Abstract
Type 2 diabetes (T2D) is a common, polygenic chronic disease with high heritability. The purpose of this whole-genome association study was to discover novel T2D-associated genes. We genotyped 500 familial cases and 497 controls with >300,000 HapMap-derived tagging single-nucleotide–polymorphism (SNP) markers. When a stringent statistical correction for multiple testing was used, the only significant SNP was at TCF7L2, which has already been discovered and confirmed as a T2D-susceptibility gene. For a replication study, we selected 10 SNPs in six chromosomal regions with the strongest association (singly or as part of a haplotype) for retesting in an independent case-control set including 2,573 T2D cases and 2,776 controls. The most significant replicated result was found at the AHI1-LOC441171 gene region.
Type 2 diabetes (T2D [MIM 222100]) is considered a major medical burden on society.1 Given the high heritability and public health importance of the trait, the identification of the underlying genes has been a high-priority mission of the scientific community for many years.2,3 Nevertheless, only a handful of genes contributing to the predisposition of T2D have been confirmed, including TCF7L2 (MIM 602228), PPARγ (MIM 601487), KCNJ11 (MIM 600937), and CAPN10 (MIM 605286).4–9 These genes, however, account for only a small fraction of the genetic variation of the disease. Although conventional linkage-based studies in affected families have had some success in providing consistent data on linkage peaks in T2D,3,10–13 it is now believed that linkage analysis may lack the statistical power and mapping resolution to identify genes affecting common complex traits such as T2D.14 Association studies are the alternative approach and are currently considered the method of choice for the identification of genes affecting common complex traits.11 Until recently, association studies have been limited to specific genes or chromosomal regions. As a result of the Human Genome Project, the SNP Consortium, and the HapMap Project, there is today a wealth of knowledge regarding human SNPs and the way they can be used for whole-genome association (WGA) studies. Furthermore, microarray technologies now allow genotyping of the genome at the necessary density.
A French Canadian consortium9 recently presented the results of the first WGA study of T2D, indicating several novel T2D candidate genes. Our study, performed with the same technology, allows a comparison of two independent WGA studies. The genotyping platform used (HumanHap300 [Illumina]) contains 317,000 SNPs, which were selected on the basis of the HapMap data.
Material and Methods
Subjects
The population studied includes 500 cases and 497 controls from four different populations, all white. Specifically, we studied 201 cases and 200 controls from eastern Finland (EF), 200 cases and 197 controls of Ashkenazi Jewish (AJ) origin from Israel, and 99 cases and 100 controls from Germany (GE) and England (UK). The Germans were from the region of Pomerania in northeastern Germany, and the English were from the Manchester region. Two of the populations represent genetically homogenous founder populations (EF and AJ), whereas the Germans and English represent the general European population. Cases and controls were sex matched so that the proportion of each sex was identical in the cases and controls within each population. The controls were selected to be older than the cases, to reduce the chance that some of the controls will develop the disease in the future. Similar study protocols and case report forms were used at all sites and were correspondingly accepted by their ethics committees. Informed consent forms were signed by all subjects participating in this study.
T2D Inclusion/Exclusion Criteria
Subjects meeting the following three criteria were defined as patients with T2D: (1) either having regular treatment with an oral hypoglycemic agent (463 of 500 cases used antidiabetic medication) or having treatment only with diet while also having either an HbA1c level ⩾7.5% or a fasting blood glucose level at least 6.7 mmol/liter, (2) with disease manifestation at age 60 years or earlier, and (3) with family history of T2D (at least one parent or sibling). Patients given diagnoses of non-T2D were excluded.
Control Inclusion/Exclusion Criteria
Subjects meeting the following two criteria were defined as controls for all populations except AJ blood donors: (1) receiving neither insulin treatment nor oral antidiabetic medication and having glucose levels <5.5 mmol/liter and (2) having no family history of diabetes (not in grandparents, parents, siblings, or offspring). Some AJ control samples were obtained from blood donors (see below) who declared not having any chronic disease.
Since it is a major risk factor and etiology of T2D, obesity was not a selection criterion for either the cases or the controls. Consequently, the mean BMI was considerably higher among the cases than the controls.
WGA Sample-Collection Process
Eastern Finland
The data and sample collection was performed in two places—by the Research Institute of Public Health, University of Kuopio (166 cases and 166 controls), and by Oy Foodfiles (35 cases and 34 controls), Kuopio, Finland. In the university collection, the sampling frame was the 17,100 examinees of a questionnaire survey mailed in 2003 to all households in a small region within the founder population, for the “North Savo Project.” In this survey, information about medical and family history and medical treatments and consent forms for further participation were collected. The subjects were reexamined in 2004 and 2005 at the institute, and blood samples were collected for DNA extraction and other measurements, in accordance with the protocol. The Foodfiles collection was performed between late 2005 and early 2006 with the use of newspaper advertisements in the surrounding communities of the city of Kuopio, with use of inclusion criteria identical to that of the university collection. On average (including both of these collections), 3.1 of the 4 grandparents were of eastern Finnish origin. A total of 201 T2D cases (99 women and 102 men) and 200 matched nondiabetic controls (99 women and 101 men) were included in the study.
