Abstract
Animal microRNAs (miRNAs) regulate gene expression through base pairing to their targets within the 3′ untranslated region (UTR) of protein-coding genes. Single-nucleotide polymorphisms (SNPs) located within such target sites can affect miRNA regulation. We mapped annotated SNPs onto a collection of experimentally supported human miRNA targets. Of the 143 experimentally supported human target sites, 9 contain 12 SNPs. We further experimentally investigated one of these target sites for hsa-miR-155, within the 3′ UTR of the human AGTR1 gene that contains SNP rs5186. Using reporter silencing assays, we show that hsa-miR-155 down-regulates the expression of only the 1166A, and not the 1166C, allele of rs5186. Remarkably, the 1166C allele has been associated with hypertension in many studies. Thus, the 1166C allele may be functionally associated with hypertension by abrogating regulation by hsa-miR-155, thereby elevating AGTR1 levels. Since hsa-miR-155 is on chromosome 21, we hypothesize that the observed lower blood pressure in trisomy 21 is partially caused by the overexpression of hsa-miR-155 leading to allele-specific underexpression of AGTR1. Indeed, we have shown in fibroblasts from monozygotic twins discordant for trisomy 21 that levels of AGTR1 protein are lower in trisomy 21.
MicroRNAs (miRNAs) are ∼21-nt small RNAs involved in posttranscriptional gene regulation. They have been shown to guide the RNA-induced silencing complex of proteins to specific target sites within mRNAs to induce immediate cleavage, localization to P-bodies, or translational repression.1 These target sites are thought to be most prevalent in the 3′ UTR of mRNAs.1
SNPs are DNA sequence variations that occur at a rate of ∼1 in every 1,000 bp in the human genome. SNPs that occur in the 3′ UTR can affect gene regulation by interfering with posttranscriptional activity, such as protein binding, polyadenylation, and miRNA binding. Two recent studies reported SNPs that alter the gene expression level by modifying miRNA targeting activity.2,3 The first showed that a 3′-UTR SNP in human SLITRK1 strengthens an existing miR-189 target site, thereby amplifying the down-regulation of SLITRK1, which is implicated in Tourette syndrome.2 The second demonstrated that a 3′-UTR SNP in the sheep Gdf8 gene creates a new illegitimate miRNA target site, which leads to significant down-regulation of Gdf8 and contributes to muscular hypertrophy.3
miRNA target sites can be categorized into two classes: 5′-dominant and 3′-compensatory. 5′-dominant target sites have perfect base pairing with at least 7 nt at the 5′ end of the miRNA, which is also referred to as the “seed” or “nucleus.”4,5 Such binding is considered in most cases to be sufficient for a functional miRNA-mRNA interaction.4,5 3′-compensatory target sites are characterized by either a <7-nt stretch of perfect base pairing or an imperfect 7-nt base pairing with the miRNA 5′ end, followed by an extended base pairing with the miRNA 3′ end.6–9 There are currently three published data sets that provide a mapping of all known SNPs onto a set of computationally predicted miRNA target sites.3,10,11 Since the introduction of computational miRNA target–prediction programs in 2003, significant progress has been made in the field.12,13 However, current algorithms still simulate only a limited part of all the biochemical processes that are responsible for a functional miRNA-mRNA interaction. As a consequence, such algorithms often lack specificity.12 Therefore, we limit our study to experimentally supported target sites.
An up-to-date and comprehensive collection of experimentally supported miRNA target sites can be found in TarBase 4.0, a manually curated database that includes >600 miRNA–target gene interactions in eight different species.14 For each supported miRNA target site, the database describes the location within the 3′ UTR where it occurs, the nature of the experiments that were conducted to validate it, and the sufficiency of the site to induce translational repression and/or cleavage. Version 4.0 of TarBase contains 143 experimentally supported human target sites for which the exact genomic location is known.
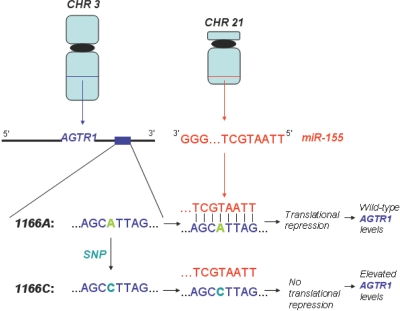
We mapped the human SNPs from dbSNP build 127 onto the experimentally supported human miRNA target sites mentioned above. This mapping revealed nine human target sites that harbor 12 SNPs (fig. 1). A recently published study employing a similar approach also reports one of these target sites (EZH2).11 For three genes (KCNH2, CAT-1, and ESR1), the target sites are of the 3′-compensatory class, and, for six genes (AGTR1, EZH2, HOXA7, SMAD1, DLL1, and GJA1), they are of the 5′-dominant class. Two of the latter six, AGTR1 and EZH2, harbor SNPs that occur in a region that interrupts 5′-dominant base pairing. The AGTR1 target site harbors three SNPs, one of which, rs5186 (1166A/C), is located in the region that interacts with the miR-155 seed and is also reported in the Allele Frequency Database (ALFRED). Because the 1166C allele interrupts the interaction with the miR-155 seed (fig. 1), we hypothesized that it may severely impair miR-155 targeting.
Figure 1. .
