Abstract
We have identified missense mutations at conserved amino acids in the PRPS1 gene on Xq22.3 in two families with a syndromic form of inherited peripheral neuropathy, one of Asian and one of European descent. The disease is inherited in an X-linked recessive manner, and the affected male patients invariably develop sensorineural hearing loss of prelingual type followed by gating disturbance and visual loss. The family of European descent was reported in 1967 as having Rosenberg-Chutorian syndrome, and recently a Korean family with the same symptom triad was identified with a novel disease locus CMTX5 on the chromosome band Xq21.32-q24. PRPS1 (phosphoribosyl pyrophosphate synthetase 1) is an isoform of the PRPS gene family and is ubiquitously expressed in human tissues, including cochlea. The enzyme mediates the biochemical step critical for purine metabolism and nucleotide biosynthesis. The mutations identified were E43D, in patients with Rosenberg-Chutorian syndrome, and M115T, in the Korean patients with CMTX5. We also showed decreased enzyme activity in patients with M115T. PRPS1 is the first CMT gene that encodes a metabolic enzyme, shedding a new light on the understanding of peripheral nerve-specific metabolism and also suggesting the potential of PRPS1 as a target for drugs in prevention and treatment of peripheral neuropathy by antimetabolite therapy.
Charcot-Marie-Tooth inherited neuropathy (CMT) represents a clinically and genetically heterogeneous group of hereditary peripheral neuropathies characterized by chronic motor and sensory impairment. It is one of the most common inherited disorders of humans and is the most common genetic cause of neuropathy, with prevalence of ∼1 in 2,500 people.1 Approximately 10%–20% of CMT is inherited in an X-linked manner (CMTX). Among the five known loci for CMTX, the causal gene has been identified only in CMTX1 (MIM 302800), in which the neuropathy is caused by mutations in the gap junction protein beta 1 gene (GJB1 [MIM 304040]) encoding the protein connexin 32 (Cx32).2 The authors previously identified a Korean family with a syndromic form of X-linked recessive CMT and localized the disease gene to a novel locus for CMTX on the chromosome band Xq21.32–24 (CMTX5 [MIM 311070]).3 The male patients with CMTX5 invariably developed a unique symptom triad of hearing loss, visual impairment, and peripheral neuropathy (table 1 and fig. 1A). The hearing loss has been characterized as sensorineural and prelingual in nature. The patients experienced progressive visual impairment from optic neuropathy at ∼10 years of age and peripheral neuropathy, at approximately the same age, that has been classified by electrophysiology as having mixed features of segmental demyelination and axonal loss. Follow-up of the youngest male patient (CMTX5:IV-11), who had been reported to have normal visual function at the time of initial presentation (at age 4 years), showed that his visual function began to be impaired at age 7 years. Pathologic examination of the sural nerve biopsy sample from the eldest living patient (CMTX5:III-18) showed loss of both large and small myelinated nerve fibers and an increase of endoneurial collagen (fig. 2A). Electron microscopic examination revealed segmental demyelination and remyelination with onion-bulb formation (fig. 2B). The symptom triad observed in CMTX5 was identical with that reported in Rosenberg-Chutorian syndrome (RCS) (table 1 and fig. 1B),4 and it has been speculated that CMTX5 and RCS are allelic disorders sharing the common disease gene. As in CMTX5, patients with RCS were not mentally retarded.
Table 1. .
Characteristics of Patients with CMTX5 and RCS, a Disease Allelic to CMTX5[Note]
| Finding (Age at Onset, in years)b |
|||||||
| Patient | Age (in years)a/Sex | Hearing Loss |
Visual Impairment |
Motor | Distal Sensory Loss |
MNCV of Median Nerve (m/sec−1) |
Uric Acid Levelc (Aged, in years) |
| CMTX5:III-18 | 28/Male | + (Since infancy) | + (11) | + (12) | + | NT | 4.48 (29) |
| CMTX5:III-21 | 25/Male | + (Since infancy) | + (13) | + (12) | − | NT | NT |
| CMTX5:III-24 | 18/Male | + (Since infancy) | + (8) | + (12) | − | NT | NT |
| CMTX5:IV-1 | 14/Male | + (Since infancy) | + (11) | + (10) | − | 51.0 | 3.36 (14) |
| CMTX5:IV-11 | 4/Male | + (Since infancy) | + (7)e | + (4) | − | 43.1 | 4.73 (5) |
| RCS:III-11 | 32/Male | + (Since infancy) | + (20) | + (5) | + | 44.8 | 5.43 (70) |
| RCS:III-13 | 29/Male | + (Since infancy) | + (19) | + (2) | + | 46.5 | 3.53 (68) |
| RCS:IV-14 | 3.5/Male | + (Since infancy) | − | Nonspecific | − | NT | NT |
Note.— MNCV = motor nerve conduction velocity; NT = Not tested.
