Abstract
A single-nucleotide variant, C/T-13910, located 14 kb upstream of the lactase gene (LCT), has been shown to be completely correlated with lactase persistence (LP) in northern Europeans. Here, we analyzed the background of the alleles carrying the critical variant in 1,611 DNA samples from 37 populations. Our data show that the T-13910 variant is found on two different, highly divergent haplotype backgrounds in the global populations. The first is the most common LP haplotype (LP H98) present in all populations analyzed, whereas the others (LP H8–H12), which originate from the same ancestral allelic haplotype, are found in geographically restricted populations living west of the Urals and north of the Caucasus. The global distribution pattern of LP T-13910 H98 supports the Caucasian origin of this allele. Age estimates based on different mathematical models show that the common LP T-13910 H98 allele (∼5,000–12,000 years old) is relatively older than the other geographically restricted LP alleles (∼1,400–3,000 years old). Our data about global allelic haplotypes of the lactose-tolerance variant imply that the T-13910 allele has been independently introduced more than once and that there is a still-ongoing process of convergent evolution of the LP alleles in humans.
The expression of the lactase enzyme (MIM 603202) in intestinal cells dramatically declines after weaning in mammals, when lactose is no longer an essential part of their diet.1 In humans, this normal mammalian condition known as “lactase nonpersistence” (LNP, also known as “adult-type hypolactasia” or “lactose intolerance” [MIM 223100]) affects most of mankind and restricts the consumption of fresh milk among adults. However, among northern Europeans and a few other ethnic populations, intestinal lactase activity persists throughout life in a substantial proportion (up to 80%–90%) of adults, a condition known as lactase persistence (LP, or lactose tolerance [MIM 223100]). The LP/LNP phenotype is genetically determined, with LP being dominant over LNP.2 We previously identified a single-nucleotide variant, C/T-13910, completely correlating with the phenotype in Finns and in a cross-sectional sample of >600 individuals from five populations.3–5 The T-13910 variant, which correlates with LP, is located 14 kb upstream of the LCT gene and has been shown to be the derived variant, compared with the C-13910 variant that represents the ancestral form of the human genome. Another variant, G/A-22018, farther upstream of LCT, was also strongly, although not completely, associated with the LP/LNP phenotype,3,5 most likely because of the substantial linkage disequilibrium (LD) in this genome region.3,6–9
Functional evidence for the C/T-13910 variant in the regulation of lactase activity has since emerged, lending additional support for this nucleotide change as the true causative variant of regulation of transcription of the lactase gene in intestinal cells.4,10,11 Adult individuals with the LP T-13910 allele show significantly higher steady-state transcript levels of LCT in their intestinal mucosa when compared with individuals with the nonpersistence C-13910 allele, which implies a transcriptional regulation of LCT.4 This is in agreement with in vitro studies demonstrating a distinct increase in the LCT promoter activity in cells transfected with the T-13910 variant.10–12 Haplotype analysis in the Finnish families demonstrated that all LP alleles among Finns originated from one common ancestor identical by descent.3 Other studies of additional European populations have also suggested the existence of one major allelic haplotype, named “haplotype A,” correlating with LP.7,13 These data indicate a single global origin for the LP T-13910 allele. In this study, we monitored the global frequencies of the LP T-13910 allele and allelic haplotype signatures of the ∼30-kb LCT locus in diverse global populations, to study the allelic background of LP in humans.
We genotyped eight SNPs and one indel polymorphism (GenBank accession number DQ109677) covering ∼30 kb of the LCT region and flanking the two LCT variants, C/T-13910 and G/A-22018, associated with LP/LNP (coverage rate of one SNP per 3.3 kb) in 1,611 samples from 37 global populations (fig. 1 and table 1). Except for the two SNPs C/T-13910 and G/A-22018, the genotyped SNPs represent common variants in all populations, with minor-allele frequencies >7% (table 2). Although this approach might not identify some rare allelic variants, especially among the LNP alleles, the most robust pattern of diversity among LP alleles—the target of our interest—will be identified in the global samples.
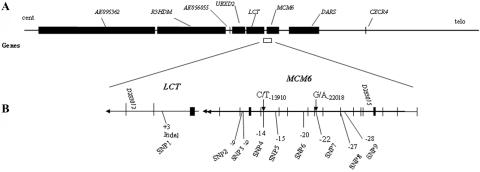
Figure 1. .
The physical map showing the analyzed genome region flanking the C/T-13910 and G/A-22018 variants associated with LP. The distance (in kb) from the first ATG of LCT is shown. A, Genes in the region studied. B, Expanded map of the 30-kb region in the LCT and MCM6 genes, showing SNPs 1–9 analyzed in the population samples.
Table 1. .
SNPs Analyzed in the DNA Samples from 37 Populations[Note]
| SNP | Type | dbSNP Accession Number | Chromosomal Position | Distance from LCT (kb) |
| 1 | Indel | DQ109677a | 136424826 | +3.64 |
| 2 | C/T | rs3754686 | 136437008 | −8.54 |
| 3 | G/C | rs3769005 | 136437098 | −8.63 |
| 4 | C/T-13910 | rs4988235 | 136442378 | −13.91 |
| 5 | C/T | rs4954493 | 136443707 | −15.239 |
| 6 | G/C | rs3099181 | 136448545 | −20.077 |
| 7 | G/A-22018 | rs182549 | 136450486 | −22.018 |
| 8 | C/T | rs4988183 | 136455779 | −27.312 |
| 9 | A/C | rs3087343 | 136456274 | −27.807 |
Table 2. .
SNP Frequencies Analyzed in 37 Population Samples[Note]
| Derived Allele 2 Frequency (SD) |
||||||||||
| Region or Population | N | SNP 1 (Indel; in=1, del=2) |
SNP 2 (C/T; T=1, G=2) |
SNP 3 (G/C; G=1, C=2) |
SNP 4 (C/T-13910; C=1, T=2) |
SNP 5 (C/T; T=1, C=2) |
SNP 6 (G/C; G=1, C=2) |
SNP 7 (G/A -22018; G=1, A=2) |
SNP 8 (A/C; C=1, A=2) |
SNP 9 (G/A; G=1, A=2) |
| South Korea | 23 | .07 (.04) | .28 (.06) | .28 (.06) | .00 (.00) | .28 (.06) | .28 (.06) | .00 (.00) | .52 (.07) | .89 (.05) |
| Han Chinese | 100 | .38 (.03) | .36 (.03) | .36 (.03) | .00 (.00) | .36 (.03) | .36 (.03) | .00 (.00) | .64 (.03) | .77 (.03) |
| Ob-Ugric speakers | 20 | .45 (.04) | .43 (.05) | .43 (.05) | .03 (.02) | .43 (.05) | .42 (.02) | .03 (.02) | .43 (.05) | .81 (.04) |
| Komi | 10 | .40 (.09) | .50 (.11) | .50 (.11) | .15 (.07) | .50 (.11) | .50 (.11) | .15 (.07) | .65 (.10) | .85 (.08) |
| Udmurts | 30 | .53 (.06) | .48 (.07) | .48 (.07) | .33 (.06) | .50 (.06) | .50 (.06) | .37 (.07) | .58 (.06) | .85 (.04) |
