Abstract
NOBOX (newborn ovary homeobox gene) is an oocyte-specific homeobox gene that plays a critical role in early folliculogenesis and represents a candidate gene for nonsyndromic ovarian failure. We investigated whether mutations in the NOBOX gene cause premature ovarian failure (POF). We sequenced the NOBOX gene in 96 white women with POF and discovered seven known single-nucleotide polymorphisms and four novel variations, two of which, p.Arg355His and p.Arg360Gln, cause missense mutations in the homeobox domain. Electrophoretic mobility shift assay (EMSA) confirmed that the missense mutation, p.Arg355His, disrupted NOBOX homeodomain binding to NOBOX DNA-binding element (NBE) and had a dominant negative effect on the binding of wild-type NOBOX to DNA. Our findings demonstrate that NOBOX mutations can cause POF.
In premature ovarian failure (POF [MIM 311360]), ovaries cease to mature oocytes before 40 years of age. The condition is characterized by secondary amenorrhea, infertility, hypoestrogenism, and elevated gonadotropin serum levels (FSH>40 IU/liter).1 POF is heritable in up to 30% of patients and is genetically heterogeneous.2,3 Mechanisms long invoked in the pathogenesis of POF include abnormalities of the X chromosome or autosomes and autoimmune, infectious, and environmental causes.4,5 For some cases, a genetic basis has been shown, and, for genes such as FMR1 (MIM 309550),6,7 FSHR (MIM 136435),8–10 POF1B (MIM 300603),11 FOXL2 (MIM 605597),12,13 and BMP15 (MIM 300247),14 functional data support causation. Because human ovaries are not easily accessible, our knowledge of ovarian development is largely derived from animal models.15 Mouse knockouts and naturally occurring mutations in sheep have been useful in identifying candidate genes for ovarian failure.16–19
Human and mouse NOBOX genes (newborn ovary homeobox [MIM 610934]) are preferentially expressed in oocytes and encode a homeobox transcriptional regulator.20,21 Nobox plays a crucial role during early folliculogenesis in the mouse.22 Disruption of the mouse Nobox gene causes nonsyndromic ovarian failure in females, whereas males are unaffected.23 NOBOX expression in adult human tissues mimics that in mice, with preferential expression in the human gonads.21 Expression within the human ovary is oocyte specific, as observed from primordial follicle through metaphase II (MII) oocytes. Mouse NOBOX homeodomain binds TAATTG,24 and we have shown elsewhere that NOBOX binds such elements in Gdf9 (MIM 601918) and Pou5f1 (MIM 164177) promoters.25 In the current study, we report two novel missense mutations in the NOBOX homeodomain in women with POF and show that p.Arg355His mutation can disrupt NOBOX homeodomain binding to DNA.
Our study subjects comprised 96 white women from the United States who had POF and had been collected at Baylor College of Medicine since 2001. Recruitment criteria comprised cessation of menstrual cycles before 40 years of age and at least two serum FSH concentrations >40 IU/liter. Women with chromosomal abnormalities were excluded. Two hundred and seventy-eight white women who denied having any medical problems were used as controls. Informed consent for molecular studies was obtained from all subjects. The study was approved by the Institutional Review Board of Baylor College of Medicine.
Peripheral blood was obtained and genomic DNA extracted. NOBOX-specific primers were designed according to the human NOBOX sequence (GenBank accession number NM_001080413 and Ensembl accession number ENST00000389325) and are presented in table 1.
Table 1. .