Nine experimentally supported miRNA target sites harbor 12 SNPs. The studies that reported the nine miRNA-target interactions shown here used one or more of the several publicly available computational target-prediction programs to compute the putative binding diagrams. Seven of the nine SNPs (rs8179014, rs17500932, rs41269629, rs17860720, rs11416, rs5185, and rs12721277) occur in a region of a 5′-dominant target site that does not affect 5′-dominant base pairing. Three SNPs (rs11541536, rs9579381, and rs9341070) occur in a region of a 3′-compensatory target site that can affect 3′-compensatory base pairing. Two SNPs (rs5186 and rs8829) occur in a region of a 5′-dominant target site that does affect 5′-dominant base pairing.
To validate our hypothesis, we proceeded with in vitro experiments. TarBase shows that this target site gained experimental support via reporter-silencing assays. More specifically, Martin et al. showed that miR-155 represses AGTR1 in lung fibroblasts via specific binding to this site.15 We inserted three different AGTR1 3′ UTRs downstream of luciferase reporter genes to create three distinct reporter gene constructs: (1) full-length AGTR1 3′ UTR with the major allele (1166A), (2) full-length AGTR1 3′ UTR with the minor allele (1166C), and (3) full-length AGTR1 3′ UTR with the miR-155 target site deleted. The AGTR1 3′ UTR with the 1166A allele was amplified by PCR with use of sense (5′-CATGTTCGAAACCTGTCCATAAAG-3′) and antisense (5′-ATAAAATTATTTTATTTTAAAGTAAAT-3′) primers. The PCR products were inserted at the XbaI site in the 3′ UTR of the luciferase gene of the pTAL-Luc vector (Promega). The pTAL-Luc AGTR1 3′ UTR with the 1166C allele was constructed similarly with genomic DNA containing the polymorphic AGTR1 1166C allele. Finally, the pTal-Luc AGTR1 deletion 1163–1169 construct was generated using the Quickchange XL site-directed mutagenesis kit (Stratagene), with use of a forward mutagenic deletion primer (5′-TCTGCAGCACTTCACTACCAAATGGCTACTTTTCAGAATTGAAGG-3′) and a complementary reverse mutagenic deletion primer (5′-CCTTCAATTCTGAAAAGTAGCCATTTGGTAGTGAAGTGCTGCAGA-3′).
293T cells were seeded in 96-well plates (104 cells/well) in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum. 293T cells were then transfected with pTAL-Luc AGTR1 3′ UTR A1166/C1166/deletion 1163–1169 (800 ng), pRL-SV40 Renilla (40 ng [Promega]), and 2.5 nM, 10 nM, and 20 nM of miR-155 precursor or 20 nM of miR-Let7c as control (double-stranded RNA oligonucleotide [Ambion]) with the use of Lipofectamine 2000 (Invitrogen), in accordance with the manufacturer’s protocol. After 48 h, firefly and Renilla luciferase activities were measured using a dual luciferase assay (Promega). The relative reporter activity was obtained by normalization to the Renilla activity.
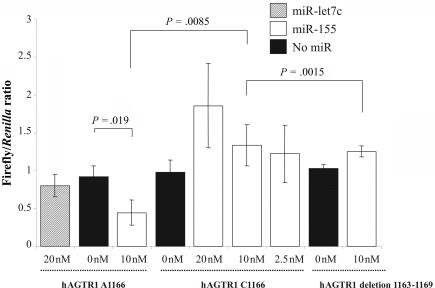
In the presence of miR-155, the expression of the luciferase containing the 1166A allele is significantly reduced (fig. 2), confirming the function of miR-155 on the AGTR1 target gene. In contrast, the expression of the luciferase containing the 1166C allele remains unchanged in the presence or absence of miR-155 (fig. 2). Furthermore, this result is similar to that obtained with the ΔmiR-155 target-site luciferase construct, implying that the 1166C allele abolishes the target site and impairs the ability of miR-155 binding, thereby elevating the levels of AGTR1. When we repeated the same experiments with a higher (20 nM) and lower (2.5 nM) amount of miR-155, the observation did not change (fig. 2).
Figure 2. .
Effects of miR-155 on the luciferase reporter genes bearing 3′ UTR segments from hAGTR1 3′-UTR. 293T cells were cotransfected with pTAL-Luc hAGTR1 3′-UTR 1166A or pTAL-Luc hAGTR1 3′ UTR 1166C, pTAL-Luc hAGTR1 3′ UTR deletion 1163–1169, and different concentrations (20, 10, or 2.5 nM) of miR-155 (white bars) or no miRNA (black bars) or 20 nM of Let-7c (unrelated miRNA [dashed lines]). Values are means of firefly/Renilla ratio from three independent experiments (each with three culture replicates). Error bars indicate 1 SD of three independent experiments. P values were calculated from a two-sided, two-sample t test.
Hsa-miR-155 maps onto human chromosome 21 and therefore is triplicated in trisomy 21. Remarkably, it has been reported that individuals with trisomy 21 have lower levels of diastolic and systolic blood pressure than do age- and sex-matched control individuals.16,17 We hypothesize that overexpression of miR-155 in trisomy 21 excessively suppresses the AGTR1 common alleles and that this may be one mechanism contributing to the lower blood pressure in individuals with trisomy 21.
