 of 1,566 bp of the coding region of the single exon of ZNF238 and found a heterozygous synonymous substitution (val74val GTG→GTA) in one patient. Sequencing of AKT3 and ZNF238 for patient LR05-101 with the t(1;13) translocation did not define any sequence changes in either the coding region or UTRs of either gene. We were therefore unable to test for monoallelic versus biallelic expression of either gene in this patient.
Developmental Expression of Akt3 and Znf238
RT-PCR and northern analysis data indicate that Akt3 and Znf238 are both expressed in the adult mouse brain23,24; however, developmental expression patterns have not been reported for either gene. We therefore performed whole-mount in situ hybridization during mouse embryogenesis, to determine whether these genes were expressed in the developing mouse cortex. In agreement with the online Brain Gene Expression Map (BGEM),25 we determined that both genes are very widely expressed in the developing CNS during embryogenesis, with high levels of expression throughout the dorsal forebrain during E9.5–E15.5. In the developing cortex, expression is observed both on the dorsal midline and laterally in the forming layers at E15.5.
Discussion
At least 30 patients with deletion of chromosome 1q42-q44 have been reported, with consistent mental retardation of variable severity, frequent seizures, and numerous congenital anomalies.19 The latter include up-slanting palpebral fissures, ear anomalies, small jaw, cleft palate, heart malformations, abnormalities of the hands and feet, hypospadias, and cryptorchidism. MIC was noted in 24 of 28 patients, although serial OFC measurements were rarely noted. At least 12 patients had ACC, most with deletion 1q42- or 1q43-qter, but one interstitial deletion of 1q42 and one small terminal deletion of 1q44 have been reported, suggesting the possibility of two separate loci.18,19,26–29 In addition, two cousins with both deletion 1q44 and duplication 12p13.3 were reported with the cortical malformation PMG.30 Two of the most consistent abnormalities in this syndrome have thus been MIC and ACC, which suggests that one or more genes involved with brain size and callosal development are found in this region. We have identified the second messenger-regulated serine-threonine kinase AKT3 as a strong candidate gene for postnatal MIC and ACC and as a probable contributor to congenital MIC, on the basis of detailed physical-mapping studies and comparison with two separate Akt3 mouse knockouts.
Physical Mapping of 1q4 Deletions and Translocation
We identified an ∼3.5-Mb critical region defined by the centromeric and telomeric boundaries of the distal deletion in subject LR02-249, who has both MIC and partial ACC. This region is not shared by the deletion of subject LP94-079, who has neither MIC nor ACC. This region extends from RP11-80B9 to RP11-241M7 (fig. 3) and contains 12 known genes: RGS7, FH, KMO, OPN3, EXO1, MLP3C_HUMAN, NP_689879, CEP170, SDCCAG8, AKT3, ZNF238, and NP_001013732. On the basis of reported expression in brain and lack of known associated phenotypes (Ensembl), only a few of these—especially OPN3, CEP171, AKT3, and ZNF238—are good candidates. We hypothesize that haploinsufficiency of one or more of these genes leads to MIC and ACC.
The 1q4 breakpoint of subject LR05-101, who also has striking postnatal MIC and ACC associated with a balanced reciprocal t(1;13) translocation, falls within this critical region. This breakpoint lies between the genes AKT3 and ZNF238 and is positioned ∼20 kb upstream of the first exon of AKT3 and ∼150 kb proximal to ZNF238. The absence of SNPs in either of these two mRNAs in subject LR05-101 precluded the investigation of monoallelic expression in this boy. Disruption of gene expression by chromosome breakpoints that are near but not within the classically defined area of a gene is a well-recognized mechanism31,32 and suggests that, of the four genes defined by deletion mapping, AKT3 and ZNF238 represent the most attractive positional candidates for a role in MIC and ACC. Our data do not suggest a single locus for CVH, since patients LP94-079 and LR02-409 do not have overlapping deletions. These data also suggest that the PMG-causative gene is likely to be located in the most distal 4.5 Mb of 1q44.