PCR Primers Used for Amplification of Human NOBOX Gene
| Primer | Sequence (5′→3′) |
Reference Sequencea |
| NOBOX 1F | ATTTAAAGACAAGCTCGAGTATC | |
| NOBOX 1R | CAAGAGTCCTCAGTGTATGGG | NM_001080413 |
| NOBOX 2F | TATCTGACCAGCCCTCCGACTTT | |
| NOBOX 2R | ACTGCTGGAATTACAGGCGTGAG | NM_001080413 |
| NOBOX 3F | ATGCTCTTGCCTCGGCTGCTGGA | |
| NOBOX 3R | AGCTTGACTATTGTGAGGATT | NM_001080413 |
| NOBOX 4F | TGCTAAGGTCAGTGGATGTTGGC | |
| NOBOX 4R | CCGGGACGAAGTGACATAC | NM_001080413 |
| NOBOX 5F | TGTAGCACCCAAGAGCGAGAA | |
| NOBOX 5R | TGCAGTGCCTTCCTCTCCTAATG | NM_001080413 |
| NOBOX 6–7F | GCAACAGCCAGGACCTAAGC | |
| NOBOX 6–7R | GTCACTCCCACCTCCATCAAACA | NM_001080413 |
| NOBOX 8F | TCTACCATTCAGCGATGCCAA | |
| NOBOX 8R | TGTGCTCCTCTCAGTTAACCC | NM_001080413 |
| NOBOX 9F | GGGACTCCGCTACTGTGGT | |
| NOBOX 9R | CGAGGGAGAAGAGCTTAATAG | ENST00000389325 |
| NOBOX 10F | GTCCTAAGCTGCGTCTATGTG | |
| NOBOX 10R | CGAGCCCAATCCTATCCCA | ENST00000389325 |
All 10 exons and exon-intron boundaries of the NOBOX gene were amplified using PCR. PCR conditions are available on request. PCR products were sequenced directly on an automated sequencer, ABI Prism Sequencer 3730XL (Applied Biosystems).
Our study revealed 11 sequence variants in the coding region of NOBOX: 7 known SNPs and 4 novel variations (fig. 1A). All sequence variants were confirmed by three independent PCR and sequencing reactions, followed by sequencing in forward and reverse directions. Details are provided in table 2.
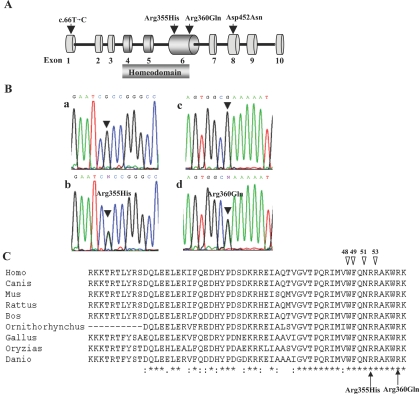
Figure 1. .
Identification of novel variants in the NOBOX gene. A, Schematic diagram of the NOBOX gene, showing three novel nonsynonymous variations (exon 6 and exon 8) and one synonymous variant (exon 1) found in the present study. Exons are a composite of human sequence NM_001080413 (GenBank) and ENST00000389325 (Ensembl). The homeodomain region is coded by exons 4–6. B, Electropherogram showing wild-type (a and c) and mutant (c.1064G→A [Arg355His] and c.1079G→A [Ar360Gln]) sequences. Arrows indicate the G→A change in POF subject 23 (b, Arg355His) and POF subject 34 (d, Ar360Gln). C, NOBOX homeodomain alignment among different species. Arrow heads indicate highly conserved amino acids highly conserved among homeodomains. The two missense mutations (p.Arg355His and p.Arg360Gln) found in this study are indicated by arrows. Asterisks (*) indicate amino acid residues that are conserved in all the species.
Table 2. .
NOBOX Sequencing Results in 96 White Women with POF
| Allele Frequency(%) |
|||||||||
| Patients with POF |
Controls |
||||||||
| Sequence Variation |
Location | Amino Acid Variation | dbSNP ID | Wild Type |
Heterozygote | Homozygote | Wild Type |
Heterozygote | Referencea |
| c.42T→C | Exon 1 | Synonymous | rs1208179 | 71.9 | 28.1 | 0 | d, e | ||
| c.66T→C | Exon 1 | Synonymous | Novel | 98.96 | 1.04 | 0 | 100 | 0 | d, e |
| c.262C→T | Exon 3 | Synonymous | rs727714 | 11.5 | 45.8 | 42.7 | d, e | ||
| c.1064G→A | Exon 6 | p.Arg355His | Novel | 99.0 | 1.01 | 0 | 100 | 0 | d, e |
| c.1079G→Ab | Exon 6 | p.Arg360Gln | Novel | 99.0 | 1.01 | 0 | 99.3 | .7 | d, e |
| c.1154+11T→C | Intron 6 | rs757388 | 11.5 | 45.8 | 42.7 | d, e | |||
| c.1155−22G→A | Intron 6 | rs11769847 | 11.5 | 45.8 | 42.7 | d, e | |||
| c.1354G→Ab | Exon 8 | p.Asp452Asn | Novel | 99.0 | 1.01 | 0 | 94.8 | 5.2 | d, e |
| c.1444G→A | Exon 8 | p.Gly482Ser | rs2525702 | 72.9 | 26.0 | 1.1 | d, e | ||
| c.603–51G→T | Intron 8 | rs11979528 | 29.2 | 44.8 | 26.0 | f, g | |||
| c.682T→C | Exon 9 | p.Phe228Leu | rs2699503 | 14.6 | 47.9 | 37.5 | f, g | ||
d = GenBank accession number NM_001080413. e = NCBI protein database accession number XP_001134420. f = Ensembl accession number ENST00000389325. g = Ensembl accession number ENSP00000373976.