To evaluate the expression of miR-155 in trisomy 21, we performed real-time quantitative PCR in fibroblasts from an MZ twin pair discordant for trisomy 21— that is, one twin was unaffected, and the other had a trisomy 21. Both twins were homozygotes for the 1166A AGTR1 allele, which is the target of miR-155. Twins were chosen so that any gene-expression differences could be attributed only to the supernumerary chromosome 21 and not to polymorphic variability in the rest of the genome. Fibroblast cells from these MZ twins were grown in DMEM with Glutamax I medium (Invitrogen) supplemented with 10% fetal calf serum and 1% penicillin and streptomycin mix (Invitrogen). Total RNA was extracted by Trizol reagent (Invitrogen). RNA quality was assessed using an Agilent 2100 BioAnalyzer with the RNA 6000 Nano LabChip. miRNA expression levels were detected by the SYBR Green I real-time PCR miRNA detection kit (mirvana [Ambion]), with the use of SuperTaq (Enzyme Technologies). Reactions were set up, by use of a Biomek 2000 robot (Beckman), in a 10-μl volume in 384-well plates. Each RNA sample was analyzed in six technical replicates and in three independent experiments, with the use of primer sets specific for miR-155 and four different miRNAs used for normalization: miR-124, miR-130b, miR-24, and miR-26a. PCRs were run in an ABI 7900 Sequence Detection System (SDS [Applied Biosystems]).
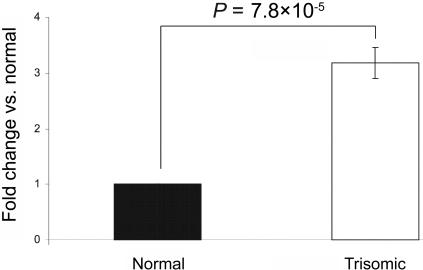
Raw cycle-threshold (CT) values were obtained using SDS 2.0 software (Applied Biosystems). From the six replicates, values outside the median ±0.5 were classified as outliers and were excluded. For all calculations, CT values were converted to quantity (q) with the formula q=2-CT. The relative expression level of mature miRNAs was normalized according to geNorm,18 with the use of miR-124, miR-130b, miR-24, and miR-26a as references to determine the normalization factor. Each miRNA was then mean normalized across the two individuals. Normalized relative expression values thus have a median of 1. The results shown in figure 3 indicate that miR-155 is indeed significantly overexpressed in fibroblasts of individuals with trisomy 21. miR-155 is conserved in mouse, and it maps within the region of mouse chromosome 16 that is triplicated in the TS65Dn partial mouse model of Down syndrome.19,20 However, this and other such mouse models could not be used to test the hypothesis, because the miR-155 target sequence in mouse AGTR1 is not conserved, and, thus, it is not expected that mmu-miR-155 interacts with mouse AGTR1.
Figure 3. .
Real-time quantitative PCR for mature miR-155 expression in fibroblast cells from MZ twins discordant for trisomy 21. Data are means (±SD) from three independent experiments analyzed in six replicates. Data are normalized with reference microRNAs, as mentioned in the text. P values were calculated from a two-sided, one-sample t test.
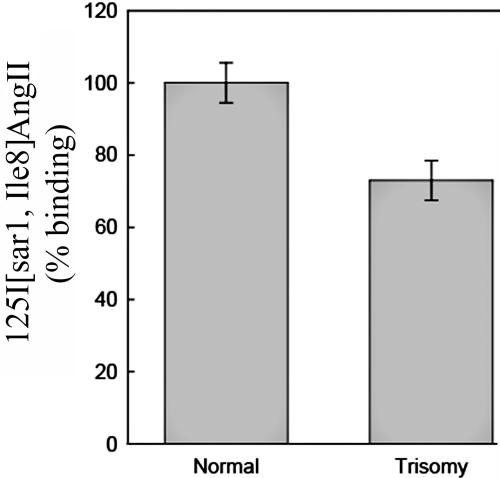
The increased levels of miR-155 expression in trisomy 21 predict that the AGTR1 protein levels in fibroblasts isolated from the MZ twins discordant for trisomy 21 should be lower in the twin with trisomy 21 than in the unaffected euploid twin. Since commercially available AGTR1 antibodies are not reliable, we performed whole-cell AGTR1 binding assays as described elsewhere.15 In brief, the fibroblasts were allowed to reach confluence, the cell medium was aspirated and was replaced with monoiodinated 125I-[Sar1,Ile8] AngII (2–3×105 cpm [Peptide Radioiodination Service]) in Hank’s Balanced Salt Solution, 20 mM Hepes, and 0.1% BSA). After incubation at room temperature for 60 min, unbound ligand was removed by washing each well twice with 1 ml ice-cold PBS. Bound ligand was recovered by dissolving the protein in each well with 1 ml 0.5-M NaOH and 0.01% SDS. Nonspecific binding was determined by performing the binding assay in the presence of 1 μM unlabeled AngII. The quantity of 125I-[Sar1,Ile8] AngII present in each sample was determined using a Cobra γ-spectrophotometer (Packard Bell). Protein content in wells was assessed using the BioRad Protein Assay dye reagent (BioRad). Values shown in figure 4 represent specific (total minus nonspecific) binding. Consistent with the prediction, the trisomy 21 fibroblasts showed reduced protein levels of AGTR1 compared with that of the euploid fibroblasts (∼30% less; P<.01).
Figure 4. .
AGTR1 radioligand binding studies in fibroblasts from MZ twins discordant for trisomy 21. There is a lower amount of AGTR1 protein in trisomy 21 fibroblasts than in the unaffected euploid twin fibroblasts.