| Mokshas | 30 | .27 (.06) | .30 (.06) | .30 (.06) | .28 (.06) | .27 (.06) | .27 (.06) | .27 (.06) | .37 (.07) | .57 (.06) |
| Erzas | 30 | .48 (.06) | .42 (.06) | .42 (.06) | .27 (.06) | .42 (.06) | .38 (.05) | .22 (.05) | .40 (.06) | .68 (.06) |
| Saami | 30 | .53 (.06) | .51 (.07) | .51 (.07) | .17 (.04) | .51 (.07) | .51 (.07) | .13 (.04) | .60 (.06) | .85 (.05) |
| Finns, eastern | 77 | .69 (.04) | .69 (.04) | .69 (.03) | .55 (.04) | .68 (.03) | .66 (.04) | .55 (.04) | .70 (.04) | .88 (.03) |
| Finns, western | 154 | .73 (.02) | .71 (.03) | .71 (.03) | .62 (.02) | .72 (.02) | .71 (.03) | .62 (.02) | .73 (.02) | .88 (.02) |
| Daghestan Druss | 17 | .23 (.07) | .23 (.07) | .18 (.07) | .12 (.06) | .21 (.07) | .21 (.07) | .12 (.06) | .26 (.07) | .62 (.09) |
| Daghestan Nog | 20 | .40 (.08) | .40 (.08) | .37 (.08) | .07 (.04) | .40 (.08) | .40 (.08) | .07 (.04) | .40 (.08) | .60 (.08) |
| Daghestan mixed | 23 | .35 (.07) | .35 (.07) | .35 (.07) | .13 (.05) | .35 (.07) | .35 (.07) | .13 (.05) | .37 (.07) | .67 (.07) |
| Pakistan Balti | 23 | .24 (.06) | .17 (.06) | .26 (.06) | .00 (.00) | .17 (.06) | .17 (.06) | .00 (.00) | .26 (.07) | .44 (.07) |
| Pakistan Burusho | 30 | .33 (.07) | .33 (.07) | .33 (.07) | .02 (.01) | .33 (.07) | .23 (.06) | .05 (.03) | .28 (.06) | .77 (.06) |
| Pakistan Kashmiri | 20 | .37 (.08) | .42 (.08) | .42 (.08) | .12 (.05) | .42 (.08) | .37 (.08) | .15 (.06) | .42 (.08) | .78 (.07) |
| Pakistan Kalash | 30 | .25 (.06) | .27 (.06) | .25 (.05) | .00 (.00) | .25 (.06) | .22 (.05) | .03 (.02) | .38 (.08) | .62 (.06) |
| Pakistan Pathan | 28 | .45 (.07) | .41 (.07) | .43 (.07) | .30 (.06) | .39 (.07) | .41 (.07) | .32 (.06) | .48 (.07) | .71 (.06) |
| Pakistan Hazara | 14 | .36 (.09) | .32 (.09) | .32 (.09) | .04 (.04) | .32 (.09) | .29 (.09) | .11 (.06) | .46 (.10) | .64 (.09) |
| Pakistan Baluch | 19 | .47 (.08) | .47 (.08) | .47 (.08) | .34 (.08) | .47 (.08) | .47 (.08) | .39 (.08) | .50 (.08) | .79 (.06) |
| Pakistan Sindi | 28 | .50 (.07) | .52 (.07) | .50 (.07) | .41 (.07) | .50 (.07) | .50 (.07) | .43 (.07) | .52 (.07) | .75 (.06) |
| Pakistan Brahui | 30 | .43 (.07) | .42 (.07) | .43 (.06) | .27 (.06) | .43 (.06) | .40 (.06) | .28 (.06) | .43 (.07) | .82 (.05) |
| Pakistan Makrani Baluch | 29 | .35 (.06) | .35 (.06) | .35 (.06) | .17 (.05) | .35 (.06) | .33 (.06) | .18 (.05) | .48 (.07) | .77 (.06) |
| Pakistan Mohannes | 29 | .38 (.06) | .38 (.06) | .36 (.07) | .28 (.06) | .36 (.07) | .36 (.06) | .28 (.06) | .36 (.06) | .67 (.07) |
| Pakistan Parsi | 29 | .21 (.05) | .19 (.05) | .19 (.05) | .14 (.05) | .22 (.06) | .17 (.05) | .03 (.02) | .33 (.06) | .55 (.07) |
| Iranians | 21 | .26 (.07) | .21 (.06) | .21 (.06) | .10 (.05) | .24 (.07) | .21 (.06) | .07 (.04) | .26 (.07) | .57 (.08) |
| Iran Qashqai | 10 | .10 (.07) | .10 (.07) | .10 (.07) | .05 (.05) | .10 (.07) | .10 (.07) | .05 (.05) | .10 (.07) | .40 (.11) |
| Arabs | 50 | .17 (.04) | .25 (.04) | .18 (.04) | .10 (.03) | .16 (.04) | .15 (.04) | .10 (.03) | .19 (.04) | .52 (.05) |
| Southern Italy | 100 | .22 (.03) | .26 (.03) | .25 (.03) | .05 (.02) | .26 (.03) | .26 (.03) | .06 (.02) | .29 (.04) | .60 (.03) |
| French | 17 | .44 (.08) | .44 (.08) | .44 (.08) | .34 (.07) | .44 (.08) | .44 (.08) | .37 (.08) | .50 (.08) | .62 (.09) |
| Basques | 85 | .71 (.03) | .70 (.03) | .73 (.03) | .66 (.04) | .72 (.04) | .69 (.03) | .64 (.04) | .70 (.03) | .86 (.03) |
| Utah | 92 | .83 (.03) | .83 (.03) | .83 (.03) | .74 (.03) | .83 (.03) | .82 (.03) | .76 (.03) | .83 (.03) | .90 (.02) |
| Somalia | 79 | .18 (.03) | .19 (.03) | .22 (.03) | .03 (.01) | .18 (.03) | .17 (.03) | .01 (.01) | .28 (.04) | .68 (.03) |
| Fulani Sudanese | 44 | .80 (.05) | .57 (.06) | .57 (.05) | .48 (.06) | .56 (.06) | .56 (.05) | .55 (.06) | .82 (.05) | .94 (.02) |
| Morocco | 90 | .35 (.03) | .33 (.04) | .33 (.03) | .18 (.03) | .35 (.03) | .33 (.03) | .16 (.03) | .41 (.04) | .64 (.03) |
| Saharawi | 57 | .36 (.05) | .36 (.04) | .37 (.04) | .26 (.04) | .36 (.05) | .36 (.04) | .29 (.04) | .36 (.04) | .72 (.04) |
| African Americans | 50 | .25 (.04) | .18 (.04) | .21 (.04) | .09 (.03) | .20 (.04) | .18 (.04) | .09 (.03) | .62 (.05) | .80 (.04) |
Note.— Alleles coded as 1 in every SNP site were the ancestral alleles, on the basis of the sequence of the primate samples, that cosegregated with the LNP phenotype, and alleles coded as 2 in every SNP site were the derived alleles that cosegregated with the LP phenotype.
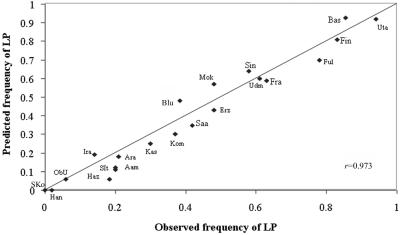
The frequency of the LP T-13910 allele in various populations was systematically correlated with the reported prevalence of LP determined elsewhere by disaccharidase activities in intestinal biopsy samples and/or lactose-tolerance tests in these populations (fig. 2 and table 3).1,2,14–17 Among the 37 populations studied (fig. 3), we identified 21 populations for which the prevalence of the LP trait was known and could establish a strong correlation (coefficient of correlation r=0.973, P<.0001) with the frequency of the T-13910 allele (fig. 2). The allele frequencies of the analyzed markers are shown in table 2, and the complete list of all observed haplotypes constructed using all nine markers with the Arlequin program18 are provided in table 4. We restricted further analysis to those haplotypes with population frequency >4% in at least one of the populations, as inferred by the Arlequin program, to avoid misleading conclusions based on rare haplotypes, which could represent artifacts of the algorithm used for the construction of the haplotypes (table 5). We identified 9 different haplotypes (H8, H9, H11, H12, H48, H49, H95, H97, and H98) with alleles carrying the T-13910 LP variant and 14 haplotypes (H1, H2, H4, H27, H34, H46, H51, H52, H54, H55, H81, H82, H84, and H87) with alleles carrying the C-13910 LNP variant (table 5). Comparison of the resulting haplotypes with the haplotypes estimated by the maximum-likelihood algorithm implemented in the PHASE program v2.1 did not reveal discrepancies (data not shown).
Figure 2. .
Correlation between the frequency of the LP trait, as measured by lactose-tolerance tests and/or disaccharidase activities, and the frequency of LP, as predicted by the frequency of the C/T-13910 allele with the assumption of Hardy-Weinberg equilibrium in different populations. A perfect correlation is not expected because the phenotype data are collected separately from the genotype data. Data about the frequency of LP were obtained from or referenced in studies reported elsewhere.1,2,14–17 Populations are coded as indicated in table 3, except for “Fin,” which includes both FiE and FiW as one group. The r coefficient was calculated using SPSS version 10.0. The two-sided test was performed using a .01 significance level.
Table 3. .