Allelic frequencies between the general population and patients with POF show no significant differences (Fisher’s exact test P>.05)
The seven known SNPs included three intronic variants (rs757388, rs11769847, and rs11979528), two synonymous variants (c.42T→C and c.262C→T), and two nonsynonymous variants (p.Gly482 Ser and p.Phe228Leu). Of the four novel variations, one was a synonymous variant (c.66T→C) and three were nonsynonymous exonic variants: c.1354G→A (p.Asp452Asn), c.1064G→A (p.Arg355His), and c.1079G→A (p.Arg360Gln). p.Asp452Asn was also found in five controls at a frequency that did not differ significantly from that in the POF sample (Fisher’s exact test P>.05). p.Arg355His and p.Arg360Gln were located in the conserved homeodomain region (fig. 1A–1C). p.Arg360Gln was also present in two controls at a frequency that was not statistically different from that in women with POF (Fisher’s exact test P>.05). By contrast, p.Arg355His was not present in the controls. p.Arg355His mutation was found in a 35-year-old woman whose FSH level was 103 IU/liter. Her menarche was at age 11 years, and she entered menopause at age 32 years. She was the mother of two healthy children and lacked overt somatic anomalies. She was the only child of a woman who conceived her at 26 years of age and who entered menopause at 42 years of age.
The protein sequence of human NOBOX is 92% identical to the corresponding homeodomain region in the mouse.20 Both mutations, p.Arg355His and p.Arg360Gln, were located within the homeodomain region that is perfectly conserved among species ranging from zebrafish to humans. Since a homeodomain is capable of mediating critical protein-DNA and protein-protein interactions, and since p.Arg355His was not present in control women, we hypothesized that this novel mutation disrupted NOBOX homeodomain binding to DNA.
We engineered p.Arg355His and p.Arg360Gln missense mutations into the mouse homeodomain to test whether these mutations will disrupt mouse NOBOX homeodomain binding to the NOBOX DNA-binding element (NBE) we identified elsewhere.25 The mouse homeobox was subcloned into the pET41b expression vector to generate a fusion protein with glutathione-S-transferase (GST-NXHD). Mouse missense mutations corresponding to human p.Arg355His and p.Arg360Gln (mouse p.Arg186His and p.Arg191Gln) were generated using the QuikChange multi site–directed mutagenesis kit (Stratagene) with oligonucleotides GTGTGGTTTCAGAACCACAGGGCAAAGTGGAGA and CGCAGGGCAAAGTGGCAGAAAGTGGAGAAACTG. We transformed BL21-pLysS Escherichia coli (Stratagene) with GST-NXHD (wild type), GST-R186H (containing mutation p.Arg186His), and GST-R191Q (containing mutation p.Arg191Gln) constructs, induced protein expression, and purified the GST-NXHD, GST-R186H, and GST-R191Q fusion proteins on GST-bind resin (Novagen). The proteins were dialyzed three times in PBS to remove excess glutathione and were quantified with BSA by use of the Lowry assay.