Ashkenazi Jews
Subjects included in the study were collected in Israel by the patients’ physicians in specialized clinics. We included 200 subjects with T2D (82 men and 118 women, mean age 64 years), each with 3 or more blood relatives of second degree or closer who had T2D. Two hundred matching nondiabetic control samples (82 men and 118 women, mean age 74 years) were collected from the Israeli Blood Bank and from elderly patients visiting general practitioners' clinics. All subjects were of AJ origin.
Germany
Cases were sampled from ∼800 inpatients with T2D from the Clinic for Diabetes and Metabolic Diseases Karlsburg and from the Department of Internal Medicine of the Ernst Moritz Arndt University Greifswald who were examined by a study physician and who fulfilled the inclusion criteria. Corresponding controls (matched by sex and age) were selected from among the 3,900 nondiabetic examinees of the population-based “Study of Health in Pomerania 1997–2001” cohort.15 Data on family history of diabetes, medication use, and comorbidity were gathered by a standardized face-to-face interview and measurement of blood pressure and BMI, and laboratory analyses were performed using standardized procedures. For the patients with T2D, additional information was drawn from the patient records. A total of 49 cases (24 women and 25 men) and 50 matched healthy controls (24 women and 26 men) from Germany were included in the study.
England
A total of 50 cases (31 women and 19 men) and 50 matched nondiabetic controls (31 women and 19 men) were included in the study. The cases were selected from among 8,300 registered patients with T2D in the city of Salford whose baseline characteristics matched those of the catchment population with T2D (n=8,300/220,000) on the basis of age, sex, and duration of diabetes. Family history data was gathered via a telephone survey with the use of a standardized questionnaire that included questions about the patients and their parents, children, siblings, and grandparents. The controls were selected from among the examinees of the Age and Cognitive Performance Research Centres volunteer panel, a group of >6,000 older adults who have been described in detail elsewhere.16 A cohort of ∼2,000 of these individuals have DNA archived in the Dyne-Steel DNA bank, and 456 of these volunteers, residents of Greater Manchester, had taken part in a research study in 2001 that included medical history, including that of T2D, and measurement of HbA1c. Of these, subjects were identified to sex match diabetic cases from Manchester.
DNA Samples
At each participating site, DNA was extracted from whole blood with the use of standard methods. The quantity and purity of each DNA sample was uniformly determined by absorbance measurements with a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies). A sample was qualified for WGA analysis if the A260:A280 ratio was ⩾1.7 and the DNA concentration was >60 ng/μl. Before WGA analysis, the samples were diluted to a concentration of 60 ng/μl in reduced Tris EDTA buffer (TEKnova).
Genotyping Platforms and SNPs
The whole-genome genotyping of DNA samples was performed using Sentrix HumanHap300 BeadChips and Infinium II genotyping assays (Illumina). The HumanHap300 BeadChip contained 317,503 tag SNPs derived from the International HapMap Project. The genotyping reactions were performed according to the Single-Sample BeadChip Manual process described in detail in the Infinium II Assay System Manual (Illumina). In brief, 750 ng of genomic DNA was subjected to whole-genome amplification at 37°C for 20–24 h. Products were fragmented, precipitated, and resuspended in hybridization buffer. The resuspended samples were denatured at 95°C for 20 min, were loaded to BeadChips, and were placed in a hybridization chamber for 16–20 h at 48°C. After hybridization, the mis- and nonhybridized DNA was washed away from the BeadChips, and an allele-specific single-base extension of the oligonucleotides on the BeadChip was performed in a 48-position Slide Chamber Rack (GenePaint [Tecan]), with the use of labeled deoxynucleotides and the captured DNA as a template. After staining of the extended DNA, BeadChips were washed and scanned with the BeadArray Reader (Illumina), and genotypes from samples were called using the BeadStudio 2.0 software (Illumina).
The Centaurus genotyping methodology uses standard PCR that uses primers to produce an amplicon of fewer than a few hundred residues. Allele-specific probes displaying a fluorescent dye at the 3′ end, a quencher at the 5′ end, and a short enhancer oligonucleotide (∼8–15 residues in length) are hybridized to the amplicon.17 The enhancer oligonucleotide hybridizes to the amplicon 3′ relative to the allele-specific probes. The uniqueness of the assay is derived through a design that produces a single-base gap between the two probes and the enhancer oligonucleotide. This gap creates a synthetic abasic site, which directs the specificity of the enzyme endonuclease IV to this specific location. This enzyme will cleave the dye on the fully complementary probe, thus separating the dye from the quencher molecule and producing a fluorescent signal. The probe that has a mismatch to the target remains uncleaved.