Age at initial clinical evaluation and electrophysiological studies.
+ = positive finding; − = negative finding.
Reference range, 3.40–7.20 mg/dL.
Age at the time of blood drawing for the determination of uric acid levels.
Follow-up examination of this patient (IV-11) showed decreased visual acuity 3 years after the initial evaluation (at age 7 years).
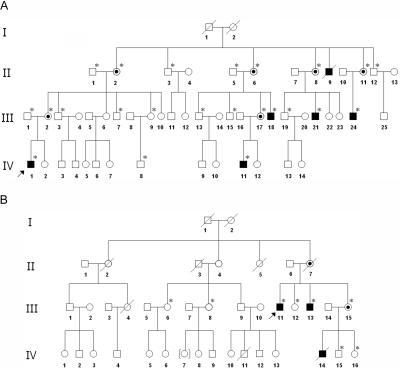
Figure 1. .
Pedigrees of the Korean family with CMTX5 (A) and the family of European descent with RCS (B). Affected male patients are indicated by blackened boxes and the obligate carrier females by circles with central dots. The individuals whose blood was drawn for the present study are marked with asterisks (*).
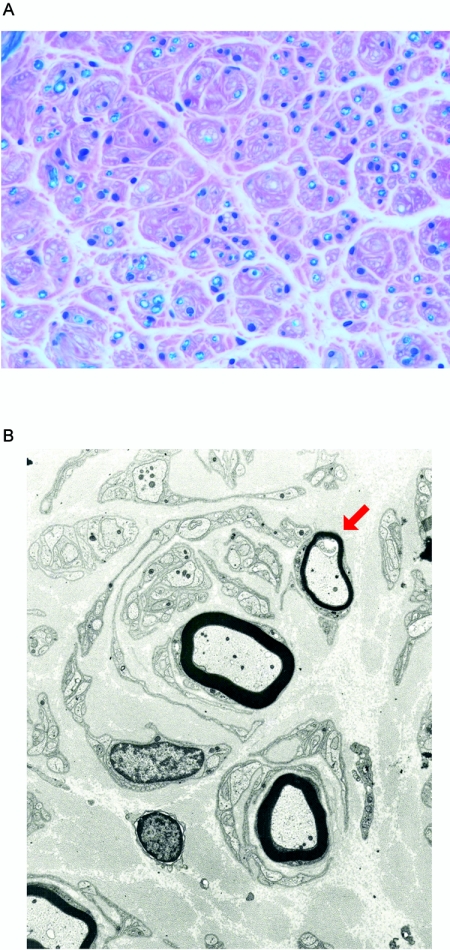
Figure 2. .
Sural nerve biopsy findings of a patient with CMTX5. A, Histological examination of the sural nerve biopsy sample revealed loss of myelinated nerve fibers and interstitial fibrosis (Luxol-fast blue, ×400). B, Electron micrograph of the sural nerve showed myelinated axons surrounded by concentrically arranged cytoplasmic processes of Schwann cells forming an “onion bulb.” The arrow indicates a remyelinating axon with an abnormally thin myelin sheath.