ALFRED reports that the minor allele, 1166C, occurs with much higher frequency in white populations (20%–30%) compared with African or Asian populations (5%–6%). We performed a literature search and identified 40 studies that investigated the association of the 1166C allele with hypertension (MIM 145500) in various populations (table A1). Of these studies, 18 reported that 1166C is a risk allele for hypertension. Type I error, ethnic admixture, and linkage disequilibrium (LD) are highly unlikely to explain the large number of studies with positive findings (table A1). Furthermore, the discrepancies between studies are usually greatly decreased in subpopulations defined by ethnic origin—for example, all six studies with Japanese subjects failed to find significant association, whereas 9 of 13 studies with white subjects showed significant association. Hirschhorn et al. indicate gene-environment factors as a likely source of variable results, and this could play a significant role in this case, since there is a strong ethnic bias in the studies with positive findings.60 Therefore, although the results must be interpreted as a function of age, sex, and ethnic origin, the literature clearly indicates an association of the 1166C allele with hypertension in several populations.
Despite this literature evidence, the molecular mechanism of the association has remained uncertain. Interestingly, it has been independently shown that elevated levels of AGTR1 contribute to cardiovascular disease.61–63 Accordingly, antagonists of AGTR1 have been developed and are now widely used in the treatment of hypertension.64 This suggests that at least one compelling mechanism for the association of 1166C with cardiovascular disease is the abrogation of miR-155 binding, which elevates AGTR1 levels (fig. 5). According to expression studies, two human organs—the spleen and kidney—concomitantly express AGTR1 and miR-155. Since the kidney is a critical organ for the regulation of blood pressure, we postulate that the abrogation of miR-155 binding to AGTR1 is most detrimental in the kidney. We note here that the mouse and rat genomes preserve only one of either miR-155 or its AGTR1 target site. Therefore, rodents are not useful model organisms for this study.
Figure 5. .
Model for molecular mechanism of 1166C association with hypertension. The 1166C allele in the 3′ UTR of AGTR1 abrogates miR-155 binding, which induces elevated levels of AGTR1.
A recent study provided convincing evidence that miRNA target sites are under negative selective pressure to harbor SNPs.10 These studies include the ∼450 experimentally verified human miRNAs in the miRBase database.65 The number of miRNAs in miRBase has grown exponentially during the past few years, and there is little expectation that this growth is complete. A recent massively parallel sequencing effort identified 447 novel miRNAs, many of which are primate specific.66 Furthermore, current computational/experimental studies conjecture that there are at least 1,000 functional miRNAs encoded within the human genome.67,68 Further characterization of these miRNAs and improvement of target-prediction programs to enable the specific prediction of nonconserved target sites will be critical for a more complete analysis of SNP abundance in miRNA target sites.
Acknowledgments
We thank Mickey M. Martin for assistance in the AGTR1 binding assays. A.G.H. thanks Nikolaus Rajewsky for helpful discussions during the early stage of this work. A.G.H. and P.S. are supported by National Science Foundation Career Award DBI-0238295. P.S. is also supported by predoctoral National Institutes of Health (NIH) training grant 5T32GM008216. T.S.E. is supported by NIH National Heart, Lung, and Blood Institute grant HL48848 and American Heart Association grant GRT00001380. S.E.A. is supported by the Swiss National Science Foundation, the National Center of Competence in Research Frontiers in Genetics, the ChildCare Foundation, and the European Union AnEUploidy project.
Appendix A: Literature Review
Of the 40 studies we examined in the literature, 18 had positive results and 22 had negative results. Discrepancies among the study results are greatly reduced in subpopulations defined by ethnic origin. Each study used a 5% significance level for type I error in the association tests. Therefore, on the basis of statistical error, we would expect, on average, only two false-positive associations among 40 studies. However, given 18 positive associations among 40 studies, the type I error alone cannot explain the discrepancy in the results, barring the unlikely possibility that there are hundreds of negative associations that have not been published. Furthermore, the discrepancy is usually greatly decreased in subpopulations defined by ethnic origin—for example, all six studies with Japanese subjects failed to find significant association, whereas 9 of 13 studies with white subjects showed significant association. Some studies on other non–ethnically defined subpopulations also produced more consistent results. For example, the three studies on pregnancy-induced hypertension all found significant association. It must be noted that some studies that used the same population type produced inconsistent results. For example, Reich et al.27 found association in males but not in females; meanwhile, Tiret et al.57 found association in females but not in males. Such discrepancies are not surprising in association testing, which is highlighted by the fact that the Hirschhorn et al.60 review found that only 6 of 166 associations that had been studied at least three times were positive >75% of the time.
The work of Hirschhorn et al.60 lists the two factors that can result in false-positive associations as ethnic admixture and LD. Ethnic admixture is unlikely to cause the observed high number of positive associations, because the studies comprised a large number of diverse populations. Furthermore, not all studies with positive results used the case-control design that is susceptible to stratification—for example, Kainulainen et al.59 used a family-based study resulting in a highly significant association. LD can result if the subjects tend to descend from a recent common ancestor. However, most of the studies used populations that were sufficiently heterogeneous to avoid this issue, and, furthermore, no markers have been found in any of the studies to be in LD with the A1166C polymorphism. Therefore, LD is unlikely to explain the large number of positive results. Hirschhorn et al.60 indicate gene-environment factors as a likely source of variable results, and this could play a significant role in our case, since there is a strong ethnic bias in the positive studies. Therefore, any association of rs5186 with cardiovascular risk must be interpreted as a function of population—in particular, age, sex, and ethnic origin.