Population Frequencies of LP Alleles C/T-1391
| No. with Genotype |
Allele Frequency(%) |
||||||||
| Designation | Region or Population |
Three-Letter Code | N | CC | CT | TT | C | T | Prevalence of LP (% [SD]) |
| 1 | South Korea | SKo | 23 | 23 | 0 | 0 | 100 | 0 | 0 (.00) |
| 2 | Han Chinese | Han | 100 | 100 | 0 | 0 | 100 | 0 | 0 (.00) |
| 3 | Ob-Ugric speakers | ObU | 62 | 58 | 4 | 0 | 96.8 | 3.2 | 6 (3.02) |
| 4 | Komi | Kom | 10 | 7 | 3 | 0 | 85 | 15 | 30 (14.50) |
| 5 | Udmurts | Udm | 30 | 12 | 16 | 2 | 66.6 | 33.4 | 60 (8.90) |
| 6 | Mokshas | Mok | 30 | 13 | 17 | 0 | 71.6 | 28.4 | 56.6 (9.01) |
| 7 | Erzas | Erz | 30 | 17 | 10 | 3 | 73.3 | 26.7 | 43.3 (9.05) |
| 8 | Saami | Saa | 30 | 20 | 10 | 0 | 83.3 | 16.7 | 33.3 (8.60) |
| 9 | Finns, eastern | FiE | 77 | 18 | 35 | 24 | 46.1 | 53.9 | 76.6 (4.75) |
| 10 | Finns, western | FiW | 154 | 25 | 68 | 61 | 38.3 | 61.7 | 83.7 (2.98) |
| 11 | Daghestan Druss | DaD | 17 | 13 | 4 | 0 | 88.2 | 11.8 | 23.5 (10.30) |
| 12 | Daghestan Nog | DaN | 20 | 15 | 5 | 0 | 87.5 | 12.5 | 25 (9.70) |
| 13 | Daghestan mixed | DaM | 23 | 19 | 3 | 1 | 89.1 | 11.9 | 17.4 (7.90) |
| 14 | Balti | Bal | 23 | 23 | 0 | 0 | 100 | 0 | 0 (.00) |
| 15 | Burusho | Bur | 30 | 29 | 1 | 0 | 98.3 | 1.7 | 3.3 (3.26) |
| 16 | Kashmiri | Kas | 20 | 15 | 5 | 0 | 87.5 | 12.5 | 25 (9.68) |
| 17 | Kalash | Kal | 30 | 30 | 0 | 0 | 100 | 0 | 0 (.00) |
| 18 | Pathan | Pat | 28 | 12 | 15 | 1 | 69.6 | 30.4 | 57.1 (9.35) |
| 19 | Hazara | Haz | 14 | 13 | 1 | 0 | 96.4 | 3.6 | 7.1 (6.86) |
| 20 | Baluch | Blu | 19 | 10 | 6 | 3 | 68.4 | 31.6 | 47.4 (11.46) |
| 21 | Sindi | Sin | 28 | 10 | 13 | 5 | 58.9 | 41.1 | 64.3 (9.11) |
| 22 | Brahui | Bra | 30 | 17 | 10 | 3 | 73.3 | 26.7 | 43.3 (9.05) |
| 23 | Makrani Baluch | MaB | 29 | 19 | 10 | 0 | 82.8 | 17.2 | 34.5 (8.83) |
| 24 | Mohannes | Moh | 29 | 16 | 10 | 3 | 72.4 | 27.6 | 44.8 (9.23) |
| 25 | Parsi | Par | 29 | 21 | 8 | 0 | 86.2 | 13.8 | 27.6 (8.30) |
| 26 | Iranians | Ira | 21 | 17 | 4 | 0 | 90.5 | 9.5 | 19 (8.56) |
| 27 | Qashqai | Qas | 10 | 9 | 1 | 0 | 95 | 5 | 10 (9.49) |
| 28 | Arabs | Ara | 51 | 42 | 8 | 1 | 90.2 | 9.8 | 17.6 (5.33) |
| 29 | Southern Italy | SIt | 100 | 89 | 11 | 0 | 94.5 | 5.5 | 11 (3.13) |
| 30 | French | Fra | 17 | 6 | 9 | 1 | 61.7 | 38.3 | 58.8 (11.94) |
| 31 | Basques | Bas | 85 | 7 | 44 | 34 | 34.1 | 65.9 | 91.7 (2.99) |
| 32 | Utah | Uta | 92 | 7 | 33 | 52 | 25.5 | 74.5 | 92.4 (2.76) |
| 33 | Somalia | Som | 79 | 74 | 5 | 0 | 96.8 | 3.2 | 6.3 (2.73) |
| 34 | Fulani Sudanese | Ful | 44 | 13 | 20 | 11 | 52 | 48 | 70.4 (6.88) |
| 35 | Saharawi | Sah | 57 | 29 | 26 | 2 | 73.7 | 26.3 | 49.1 (6.62) |
| 36 | Morocco | Mor | 90 | 62 | 25 | 3 | 82.7 | 17.3 | 31.1 (4.88) |
| 37 | African Americans | Aam | 50 | 44 | 3 | 3 | 91 | 9 | 12 (4.60) |
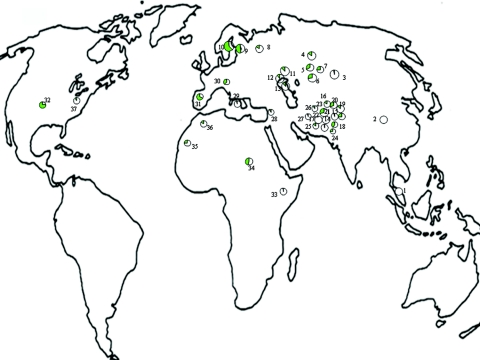
Figure 3. .
Population frequencies for the T-13910 allele associated with LP in worldwide populations. For each population, the pie chart denotes the frequency of the T-13910 allele (green shading). Populations and frequency details are shown in table 3.
Table 4. .
The Complete List of the 30-kb Haplotype Frequencies in 37 Populations[Note]
| Population Frequency(SD) |
||||||||||||||||||||||||||||||||||||||
| Haplotypea | SKo (N=46) | Han (N=200) | ObU (N=124) | Kom (N=20) | Udm (N=60) | Mok (N=60) | Erz (N=60) |
Saa (N=60) | FiW (N=306) | FiE (N=154) | DaD (N=34) | DaN (N=40) | DaM (N=46) | Bal (N=46) | Bur (N=60) | Kas (N=40) | Kal (N=60) | Pat (N=56) | Haz (N=28) | Blu (N=38) | Sin (N=56) | Bra (N=60) | MaB (N=58) | Moh (N=58) | Par (N=58) | Ira (N=42) | Qas (N=20) | Ara (N=102) | SIt (N=200) | Fra (N=34) | Bas (N=170) | Uta (N=184) | Som (N=158) | Mor (N=180) | Sah (N=114) | Aam (N=100) | Ful (N=88) | |
| H1 | 111111111 | .11 (.05) | .21 (.03) | .19 (.04) | .15 (.09) | .04 (.03) | .32 (.06) | .17 (.05) | .12 (.04) | .10 (.02) | .11 (.02) | .23 (.07) | .40 (.07) | .33 (.07) | .54 (.07) | .18 (.04) | .18 (.07) | .37 (.08) | .21 (.07) | .36 (.09) | .17 (.06) | .21 (.06) | .17 (.05) | .22 (.06) | .29 (.07) | .39 (.07) | .38 (.09) | .60 (.12) | .41 (.05) | .32 (.04) | .34 (.09) | .12 (.03) | .10 (.02) | .27 (.04) | .31 (.03) | .25 (.05) | .19 (.04) | .01 (.01) |
| H2 | 111111112 | .35 (.08) | .12 (.02) | .31 (.04) | .10 (.08) | .19 (.06) | .18 (.05) | .25 (.07) | .25 (.05) | .12 (.02) | .16 (.03) | .50 (.09) | .15 (.05) | .26 (.07) | .13 (.07) | .44 (.07) | .31 (.08) | .22 (.05) | .27 (.06) | .11 (.06) | .33 (.07) | .23 (.06) | .33 (.06) | .30 (.06) | .31 (.06) | .21 (.06) | .26 (.08) | .30 (.11) | .26 (.05) | .21 (.03) | .13 (.06) | .09 (.02) | .05 (.02) | .38 (.04) | .20 (.03) | .34 (.05) | .17 (.04) | .07 (.04) |
| H3 | 111111121 | .01 (.01) | .01 (.01) | .01 (.01) | ||||||||||||||||||||||||||||||||||
| H4 | 111111122 | .26 (.08) | .28 (.03) | .02 (.01) | .25 (.12) | .07 (.04) | .07 (.04) | .03 (.01) | .05 (.01) | .03 (.03) | .05 (.04) | .07 (.04) | .04 (.03) | .03 (.03) | .15 (.05) | .07 (.03) | .18 (.08) | .03 (.02) | .02 (.02) | .03 (.02) | .13 (.04) | .12 (.04) | .02 (.02) | .03 (.01) | .06 (.05) | .01 (.01) | .02 (.01) | .04 (.02) | .06 (.02) | .01 (.01) | .32 (.05) | .05 (.02) | ||||||
| H5 | 111111211 | .02 (.02) | .01 (.01) | .01 (.01) | ||||||||||||||||||||||||||||||||||
| H6 | 111111212 | .02 (.02) | .05 (.03) | .01 (.01) | .02 (.01) | |||||||||||||||||||||||||||||||||
| H7 | 111111222 | .02 (.02) | .01 (.01) | |||||||||||||||||||||||||||||||||||
| H8 | 111211111 | .02 (.02) | .05 (.03) | |||||||||||||||||||||||||||||||||||
| H9 | 111211112 | .02 (.02) | .05 (.04) | .01 (.01) | .01 (.00) | .01 (.01) | .02 (.01) | .01 (.01) | ||||||||||||||||||||||||||||||
| H10 | 111211122 | .02 (.01) | .00 (.00) | |||||||||||||||||||||||||||||||||||
| H11 | 111211211 | .07 (.04) | .11 (04) | .02 (.02) | .02 (.02) | |||||||||||||||||||||||||||||||||
| H12 | 111211212 | .06 (.05) | ||||||||||||||||||||||||||||||||||||
| H13 | 111211222 | .03 (.03) | ||||||||||||||||||||||||||||||||||||
| H14 | 111212222 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H15 | 111222212 | .01 (01) | ||||||||||||||||||||||||||||||||||||
| H16 | 111221111 | .02 (.02) | ||||||||||||||||||||||||||||||||||||
| H17 | 111221122 | .02 (02) | .01 (.01) | |||||||||||||||||||||||||||||||||||
| H18 | 111121111 | .00 (.00) | .02 (.02) | .02 (.01) | ||||||||||||||||||||||||||||||||||
| H19 | 111121112 | .00 (.00) | .01 (.01) | .01 (.01) | .01 (.01) | .01 (.01) | .02 (.01) | |||||||||||||||||||||||||||||||
| H20 | 111121122 | .00 (.00) | .01 (.01) | .01 (.00) | ||||||||||||||||||||||||||||||||||
| H21 | 111122111 | .00 (.00) | ||||||||||||||||||||||||||||||||||||
| H22 | 111112111 | .01 (.01) | .01 (.01) | |||||||||||||||||||||||||||||||||||
| H23 | 111112112 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H24 | 121111111 | .02 (.02) | .02 (.02 | .01 (.00) | .01 (.01) | |||||||||||||||||||||||||||||||||
| H25 | 121111121 | .02 (.01) | ||||||||||||||||||||||||||||||||||||
| H26 | 121111112 | .01 (.00) | .02 (.01) | .01 (.01) | ||||||||||||||||||||||||||||||||||
| H27 | 121111122 | .07 (.03) | .04 (.02) | .02 (.01) | .01 (.01) | .05 (02) | ||||||||||||||||||||||||||||||||