Initial SNP Selection for Statistical Analysis
To check that the same individual was not included twice in the study, a set of 23 SNPs (one polymorphic SNP from each chromosome) was compared across all subjects before the statistical analysis. SNP quality was also assessed on the basis of three criteria: the call rate (CR), minor-allele frequency (MAF), and Hardy-Weinberg (H-W) equilibrium. The CR is the proportion of samples genotyped successfully, regardless of whether the genotypes are correct. The MAF is the frequency of the allele that is less frequent in the study sample. H-W equilibrium was tested for controls with use of the standard χ2 test of goodness of fit with 1 df. A SNP not in H-W equilibrium can be due to genotyping error or to some unknown population dynamics (e.g., random drift or selection). Only the SNPs that had CR >90% and MAF >1% and were in H-W equilibrium (χ2 test statistic <27.5 in controls) were used in the statistical analysis. A total of 315,917 of 317,503 SNPs available in the Illumina HumanHap300 assay fulfilled the above criteria and were included in the statistical analysis.
Single-Point Analysis
The single-point analysis was performed using Cochran-Mantel-Haenszel χ2 tests to detect association between disease status and the allele frequency of each SNP, with stratification of the samples by population. Our sample set consists of four different populations. Cases and controls were carefully matched within each population by sex and ethnicity, to avoid spurious associations. Accounting for multiple testing, we computed the significance threshold, using 10,000 permutations. It should be noted that the number of permuted P values is 10,000×315,917 SNPs and is thus appropriate for obtaining a very accurate threshold. The disease status was randomly reassigned to subjects within each population, and the same protocol of testing was applied to the permuted samples. The statistical analysis was performed using the R programming environment. We used the specifications of the function “mantelhaen.test” in the implementation of the test.
In addition, the associations of genotypes with T2D were analyzed with an adjustment for age, sex, and population by use of binary logistic modeling. The population was entered as a categorical variable, so each population had its own correction for the dependent variable.
Haplotype Analysis
Haplotypes of large chromosomal segments (e.g., 400 SNPs) were estimated first with the HaploRec program, and then the segments were joined to create fully phased chromosomes.18 The HaploRec program was used because it has recently been found to be extremely efficient and powerful compared with popular alternatives.19 Each population was haplotyped separately because population haplotype frequencies differ; thus, estimation of haplotypes within a population is more accurate than use of the pooled data. Populations were combined, and haplotype frequencies were estimated with the HPM program.20 The program was used to test haplotypes of length 2, 3, 4, and 5 SNPs. For each haplotype, HPM calculates how many times the haplotype is present in case and control chromosomes and how many times it is not present in case and control chromosomes. This results in a 2×2 table for each single haplotype tested against all other haplotypes. To assess statistical significance, 10,000 permutations were performed similar to the single-point analysis.
Replication Study
The replication study included 2,573 T2D cases and 2,776 normoglycemic control subjects given diagnoses in accordance with the 1997 American Diabetes Association criteria. The T2D cases were recruited at the Sud Francilien Hospital or at the Centre National de la Recherche Scientifique UMR8090, Lille. All cases had family history of T2D (i.e., the proband had at least one first-degree relative with T2D). Individuals with maturity-onset diabetes of the young or neonatal or mitochondrial diabetes were excluded. The control subjects were obtained from a prospective population-based cohort of middle-aged subjects (N=5,153 at baseline).21 They had fasting blood glucose level <6.1 mmol/liter at baseline and during a 9-year follow-up (measurements at 0, 3, 6, and 9 years), and their family histories were free of T2D. Genotyping of the 10 SNPs in these samples was performed with the TaqMan technology (Applied Biosystems).
Results
In the initial WGA study, we genotyped 500 cases and 497 controls, producing >300 million genotype results. The cases and controls were selected from four populations (EF, AJ, GE, and UK). All populations are of white origin but are distinct, thus allowing the examination of the common basis of any finding across different populations. Sex was matched between cases and controls within each population. The age distribution was higher in controls, to include as few as possible nondiabetic individuals who may develop the disease in the future.
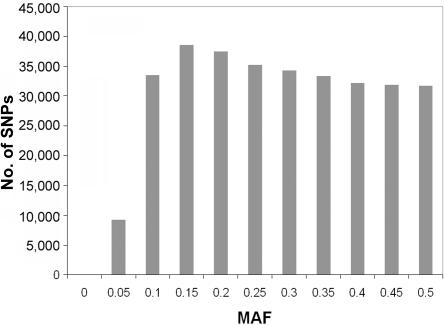
The average CR for all SNPs was very impressive (99.5%), with only 1,341 (0.42%) of the 317,503 SNPs tested having a CR <90%. Four subjects chosen randomly were genotyped twice for all SNPs. Reproducibility of genotypes was >99.9%. In addition, to study the validity of the Infinium method, 12 HumanHap300 SNPs were also genotyped for 393 samples with the Centaurus methodology.17 The concordance of the genotypes was >99.9%. Figure 1 presents the distribution of the MAF of all SNPs. As expected, the MAF was found to be quite uniformly distributed between 0.1 and 0.5. Of the 317,503 SNPs available in the Illumina HumanHap300 assay, 315,917 fulfilled our criteria of having CR >90% and MAF >1% and not being in extreme H-W disequilibrium (P>1.5×10-7) and were included in the statistical analysis.