We performed electrophoretic mobility shift assay (EMSA) as described elsewhere.25 In brief, NBE (containing NOBOX-binding consensus sequence TAATTG) was labeled by end-filling annealed primers with [α-32P] dCTP and Klenow polymerase (Invitrogen) at room temperature. The top and bottom strand oligonucleotides were 5′-ACGAGCTACCTTACTTAATTGGACGT-3′ and 5′-ACAGTACGCGTTCAACGTC-3′, respectively. Binding reactions were conducted by incubating 32P-labeled probe (250,000 cpm/reaction) individually with 50 ng of purified GST-NXHD, GST-R186H, and GST-R191Q. Polyclonal anti-GST antibodies (Amersham Biosciences) were used to supershift DNA-protein complexes. Binding reactions were resolved on a 4% polyacrylamide gel. The gel was then fixed, dried, and exposed to Kodak BioMax XAR Film.
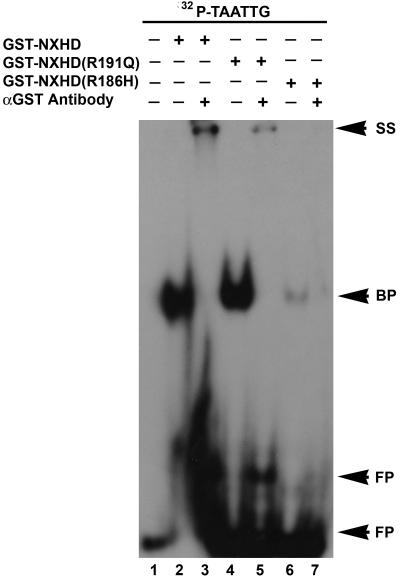
As shown in figure 2 (lanes 2 and 4), GST-NXHD or GST-R191Q protein bound radiolabeled NBE with similar affinity. In contrast, GST-R186H protein showed a dramatic decrease in the NBE-binding capacity (fig. 2, lane 6). In the supershift assay, addition of anti-GST antibodies decreased the mobility of the GST-NXHD-NBE and GST-R191Q-NBE complex and, as expected, diminished the total amount of the DNA-protein complex (fig. 2, lanes 3, 5, and 7). These results show that R186H mutation disrupted NOBOX homeodomain binding, whereas R191Q did not exhibit a similar effect.
Figure 2. .
GST-R186H binds to the TAATTG sequences with low affinity. 32P-labeled TAATTG was incubated with purified recombinant GST-NXHD, GST-R191Q, or GST-R186H. 32P-labeled TAATTG formed a strong DNA-protein complex with the recombinant GST-NXHD (lane 2) and GST-R191Q protein (lane 4), but not with GST-R186H (lane 6). The DNA-protein complex was supershifted when incubated with polyclonal antibodies against GST (lanes 3, 5 and 7). The presence or absence of recombinant protein and/or antibody is indicated above each lane with a plus (+) or minus (−) sign, respectively. Arrows indicate DNA-protein complex (BP), supershifted complex (SS) and free probe (FP). Oligonucleotide sequence containing TAATTG is 5′-ACG AGC TAC CTT ACT TAA TTG GAC GTT GAA CGC GTA CTG T-3′.
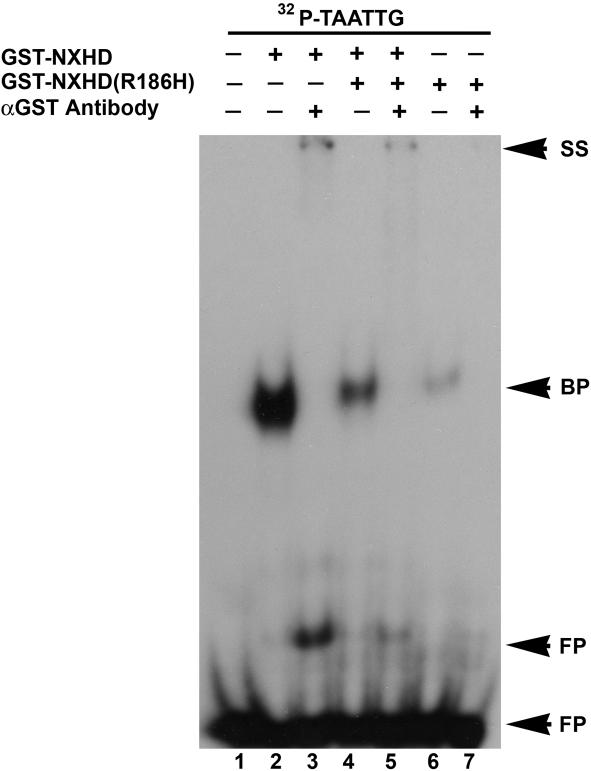
We also examined whether the R186H mutation interfered with the DNA-binding capacity of the wild-type NOBOX homeodomain protein. Equivalent amounts of GST-NXHD and GST-R186H (50 ng) were coincubated (fig. 3), and the mixture of wild-type and R186H homedomains exhibited weaker binding to NBE than did wild type alone (fig. 3, lanes 4 and 2). These experiments indicate that the R186H mutation has a dominant negative effect on the wild-type NOBOX homeodomain and that it causes diminished binding affinity for the NBE. This effect was not observed with the R191Q missense mutation.