Akt3 and Znf238 Are Widely Expressed in Brain
We have demonstrated through whole-mount in situ hybridization in mouse that both Akt3 and Znf238 are widely expressed in the CNS during embryogenesis. We found high levels of expression throughout the dorsal forebrain (E9.5-E15.5) and in the developing cortex (E15.5), consistent with the findings of others. Becker et al.24 studied the expression pattern of Znf238 by northern blot and, in adult mice, found transcription only in the brain with the highest levels in the cerebellum. The gene was also expressed in the head in mouse embryos, which suggests a role in both the developing and mature CNS. Akt3 is more highly expressed in brain than are its protein family members Akt1 and Akt2 and is the predominant Akt protein in cortex and hippocampus.23 In humans, AKT3 is expressed in brain, lungs, and kidneys in adults and in fetal brain, heart and liver. These data suggest a role for both AKT3 and ZNF238 in brain development and do not allow further prioritization of these genes as ACC or MIC candidates on the basis of expression data alone.
ZNF238 is a C2H2-type zinc-finger protein for which little functional information is available. It contains a Pox virus and zinc finger (POZ) domain that functions as a sequence-specific DNA-binding transcriptional repressor.24,33,34 No animal models have been reported. A single highly conserved noncoding sequence, designated “UC.43,” is located in intron 1 of AKT3 but may regulate ZNF238 expression, since such noncoding sequences are preferentially found near transcription factors.16
AKT3 Functions Downstream of PI3K and Regulates Brain Size in Mice
Substantial functional data exist that support AKT3 as a causative gene for MIC. The AKT or protein kinase B family consists of three proteins that contain a pleckstrin-homology domain (a phosphorylation substrate), a kinase domain, and a regulator domain, with the last identical in all three AKT proteins (reviewed by Bellacosa et al.35). All three AKT proteins function in a signaling pathway with insulin or insulinlike growth factor 1 (IGF1), insulin receptor substrate (IRS), phosphoinositide-3-kinases (PI3K), phophoinositide-3,4,5-triphosphate (PIP3), and phosphoinositide-dependent protein kinases (PDPK1 and PDPK2), to regulate many important cellular processes. These include transcription, cell proliferation, apoptosis, migration, glucose metabolism, and growth, including maintenance of both cell size and cell number.36–39 Overexpression of AKT proteins results in an increase in size of various tissues, whereas deficiency results in defects in growth and glucose regulation. For example, the Akt1−/− mouse knockout has impaired overall growth (including small brain size), whereas the Akt2−/− mouse has insulin intolerance and a diabetes-like syndrome.38,39
Two Akt3 mouse knockouts were recently reported, one targeting exon 3 in an unstated strain of 129 mice that we here designate “Akt3(ex3)−/−,”23 and the other targeting exon 4 in 129 Ola mice that we designate “Akt3(ex4)−/−.”40 Body size and size of all organs other than the brain were normal in both lines. In newborn mice, brain size was normal in Akt3(ex4)−/− mutants or was reduced by ∼15% in Akt3(ex3)−/− mutants. But postnatal growth was significantly reduced in both lines, with brain size reduced by ∼25% by age 3 mo, as measured by weight or volume. The small size was caused by reductions in both cell size and cell number. The cause of the reduced cell number may be related to increased cell death, since exposure of primary hippocampal cell cultures from Akt3(ex4)−/− mutants to either glutamate or staurosporine increased the rate of apoptosis.40 Akt3 also appears to have a role in callosal development. One of the two mutant strains, Akt3(ex4)−/−, had callosal hypoplasia, as assessed by both brain imaging and pathological analysis,40 although the other strain did not.23 This suggests a strain-specific effect on the phenotype. The same phenomenon was observed in our five patients with deletion or disruption of AKT3: three had total ACC, whereas two had partial ACC consisting of overall reduced thickness and absent splenium (fig. 1). Thus, substantial functional data support a role for AKT3 in both prenatal and postnatal brain growth, as well as corpus callosum development.