| H28 | 121111222 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H29 | 121122122 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H30 | 121122211 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H31 | 121211112 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H32 | 121211122 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H33 | 121222222 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H34 | 112111111 | .02 (.01) | .02 (.02) | .06 (.02) | .01 (.01) | .01 (.01) | .02 (.01) | |||||||||||||||||||||||||||||||
| H35 | 112111121 | .02 (.02) | ||||||||||||||||||||||||||||||||||||
| H36 | 112111112 | .02 (.02) | ||||||||||||||||||||||||||||||||||||
| H37 | 112111122 | .01 (.01) | .01 (.01) | .01 (.00) | ||||||||||||||||||||||||||||||||||
| H38 | 112122112 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H39 | 112122122 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H40 | 112212212 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H41 | 112222122 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H42 | 111112111 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H43 | 122111111 | .00 (.00) | ||||||||||||||||||||||||||||||||||||
| H44 | 122111112 | .02(.02) | .01 (.01) | |||||||||||||||||||||||||||||||||||
| H45 | 122111122 | .02 (.02) | .00 (.00) | .04 (.03) | ||||||||||||||||||||||||||||||||||
| H46 | 122122122 | .22 (.07) | .03 (.02) | .02 (.02) | .02 (.02) | .02 (.02) | .05 (.04) | .12 (.03) | .01 (.01) | |||||||||||||||||||||||||||||
| H47 | 122122222 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H48 | 122222212 | .10 (.7) | ||||||||||||||||||||||||||||||||||||
| H49 | 122222222 | .02 (.02) | .06 (.04) | .03 (.01) | .03 (.03) | |||||||||||||||||||||||||||||||||
| H50 | 122211111 | .00 (.00) | ||||||||||||||||||||||||||||||||||||
| H51 | 211111111 | .08 (.04) | .03 (.02) | .01 (.01) | .04 (.03) | .02 (.02) | .02 (.02) | .02 (.02) | .06 (.02) | .01 (.01) | ||||||||||||||||||||||||||||
| H52 | 211111112 | .03 (.02) | .00 (.00) | .03 (.03) | ||||||||||||||||||||||||||||||||||
| H53 | 211111211 | .04 (.03) | .01 (.01) | .01 (.01) | .01 (.01) | .07 (.02) | .14 (.05) | |||||||||||||||||||||||||||||||
| H54 | 211111122 | .02 (.01) | .01 (.01) | .02 (.02) | .03 (.02) | .06 (.03) | ||||||||||||||||||||||||||||||||
| H55 | 211111222 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H56 | 211121111 | |||||||||||||||||||||||||||||||||||||
| H57 | 211112112 | .00 (.00) | ||||||||||||||||||||||||||||||||||||
| H58 | 211122111 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H59 | 211122112 | .00 (.00) | ||||||||||||||||||||||||||||||||||||
| H60 | 211122121 | .03 (.03) | .01 (.00) | |||||||||||||||||||||||||||||||||||
| H61 | 211122122 | .02 (.02) | .02 (.01) | .02 (.02) | ||||||||||||||||||||||||||||||||||
| H62 | 211122212 | |||||||||||||||||||||||||||||||||||||
| H63 | 211211222 | .02 (.02) | .02 (.02) | |||||||||||||||||||||||||||||||||||
| H64 | 211222111 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H65 | 211222222 | .02 (.02) | .02 (.03) | |||||||||||||||||||||||||||||||||||
| H66 | 211211222 | .03 (.03) | .00 (.00) | |||||||||||||||||||||||||||||||||||
| H67 | 212111111 | .01 (.01) | .00 (.00) | .01 (.01) | .01 (.01) | |||||||||||||||||||||||||||||||||
| H68 | 212122122 | .02 (.02) | .01 (.01) | |||||||||||||||||||||||||||||||||||
| H69 | 212222211 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H70 | 212212222 | .02 (.02) | .02 (.02) | .03 (.02) | ||||||||||||||||||||||||||||||||||
| H71 | 212222222 | .02 (.02) | .02 (.02) | |||||||||||||||||||||||||||||||||||
| H72 | 221122122 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H73 | 221122222 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H74 | 221222211 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H75 | 221222222 | .02 (.02) | ||||||||||||||||||||||||||||||||||||
| H76 | 222111111 | .00 (.00) | .01 (.01) | .01 (.00) | ||||||||||||||||||||||||||||||||||
| H77 | 222111112 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H78 | 222112122 | .02 (.02) | .00 (.00) | .03 (.03) | ||||||||||||||||||||||||||||||||||
| H79 | 222121122 | .02 (.02) | .01 (.01) | .02 (.02) | .00 (.00) | .02 (.01) | ||||||||||||||||||||||||||||||||
| H80 | 222121212 | .02 (.02) | ||||||||||||||||||||||||||||||||||||
| H81 | 222121112 | .02 (.01) | .00 (.00) | .03 (.01) | .09 (.04) | .05 (.04) | .03 (.02) | .04 (.03) | .02 (.01) | .01 (.01) | .02 (.01) | .01 (.01) | .02 (.01) | |||||||||||||||||||||||||
| H82 | 222122112 | .02 (.03) | .02 (.01) | .05 (.04) | .05 (.03) | .02 (.02) | .01 (.01) | |||||||||||||||||||||||||||||||
| H83 | 222122121 | .03 (.03) | .04 (.04) | |||||||||||||||||||||||||||||||||||
| H84 | 222122122 | .04 (.03) | 36 (.03) | .33 (.04) | .35 (.12) | .35 (.07) | .09 (.04) | .13 (.05) | .33 (.08) | .06 (.01) | .11 (.02) | .05 (.05) | .08 (.04) | .19 (.06) | .15 (.06) | .18 (.05) | .20 (.08) | .18 (.05) | .07 (.04) | .18 (.10) | .04 (.03) | .09 (.04) | .08 (.04) | .17 (.06) | .09 (.04) | .11 (.04) | .17 (.07) | .05 (.04) | .04 (.02) | .04 (.02) | .09 (.06) | .03 (.01) | .04 (.02) | .15 (.03) | .15 (.03) | .06 (.02) | .09 (.03) | .06 (.03) |
| H85 | 222122212 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H86 | 222122221 | .03 (.02) | .01 (.01) | |||||||||||||||||||||||||||||||||||
| H87 | 222122222 | .00 (.00) | .08 (.04) | .03 (.02) | .02 (.02) | .07 (.06) | .05 (.04) | .02 (.02) | .01 (.02) | .02 (.01) | .03 (.03) | .02 (.01) | .02 (.01) | |||||||||||||||||||||||||
| H88 | 222211221 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H89 | 222212221 | .02 (.02) | ||||||||||||||||||||||||||||||||||||
| H90 | 222212222 | .01 (.01) | .00 (.00) | .02 (.01) | .02 (.01) | |||||||||||||||||||||||||||||||||
| H91 | 222221221 | .02 (.01) | ||||||||||||||||||||||||||||||||||||
| H92 | 222221222 | .01 (.00) | ||||||||||||||||||||||||||||||||||||
| H93 | 222221112 | .00 (.00) | .03 (.03) | |||||||||||||||||||||||||||||||||||
| H94 | 222222121 | .01 (.01) | ||||||||||||||||||||||||||||||||||||
| H95 | 222222122 | .03 (.02) | .05 (.03) | .02 (.01) | .01 (.01) | .01 (.01) | .01 (.01) | |||||||||||||||||||||||||||||||
| H96 | 222222212 | .01 (.01) | .01 (.01) | |||||||||||||||||||||||||||||||||||
| H97 | 222222221 | .09 (.05) | .02 (.02) | .01 (.01) | .01 (.01) | |||||||||||||||||||||||||||||||||
| H98 | 222222222 | .03 (.02) | .05 (.06) | .14 (.05) | .14 (.05) | .20 (.06) | .12 (.04) | .61 (03) | .53 (.04) | .13 (.06) | .11 (.05) | .06 (.05) | .27 (.06) | .04 (.03) | .34 (.08) | .39 (.07) | .25 (.06) | .15 (.05) | .28 (.06) | .01 (.01) | .02 (.02) | .05 (.05) | .05 (.02) | .01 (.01) | .31 (.09) | .56 (.04) | .74 (.03) | .01 (.01) | .12 (.02) | .26 (.04) | .06 (.03) | .47 (.06) | ||||||
| H103 | 212112110 | .02 (.02) | ||||||||||||||||||||||||||||||||||||
| H104 | 221221220 | .02 (.02) | ||||||||||||||||||||||||||||||||||||
Note.— Haplotypes >4% in any population have been included in the table. Alleles coded as 1 are based on the sequence of the primate samples and were cosegregated with the LNP phenotype, and alleles coded as 2 represent the alleles cosegregated with the LP phenotype. The SNPs used in constructing the haplotypes are shown in table 2. The three-letter codes for the populations are used; the complete names are given in table 3.