Hirschhorn et al.60 also discuss the likelihood of obtaining false-negative results when the genetic effect is relatively weak and there is a lack of power in the study to detect it. Given that the association is real, this is likely the cause of some of the observed false-negative results, because the A1166C polymorphism has a fairly low allele frequency in the population (∼10% to ∼20% in the 40 studies considered). Moreover, when an association is found, it is not at an extreme level in any case—for example, an allele frequency of 28% versus 16% in cases versus controls in the work of Kainulainen et al.59 Indeed, a number of the studies found positive trends or P values that are on the threshold of significance. Therefore, we conclude that the literature provides a very strong case for a true association of the A1166C polymorphism and hypertension in several major subpopulations.
Table A1. .
Results of 40 Studies that Tested the Association of AGTR1 1166C with Hypertension
| Study | Association |
| Ono et al.21 | None in Japanese |
| Takami et al.22 | None in Japanese |
| Sugimoto et al.23 | None in Japanese |
| Kato et al.24 | None in Japanese |
| Katsuya et al.25 | None in elderly Japanese |
| Bonnardeaux et al.26 | In whites |
| Reich et al.27 | In white males |
| Castellano et al.28 | In Italians with “clinical” but not “ambulatory” blood pressure and homozygosity |
| Schmidt et al.29 | None in whites |
| Thomas et al.30 | None in Chinese |
| Thomas et al.31 | None in Chinese |
| Zhu et al.32 | In Chinese |
| Barbalic et al.33 | None in Croatians |
| Kaidashev et al.34 | In Ukrainians |
| Zhang et al.35 | None in Chinese (Han) |
| Spiering et al.36 | None in whites |
| Abdollahi et al.37 | None |
| Henskens et al.38 | Association |
| Gardier et al.39 | None |
| Kobashi et al.40 | With pregnancy hypertension |
| Agachan et al.41 | In Turks |
| Liu et al.42 | In Tibetans but not in Han or Yi |
| Porto et al.43 | None with young-onset hypertension |
| Petrovic et al.44 | None with young-onset hypertension |
| Stankovic et al.45 | In Serbian males but not in females |
| Hindorff et al.46 | In whites but not in large white and black mixed study sample |
| Jiang et al.47 | In Chinese (Han) |
| Giner et al.48 | None in Spanish |
| Dzida et al.49 | In Polish |
| Davis et al.50 | None |
| Li et al.51 | None in Chinese |
| Xiang et al.52 | None in Chinese |
| Nalogowska et al.53 | With pregnancy hypertension |
| Seremak-Mrozikiewicz et al.54 | With pregnancy hypertension in Polish |
| Liyou et al.55 | None in Australian elderly |
| Berge et al.56 | Trend seen in three Norwegian populations but not to level of statistical significance |
| Tiret et al.57 | In French females but not in males |
| Wang et al.58 | In whites |
| Kainulainen et al.59 | In Finns |
Web Resources
The URLs for data presented herein are as follows:
- ALFRED, http://alfred.med.yale.edu/alfred/
- dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for hypertension)
- TarBase, http://diana.pcbi.upenn.edu/tarbase (for the database of experimentally supported targets)
References
- 1.Kloosterman WP, Plasterk RH (2006) The diverse functions of microRNAs in animal development and disease. Dev Cell 11:441–450 10.1016/j.devcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 2.Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al (2005) Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science 310:317–320 10.1126/science.1116502 [DOI] [PubMed] [Google Scholar]
- 3.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, et al (2006) A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet 38:813–818 10.1038/ng1810 [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 5.Krek A, Gruen D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al (2005) Combinatorial microRNA target predictions. Nature Genetics 37:495–500 10.1038/ng1536 [DOI] [PubMed] [Google Scholar]
- 6.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioykos C, Mourelatos Z, Hatzigeorgiou AG (2004) A combined computational-experimental approach predicts human microRNA targets. Genes Dev 18:1165–1178 10.1101/gad.1184704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of microRNA-target recognition. PLoS Biol 3:e85 10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME (2006) A brain-specific microRNA regulates dendritic spine development. Nature 439:283–289 10.1038/nature04367 [DOI] [PubMed] [Google Scholar]
- 9.Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, MacMenamin P, et al (2006) A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol 16:460–471 10.1016/j.cub.2006.01.050 [DOI] [PubMed] [Google Scholar]
- 10.Chen K, Rajewsky N (2006) Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet 38:1452–1456 10.1038/ng1910 [DOI] [PubMed] [Google Scholar]