Figure 3. .
Dominant negative effect of GST-R186H. Radiolabeled NBE (32P-TAATTG) was incubated individually with purified recombinant GST-NXHD, GST-R186H, or a 50/50 mixture of both homeodomains. GST-R186H reduced GST-NXHD binding affinity to the 32P-labeled TAATTG (lane 4). The specificity of the DNA-protein complexes was confirmed by polyclonal antibodies against GST. The presence or absence of recombinant protein and/or antibody is indicated above each lane with a plus (+) or minus (−) sign, respectively. Arrows indicate DNA-protein complex (BP), supershifted complex (SS) and free probe (FP).
In this study of 96 white women with POF, we found two missense mutations in the NOBOX homeodomain, one of which, p.Arg355His, is absent in the control population and disrupts NOBOX homeodomain binding to NBE in a dominant negative fashion. Mice that are null for Nobox are infertile and lose oocytes rapidly during ovarian development. Nobox is therefore a critical oocyte prosurvival factor. Partial deficiency in NOBOX function in humans can conceivably lead to a more gradual loss of oocytes in women and can cause POF after the onset of puberty.
Homeodomains are generally composed of 60 aa, as is predicted for NOBOX. The p.Arg355His missense mutation is located within the highly conserved portion of the NOBOX homeodomain at position 52 (Arg52) (fig. 1C). Several amino acids are conserved in the majority of homeodomains. These include Trp48 (W), Phe49 (F), Asn51 (N), and Arg53 (R) within the homeodomain (fig. 1C); all reside in helix III, which has critical implications for DNA binding and overall stability of the tertiary structure of the homeodomains.24 Prior studies have demonstrated that substitution of glutamine for the conserved asparagine residue at position 51 abolished NOBOX homeodomain binding to NBEs.25,26
Despite histidine being a polar amino acid like arginine, p.Arg355His mutation, when engineered into a mouse homeodomain, disrupts NOBOX binding to DNA. Arg355 in the NOBOX homeodomain is conserved from zebrafish to humans and is a critical residue for NOBOX homeodomain binding to the DNA. Homeodomains are also known to be critical in protein-protein interactions, involving either homeodomains or other protein domains.27 Direct protein-protein interactions are necessary for transcriptional activity.28,29 The dominant negative effect also suggests that the NOBOX homeodomain may function as a dimer. The p.Arg360Gln missense mutation, when engineered into a mouse homeodomain, does not disrupt protein-DNA interaction in the EMSA assay, was also found in the control population, and is consistent with being a novel SNP.
Our study shows that mutations within NOBOX can cause human POF. The only previous study involved 35 Japanese women with POF and did not find mutations in the NOBOX homeodomain.30 By contrast, our results indicate that mutations within NOBOX likely account for POF in a small subset of women. There are numerous oocyte-specific genes, each of which may be responsible for some cases of POF.31 Future high-throughput sequencing of such genes in women with idiopathic ovarian failure should provide a better idea of the contribution that oocyte-specific genes make to nonsyndromic POF.