Figure 1. .
Distribution of MAFs in 317,503 SNPs
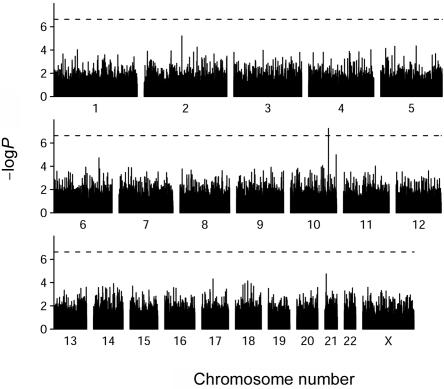
Because we analyzed four different populations under the same null hypothesis and model, the statistical inferences for single SNPs were based on the Cochrane-Mantel-Haenszel statistic. Figure 2 presents the nominal P value of each of the SNPs tested across the entire genome. Except for one SNP on chromosome 10 at the TCF7L2 gene (rs7903146), no other SNP crossed the stringent corrected-statistical-significance threshold (fig. 2, dashed lines at -log(P)=6.60, P=2.5×10-7, based on 10,000 permutations). Table 1 presents the detailed results for the 10 most significant SNPs with MAF >0.05. The effect of statistical adjustment for age, sex, and population was small (data not shown).
Figure 2. .
Vertical lines plotted for each SNP analyzed, placed according to their chromosomal locations. The height of the lines correspond to the nominal P value (plotted as −logP) of each SNP as obtained from a single-point analysis with use of a Cochran-Mantel-Haenszel χ2 test (see the “Material and Methods” section). The horizontal dashed lines (at -logP=6.64, P=2.3×10-7) correspond to the statistical threshold adjusted for multiple testing and an overall statistical significance of .05, after 1,000 permutations.
Table 1. .
SNPs with the Strongest Association with T2D in the WGA Study
| SNP Markera | P | Allelic ORb (95% CI) | Minor Allele | Chromosome | Gene |
| rs7903146 | 5.52×10−8 | 1.71 (1.41–2.08) | A | 10 | TCF7L2 |
| rs7901695 | 3.42×10−7 | 1.66 (1.37–2.01) | G | 10 | TCF7L2 |
| rs12255372 | 5.31×10−7 | 1.64 (1.35–2.00) | A | 10 | TCF7L2 |
| rs6712932 | 6.25×10−6 | .66 (.55–.79) | G | 2 | None |
| rs7910485 | 1.02×10−5 | 1.58 (1.29–1.94) | A | 10 | STK32C |
| rs200801 | 1.78×10−5 | 1.48 (1.24–1.77) | A | 21 | None |
| rs1535435 | 1.86×10−5 | 2.34 (1.58–3.46) | A | 6 | AHI1 |
| rs158081 | 2.36×10−5 | .66 (.54–.80) | G | 21 | None |
| rs9494266 | 2.67×10−5 | 2.31 (1.56–3.42) | A | 6 | LOC441171 |
| rs2254434 | 3.86×10−5 | .68 (.56–.81) | A | 21 | None |
Markers selected for the replication study are highlighted in bold.
ORs presented are for all four populations pooled.
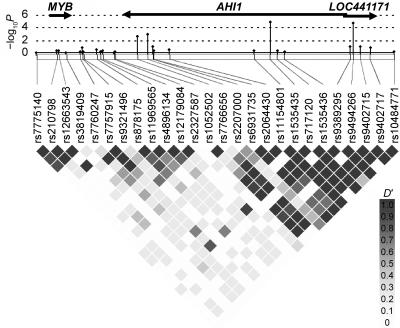
The intragenic SNPs with the next strongest univariate association were in STK32C, AHI1 (Abelson helper integration site 1 [MIM 608894]), and LOC441171, which flanks and overlaps AHI1. As shown in figure 2, the AHI1 and LOC441171 markers were in complete linkage disequilibrium (LD) (D′=1). Since haplotype analysis may have higher statistical power, the data were also analyzed using haplotypes for all possible sets of two to five consecutive SNPs (table 2). The next strongest haplotypes, after those in TCF7L2, were in the genes MYO10 (MIM 601481) and TTC7B. With use of a stringent correction for multiple testing, only a P value of 2.02×10−8 (based on 10,000 permutations) can be considered significant in its own right; no haplotype crossed this value.
Table 2. .