- 11.Saunders MA, Liang H, Wen-Hsuing L (2007) Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA 104:3300–3305 10.1073/pnas.0611347104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sethupathy P, Megraw M, Hatzigeorgiou AG (2006) A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods 3:881–886 10.1038/nmeth954 [DOI] [PubMed] [Google Scholar]
- 13.Rajewsky N (2006) microRNA target predictions in animals. Nat Genet 38:S8–S13 10.1038/ng1798 [DOI] [PubMed] [Google Scholar]
- 14.Sethupathy P, Corda B, Hatzigeorgiou AG (2006) TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA 12:192–197 10.1261/rna.2239606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS (2006) MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J Biol Chem 281:18277–18284 10.1074/jbc.M601496200 [DOI] [PubMed] [Google Scholar]
- 16.Morrison RA, McGrath A, Davidson G, Brown JJ, Murray GD, Lever AF (1996) Low blood pressure in Down’s syndrome: a link with Alzheimer’s disease? Hypertension 28:569–575 [DOI] [PubMed] [Google Scholar]
- 17.Draheim CC, McCubbin JA, Williams DP (2002) Differences in cardiovascular disease risk between nondiabetic adults with mental retardation with and without Down syndrome. Am J Ment Retard 107:201–211 [DOI] [PubMed] [Google Scholar]
- 18.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Spelman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, Schmidt C, Bronson RT, Davisson MT (1995) A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet 11:177–184 10.1038/ng1095-177 [DOI] [PubMed] [Google Scholar]
- 20.Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S (2004) Chromosome 21 and Down syndrome: from genomics to pathophysiology. Nat Rev Genet 5:725–738 10.1038/nrg1448 [DOI] [PubMed] [Google Scholar]
- 21.Ono K, Mannami T, Baba S, Yasui N, Ogihara T, Iwai N (2003) Lack of association between angiotensin II type 1 receptor gene polymorphism and hypertension in Japanese. Hypertens Res 26:131–134 10.1291/hypres.26.131 [DOI] [PubMed] [Google Scholar]
- 22.Takami S, Katsuya T, Rakugi H, Sato N, Nakata Y, Kamitani A, Miki T, Higaki J, Ogihara T (1998) Angiotensin II type 1 receptor gene polymorphism is associated with increase of left ventricular mass but not with hypertension. Am J Hypertens 11:316–321 10.1016/S0895-7061(97)00457-3 [DOI] [PubMed] [Google Scholar]
- 23.Sugimoto K, Katsuya T, Ohkubo T, Hozawa A, Yamamoto K, Matsuo A, Rakugi H, Tsuji I, Imai Y, Ogihara T (2004) Association between angiotensin II type 1 receptor gene polymorphism and essential hypertension: the Ohasama Study. Hypertens Res 27:551–556 10.1291/hypres.27.551 [DOI] [PubMed] [Google Scholar]
- 24.Kato N, Sugiyama T, Morita H, Kurihara H, Furukawa T, Isshiki T, Sato T, Yamori Y, Yazaki Y (2000) Comprehensive analysis of the renin-angiotensin gene polymorphisms with relation to hypertension in the Japanese. J Hypertens 18:1025–1032 10.1097/00004872-200018080-00006 [DOI] [PubMed] [Google Scholar]
- 25.Katsuya T, Higaki J, Ishikawa K, Sato N, Ogihara T (1999) Genetic analysis of candidate gene polymorphisms in elderly hypertension. Nippon Ronen Igakkai Zasshi 36:547–552 [DOI] [PubMed] [Google Scholar]
- 26.Bonnardeaux A, Davies E, Jeunemaitre X, Fery I, Charru A, Clauser E, Tiret L, Cambien F, Corvol P, Soubrier F (1994) Angiotensin II type 1 receptor gene polymorphisms in human essential hypertension. Hypertension 24:63–69 [DOI] [PubMed] [Google Scholar]
- 27.Reich H, Duncan JA, Weinstein J, Cattran DC, Scholey JW, Miller JA (2003) Interactions between gender and the angiotensin type 1 receptor gene polymorphism. Kidney Int 63:1443–1449 10.1046/j.1523-1755.2003.00867.x [DOI] [PubMed] [Google Scholar]
- 28.Castellano M, Muiesan ML, Beschi M, Rizzoni D, Cinelli A, Salvetti M, Pasini G, Porteri E, Bettoni G, Zulli R, et al (1996) Angiotensin II type 1 receptor A/C1166 polymorphism relationships with blood pressure and cardiovascular structure. Hypertension 28:1076–1080 [DOI] [PubMed] [Google Scholar]
- 29.Schmidt S, Beige J, Walla-Friedel M, Michel MC, Sharma AM, Ritz E (1997) A polymorphism in the gene for the angiotensin II type 1 receptor is not associated with hypertension. J Hypertens 15:1385–1388 10.1097/00004872-199715120-00003 [DOI] [PubMed] [Google Scholar]
- 30.Thomas GN, Tomlinson B, Chan JC, Sanderson JE, Cockram CS, Critchley JA (2001) Renin-angiotensin system gene polymorphisms, blood pressure, dyslipidemia, and diabetes in Hong Kong Chinese: a significant association of the ACE insertion/deletion polymorphism with type 2 diabetes. Diabetes Care 24:356–361 10.2337/diacare.24.2.356 [DOI] [PubMed] [Google Scholar]
- 31.Thomas GN, Young RP, Tomlinson B, Woo KS, Sanderson JE, Critchley JA (2000) Renin-angiotensin-aldosterone system gene polymorphisms and hypertension in Hong Kong Chinese. Clin Exp Hypertens 22:87–97 10.1081/CEH-100100064 [DOI] [PubMed] [Google Scholar]