Deletion 1q4 Is Also Associated with CVH and PMG
All six patients also had CVH, but our data are difficult to reconcile with a single locus causing this phenotype. Among the five patients with deletion 1q4 and CVH, only BAC RP11-518L10 was deleted in all five patients. This BAC contains all or portions of three known or potential genes: cen-C1orf100-TGIF2P1-ADSS-tel. TGIF2P1 is a pseudogene, ADSS (or adenylosuccinate synthetase) is involved in purine biosynthesis, and C1orf100 is a predicted gene of unknown function with no expression data available. C1orf100 is not disrupted by the t(1;13) translocation in patient LR05-101, although the CVH in this boy is minimal. The next gene toward the centromere is ZNF238, discussed above, but it is not contained in RP11-518L10. Although it is possible that deletion of C1orf100 or the adjacent ZNF238 is associated with CVH, the data are not compelling. We think our data are more consistent with the hypothesis that at least two genes contribute to CVH. Patient LP94-079 has severe CVH with an occipital cephalocele and has a deletion from RP11-518L10 to the 1q telomere. We propose that one gene for CVH and another causing PMG are located in this region. Alternatively, both phenotypes could be caused by deletion of a single gene. The remaining four patients have deletions for the same 3.5-Mb region between RP11-80B9 and RP11-241M7 (fig. 3), discussed in regard to MIC and ACC. We propose that a second gene causing CVH is located in this region. This is not likely to be AKT3, since the reduced size of the cerebellum in Akt3−/− mutants is proportional to the reduced size of the cerebrum and also affects both vermis and hemispheres similarly,40 in contrast to human CVH.
Deletion 1q4 is Consistently Associated with MIC and ACC
Our observations of both MIC and ACC in the children reported here with deletion 1q4 support a role for AKT3 (less likely ZNF238) in human brain growth and callosal development (table 1). There are two further reports of patients with large 1q4 deletions that included AKT3 and ZNF238. The first had an ∼15-Mb subtelomeric deletion,18 whereas the second had an ∼8.5-Mb subtelomeric deletion involving the RGS7 gene to the telomere.19 Both had MIC and partial ACC, as determined by cranial CT scan. The former consisted of congenital MIC (−2 or −3 SD) in both children and postnatal MIC (−4 SD) at age 8 mo in the child who survived. Importantly, the boy with putative disruption of AKT3 expression by the nearby t(1;13) breakpoint had normal (mean) OFC at birth, but it was −2 SD by age 1 year and −2.5 SD by age 2 years. This is comparable to data in the mouse Akt3−/− mutants. When our data are combined with the literature, at least four patients with deletion 1q42-q44 that include AKT3 have had congenital MIC (−2 to −5 SD), with at least two more demonstrating additional postnatal slowing of head growth (table 1). By contrast, another patient with a 1q4 deletion distal to AKT3 had normal OFC at age 15 years (table 1). In summary, we believe that several complementary lines of evidence support AKT3 as an excellent candidate gene for both MIC and ACC in humans. We suggest that haploinsufficiency of AKT3 causes postnatal MIC in humans and may contribute to congenital MIC as well.
Acknowledgments
E.B. was funded by the Health Foundation. G.B. is a Wellcome Trust Senior Clinical Research Fellow. We thank the staff of the Northwest Regional Cytogenetic Department, St Mary’s Hospital, Manchester, and Northern Ireland Regional Cytogenetic Laboratory, Belfast City Hospital, for initial karyotyping of patients. We thank the staff of the DNA Sequencing Facility of the University of Chicago Cancer Research Center and the Sanger Centre Clone Supply Unit for providing BAC and fosmid clones. We also thank the staff and students of the Millen and Dobyns Labs at the University of Chicago for much advice and assistance. This work was supported in part by grants from the National Institutes of Health (NIH) to E.H.S. and to W.B.D. and K.J.M. (R01-NS050375) and from the March of Dimes. This publication was made possible in part by grant UL RR024131-01 from the National Center for Research Resources (NCRR) (a component of the NIH) and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NCRR or the NIH.