The fourth position in each haplotype is the common LNP/LP variant, C/T-13910, where 1 denotes C-13910 and 2 denotes T-13910. The seventh position denotes the SNP G/A-22018, where 1 denotes G-22018 and 2 denotes A-22018.
Table 5. .
| Population Frequency (% [SD]) |
|||||||||||||||||||||||||||||||||||||
| Haplotype | SKo | Han | ObU | Kom | Udm | Mok | Erz | Saa | FiW | FiE | DaD | DaN | DaM | Bal | Bur | Kas | Kal | Pat | Haz | Blu | Sin | Bra | MaB | Moh | Par | Ira | Qas | Ara | SIt | Fra | Bas | Uta | Som | Mor | Sah | Aam | Ful |
| N | 46 | 200 | 124 | 20 | 60 | 60 | 60 | 60 | 154 | 306 | 34 | 40 | 46 | 46 | 60 | 40 | 60 | 56 | 28 | 38 | 56 | 60 | 58 | 58 | 58 | 42 | 20 | 102 | 200 | 34 | 170 | 184 | 158 | 180 | 114 | 100 | 88 |
| LNP: | |||||||||||||||||||||||||||||||||||||
| H1 | 11 (5) | 21 (3) | 19 (4) | 15 (9) | 4 (3) | 32 (6) | 17 (5) | 12 (4) | 10 (2) | 11 (2) | 23 (7) | 40 (7) | 33 (7) | 54 (7) | 18 (4) | 18 (7) | 37 (8) | 21 (7) | 36 (9) | 17 (6) | 21 (6) | 17 (5) | 22 (6) | 29 (7) | 39 (7) | 38 (9) | 60 (12) | 41 (5) | 32 (4) | 34 (9) | 12 (3) | 10 (2) | 27 (4) | 31 (3) | 25 (5) | 19 (4) | 1 (1) |
| H2 | 35 (8) | 12 (2) | 31 (4) | 10 (8) | 19 (6) | 18 (5) | 25 (7) | 25 (5) | 12 (2) | 16 (2) | 50 (9) | 15 (5) | 26 (7) | 13 (7) | 44 (7) | 31 (8) | 22 (5) | 27 (6) | 11 (6) | 33 (7) | 23 (6) | 33 (6) | 30 (6) | 31 (6) | 21 (6) | 26 (8) | 30 (11) | 26 (5) | 21 (3) | 13 (6) | 9 (2) | 5 (2) | 38 (4) | 20 (3) | 34 (5) | 17 (4) | 7 (3) |
| H4 | 26 (8) | 28 (3) | 2 (1) | 25 (12) | … | 7 (4) | … | 7 (4) | 3 (1) | 5 (1) | 3 (3) | 5 (4) | 7 (4) | … | 4 (3) | 3 (3) | 15 (5) | 7 (3) | 18 (8) | 3 (2) | 2 (2) | 3 (2) | 13 (4) | … | 12 (4) | 2 (2) | … | … | 3 (1) | 6 (5) | 1 (1) | 2 (1) | 4 (2) | 6 (2) | 1 (1) | 32 (5) | 5 (2) |
| H27 | … | … | … | … | … | … | … | … | … | … | … | … | … | 7 (3) | … | … | … | … | … | … | … | … | … | … | … | … | … | 4 (2) | … | … | … | … | 2 (1) | 1 (1) | … | 5 (2) | … |
| H34 | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | 2 (1) | … | … | 2 (2) | … | … | … | 6 (2) | 1 (1) | … | 1 (1) | … | 2 (1) | … | … | … | … |
| H46 | 22 (7) | … | 3 (2) | … | … | 2 (2) | 2 (2) | 2 (2) | … | … | … | … | … | … | … | 5 (4) | … | … | … | … | … | … | … | … | … | … | … | … | 12 (3) | … | … | 1 (1) | … | … | … | … | … |
| H51 | … | … | … | … | … | … | 8 (4) | 3 (2) | 1 (1) | … | … | … | … | … | … | 4 (3) | … | … | … | … | … | … | … | 2 (2) | 2 (2) | 2 (2) | … | … | 6 (2) | … | … | 1 (1) | … | … | … | … | … |
| H52 | … | … | 3 (2) | … | … | … | … | … | 0 (0) | … | … | … | … | 2 (2) | … | … | … | … | 4 (3) | … | … | … | … | … | … | … | … | … | 7 (3) | … | … | … | … | … | … | … | 5 (2) |
| H54 | … | 2 (1) | 1 (1) | … | … | … | … | 2 (2) | … | … | … | … | … | 4 (3) | … | … | … | … | … | … | … | … | … | … | … | … | … | … | 1 (1) | … | … | … | 1 (1) | 1 (1) | … | 7 (2) | 14 (5) |
| H55 | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | 3 (2) | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | 6 (3) |
| H81 | … | … | … | … | … | … | 2 (1) | … | 0 (0) | 3 (1) | … | … | … | … | 9 (4) | 5 (4) | 3 (2) | … | 4 (3) | … | … | … | … | … | … | … | … | 2 (1) | 1 (1) | … | 2 (1) | 1 (1) | … | 2 (1) | … | … | … |
| H82 | 2 (3) | … | 2 (1) | … | … | … | … | … | … | … | … | 5 (4) | 5 (3) | 2 (2) | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | 1 (1) | … | … | … | … | … | … |
| H84 | 4 (3) | 36 (3) | 33 (4) | 35 (12) | 35 (7) | 9 (4) | 13 (5) | 33 (8) | 6 (1) | 11 (2) | 5 (5) | 8 (4) | 19 (6) | 15 (6) | 18 (5) | 20 (8) | 18 (5) | 7 (4) | 18 (10) | 4 (3) | 9 (4) | 8 (4) | 17 (6) | 9 (4) | 11 (4) | 17 (7) | 5 (4) | 4 (2) | 4 (2) | 9 (6) | 3 (1) | 4 (2) | 15 (3) | 15 (3) | 6 (2) | 9 (3) | 6 (3) |
| H87 | … | … | … | … | … | … | … | … | 0 (0) | … | … | 8 (4) | … | … | … | … | 3 (2) | 2 (2) | 7 (6) | 5 (4) | 2 (2) | 1 (2) | 2 (1) | … | … | 3 (3) | … | … | … | … | 2 (1) | 2 (1) | … | … | … | … | … |
| LP: | |||||||||||||||||||||||||||||||||||||
| H8 | … | … | … | … | 2 (2) | … | 5 (3) | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| H9 | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | 2 (2) | 5 (4) | … | 1 (1) | 1 (1) | … | 1 (1) | … | 2 (1) | 1 (1) | … | … | … |
| H11 | … | … | … | … | 7 (4) | 11 (4) | … | … | … | … | … | … | … | … | … | … | … | … | … | 2 (2) | … | … | … | … | 2 (2) | … | … | … | … | … | … | … | … | … | … | … | … |
| H12 | … | … | … | … | 6 (5) | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| H48 | … | … | … | 10 (7) | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| H49 | … | … | … | … | … | … | … | 2 (2) | … | … | … | … | … | … | … | 6 (4) | … | … | … | … | … | … | … | … | … | … | … | … | 3 (1) | 3 (3) | … | … | … | … | … | … | … |
| H95 | … | … | … | … | … | … | … | 3 (2) | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | 5 (3) | … | … | … | … | … | 2 (1) | 1 (1) | … | 1 (1) | … | … | 1 (1) |
| H97 | … | … | … | … | … | … | … | … | … | … | 9 (5) | … | … | … | 2 (2) | 1 (1) | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | 1 (1) | … |
| H98 | … | … | 3 (2) | 5 (6) | 14 (5) | 14 (5) | 20 (6) | 12 (4) | 61 (4) | 53 (3) | … | 13 (6) | 11 (5) | … | … | 6 (5) | … | 27 (6) | 4 (3) | 34 (8) | 39 (7) | 25 (6) | 15 (5) | 28 (6) | 1 (1) | 2 (2) | 5 (5) | 5 (2) | 1 (1) | 31 (9) | 56 (4) | 74 (3) | 1 (1) | 12 (2) | 26 (4) | 6 (3) | 47 (6) |
One of the nine haplotypes (H98) distinctly dominated in LP alleles in most study populations, with only a few exceptions: in populations of Udmurts, Erzas, and Mokshas, five other LP haplotypes (H8–H12) were observed at the reasonable frequency (table 5). Among these “other” LP alleles, the frequency of H8 was highest (5%) among Erzas, whereas H11 was present at the frequency of 11% and 7% among Mokshas and Udmurts, respectively (table 5). Of the 14 identified LNP haplotypes listed in table 5, 3 were found to be present in all populations (H1, H2, and H84). Interestingly, when we monitored the structure of these global LCT alleles, we saw that the major LP H98 allele diverges the most from the major LNP H1 allele; these two haplotypes differ at every SNP. Another common LNP H84 allele differs from the major LP H98 allele only at the positions of the two critical variants (C/T-13910 and G/A-22018) that correlate with LP (table 5). Thus, two common LNP alleles in H1 and H84 show a highly divergent allelic background, and the frequencies of intervening haplotypes between them are low, which are most probably lost because of recombinations and/or genetic drift (table 5). The sequence identity between H84 (LNP) and H98 (LP) not only covers the 30-kb region thoroughly analyzed in all populations but actually spans 700 kb in some tested populations (e.g., Finns; data not shown), underlining their close relationship in the evolution.