Haplotypes with the Strongest Association with T2D in the WGA
| Haplotype | SNP Markersa | P | Haplotype ORb (95% CI) | Haplotype | Chromosome | Gene |
| 1 | rs7895307, rs7901695, and rs7903146 | 3.47×10−8 | 1.76 (1.44–2.16) | GGA | 10 | TCF7L2 |
| 2 | rs3936203, rs10933514, and rs4630763 | 5.02×10−7 | 1.77 (1.41–2.21) | GGC | 2 | None |
| 3 | rs2824577 and rs200801 | 1.28×10−6 | .64 (.54–.77) | AG | 21 | None |
| 4 | rs2288440, rs31299, rs40979, rs173738, and rs1445946 | 1.30×10−6 | 1.61 (1.33–1.96) | AACGG | 5 | MYO10 |
| 5 | rs1749718, rs1742083, and rs8018904 | 3.02×10−6 | .64 (.53–.77) | GAC | 14 | TTC7B |
| 6 | rs10491118, rs854685, and rs854692 | 3.97×10−6 | .34 (.21–.55) | CAA | 17 | CCL14-CCL15 |
| 7 | rs4495839, rs4466762, rs11596854, rs4128664, and rs925587 | 4.13×10−6 | .46 (.33–.64) | AAGAG | 10 | ANTXRL |
| 8 | rs1042778, rs237887, rs2268490, rs2268491, and rs237889 | 5.82×10−6 | .04 (.01–.31) | AGAAG | 3 | OXTR |
| 9 | rs930306, rs13136503, rs7682282, rs13106255, and rs7695711 | 5.86×10−6 | .28 (.16–.50) | AGAGG | 4 | None |
| 10 | rs6712932 and rs1545122 | 6.86×10−6 | .66 (.54–.79) | GG | 2 | None |
Markers selected for the replication study are highlighted in bold.
OR for the defined haplotype versus all other haplotypes (per increment of the number of copies). Haplotypes 1, 4, 5, 6, and 8 are intragenic.
Ten SNPs (table 3)—including the most promising ones from tables 1 and 2, two additional SNPs in MYO10 (rs31313 and rs253336), and one additional SNP in TTC7B (rs942740)—were retested in a case-control set that was independent of the WGA subjects: 2,573 cases and 2,776 controls from France. These cases and controls have already been found to be well matched and appropriate for association studies of T2D.9 SNPs in TCF7L2 were not included in the replication study, since this gene has been well established as a T2D-susceptibility gene, including in the samples used here for the replication study.9 Table 3 presents the detailed results of these 10 SNPs in the initial WGA samples, and table 4 presents the results of these SNPs in the replication samples. Five of these 10 SNPs reached a nominal statistical significance of P<.01. However, only two SNPs (rs1535435 and rs9494266) in the same haplotype reached the Bonferroni-corrected P value <.005 for allelic association in the replication study (P=.0002 and P=.00005, respectively) and exhibited odds ratios (ORs) in the same direction as those found in the initial WGA study. (SNPs in the TTC7B gene also reached a P value <.005; however, ORs were in opposite directions.) The respective P values for rs1535435 and rs9494266 in a pooled Cochran-Mantel-Haenszel analysis of the WGA together with the replication study were 10−6 and 2.28×10−7. Figure 3 presents the haplotype structure around the significant region. The most significant finding was on chromosome 6, located within an LD block of 178 kb encompassing 19 introns of the AHI1 gene and the non–protein-coding LOC441171 on chromosome 6 (fig. 3). This region has been identified elsewhere as a T2D-susceptibility locus in several linkage studies.22,23 LOC441171 (or C6orf217) is a primate-specific gene expressed in brain, pancreas, and liver, and it does not seem to encode a functional protein. Therefore, AHI1 might be a preferred candidate gene, although a functional effect of LOC441171 cannot be entirely discarded.
Table 3. .
Detailed Results of the 10 SNPs Selected for Replication
| EFa |
AJb |
GE and UKc |
Four PopulationsCombinedd |
|||||||||||
| SNP | Gene | Minor Allele | MAF | OR | P | MAF | OR | P | MAF | OR | P | MAF | OR | P |
| rs6712932 | None | G | .294 | .568 | .0003 | .448 | .702 | .0135 | .317 | .699 | .0977 | .360 | .657 | .000008 |
| rs7910485 | STK32C | A | .287 | 1.750 | .0004 | .246 | 1.457 | .0232 | .236 | 1.500 | .0876 | .260 | 1.583 | .000008 |
| rs200801 | None | A | .365 | 1.479 | .0078 | .504 | .661 | .0036 | .399 | 1.454 | .0685 | .427 | 1.481 | .000016 |
| rs1535435 | AHI1 | A | .039 | 3.565 | .0019 | .065 | 1.626 | .0959 | .101 | 2.948 | .0024 | .062 | 2.337 | .00001 |
| rs9494266 | LOC441171 | A | .039 | 3.565 | .0019 | .065 | 1.626 | .0959 | .098 | 2.830 | .0037 | .061 | 2.307 | .00002 |
| rs253336 | MYO10 | A | .288 | 1.221 | .2003 | .198 | 1.668 | .0046 | .317 | 1.545 | .0446 | .258 | 1.419 | .00067 |
| rs173738 | MYO10 | A | .307 | .697 | .0191 | .402 | .719 | .0230 | .359 | .732 | .1357 | .355 | .716 | .00037 |
| rs31313 | MYO10 | A | .268 | 1.595 | .0037 | .181 | 1.668 | .0063 | .178 | .903 | .6989 | .215 | 1.46 | .00057 |
| rs942740 | TTC7B | A | .187 | 1.691 | .0042 | .194 | 1.271 | .1830 | .121 | 2.217 | .0124 | .177 | 1.55 | .00022 |
| rs1749718 | TTC7B | A | .459 | 1.513 | .0036 | .364 | 1.228 | .1652 | .490 | 1.497 | .0451 | .427 | 1.386 | .00032 |
201 cases and 200 controls.