Web Resources
The URLs for data presented herein are as follows:
- BGEM, http://www.stjudebgem.org/
- Ensembl, http://www.ensembl.org/
- National Center for Bioinformatics, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ACC) [PubMed]
- Primer3, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
- UCSF Comprehensive Cancer Center, http://cancer.ucsf.edu/array/index.php
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/
References
- 1.Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, Mitchell HF, Donis-Keller H, Helms C, Hing AV, et al (1996) Identification of sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat Genet 14:353–356 10.1038/ng1196-353 [DOI] [PubMed] [Google Scholar]
- 2.Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC, Muenke M (1996) Mutations in the human sonic hedgehog gene cause holoprosencephaly. Nat Genet 14:357–360 10.1038/ng1196-357 [DOI] [PubMed] [Google Scholar]
- 3.Gripp KW, Wotton D, Edwards MC, Roessler E, Ades L, Meinecke P, Richieri-Costa A, Zackai EH, Massague J, Muenke M, et al (2000) Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat Genet 25:205–208 10.1038/76074 [DOI] [PubMed] [Google Scholar]
- 4.Wallis DE, Roessler E, Hehr U, Nanni L, Wiltshire T, Richieri-Costa A, Gillessen-Kaesbach G, Zackai EH, Rommens J, Muenke M (1999) Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat Genet 22:196–198 10.1038/9718 [DOI] [PubMed] [Google Scholar]
- 5.Hamilton G, Brown N, Oseroff V, Huey B, Segraves R, Sudar D, Kumler J, Albertson D, Pinkel D (2006) A large field CCD system for quantitative imaging of microarrays. Nucleic Acids Res 34:e58 10.1093/nar/gkl160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH (1993) Isolation of a Miller-Dieker lissencephaly gene containing G protein β-subunit-like repeats. Nature 364:717–721 10.1038/364717a0 [DOI] [PubMed] [Google Scholar]
- 7.Chong SS, Pack SD, Roschke AV, Tanigami A, Carrozzo R, Smith ACM, Dobyns WB, Ledbetter DH (1997) A revision of the lissencephaly and Miller-Dieker syndrome critical regions in chromosome 17p13.3. Hum Molec Genet 6:147–155 10.1093/hmg/6.2.147 [DOI] [PubMed] [Google Scholar]
- 8.Grinberg I, Northrup H, Ardinger H, Prasad C, Dobyns WB, Millen KJ (2004) Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat Genet 36:1053–1055 10.1038/ng1420 [DOI] [PubMed] [Google Scholar]
- 9.Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, et al (2003) Mutations in SOX2 cause anophthalmia. Nat Genet 33:461–463 10.1038/ng1120 [DOI] [PubMed] [Google Scholar]
- 10.Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo W-L, Collins C, et al (1998) Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet 63:1316–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaser T, Walton DS, Maas RL (1992) Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat Genet 2:232–239 10.1038/ng1192-232 [DOI] [PubMed] [Google Scholar]
- 12.Jordan T, Hanson I, Zaletayev D, Hodgson S, Prosser J, Seawright A, Hastie N, van Heyningen V (1992) The human PAX6 gene is mutated in two patients with aniridia. Nat Genet 1:328–332 10.1038/ng0892-328 [DOI] [PubMed] [Google Scholar]
- 13.Gleeson JG, Allen KM, Fox JW, Lamperti ED, Berkovic S, Scheffer I, Cooper EC, Dobyns WB, Minnerath SR, Ross ME, et al (1998) Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell 92:63–72 10.1016/S0092-8674(00)80899-5 [DOI] [PubMed] [Google Scholar]