The disease locus of CMTX5 spans 15.2 cM and harbors >170 known genes and predicted genes. Because of the large number of positional candidate genes, we began by evaluating those candidate genes that were known to be expressed in the inner ear according to the cochlear expression database (Morton cochlear expression database)5 within the locus, since sensorineural hearing loss has been the earliest symptom and sign of both CMTX5 and RCS. The study was approved by the Institutional Review Board of the Samsung Medical Center, and written informed consent was obtained from all adult subjects and from parents on behalf of their children. Blood samples were collected and genomic DNA was isolated from peripheral blood leukocytes by use of the Wizard genomic DNA purification kit, according to the manufacturer’s instructions (Promega). All coding exons and their flanking intronic sequences of the candidate genes were amplified by PCR using the primers designed by the authors with the Primer3 program. Cycle sequencing was performed with the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) on the ABI 3100 Genetic Analyzer (Applied Biosystems). In the course of screening the positional candidate genes, including those expressed in the inner ear (number of genes screened = 71), the authors identified a 344T→C transition mutation in exon 3 of the phosphoribosyl pyrophosphate synthetase gene (PRPS1 [MIM 311850; Genbank accession number NM_002764]) on Xq22.3 that generates an amino acid substitution M115T (figs. 3 and 4A). This sequence variation was not observed in 1,045 unrelated control chromosomes of Korean origin. On the basis of our hypothesis that the disease-causing gene would be same in CMTX5 and RCS, we obtained DNA samples from patients and family members with RCS. Direct sequencing analyses of the PRPS1 gene revealed a 129A→C transversion mutation in exon 2 of PRPS1 giving rise to E43D in the two clinically affected patients with RCS and confirmed the X-linked recessive inheritance in the family (fig. 4A). Again, we confirmed that E43D was not observed in 1,103 unrelated control chromosomes of Korean origin. We also performed direct sequencing of exon 2 and exon 3 of PRPS1 on DNA samples from 50 unrelated individuals of white origin (Coriell Cell Repositories catalogue ID HD50CAU) and confirmed that no sequence variations were observed. The primer sequences for the PRPS1 gene are available on request. The mutated amino acids in the Korean patients with CMTX5 and the patients with RCS both showed perfect conservation from zebrafish to human (fig. 4B). To further investigate the frequency of PRPS1 mutations in CMT, we performed direct sequencing of all coding exons and exon-intron boundary sequences of PRPS1 in 101 patients with CMT whose previous genetic test results had ruled out CMT1A (MIM 118220), the most common subtype of CMT (57 male patients and 44 female patients). We observed a single patient with a sequence variation in exon 4 (420G→A), which was considered to be a rare synonymous polymorphism (P140P) on the basis of our further observation of its occurrence in control chromosomes (1.1%; 2/185), and no patient had a mutation that altered the amino acid sequence of PRPS1. This indicates, first, that PRPS1 is specifically mutated in CMTX5, causing the unique symptom triad, and, second, that the gene is highly conserved even within the human species.
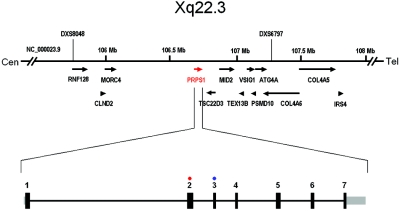
Figure 3. .
The transcript map of the region on chromosome band Xq22.3 within the CMTX5 locus and the genomic structure of PRPS1 with seven coding exons. Black bars represent coding sequences, and gray bars represent UTRs. All seven coding exons of PRPS1 were amplified and directly sequenced, according to standard procedures. The patients with CMTX5 had a missense mutation in exon 3 (blue circle), and the patients with RCS had a missense mutation in exon 2 (red circle). Tel = telomeric; Cen = centromeric.
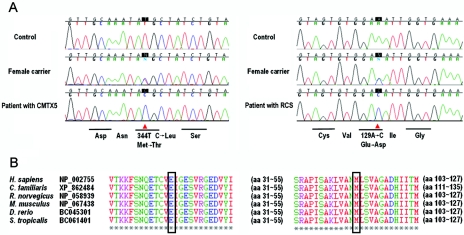
Figure 4. .
A, The chromatograms of the PRPS1 mutations detected by direct sequencing. M115T in patients with CMTX5 (left panel) and E43D in patients with RCS (right panel). B, The affected amino acids are perfectly conserved across different species.