Acknowledgments
This study was supported in part by a National Institutes of Health grant (HD44858) and a March of Dimes Basil O’Connor Award (5-FY02-266) to A.R. and by grants from the National Natural Science Foundation Committee (30470703 and 30670777) of the People’s Republic of China and National Basic Research Program of China “973 Program” (2006CB944004) to Z. Chen.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Ensembl, http://www.ensembl.org/(for NOBOX [accession number ENST00000389325])
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for IDs shown in table 2)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for NOBOX [accession number NM_001080413])
- NCBI Protein Database, http://www.ncbi.nlm.nih.gov/sites/entrez?db=protein&cmd=search&term= (accession number XP_001134420)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nim.nih.gov/Omim (for POF, NOBOX, FMR1, POF1B, FSHR, FOXL2, BMP15, GDF9, and POU5F1)
Reference
- 1.Coulam CB, Adamson SC, Annegers JF (1986) Incidence of premature ovarian failure. Obstet Gynecol 67:604–606 [PubMed] [Google Scholar]
- 2.Woad KJ, Watkins WJ, Prendergast D, Shelling AN (2006) The genetic basis of premature ovarian failure. Aust N Z J Obstet Gynaecol 46:242–244 10.1111/j.1479-828X.2006.00585.x [DOI] [PubMed] [Google Scholar]
- 3.Vegetti W, Grazia Tibiletti M, Testa G, de Lauretis Y, Alagna F, Castoldi E, Taborelli M, Motta T, Bolis PF, Dalpra L, et al (1998) Inheritance in idiopathic premature ovarian failure: analysis of 71 cases. Hum Reprod 13:1796–1800 10.1093/humrep/13.7.1796 [DOI] [PubMed] [Google Scholar]
- 4.Simpson JL, Rajkovic A (1999) Ovarian differentiation and gonadal failure. Am J Med Genet 89:186–200 [DOI] [PubMed] [Google Scholar]
- 5.Simpson JL, Rajkovic A (2004) Germ cell failure and ovarian resistance: human genes and disorders. In: Leung PCK, Adashi EY (eds) The ovary, 2nd ed. Academic Press, San Diego, pp 541–557 [Google Scholar]
- 6.Wittenberger, Hagerman RJ, Sherman SL, McConkie-Rosell A, Welt CK, Rebar RW, Corrigan EC, Simpson JL, Nelson LM (2006) The FMR1 premutation and reproduction. Fertil Steril 87:456–465 10.1016/j.fertnstert.2006.09.004 [DOI] [PubMed] [Google Scholar]
- 7.Sullivan AK, Marcus M, Epstein MP, Allen EG, Anido AE, Paquin JJ, Yadav-Shah M, Sherman SL (2005) Association of FMR1 repeat size with ovarian dysfunction. Hum Reprod 20:402–412 10.1093/humrep/deh635 [DOI] [PubMed] [Google Scholar]
- 8.Aittomaki K, Lucena JL, Pakarinen P, Sistonen P, Tapanainen J, Gromoll J, Kaskikari R, Sankila EM, Lehvaslaiho H, Engel AR, et al (1995) Mutation in the follicle-stimulating hormone receptor gene causes hereditary hypergonadotropic ovarian failure. Cell 82:959–968 10.1016/0092-8674(95)90275-9 [DOI] [PubMed] [Google Scholar]
- 9.Touraine P, Beau I, Gougeon A, Meduri G, Desroches A, Pichard C, Detoeuf M, Paniel B, Prieur M, Zorn JR, et al (1999) New natural inactivating mutations of the follicle-stimulating hormone receptor: correlations between receptor function and phenotype. Mol Endocrinol 13:1844–1854 10.1210/me.13.11.1844 [DOI] [PubMed] [Google Scholar]
- 10.Rannikko A, Pakarinen P, Manna PR, Beau I, Misrahi M, Aittomaki K, Huhtaniemi I (2002) Functional characterization of the human FSH receptor with an inactivating Ala189Val mutation. Mol Hum Reprod 8:311–317 10.1093/molehr/8.4.311 [DOI] [PubMed] [Google Scholar]
- 11.Lacombe A, Lee H, Zahed L, Coucair M, Muller JM, Nelson SF, Salameh W, Vilain E (2006) Disruption of POF1B binding to nonmuscle actin filaments is associated with premature ovarian failure. Am J Hum Genet 79:113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prueitt RL, Zinn AR (2001) A fork in the road to fertility. Nat Genet 27:159–166 10.1038/84735 [DOI] [PubMed] [Google Scholar]