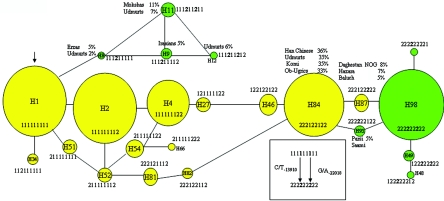
To explore the relationship between different haplotypes of the LCT alleles, we constructed a median-joining (MJ) haplotype network of the 30-kb LCT region in the global samples, using a total of 23 haplotypes showing frequency >4% in at least one population. The network was constructed using the NETWORK software under the default parameters. The population frequencies of the relevant haplotypes are shown in figure 4. Comparisons with primate (i.e., chimpanzee, orangutan, gorilla, and rhesus monkey) sequences revealed that H1 represents the ancestral haplotype for the human LCT gene; therefore, it was used as the rooted haplotype in the MJ network (fig. 4). The MJ haplotype network further exposes two distinct clusters of LP haplotypes carrying the T-13910 variant. These clusters are separated by more than five mutational steps (fig. 4). The first cluster of LP haplotypes consists of H8, H9, H11, and H12, and the second cluster consists of H48, H49, H95, H97, and H98, of which LP H98 is the most common among all populations tested (fig. 4). The first cluster (H8–H12), which is relatively common among the populations of Udmurts, Mokshas, Erzas, and Iranians, cannot represent an outcome of simple recombination events among the common LNP haplotypes H1, H2, H4, and H84 and the major LP allele H98. This observation could reflect multiple recombination events in history or could actually report two different origins of the LP T-13910 allele in the populations living north of the Caucasus and west of the Urals, which we consider the most probable option.
Figure 4. .
MJ haplotype network for eight SNPs and one indel marker in the 30-kb LCT region among 37 populations, constructed using NETWORK version 4.1.1.2. The analysis includes all haplotypes with an estimated population frequency >4% in at least one population. The arrow denotes the rooted haplotype LNP H1 of the network. LNP haplotypes are shown as yellow circles, and LP haplotypes are shown as green circles. The size of the circles corresponds to the estimated haplotype frequency in the global sample, and the haplotypes are shown inside or above the circles. The population frequencies for some haplotypes relevant to the origin of the LP alleles and discussed in the text are shown. The positions of the C/T-13910 and G/A-22018 alleles within the haplotype are shown in the enclosed box. The SNPs have been coded for each site as 1 for the ancestral SNP and 2 for the derived SNP. The haplotype details are shown in tables 2 and 5.
The haplotype network also shows that the two haplotypes representing different phenotypes, LNP H87 and LP H95, are both connected to LP H98. We wanted to assess the possibility that these haplotypes represent recombinants, by genotyping 14 of 19 individuals carrying H87 for more-distant flanking markers. This analysis revealed one major haplotype block covering >800 kb flanking C/T-13910 (data not shown). Further, we sequenced the entire 3,435-bp region of intron 13 of the MCM6 gene (where C/T-13910 resides) of the H87 haplotype and established that the SNPs flanking the C-13910 allele in H87 are all part of the same 800-kb haplotype block. Thus, we were not able to obtain any evidence that the C-13910 allele of H87 was generated by a recombination event, and we concluded that H87 represents the allelic background on which LP T-13910 occurred, resulting in LP H98. For H95, in three of the six individuals genotyped for more-distant flanking SNP markers, the haplotype is broken at 450 kb, 3′ of C/T-13910 (data not shown), and we were not able to obtain any evidence that H95 was generated by a recombination event from other haplotypes (H84 and H98). This prompted us to assume a different origin for LP T-13910 on H95 than for the mutation on H98, which implies that the origin of LP T-13910 has occurred more than once in recent human history.
The MJ network suggested that the common ancestral LNP haplotype background on which the major LP H98 variants occurred was LNP H84. Therefore, we monitored the prevalence pattern of the common LNP H84 haplotype in our samples from global populations, to assess the distribution of this allele, which might help in the elucidation of the historical origin of LP H98. A high prevalence of H84 is characteristic to the eastern part of the Ural Mountains, among Ob-Ugric speakers, where the prevalence reaches as high as 33%. The high prevalence of H84 extends east to the populations totally lacking the LP mutation, like Han Chinese (36%) (table 5). The high population frequency of this particular allele can be seen also in South Korea, where H46, the haplotype deviating from H84 by one mutational step, can be observed at 22% frequency (table 5 and fig. 4). Among the populations living west of the Urals on the European side of Russia (e.g., Komi and Udmurts), as well as among Saami, the frequencies of this haplotype are 33%–35%. These prevalence figures imply that the ancestral H84 allele, the target of the most common LP H98 mutation(s), originates from Asian populations. On the basis of population frequencies, we can actually monitor the western migration of this allele. We recognize that this interpretation could be reversed if the common LNP H84 arose via a gene-conversion event from the common LP H98 and not vice versa. We consider this unlikely, given the relatively recent age of LP H98 and the fact that the common LNP H84 haplotype was found in all 37 populations, which indicates introduction into global populations earlier than predicted for LP H98.
We also monitored the prevalence pattern of the less common LNP H87 haplotype that, on the basis of the MJ network, represents the immediate allelic haplotype on which the LP H98 mutation occurred. The highest frequencies of H87 alleles were observed among Daghestan Nogais (8%) and Hazara (7%). This allele was detected in Daghestan Nogais, Hazara, Baluch, Sindi, Brahui, Makrani Baluch, Iranians, Basques, individuals from Utah, and Finns (eastern region). From this distribution of H87, we were able to propose that the ancestral population in which the LP T-13910 H98 mutation occurred is of Caucasian origin.
We recognize the role of selection in shaping the present-day frequencies of LP alleles2,9,13,14,16,17,19 and other demographic processes such as genetic drift, which could have a major effect on the frequencies in some populations and could result in a biased interpretation of the global history of the LP trait. For example, the wide LD interval providing a strong signal for selection of the LCT region could interfere with our interpretation based on the population frequencies.9,20,21 Although MJ networks can be used to analyze large data sets and multistate characters, we recognize that the algorithm on which the MJ haplotype network construction is based requires a recombination-free region, such as the mtDNA region.22 We tried to minimize the recombination events in the critical LCT region and analyzed the variants in a very restricted DNA region (30 kb); we used only haplotypes that exceeded 4% frequency in any population. We recognize that some recombinants still could have taken place and could have interfered with the interpretation of the results. Despite these limitations, we think our data provide a solid basis for a hypothesis of more than one allelic origin of the LP T-13910 mutations and the evolutionary history of the LP trait. Importantly, we base our conclusion on the frequency of the critical background alleles defined by haplotypes (like LNP H84 and H87) not directly affected by selection. Further, we base our interpretation on the analyses of reasonably large study samples from diverse populations, and, although the DNA samples analyzed here do not provide complete global covering, they do cover the critical regional populations in Eurasia.
To further address the issue of the historical origin of the common LP mutation in two diverse populations—Finns and Fulanis—we first estimated the most recent common ancestor (TMRCA) of the LP H98 T-13910 alleles in the Finns, using LD-decay analysis for marker D2S3014, which shows the highest LD with the LP phenotype in the Finns.3 Using a generation time of 25 years and the algorithm by Risch et al.,23,24 we found an age estimate of 5,275 years (95% CI 4,875–5,640) for the Finnish alleles. Use of the same marker, D2S3014, in the Fulani Sudanese population in the LD-decay analysis gave an age estimate of 6,475 years (95% CI 5,875–7,100). With three flanking markers (D2S3013, D2S3015, and D2S3016) that show less LD in LP alleles,3 the average square distance (ASD) method that used the Ytime program19 gave an age estimate of 9,252 years (95% CI 100–34,000) in this population (table 6).
Table 6. .