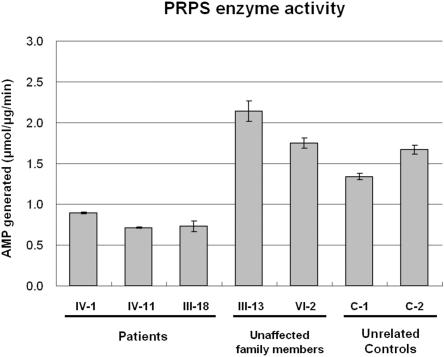
PRPS1 is a ubiquitously expressed gene and is the major isoform of the human phosphoribosyl pyrophosphate (PRPP) synthetase gene family.6 The gene gives rise to a 2-kb transcript and encodes 318 amino acids (including the first Met). The whole protein sequence of PRPS1 shows an exceptionally high degree of conservation, with homologies >95% across different species from zebrafish to human (fig. 5). PRPS1 is an enzyme that catalyzes the synthesis of PRPP, a substrate common to the biosynthesis of purines and pyrimidines, from ATP and Rib-5-P (EC 2.7.6.1). To see the functional outcome of the mutation, the authors determined the PRPS1 enzyme activity from fibroblasts obtained from skin biopsy samples from three patients with CMTX5 (CMTX5:IV-1, IV-11, and III-18), two nonaffected individuals in the CMTX5 family (CMTX5:III-13 and IV-2), and two unrelated adult control individuals (C-1 and C-2). The PRPS activity was determined as previously described.7 Briefly, 100 μl of cell extract was incubated for 60 min at 37°C, with 100 μl of a pH 7.4 reaction mixture containing: TrisHCl 50 mM, MgCl2 5 mM, EDTA 1 mM, DTT 1 mM, NaPi 32 mM, saturating concentrations of the substrates ATP (0.5 mM) and ribose 5-phosphate (0.15 mM), and P1 P5 diadenosine pentaphosphate (Ap5A, an inhibitor of adenylate kinase, 0.25 mM). On completion of the incubation, the reaction was stopped by heating at 70°C, and samples were centrifuged in Amicon cones (30,000 MW, Centricon30). To determine the level of AMP generated from ATP, the filtrate was injected into the high-performance liquid chromatography (HPLC) system with use of Waters prepacked μBondapak C18 10-μm column and was eluted at room temperature with 20 mM phosphate buffer (pH 2.5) at a flow rate of 1 ml/min. Nucleotide peaks were identified by coinjection with 25 μM AMP and GMP as standard. The instrument for HPLC and ultraviolet spectroscopy was the Waters Breeze HPLC system, and all reagents were obtained form Sigma-Aldrich. As a result, the authors observed that the mutation causing CMTX5 results in a partial loss of normal function, because we found decreased PRPS enzyme activity in patients compared with unaffected family members (i.e., those without the mutation) and unrelated control individuals (fig. 6).
Figure 5. .
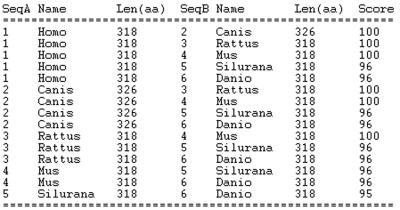
Multiple sequence alignment of PRPS1 by ClustalW, showing the degree of conservation of the amino acid sequences of the PRPS1 protein across different species. Homo = Homo sapiens; Canis = Canis familiaris; Rattus = Rattus norvegicus; Mus = Mus musculus; Danio = Danio rerio; Silurana = Silurana tropicalis. The reference amino acid sequences for alignment are same as in figure 4B.
Figure 6. .
PRPS enzyme activity, determined by measuring the degree of generation of AMP, the end product of PRPS enzyme, in cultured fibroblasts. The activity was decreased in patients with CMTX5 (CMTX5:IV-1, IV-11, and III-18) compared with unaffected family members and unrelated control individuals (CMTX5:III-13, VI-2 and C-1, C-2).