200 cases and 197 controls.
99 cases and 100 controls.
500 cases and 497 controls.
Table 4. .
Association Results of the Replication Study in 2,573 T2D Cases and 2,776 Controls
| SNP | Gene | Minor Allele | MAF | ORa (95% CI) | P |
| rs6712932 | None | G | .351 | 1.117 (1.030–1.212) | .008 |
| rs7910485 | STK32C | A | .247 | .980 (.896–1.072) | .657 |
| rs200801 | None | A | .477 | 1.031 (.955–1.113) | .433 |
| rs1535435 | AHI1 | A | .096 | 1.287 (1.126–1.470) | .0002 |
| rs9494266 | LOC441171 | A | .100 | 1.308 (1.149–1.489) | .00005 |
| rs253336 | MYO10 | A | .300 | .986 (.906–1.073) | .746 |
| rs173738 | MYO10 | A | .357 | 1.060 (.976–1.151) | .167 |
| rs31313 | MYO10 | A | .193 | 1.024 (.928–1.130) | .634 |
| rs942740 | TTC7B | A | .185 | .858 (.777–.948) | .003 |
| rs1749718 | TTC7B | A | .499 | .878 (.812–.950) | .001 |
ORs are presented for the minor allele versus the major allele.
Figure 3. .
Pairwise LD diagram for SNPs in the AHI1 gene region. The LD between pairs of SNPs is presented using D′, which was calculated from the DiaGen genotyping data with Haploview software. The bar graph indicates the negative logarithm of the P value for each SNP from single-point analysis.
AHI1 encodes a unique AHI1 (Jouberin) protein, the only known protein containing an SH3 motif, seven WD40-repeat domains, and multiple SH3-binding domains (PXXP motifs).24,25 WD motifs are evolutionarily conserved across the species and form a stable β-propeller structure that serves as a platform for protein-protein interactions.26 Specific functions of WD-repeat proteins seem to be determined by other motifs and domains represented in the protein.26 Therefore, the domain structure suggests that AHI1 is likely to act as a scaffold protein and to participate in a variety of processes such as signal transduction, cytoskeleton organization, cell cycle, and cell-cell communication.26,27 However, the exact function of AHI1 is not clarified yet, and additional studies will be required to determine its possible involvement in T2D.
The SNPs rs1535435 and rs9494266 may directly affect susceptibility to T2D or may predispose an individual to obesity (MIM 601665) and thus indirectly affect susceptibility to T2D. For 1,084 of the cases in the replication set and 1,084 of the controls, we have BMI information. In this restricted set, the allelic P values of rs1535435 and rs9494266 were .005 and .003, respectively. Analyzing these SNPs with BMI as a covariate resulted in allelic P values of .019 and .011, respectively. Therefore, although the observed effect on T2D may be partially mediated through obesity, it seems that a direct and independent effect is also present.
Discussion
In the current study, we aimed to assess the potential value of WGA studies on the basis of HapMap-derived high-density SNP arrays. To increase statistical power, we primarily used samples from homogeneous founder populations (801 of the 997 subjects included in this study). In addition, we included disease cases that have a high familial prevalence of the disease—on average, more than two affected relatives in addition to the proband. In spite of our effort to carefully select samples in a manner that may increase statistical power and the use of the most advanced technology available today, a sample of ∼500 cases and ∼500 controls was powerful enough to identify only one known gene and to provide suggestive evidence of the identification of other genes affecting T2D. This is similar to the findings of another WGA study of T2D.9 Following a genomewide statistical correction, Sladek et al.9 also found only the TCF7L2 gene to be associated with T2D. They also performed a replication study in which eight additional SNPs were replicated.9 Of these eight SNPs, five are present in the HumanHap300 microarray that we used. Interestingly, none of these five SNPs reached a nominal statistical significance of .01 in our WGA study. It should be noted that the samples used for replication by Sladek et al.9 are the same ones used for replication in the current study.