Age Estimates of TMRCA for LP H98 T-13910 in Global Populations
| Selection Methodb(years [SD]) |
Rho Methodc(years [±SD]) |
||||
| Populationa | s=.04 | s=.05 | LD or ASD Method (years [95% CI]) |
Lower Boundd | Upper Bounde |
| Par | 1,425 (0–1,875) | 1,150 (0–1,500) | … | 262 (±29) | 459 (±51) |
| Som | 1,425 (0–1,875) | 1,150 (0–1,500) | … | 280 (±44) | 490 (±77) |
| Ira | 1,875 (0–2,325) | 1,500 (0–1,850) | … | 371 (±82) | 649 (±144) |
| Qas | 2,475 (0–2,975) | 1,975 (0–2,350) | … | 485 (±76) | 848 (±133) |
| Haz | 2,325 (1,425–2,700) | 1,850 (1,150–2,150) | … | 573 (±63) | 1,002 (±111) |
| Ara | 2,475 (2,125–2,700) | 1,975 (1,700–2,150) | … | 670 (±105) | 1,173 (±184) |
| SIt | 1,425 (0–1,875) | 1,150 (0–1,500) | … | 795 (±272) | 1,391 (±476) |
| ObU | 2,125 (1,875–2,475) | 1,700 (1,150–1,975) | … | 900 (±141) | 1,575 (±247) |
| Kas | 2,600 (1,425–3,025) | 2,075 (1,150–2,425) | … | 1,400 (±220) | 2,450 (±385) |
| DaN | 3,175 (2,700–3,500) | 2,525 (2,150–2,775) | … | 1,500 (±167) | 2,625 (±292) |
| Aam | 2,600 (2,115–2,875) | 2,075 (1,700–2,300) | … | 1,512 (±238) | 2,646 (±416) |
| DaDf | 2,875 (2,325–3,225) | 2,300 (1,850–2,575) | … | 1,527 (±270) | 2,673 (±472) |
| Kom | 2,475 (0–2,975) | 1,975 (0–2,350) | … | 1,575 (±247) | 2,756 (±433) |
| DaM | 3,025 (2,600–3,350) | 2,425 (2,075–2,675) | … | 1,575 (±247) | 2,756 (±433) |
| Mor | 3,110 (2,950–3,225) | 2,475 (2,350–2,575) | … | 1,777 (±198) | 3,110 (±346) |
| Mok | 3,225 (2,875–3,500) | 2,600 (2,300–2,775) | … | 1,867 (±293) | 3,267 (±513) |
| MaB | 3,275 (2,950–3,550) | 2,625 (2,350–2,825) | … | 2,577 (±286) | 4,510 (±501) |
| Fra | 4,025 (3,625–4,400) | 3,200 (2,900–3,500) | … | 3,013 (±474) | 5,272 (±829) |
| Moh | 3,900 (3,625–4,150) | 3,100 (2,900–3,300) | … | 3,054 (±480) | 5,345 (±840) |
| Saa | 3,100 (2,800–3,350) | 2,475 (2,225–2,675) | … | 3,150 (±350) | 5,513 (±613) |
| Sah | 3,825 (3,625–3,975) | 3,025 (2,900–3,175) | … | 3,203 (±356) | 5,606 (±623) |
| Erz | 3,550 (3,225–3,825) | 2,475 (2,225–2,650) | … | 3,437 (±540) | 6,014 (±945) |
| Pat | 3,850 (3,575–4,100 | 3,075 (2,850–3,275) | … | 3,500 (±550) | 6,125 (±962) |
| Bra | 3,750 (3,475–4,025) | 3,000 (2,775–3,200) | … | 3,780 (±420) | 6,615 (±735) |
| Sin | 4,350 (4,050–4,650) | 3,475 (3,225–3,725) | … | 4,077 (±453) | 7,134 (±793) |
| Blu | 4,150 (3,800–4,475) | 3,300 (3,025–3,575) | … | 4,310 (±479) | 7,543 (±838) |
| Udm | 3,225 (2,875–3,500) | 2,575 (2,300–2,775) | … | 5,040 (±792) | 8,820 (±1,386) |
| Bas | 5,150 (4,950–5,425) | 4,125 (3,950–4,300) | … | 5,205 (±578) | 9,108 (±1,012) |
| FiW | 5,000 (4,800–5,225) | 3,975 (3,825–4,175) | 5,275 (4,875–5,640)g | 5,207 (±579) | 9,113 (±1,013) |
| FiE | 5,475 (5,275–5,675) | 4,350 (4,200–4,525) | 9,252 (100–34,000)h | 5,433 (±854) | 9,508 (±1,494) |
| Uta | 6,625 (6,275–7,050) | 5,275 (5,000–5,625) | … | 5,563 (±618) | 9,736 (±1,082) |
| Ful | 4,700 (4,475–4,950) | 3,750 (3,575–3,950) | 6,475 (5,875–7,100)i | 6,134 (±682) | 10,735 (±1,193) |
The three-letter codes for the populations are used; the complete names are given in table 3.
P=.001. The SD of the estimate is based on the SD of the current allele frequencies (p).
The calculations for the Rho method were performed using the NETWORK program, version 4.1.1.2.
The lower boundary for mutation-rate calibration was based on the LD method and translated into one mutation per 700 years.
The upper boundary for the mutation-rate calibration was based on the ASD method and translated into one mutation per 1,225 years.
The LP allele detected in this population was LP 97.
The estimate was based on the LD method in the Finnish families.
The estimate was based on the ASD method in the Finnish families.
The estimate was based on the LD method.
For other populations, we applied two different methods to estimate the age of the LP mutation on the basis of the obtained haplotype frequencies. In the first method, we tried to take advantage of the role of selection that shaped the LCT region, to estimate the age of the LP T-13910 alleles among different populations. Previous studies have shown the selection coefficient, s, which measures the proportional excess of fitness of LP allele in relation to LNP allele, to range from 0.02 to 0.19.2,9,25,26 With the assumption of a dominant model for LP, s is proposed to be 0.04–0.05, and initial allele frequency p0 to be 0.001. We applied the general selection formula (ln(p/q)+1/q=ln(p0/q0)+1/q0+st) to roughly estimate the age of the selected allele, using the current allele frequencies (p) in every population.27 In the second, phylogeny-based method, we specifically analyzed the sequence of the critical 30-kb region. The age estimates were obtained by constructing the MJ network of 30-kb LCT haplotypes in each population separately, and, from these networks, we measured the rho statistic (ρ)—the average number of mutations from the root haplotype, LNP H1—in these populations. We included the SDs and a generation time of 25 years, to estimate TMRCA of the LP T-13910 alleles, using the NETWORK 4.1.1.2 program, which applied the formula t=ρ/μ (where t is the time since TMRCA and μ is the mutation rate for the region per year).22 This method needs a calibration point to estimate the mutation rate of the region. We chose our previous age estimations on the basis of LD decay in Finns and Fulani and used the ASD in Finns as the internal calibration point to estimate the mutation rate of the region. The LD-decay method used here is considered to represent the lower boundary for mutation-rate calibration—4.54×10−8 bp/year, which translates into one mutation per 700 years. The second, ASD-based method is considered to represent the upper boundary for mutation-rate calibration—2.59×10−8 bp/year, which translates into one mutation per 1,225 years.
Although mutation-dating methods involve many assumptions and uncertainties, the results clearly indicate that our age estimate of the first cluster of LP haplotypes (H8–H12) indicates a substantially more recent introduction of the LP variants (1,400–3,000 years) than does the age estimate of the second cluster, representing the LP H98 haplotype (5,000–12,000 years). This supports the concept of two different origins of the LP T-13910 allele (tables 6 and 7). The oldest age estimates for the LP H98 T-13910 allele are obtained within the populations from widely divergent regions, such as African Fulani Sudanese, individuals from Utah, Finns, Basques, and Udmurts, in the age ±SD range of 5,040±792 to 10,735±1,193 years (table 6). Interestingly, if we take into account the high prevalence of LP and the LP T-13910 allele in the Fulani and northern Europeans, as well as the almost-identical LP H98 allelic haplotype carrying the T-13910 allele (not only in the 30-kb region studied here but also in an 800-kb region in some populations [data not shown]), similar age estimates emerge for the T-13910 allele in both populations. This would indicate that the African Fulani and northern Europeans probably share the origin of this mutation and perhaps also share a dairy culture. Previous studies in Fulani have also suggested a degree of Caucasian admixture in their gene pool, a finding that supports the Caucasian origin of the LP H98 T-13910 allele.28
Table 7. .
Age Estimates Using TMRCA for the “Less Common” LP Alleles (H8–H12) in Various Populations[Note]
| Age Estimate(years [SD]) |
||||
| Population or Region |
H8 | H9 | H11 | H12 |
| Udmurts | 1,850 (0–2,300) | … | 2,675 (2,100–3,025) | 2,575 (1,425–3,050) |
| Erza | 2,450 (1,850–2,775) | … | … | … |
| Mokshas | … | … | 3,025 (2,675–3,275) | … |
| Baluch | … | … | 1,850 (0–2,300) | … |
| Parsi | … | 1,850 (0–2,300) | 1,850 (0–2,300) | … |
| Iranians | … | 2,450 (1,425–2,875) | … | … |
| Arabs | … | 1,425 (0–1,850) | … | … |
| Southern Italy | … | 1,425 (0–1,850) | … | … |
| Basques | … | 1,425 (0–1,850) | … | … |
| Somalia | … | 1,850 (1,425–2,125) | … | … |
| Morocco | … | 1,425 (0–1,850) | … | … |
Note.— The selection method was used, with P=.001 and s=.04. The SD of the estimate is based on the SD in the current allele frequencies (p).
Although it is unlikely that all the populations exhibit the same initial allele frequency or would have experienced the same selection pressures throughout history, the selection method gives very reasonable estimates for the majority of the populations analyzed when compared with the other methods (table 6). It is interesting that, for many populations analyzed here, the age estimates obtained correlate very well with the dates estimated for the age of the LP H98 T-13910 allele in populations reported in other studies, such as northern Europeans.9,19 An interesting, recent report by Burger et al.29 showed the relative absence of the LP T-13910 allele in human remains in Europe (dated 7,000–7,800 years ago), implying that LP was rare in early Neolithic European farmers. This finding provides further support for our age estimates of the introduction of the LP T-13910 allele to global populations.29
The presence of the same LP allelic haplotype, H98, in dramatically diverse populations observed here, including Europeans, Asians, Arabs, some Sub-Saharan Africans, and North Africans (fig. 3), supports the concept of a single and relatively ancient global origin for the LP T-13910 H98 allele.3,7,13 Recently, Myles et al. interpreted the presence of the LP T-13910 allele among three North African Berber populations (from Morocco and Algeria) as genetic evidence of a shared origin of the dairy culture among those populations from Europe and Asia that show the presence of the LP T-13910 allele.30 More-recent data indicated the lack of the T-13910 variant among most Sub-Saharan African populations known to show high prevalence of LP, implying that other LP mutations must exist globally.31 Interestingly, two new reports have shown the presence of more than three variants that have risen independently in the close vicinity of the C/T-13910 variant correlating with LP in Africa.25,32 Taken together, these data and our results show that the LP T-13910 variant is of Caucasian origin and was most probably introduced independently more than once in human history. The accumulating data also imply the critical functional role of the −13910 region, as indicated by the recent reports of other mutations at or near this site: −13907, −13915, −13913, −13914, and −14010 variants, shown to correlate with LP in different populations. Some of them are driven to high population frequencies, whereas others still show low frequencies. These data lend strong support to the concept of convergent and still-ongoing adaptation of LP evolution in response to adult milk consumption in different human populations.