- 32.Zhu S, Meng QH (2006) Association of angiotensin II type 1 receptor gene polymorphism with carotid atherosclerosis. Clin Chem Lab Med 44:282–284 10.1515/CCLM.2006.048 [DOI] [PubMed] [Google Scholar]
- 33.Barbalic M, Skaric-Juric T, Cambien F, Barbaux S, Poirier O, Turek S, Vrhovski-Hebrang D, Cubrilo-Turek M, Rudan I, Rudan P, et al (2006) Gene polymorphisms of the renin-angiotensin system and early development of hypertension. Am J Hypertens 19:837–842 10.1016/j.amjhyper.2006.01.004 [DOI] [PubMed] [Google Scholar]
- 34.Kaidashev IP, Rasin MS, Savchenko LG, Shlykova OA, Iakimishina LI (2005) Clinical efficiency of candesatran depends on angiotension II receptor, type 1 gene polymorphism. Lik Sprava 8:66–71 [PubMed] [Google Scholar]
- 35.Zhang KX, Liu TB, Xu QX, Zhu DL, Huang W (2005) Association of angiotensin II receptor type 1 gene single nucleotide polymorphism with Chinese essential hypertension complicated with coronary heart disease. Zhonghua Xin Xue Guan Bing Za Zhi 33:720–723 [PubMed] [Google Scholar]
- 36.Spiering W, Zwaan IM, Kroon AA, de Leeuw PW (2005) Genetic influences on 24 h blood pressure profiles in a hypertensive population: role of the angiotensin-converting enzyme insertion/deletion and angiotensin II type 1 receptor A1166C gene polymorphisms. Blood Press Monit 10:135–141 10.1097/00126097-200506000-00004 [DOI] [PubMed] [Google Scholar]
- 37.Abdollahi MR, Gaunt TR, Syddall HE, Cooper C, Phillips DI, Ye S, Day IN (2005) Angiotensin II type I receptor gene polymorphism: anthropometric and metabolic syndrome traits. J Med Genet 42:396–401 10.1136/jmg.2004.026716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henskens LH, Spiering W, Stoffers HE, Soomers FL, Vlietinck RF, de Leeuw PW, Kroon AA (2003) Effects of ACE I/D and AT1R-A1166C polymorphisms on blood pressure in a healthy normotensive primary care population: first results of the Hippocates study. J Hypertens 21:81–86 10.1097/00004872-200301000-00017 [DOI] [PubMed] [Google Scholar]
- 39.Gardier S, Vincent M, Lantelme P, Rial MO, Bricca G, Milon H (2004) A1166C polymorphism of angiotensin II type 1 receptor, blood pressure and arterial stiffness in hypertension. J Hypertens 22:2135–2142 10.1097/00004872-200411000-00016 [DOI] [PubMed] [Google Scholar]
- 40.Kobashi G, Hata A, Ohta K, Yamada H, Kato EH, Minakami H, Fujimoto S, Kondo K (2004) A1166C variant of angiotensin II type 1 receptor gene is associated with severe hypertension in pregnancy independently of T235 variant of angiotensinogen gene. J Hum Genet 49:182–186 (http://www.springerlink.com/content/9tgdfpqexvxgpkmj/fulltext.html) (electronically published March 23, 2004; accessed June 29, 2007) 10.1007/s10038-004-0129-4 [DOI] [PubMed] [Google Scholar]
- 41.Agachan B, Isbir T, Yilmaz H, Akoglu E (2003) Angiotensin converting enzyme I/D, angiotensinogen T174M-M235T and angiotensin II type 1 receptor A1166C gene polymorphisms in Turkish hypertensive patients. Exp Mol Med 35:545–549 [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Shan GL, Cui CY, Hou SQ, Zhuoma C, Cen WJ, Cai D, Zheng HQ, Xiao ZS, Wu ZL, Zhou WY, Qiu CC (2003) A1166C polymorphism of the angiotensin II type 1 receptor gene and essential hypertension in Han, Tibetan and Yi populations. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 20:220–224 [PubMed] [Google Scholar]
- 43.Porto PI, Garcia SI, Dieuzeide G, Gonzalez C, Pirola CJ (2003) Renin-angiotensin-aldosterone system loci and multilocus interactions in young-onset essential hypertension. Clin Exp Hypertens 25:117–130 10.1081/CEH-120017932 [DOI] [PubMed] [Google Scholar]
- 44.Petrovic D, Bidovec M, Peterlin B (2002) Gene polymorphisms of the renin-angiotensin-aldosterone system and essential arterial hypertension in childhood. Folia Biol (Krakow) 50:53–56 [PubMed] [Google Scholar]
- 45.Stankovic A, Zivkovic M, Glisic S, Alavantic D (2003) Angiotensin II type 1 receptor gene polymorphism and essential hypertension in Serbian population. Clin Chim Acta 327:181–185 10.1016/S0009-8981(02)00340-6 [DOI] [PubMed] [Google Scholar]
- 46.Hindorff LA, Heckbert SR, Tracy R, Tang Z, Psaty BM, Edwards KL, Siscovick DS, Kronmal RA, Nazar-Stewart V (2002) Angiotensin II type 1 receptor polymorphisms in the cardiovascular health study: relation to blood pressure, ethnicity, and cardiovascular events. Am J Hypertens 15:1050–1056 10.1016/S0895-7061(02)03063-7 [DOI] [PubMed] [Google Scholar]
- 47.Jiang Z, Zhao W, Yu F, Xu G (2001) Association of angiotensin II type 1 receptor gene polymorphism with essential hypertension. Chin Med J (Engl) 114:1249–1251 [PubMed] [Google Scholar]