- 14.Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, Snijders A, Albertson DG, Pinkel D, Marra MA, et al (2004) A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet 36:299–303 10.1038/ng1307 [DOI] [PubMed] [Google Scholar]
- 15.Jain AN, Tokuyasu TA, Snijders AM, Segraves R, Albertson DG, Pinkel D (2002) Fully automatic quantification of microarray image data. Genome Res 12:325–332 10.1101/gr.210902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D (2004) Ultraconserved elements in the human genome. Science 304:1321–1325 10.1126/science.1098119 [DOI] [PubMed] [Google Scholar]
- 17.Chizhikov VV, Millen KJ (2004) Control of roof plate formation by Lmx1a in the developing spinal cord. Development 131:2693–2705 10.1242/dev.01139 [DOI] [PubMed] [Google Scholar]
- 18.Gentile M, Di Carlo A, Volpe P, Pansini A, Nanna P, Valenzano MC, Buonadonna AL (2003) FISH and cytogenetic characterization of a terminal chromosome 1q deletion: clinical case report and phenotypic implications. Am J Med Genet A 117:251–254 10.1002/ajmg.a.10018 [DOI] [PubMed] [Google Scholar]
- 19.van Bever Y, Rooms L, Laridon A, Reyniers E, van Luijk R, Scheers S, Wauters J, Kooy RF (2005) Clinical report of a pure subtelomeric 1qter deletion in a boy with mental retardation and multiple anomalies adds further evidence for a specific phenotype. Am J Med Genet A 135:91–95 [DOI] [PubMed] [Google Scholar]
- 20.de Vries BB, Knight SJ, Homfray T, Smithson SF, Flint J, Winter RM (2001) Submicroscopic subtelomeric 1qter deletions: a recognisable phenotype? J Med Genet 38:175–178 10.1136/jmg.38.3.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Vries BB, White SM, Knight SJ, Regan R, Homfray T, Young ID, Super M, McKeown C, Splitt M, Quarrell OW, et al (2001) Clinical studies on submicroscopic subtelomeric rearrangements: a checklist. J Med Genet 38:145–150 10.1136/jmg.38.3.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniel A, Baker E, Chia N, Haan E, Malafiej P, Hinton L, Clarke N, Adès L, Darmanian A, Callen D (2003) Recombinants of intrachromosomal transposition of subtelomeres in chromosomes 1 and 2: a cause of minute terminal chromosomal imbalances. Am J Med Genet A 117:57–64 10.1002/ajmg.a.10048 [DOI] [PubMed] [Google Scholar]
- 23.Easton RM, Cho H, Roovers K, Shineman DW, Mizrahi M, Forman MS, Lee VM, Szabolcs M, de Jong R, Oltersdorf T, et al (2005) Role for Akt3/protein kinase Bγ in attainment of normal brain size. Mol Cell Biol 25:1869–1878 10.1128/MCB.25.5.1869-1878.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becker KG, Lee IJ, Nagle JW, Canning RD, Gado AM, Torres R, Polymeropoulos MH, Massa PT, Biddison WE, Drew PD (1997) C2H2-171: a novel human cDNA representing a developmentally regulated POZ domain/zinc finger protein preferentially expressed in brain. Int J Dev Neurosci 15:891–899 10.1016/S0736-5748(97)00034-8 [DOI] [PubMed] [Google Scholar]
- 25.Magdaleno S, Jensen P, Brumwell CL, Seal A, Lehman K, Asbury A, Cheung T, Cornelius T, Batten DM, Eden C, et al (2006) BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol 4:e86 10.1371/journal.pbio.0040086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Rosa G, Pardeo M, Bria S, Caresta E, Vasta I, Zampino G, Zollino M, Zuppa AA, Piastra M (2005) Isolated myocardial non-compaction in an infant with distal 4q trisomy and distal 1q monosomy. Eur J Pediatr 164:255–256 10.1007/s00431-005-1620-4 [DOI] [PubMed] [Google Scholar]