Acknowledgments
We are grateful to the participants for providing their samples for this study and to the following institutions for providing their financial support: The Emil Aaltonen Foundation (Tampere, Finland), The Center of Excellence in Complex Disease Genetics of the Academy of Finland, Biocentrum Helsinki, Research and Science Foundation of Farmos, The Sigrid Jusélius Foundation (Helsinki), and The Helsinki University Hospital Research Funding.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Arlequin, http://lgb.unige.ch/arlequin/
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for SNPs 2 [rs3754686], 3 [rs3769005], 4 [rs4988235], 5 [rs4954493], 6 [rs3099181]), 7 [rs182549], 8 [rs4988183], and 9 [rs3087343])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for indel polymorphism sequence within intron 1 of LCT [accession number DQ109677])
- NETWORK version 4.1.1.2, http://www.fluxus-engineering.com/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for lactase, LNP, and LP)
References
- 1.Sahi, T, Isokoski M, Jussila J, Launiala K, Pyorala K (1973) Recessive inheritance of adult-type lactose malabsorption. Lancet 2:823–826 10.1016/S0140-6736(73)90862-3 [DOI] [PubMed] [Google Scholar]
- 2.Sahi T (1994) Genetics and epidemiology of adult-type hypolactasia. Scand J Gastroenterol Suppl 202:7–20 [DOI] [PubMed] [Google Scholar]
- 3.Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvelä I (2002) Identification of a variant associated with adult-type hypolactasia. Nat Genet 30:233–237 10.1038/ng826 [DOI] [PubMed] [Google Scholar]
- 4.Kuokkanen M, Enattah NS, Oksanen A, Savilahti E, Orpana A, Jarvela I (2003) Transcriptional regulation of the lactase-phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut 52:647–652 10.1136/gut.52.5.647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasinpera H, Savilahti E, Enattah NS, Kuokkanen M, Totterman N, Lindhal H, Jarvela I, Kolho K-L (2004) A genetic test which can be used to diagnose adult-type hypolactasia in children. Gut 53:1571–1576 10.1136/gut.2004.040048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey CB, Pratt WS, Islam I, Whitehouse DB, Swallow DM (1995) DNA polymorphisms in the lactase gene: linkage disequilibrium across the 70-kb region. Eur J Hum Genet 3:27–41 [DOI] [PubMed] [Google Scholar]
- 7.Harvey CB, Hollox EJ, Poulter M, Wang Y, Rossi M, Auricchio S, Iqbal TH, Cooper BT, Barton R, Sarner M, et al (1998) Lactase haplotype frequencies in Caucasians: association with the lactase persistence/non-persistence polymorphism. Ann Hum Genet 62:215–223 10.1046/j.1469-1809.1998.6230215.x [DOI] [PubMed] [Google Scholar]
- 8.Poulter M, Hollox E, Harvey CB, Mulcare C, Peuhkuri K, Kajander K, Sarner M, Korpela R, Swallow DM (2003) The causal element for the lactase persistence/non-persistence polymorphism is located in a 1 Mb region of linkage disequilibrium in Europeans. Ann Hum Genet 67:298–311 10.1046/j.1469-1809.2003.00048.x [DOI] [PubMed] [Google Scholar]
- 9.Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, Hirschhorn JN (2004) Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet 74:1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olds LC, Sibley E (2003) Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet 12:2333–2340 10.1093/hmg/ddg244 [DOI] [PubMed] [Google Scholar]
- 11.Troelsen JT, Olsen J, Moller J, Sjostrom H (2003) An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology 125:1686–1694 10.1053/j.gastro.2003.09.031 [DOI] [PubMed] [Google Scholar]
- 12.Lewinsky RH, Jensen TG, Moller J, Stensballe A, Olsen J, Troelsen JT (2005) T-13910 DNA variant associated with lactase persistence interacts with Oct-1 and stimulates lactase promoter activity in vitro. Hum Mol Genet 14:3945–3953 10.1093/hmg/ddi418 [DOI] [PubMed] [Google Scholar]
- 13.Hollox EJ, Poulter M, Zvarik M, Ferak V, Krause A, Jenkins T, Saha N, Kozlov AI, Swallow DM (2001) Lactase haplotype diversity in the Old World. Am J Hum Genet 68:160–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simoons FJ (1978) The geographic hypothesis and lactose malabsorption: a weighing of the evidence. Am J Dig Dis 23:963–980 10.1007/BF01263095 [DOI] [PubMed] [Google Scholar]
- 15.Swallow DM (2003) Genetics of lactase persistence and lactose intolerance. Annu Rev Genet 37:197–219 10.1146/annurev.genet.37.110801.143820 [DOI] [PubMed] [Google Scholar]
- 16.Holden C, Mace R (1997) Phylogenetic analysis of the evolution of lactose digestion in adults. Hum Biol 69:605–628 [PubMed] [Google Scholar]
- 17.Flatz G, Rotthauwe HW (1977) The human lactase polymorphism: physiology and genetics of lactose absorption and malabsorption. Prog Med Genet 2:205–249 [PubMed] [Google Scholar]
- 18.Schneider S, Roessli D, Excoffier L (2000) Arlequin version 2.000: a software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva [Google Scholar]
- 19.Coelho M, Luiselli D, Bertorelle G, Lopes AI, Seixas S, Destro-Bisol G, Rocha J (2005) Microsatellite variation and evolution of human lactase persistence. Hum Genet 117:329–339 10.1007/s00439-005-1322-z [DOI] [PubMed] [Google Scholar]
- 20.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P (2005) A haplotype map of the human genome. Nature 437:1299–1320 10.1038/nature04226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AJ, Bustamante C (2005) Genomic scans for selective sweeps using SNP data. Genome Res 15:1566–1575 10.1101/gr.4252305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 [DOI] [PubMed] [Google Scholar]
- 23.Risch N, De Leon D, Ozelius L, Kramer P, Almasy L, Singer B, Fahn S, Breakefield X, Bressman S (1995) Genetic analysis of idiopathic torsion dystonia in Ashkenazi Jews and their recent descent from a small founder population. Nat Genet 9:152–159 10.1038/ng0295-152 [DOI] [PubMed] [Google Scholar]
- 24.Labuda M, Labuda D, Korab-Laskowska M, Cole DE, Zietkiewicz E, Wiessenbach J, Popowska E, Pronicka E, Root AW, Glorieux FH (1996) Linkage disequilibrium analysis in young populations: pseudo-vitamin D-deficiency rickets and the founder effect in French Canadians. Am J Hum Genet 59:633–643 [PMC free article] [PubMed] [Google Scholar]
- 25.Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M, et al (2007) Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet 39:31–40 10.1038/ng1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- 27.Hartl DL, Clark AG (1997) Principles of population genetics, 3rd ed. Sinauer, Sunderland, United Kingdom [Google Scholar]
- 28.Modiano D, Luoni G, Petrarca V, Sodiomon-Sirima B, De Luca M, Simpore J, Coluzzi M, Bodmer JG, Modiano G (2001) HLA class I in three West African ethnic groups: genetic distances from sub-Saharan and Caucasoid populations. Tissue Antigens 57:128–137 10.1034/j.1399-0039.2001.057002128.x [DOI] [PubMed] [Google Scholar]
- 29.Burger M, Kirchner M, Bramanti B, Haak W, Thomas MG (2007) Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc Natl Acad Sci USA 104:3736–3741 10.1073/pnas.0607187104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myles S, Bouzekri N, Haverfield E, Cherkaoui M, Dugoujon JM, Ward R (2005) Genetic evidence in support of a shared Eurasian-North African dairying origin. Hum Genet 117:34–42 10.1007/s00439-005-1266-3 [DOI] [PubMed] [Google Scholar]
- 31.Mulcare CA, Weale ME, Jones AL, Connell B, Zeitlyn D, Tarekegn A, Swallow D, Bradman M, Thomas MG (2004) The T allele of a single-nucleotide polymorphism 13.9 kb upstream of the lactase gene (LCT) (C-13.9kbT) does not predict or cause the lactase-persistence phenotype in Africans. Am J Hum Genet 74:1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingram CJ, Elamin MF, Mulcare CA, Weale ME, Tarekegn A, Raga TO, Bekele E, Elamin FM, Thomas MG, Bradman N, et al (2007) A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence? Hum Genet 120:779–788 10.1007/s00439-006-0291-1 [DOI] [PubMed] [Google Scholar]