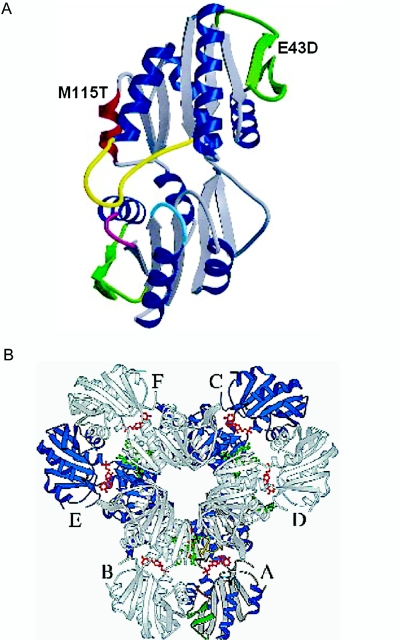
It is of note that previous reports of missense mutations in PRPS1 have demonstrated increased rather than decreased enzyme activity, with “superactive” enzymes resulting in hyperuricemia and gout (MIM 311850).8,9 One such mutation also caused sensorineural hearing loss and developmental delay.10 We therefore measured serum uric acid levels in patients with CMTX5 and RCS, all of whom were shown to have normal values (table 1). No patient with CMTX5 or RCS had been diagnosed with gout or had other hyperuricemia-related symptoms or signs. These results are consistent with our observation that the PRPS1 activity was decreased in our patients and also demonstrate that mutations resulting in a decreased enzymatic activity can damage the nervous system without elevating uric acid levels. In addition, PRPS1 is considered to be another rare example of a human disease gene in which activating and inactivating mutations cause distinct hereditary disorders. Why some PRPS1 mutations cause distinct phenotypes associated with increased or decreased enzyme activity is not known but is likely related to the site or nature of the mutation. The functional form of the enzyme was shown to have a hexameric structure. The PRPS1 monomer has five-stranded parallel β sheets and four α-helices on each of the N- and C-terminal domains, flanked by a short antiparallel β sheet protruding from the central core (a “flag” region).11 In addition to catalytic and regulatory binding sites, PRPS1 has functional residues involved in intersubunit interactions and maintaining the stability of the enzyme. The mutation in CMTX5 occurred at the Met 115 residue of α-helix of N-terminal domain, and the mutation in RCS occurred at the Glu 43 residue of the “flag” region of the N-terminal domain11 (fig. 7A). It was speculated that the increased enzyme activity was caused by mutations that alter the transmission of allosteric effects on the active sites of PRPS1, resulting in decreased inhibition.8 Intriguingly, the PRPS1 mutations causing decreased enzyme activity (CMTX5 and RCS) were occurring in the interface between the A and D subunits, whereas those causing increased enzyme activity were found in the interface between the A and C subunits, indicating that the different regulatory behaviors of mutations are from altered interactions between subunits (fig. 7B and S. Larsen, personal communication11).
Figure 7. .
A, The locations of the mutations on the ribbon diagram of the PRPS1 monomer.11 M115T in CMTX5 on the α-helix of N-terminal domain (blue) and E43D in RCS on the “flag” region of the N-terminal domain (green). B, Hexameric structure and subunits of PRPS1. Reprinted with permission from Nature Publishing Group.11
Lastly, acquired peripheral neuropathy from antimetabolite medications is one of the major side effects of cancer chemotherapy and of other medications, such as statins and nucleoside reverse transcriptase inhibitors utilized to treat patients with HIV (drug-/chemotherapy-induced peripheral neuropathy).12,13 Some of these medications also cause sensorineural hearing loss and optic neuropathy, as in the patients with CMTX5 and RCS. Several agents have been developed for potential use as chemoprotectants to reduce these side effects, but none have proven effective.14 As the metabolic enzyme critical for nucleotide biosynthesis, our findings provide a clue to the pathophysiology of acquired form of peripheral neuropathy and also raise the potential of the PRPS1 gene as a therapeutic target to prevent or treat medication-induced acquired peripheral neuropathy, sensorineural hearing loss, and/or optic neuropathy.14,15 Indeed, the PRPS1 gene, along with other PRPS family genes, has been included in the part of the human genome thought to offer opportunities for drug therapy.16
In summary, we found that missense mutations in the PRPS1 gene cause a syndromic form of peripheral neuropathy associated with sensorineural hearing loss and optic neuropathy with the help of the cochlear expression database. This is the first report of inherited peripheral neuropathy from decreased activity of a metabolic enzyme critical for purine metabolism and nucleotide biosynthesis, shedding a new light on the understanding of peripheral nerve-specific metabolism and the pathophysiology of antimetabolite-induced acquired peripheral neuropathy.