- 27.Puthuran MJ, Rowland-Hill CA, Simpson J, Pairaudeau PW, Mabbott JL, Morris SM, Crow YJ (2005) Chromosome 1q42 deletion and agenesis of the corpus callosum. Am J Med Genet A 138:68–69 [DOI] [PubMed] [Google Scholar]
- 28.Hathout EH, Thompson K, Baum M, Dumars KW (1998) Association of terminal chromosome 1 deletion with sertoli cell-only syndrome. Am J Med Genet 80:396–398 [DOI] [PubMed] [Google Scholar]
- 29.Zollino M, Colosimo C, Zuffardi O, Rossi E, Tosolini A, Walsh CA, Neri G (2003) Cryptic t(1;12)(q44;p13.3) translocation in a previously described syndrome with polymicrogyria, segregating as an apparently X-linked trait. Am J Med Genet A 117:65–71 10.1002/ajmg.a.10068 [DOI] [PubMed] [Google Scholar]
- 30.Meinecke P, Vogtel D (1987) A specific syndrome due to deletion of the distal long arm of chromosome 1. Am J Med Genet 28:371–376 10.1002/ajmg.1320280215 [DOI] [PubMed] [Google Scholar]
- 31.Kleinjan DA, van Heyningen V (2005) Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 76:8–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleinjan DJ, van Heyningen V (1998) Position effect in human genetic disease. Hum Mol Genet 7:1611–1618 10.1093/hmg/7.10.1611 [DOI] [PubMed] [Google Scholar]
- 33.Meng G, Inazawa J, Ishida R, Tokura K, Nakahara K, Aoki K, Kasai M (2000) Structural analysis of the gene encoding RP58, a sequence-specific transrepressor associated with heterochromatin. Gene 242:59–64 10.1016/S0378-1119(99)00477-1 [DOI] [PubMed] [Google Scholar]
- 34.Kang JS, Yang YC, Liu HL, Li YH, Du YC, Li RX (2001) Cloning and distribution of rRP58, a novel neuronal gene. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 33:563–568 [PubMed] [Google Scholar]
- 35.Bellacosa A, Testa JR, Moore R, Larue L (2004) A portrait of AKT kinases: human cancer and animal models depict a family with strong individualities. Cancer Biol Ther 3:268–275 [DOI] [PubMed] [Google Scholar]
- 36.Yang ZZ, Tschopp O, Hemmings-Mieszczak M, Feng J, Brodbeck D, Perentes E, Hemmings BA (2003) Protein kinase B α/Akt1 regulates placental development and fetal growth. J Biol Chem 278:32124–32131 10.1074/jbc.M302847200 [DOI] [PubMed] [Google Scholar]
- 37.Brodbeck D, Cron P, Hemmings BA (1999) A human protein kinase Bγ with regulatory phosphorylation sites in the activation loop and in the C-terminal hydrophobic domain. J Biol Chem 274:9133–9136 10.1074/jbc.274.14.9133 [DOI] [PubMed] [Google Scholar]
- 38.Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ (2001) Akt1/PKBα is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem 276:38349–38352 10.1074/jbc.C100462200 [DOI] [PubMed] [Google Scholar]
- 39.Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB 3rd, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ (2001) Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB β). Science 292:1728–1731 10.1126/science.292.5522.1728 [DOI] [PubMed] [Google Scholar]
- 40.Tschopp O, Yang ZZ, Brodbeck D, Dummler BA, Hemmings-Mieszczak M, Watanabe T, Michaelis T, Frahm J, Hemmings BA (2005) Essential role of protein kinase Bγ (PKBγ/Akt3) in postnatal brain development but not in glucose homeostasis. Development 132:2943–2954 10.1242/dev.01864 [DOI] [PubMed] [Google Scholar]