In the current study, we selected the 10 most promising SNPs and genotyped them in the replication sample set. The most promising result was found at the AHI1 gene region. The selection of only 10 SNPs is somewhat arbitrary, and there might be value in testing a significantly larger number of SNPs. The partial inconsistency of the WGA findings between populations and studies deserves further research, since it may suggest either population specificity of some of the observed associations, insufficient statistical power, or possible false-positive results in either study. Population specificity could be due to differences in LD blocks, population-specific interactions between genes and nongenetic factors, population-specific epigenetic effects, or the different inclusion criteria used (specifically, BMI <30 for the cases in the study by Sladek et al.,9 whereas BMI was not a selection criterion in our study). The possibility of false-positive results should be carefully considered, particularly in light of the unexpectedly high proportion of SNPs in the replication study that exhibit a significant effect (5 of 10), 3 of them with ORs in a direction opposite to the initial findings. Population stratification is unlikely to be a source of false-positive results in the initial WGA study, since samples were carefully matched by ethnicity and sex. More importantly, the only significant result in the initial WGA study was found in a gene known to affect T2D, providing strong evidence of the lack of population stratification in these samples. Nevertheless, population stratification as a source of false-positive results in the replication set cannot be entirely discarded. As in any association study of this kind, further replication is necessary. On the positive side, the replication of the TCF7L2 finding provides strong evidence of the robustness of the WGA-study approach, at least for the identification of genes with allelic ORs >1.7. Our study also indicates, as has been suggested on a theoretical basis,28 that larger sample sizes will be necessary to unequivocally establish the involvement of genes with modest ORs (i.e., <1.7) across different populations.
Acknowledgments
We thank the personnel of Oy Jurilab, all participating institutes, the former employees and clinical collaborators of IDgene Pharmaceuticals, Stephane Lobbens for technical assistance, and the subjects who donated samples for this study. This work was partially funded by support from the Finnish Funding Agency for Technology and Innovation (TEKES), to Oy Jurilab. The Study of Health in Pomerania is funded by grant 01ZZ96030 from the German Ministry for Education and Research and by grants from the Ministry for Education, Research, and Cultural Affairs and the Ministry for Social Affairs of the Federal State of Mecklenburg, West Pomerania. The collection of the Dyne-Steel DNA bank for cognitive genetic studies was partially funded by Research Into Ageing. The materials, anonymized data, and associated protocols are available on request for the SNPs and haplotypes presented in this article. Several of the coauthors (J.T.S., P.U., J.-M.A., M.P., J.K., B.T., and C.D.) are inventors involved in one to four patent applications (U.S. application numbers 11/325,330, 60/798,706, 60/805,522, and 60/863,438) related to the findings of this study.
Web Resources
The URLs for data presented herein are as follows:
- HapMap Project and SNP Consortium, http://www.hapmap.org/
- Human Genome Project, http://www.ornl.gov/sci/techresources/Human_Genome/home.shtml
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for T2D, TCF7L2, PPARγ, KCNJ11, CAPN10, AHI1, MYO10, and obesity)
- The R Project for Statistical Computing, http://www.r-project.org/
References
- 1.Narayan KM, Boyle JP, Geiss LS, Saaddine JB, Thompson TJ (2006) Impact of recent increase in incidence on future diabetes burden: U.S., 2005–2050. Diabetes Care 29:2114–2116 10.2337/dc06-1136 [DOI] [PubMed] [Google Scholar]
- 2.Permutt MA, Wasson J, Cox N (2005) Genetic epidemiology of diabetes. J Clin Invest 115:1431–1439 10.1172/JCI24758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barroso I (2005) Genetics of type 2 diabetes. Diabet Med 22:517–535 10.1111/j.1464-5491.2005.01550.x [DOI] [PubMed] [Google Scholar]
- 4.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al (2006) Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38:320–232 10.1038/ng1732 [DOI] [PubMed] [Google Scholar]
- 5.Saxena R, Gianniny L, Burtt NP, Lyssenko V, Giuducci C, Sjogren M, Florez C, Almgren P, Isomaa B, Orho-Melander M, et al (2006) Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes 55:2890–2895 10.2337/db06-0381 [DOI] [PubMed] [Google Scholar]
- 6.Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, et al (2000) The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26:76–80 10.1038/79839 [DOI] [PubMed] [Google Scholar]