Acknowledgments
This work was supported by grants from the Korean HapMap Project of MOST (Ministry of Science and Technology of Korea) and from the National Institutes of Health (R01 NS43168-01A1). We thank the patients and their family members who participated in this study for their cooperation.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- ClustalW, http://www.ebi.ac.uk/clustalw/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for PRPS1 [accession number NM_002764])
- Morton Lab Web site, http://www.brighamandwomens.org/bwh_hearing/ (for cochlear expression database)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- Primer3, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
References
- 1.Skre H (1974) Genetic and clinical aspects of Charcot-Marie-Tooth’s disease. Clin Genet 6:98–118 [DOI] [PubMed] [Google Scholar]
- 2.Bergoffen J, Scherer SS, Wang S, Scott MO, Bone LJ, Paul DL, Chen K, Lensch MW, Chance PF, Fischbeck KH (1993) Connexin mutations in X-linked Charcot-Marie-Tooth disease. Science 262:2039–2042 10.1126/science.8266101 [DOI] [PubMed] [Google Scholar]
- 3.Kim HJ, Hong SH, Ki CS, Kim BJ, Shim JS, Cho SH, Park JH, Kim JW (2005) A novel locus for X-linked recessive CMT with deafness and optic neuropathy maps to Xq21.32-q24. Neurology 64:1964–1967 10.1212/01.WNL.0000163768.58168.3A [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg RN, Chutorian A (1967) Familial opticoacoustic nerve degeneration and polyneuropathy. Neurology 17:827–832 [DOI] [PubMed] [Google Scholar]
- 5.Resendes BL, Robertson NG, Szustakowski JD, Resendes RJ, Weng Z, Morton CC (2002) Gene discovery in the auditory system: characterization of additional cochlear-expressed sequences. J Assoc Res Otolaryngol 3:45–53 10.1007/s101620020005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taira M, Iizasa T, Yamada K, Shimada H, Tatibana M (1989) Tissue-differential expression of two distinct genes for phosphoribosyl pyrophosphate synthetase and existence of the testis-specific transcript. Biochim Biophys Acta 1007:203–208 [DOI] [PubMed] [Google Scholar]
- 7.Torres RJ, Mateos FA, Puig JG, Becker MA (1996) Determination of phosphoribosylpyrophosphate synthetase activity in human cells by a non-isotopic, one step method. Clin Chim Acta 245:105–112 10.1016/0009-8981(95)06178-9 [DOI] [PubMed] [Google Scholar]
- 8.Becker MA, Smith PR, Taylor W, Mustafi R, Switzer RL (1995) The genetic and functional basis of purine nucleotide feedback-resistant phosphoribosylpyrophosphate synthetase superactivity. J Clin Invest 96:2133–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker MA, Taylor W, Smith PR, Ahmed M (1996) Overexpression of the normal phosphoribosylpyrophosphate synthetase 1 isoform underlies catalytic superactivity of human phosphoribosylpyrophosphate synthetase. J Biol Chem 271:19894–19899 10.1074/jbc.271.33.19894 [DOI] [PubMed] [Google Scholar]
- 10.Simmonds HA, Webster DR, Lingam S, Wilson J (1985) An inborn error of purine metabolism, deafness and neurodevelopmental abnormality. Neuropediatrics 16:106–108 [DOI] [PubMed] [Google Scholar]
- 11.Eriksen TA, Kadziola A, Bentsen AK, Harlow KW, Larsen S (2000) Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase. Nat Struct Biol 7:303–308 10.1038/74069 [DOI] [PubMed] [Google Scholar]
- 12.Peltier AC, Russell JW (2006) Advances in understanding drug-induced neuropathies. Drug Saf 29:23–30 10.2165/00002018-200629010-00002 [DOI] [PubMed] [Google Scholar]
- 13.Sul JK, Deangelis LM (2006) Neurologic complications of cancer chemotherapy. Semin Oncol 33:324–332 10.1053/j.seminoncol.2006.03.006 [DOI] [PubMed] [Google Scholar]
- 14.Hausheer FH, Schilsky RL, Bain S, Berghorn EJ, Lieberman F (2006) Diagnosis, management, and evaluation of chemotherapy-induced peripheral neuropathy. Semin Oncol 33:15–49 10.1053/j.seminoncol.2005.12.010 [DOI] [PubMed] [Google Scholar]
- 15.Brinkman RR, Dube MP, Rouleau GA, Orr AC, Samuels ME (2006) Human monogenic disorders - a source of novel drug targets. Nat Rev Genet 7:249–260 10.1038/nrg1828 [DOI] [PubMed] [Google Scholar]
- 16.Hopkins AL, Groom CR (2002) The druggable genome. Nat Rev Drug Discov 1:727–730 10.1038/nrd892 [DOI] [PubMed] [Google Scholar]