- 48.Giner V, Corella D, Chaves FJ, Pascual JM, Portoles O, Marin P, Lozano JV, Armengod ME, Redon J (2001) Renin-angiotensin system genetic polymorphisms and essential hypertension in the Spanish population. Med Clin (Barc) 117:525–529 [DOI] [PubMed] [Google Scholar]
- 49.Dzida G, Sobstyl J, Puzniak A, Golon P, Mosiewicz J, Hanzlik J (2001) Polymorphisms of angiotensin-converting enzyme and angiotensin II receptor type 1 genes in essential hypertension in a Polish population. Med Sci Monit 7:1236–1241 [PubMed] [Google Scholar]
- 50.Davis D, Liyou N, Johnson A (2001) The ACE gene I/D polymorphism, but not the angiotensin II type I receptor gene A1166C polymorphism is associated with isolated systolic hypertension. J Hum Hypertens 15:653–654 10.1038/sj.jhh.1001221 [DOI] [PubMed] [Google Scholar]
- 51.Li X, Wang L, Han X (2001) Association between angiotensin system gene polymorphism and essential hypertension. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 18:292–295 [PubMed] [Google Scholar]
- 52.Xiang K, Zheng T, Sun D, Li J (1998) The relationship between angiotensin II type 1 receptor gene and coronary heart disease, hypertension and diabetes mellitus in Chinese. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 15:9–12 [PubMed] [Google Scholar]
- 53.Nalogowska-Glosnicka K, Lacka BI, Zychma MJ, Grzeszczak W, Zukowska-Szczechowska E, Poreba R, Michalski B, Kniazewski B, Rzempoluch J (2000) Angiotensin II type 1 receptor gene A1166C polymorphism is associated with the increased risk of pregnancy-induced hypertension. Med Sci Monit 6:523–529 [PubMed] [Google Scholar]
- 54.Seremak-Mrozikiewicz A, Drews K, Chmara E, Mrozikiewicz PM, Slomko Z (2000) Gestational hypertension (GH) and a1166c polymorphism of angiotensin II type 1 receptor. Ginekol Pol 71:783–788 [PubMed] [Google Scholar]
- 55.Liyou N, Davis D, James K, Simons L, Friedlander Y, Simons J, McCallum J, Johnson A (1999) The A1166C mutation in the angiotensin II type I receptor and hypertension in the elderly. Clin Exp Pharmacol Physiol 26:525–526 10.1046/j.1440-1681.1999.03066.x [DOI] [PubMed] [Google Scholar]
- 56.Berge KE, Berg K (1998) Polymorphisms at the angiotensinogen (AGT) and angiotensin II type 1 receptor (AT1R) loci and normal blood pressure. Clin Genet 53:214–219 [DOI] [PubMed] [Google Scholar]
- 57.Tiret L, Blanc H, Ruidavets JB, Arveiler D, Luc G, Jeunemaitre X, Tichet J, Mallet C, Poirier O, Plouin PF, Cambien F (1998) Gene polymorphisms of the renin-angiotensin system in relation to hypertension and parental history of myocardial infarction and stroke: the PEGASE study. J Hypertens 16:37–44 10.1097/00004872-199816010-00007 [DOI] [PubMed] [Google Scholar]
- 58.Wang WY, Zee RY, Morris BJ (1997) Association of angiotensin II type 1 receptor gene polymorphism with essential hypertension. Clin Genet 51:31–34 [DOI] [PubMed] [Google Scholar]
- 59.Kainulainen K, Perola M, Terwilliger J, Kaprio J, Koskenvuo M, Syvanen A, Vartiainen E, Peltonen L, Kontula K (1999) Evidence for involvement of the type 1 angiotensin II receptor locus in essential hypertension. Hypertension 33:844–849 [DOI] [PubMed] [Google Scholar]
- 60.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K (2002) A comprehensive review of genetic association studies. Genet Med 4:45–61 [DOI] [PubMed] [Google Scholar]
- 61.van Geel PP, Pinto YM, Voors AA, Buikema H, Oosterga M, Crijns HJ, van Gilst WH (2000) Angiotensin II type 1 receptor A1166C gene polymorphism is associated with an increased response to angiotensin II in human arteries. Hypertension 35:717–721 [DOI] [PubMed] [Google Scholar]
- 62.Carswell CI, Goa KL (2003) Losartan in diabetic nephropathy. Drugs 63:407–414 10.2165/00003495-200363040-00006 [DOI] [PubMed] [Google Scholar]
- 63.Song JC, White CM (2002) Clinical pharmacokinetics and selective pharmacodynamics of new angiotensin converting enzyme inhibitors: an update. Clin Pharmacokinet 41:207–224 10.2165/00003088-200241030-00005 [DOI] [PubMed] [Google Scholar]
- 64.Burnier M, Brunner HR (2000) Angiotensin II receptor antagonists. Lancet 355:637–645 10.1016/S0140-6736(99)10365-9 [DOI] [PubMed] [Google Scholar]
- 65.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144 10.1093/nar/gkj112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH (2006) Diversity of microRNAs in human and chimpanzee brain. Nat Genet 38:1375–1377 10.1038/ng1914 [DOI] [PubMed] [Google Scholar]
- 67.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E (2005) Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120:21–24 10.1016/j.cell.2004.12.031 [DOI] [PubMed] [Google Scholar]
- 68.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einat U, Meiri E, et al (2005) Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37:766–770 10.1038/ng1590 [DOI] [PubMed] [Google Scholar]