- 7.Gloyn AL, Weedon MN, Owen KR, Turner MJ, Knight BA, Hitman G, Walker M, Levy JC, Sampson M, Halford S, et al (2003) Large-scale association studies of variants in genes encoding the pancreatic β-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes 52:568–572 10.2337/diabetes.52.2.568 [DOI] [PubMed] [Google Scholar]
- 8.Tsuchiya T, Schwarz PE, Bosque-Plata LD, Geoffrey Hayes M, Dina C, Froguel P, Wayne Towers G, Fischer S, Temelkova-Kurktschiev T, Rietzsch H, et al (2006) Association of the calpain-10 gene with type 2 diabetes in Europeans: results of pooled and meta-analyses. Mol Genet Metab 89:174–184 10.1016/j.ymgme.2006.05.013 [DOI] [PubMed] [Google Scholar]
- 9.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al (2007) A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445:881–885 10.1038/nature05616 [DOI] [PubMed] [Google Scholar]
- 10.Permutt MA (1991) Use of DNA polymorphisms for genetic analysis of non-insulin dependent diabetes mellitus. Baillieres Clin Endocrinol Metab 5:495–526 10.1016/S0950-351X(05)80144-2 [DOI] [PubMed] [Google Scholar]
- 11.Laird NM, Lange C (2006) Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet 7:385–394 10.1038/nrg1839 [DOI] [PubMed] [Google Scholar]
- 12.McCarthy MI (2003) Growing evidence for diabetes susceptibility genes from genome scan data. Curr Diab Rep 3:159–167 10.1007/s11892-003-0040-y [DOI] [PubMed] [Google Scholar]
- 13.Hansen L, Pedersen O (2005) Genetics of type 2 diabetes mellitus: status and perspectives. Diabetes Obes Metab 7:122–135 10.1111/j.1463-1326.2004.00396.x [DOI] [PubMed] [Google Scholar]
- 14.Balding DJ (2006) A tutorial on statistical methods for population association studies. Nat Rev Genet 7:781–791 10.1038/nrg1916 [DOI] [PubMed] [Google Scholar]
- 15.Luedemann J, Schminke U, Berger K, Piek M, Willich SN, Doring A, John U, Kessler C (2002) The association between behavior dependent cardiovascular risk factors and asymptomatic carotid atherosclerosis in a general population. Stroke 33:2929–2935 10.1161/01.STR.0000038422.57919.7F [DOI] [PubMed] [Google Scholar]
- 16.Rabbitt PMA, McInnes L, Diggle P, Holland F, Bent N, Abson V, Pendleton N, Horan M (2004) The University of Manchester Longitudinal Study of Cognition in Normal Healthy Old Age, 1983 through 2003. Neuropsychol Dev Cogn B Aging Neuropsychol Cogn 11:245–279 10.1080/13825580490511116 [DOI] [Google Scholar]
- 17.Kutyavin IV, Milesi D, Belousov Y, Podyminogin M, Vorobiev A, Gorn V, Lukhtanov EA, Vermeulen NM, Mahoney W (2006) A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res 34:e128 10.1093/nar/gkl679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eronen L, Geerts F, Toivonen H (2004) A Markov chain approach to reconstruction of long haplotypes. Pac Symp Biocomput 9:104–115 [DOI] [PubMed] [Google Scholar]
- 19.Eronen L, Geerts F, Toivonen H (2006) HaploRec: efficient and accurate large-scale reconstruction of haplotypes. BMC Bioinformatics 7:542 10.1186/1471-2105-7-542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toivonen HT, Onkamo P, Vasko K, Ollikainen V, Sevon P, Mannila H, Herr M, Kere J (2000) Data mining applied to linkage disequilibrium mapping. Am J Hum Genet 67:133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balkau B (1996) An epidemiologic survey from a network of French Health Examination Centres, (D.E.S.I.R.): epidemiologic data on the insulin resistance syndrome. Rev Epidemiol Sante Publique 44:373–375 [PubMed] [Google Scholar]
- 22.Xiang K, Wang Y, Zheng T, Jia W, Li J, Chen L, Shen K, Wu S, Lin X, Zhang G, et al (2004) Genome-wide search for type 2 diabetes/impaired glucose homeostasis susceptibility genes in the Chinese: significant linkage to chromosome 6q21-q23 and chromosome 1q21-q24. Diabetes 53:228–234 10.2337/diabetes.53.1.228 [DOI] [PubMed] [Google Scholar]
- 23.Hanson LR, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Foroud T, Kobes S, Baier L, Burns DK, et al (1998) An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am J Hum Genet 63:1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon-Salazar T, Silhavy JL, Marsh SE, Louie CM, Scott LC, Gururaj A, Al-Gazali L, Al-Tawari AA, Kayserili H, Sztriha L, et al (2004) Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am J Hum Genet 75:979–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louie CM, Gleeson JG (2005) Genetic basis of Joubert syndrome and related disorders of cerebellar development. Hum Mol Genet 14:R235–R242 10.1093/hmg/ddi264 [DOI] [PubMed] [Google Scholar]
- 26.Li D, Roberts R (2001) WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci 58:2085–2097 10.1007/PL00000838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer BJ (2001) SH3 domains: complexity in moderation. J Cell Sci 114:1253–1263 [DOI] [PubMed] [Google Scholar]
- 28.Wang WY, Barratt BJ, Clayton DG, Todd JA (2005) Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet 6:109–118 10.1038/nrg1522 [DOI] [PubMed] [Google Scholar]