- 13.Nallathambi J, Moumne L, De Baere E, Beysen D, Usha K, Sundaresan P, Veitia RA (2007) A novel polyalanine expansion in FOXL2: the first evidence for a recessive form of the blepharophimosis syndrome (BPES) associated with ovarian dysfunction. Hum Genet 121:107–112 10.1007/s00439-006-0276-0 [DOI] [PubMed] [Google Scholar]
- 14.Di Pasquale E, Beck-Peccoz P, Persani L (2004) Hypergonadotropic ovarian failure associated with an inherited mutation of human bone morphogenetic protein-15 (BMP15) gene. Am J Hum Genet 75:106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pangas SA, Rajkovic A (2006) Transcriptional regulation of early oogenesis: in search of masters. Hum Reprod Update 12:65–76 10.1093/humupd/dmi033 [DOI] [PubMed] [Google Scholar]
- 16.Pangas SA, Li X, Robertson EJ, Matzuk MM (2006) Premature luteinization and cumulus cell defects in ovarian-specific Smad4 knockout mice. Mol Endocrinol 20:1406–1422 10.1210/me.2005-0462 [DOI] [PubMed] [Google Scholar]
- 17.Racki WJ, Richter JD (2006) CPEB controls oocyte growth and follicle development in the mouse. Development 133:4527–4537 10.1242/dev.02651 [DOI] [PubMed] [Google Scholar]
- 18.Shiina H, Matsumoto T, Sato T, Igarashi K, Miyamoto J, Takemasa S, Sakari M, Takada I, Nakamura T, Metzger D, et al (2006) Premature ovarian failure in androgen receptor-deficient mice. Proc Natl Acad Sci USA 103:224–229 10.1073/pnas.0506736102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodin L, Di Pasquale E, Fabre S, Bontoux M, Monget P, Persani L, Mulsant P (2007) A novel mutation in the bone morphogenetic protein 15 gene causing defective protein secretion is associated with both increased ovulation rate and sterility in Lacaune sheep. Endocrinology 148:393–400 10.1210/en.2006-0764 [DOI] [PubMed] [Google Scholar]
- 20.Suzumori N, Yan C, Matzuk MM, Rajkovic A (2002) Nobox is a homeobox-encoding gene preferentially expressed in primordial and growing oocytes. Mech Dev 111:137–141 10.1016/S0925-4773(01)00620-7 [DOI] [PubMed] [Google Scholar]
- 21.Huntriss J, Hinkins M, Picton HM (2006) cDNA cloning and expression of the human NOBOX gene in oocytes and ovarian follicles. Mol Hum Reprod 12:283–289 10.1093/molehr/gal035 [DOI] [PubMed] [Google Scholar]
- 22.Choi Y, Rajkovic A (2006) Genetics of early mammalian folliculogenesis. Cell Mol Life Sci 63:579–590 10.1007/s00018-005-5394-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM (2004) NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science 305:1157–1159 10.1126/science.1099755 [DOI] [PubMed] [Google Scholar]
- 24.Banerjee-Basu S, Baxevanis AD (2001) Molecular evolution of the homeodomain family of transcription factors. Nucleic Acids Res 29:3258–3269 10.1093/nar/29.15.3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi Y, Rajkovic A (2006) Characterization of NOBOX DNA binding specificity and its regulation of Gdf9 and Pou5f1 promoters. J Biol Chem 281:35747–3756 10.1074/jbc.M604008200 [DOI] [PubMed] [Google Scholar]
- 26.Kim DW, Kempf H, Chen RE, Lassar AB (2003) Characterization of Nkx3.2 DNA binding specificity and its requirement for somitic chondrogenesis. J Biol Chem 278:27532–27539 10.1074/jbc.M301461200 [DOI] [PubMed] [Google Scholar]
- 27.Bruun JA, Thomassen EI, Kristiansen K, Tylden G, Holm T, Mikkola I, Bjorkoy G, Johansen T (2005) The third helix of the homeodomain of paired class homeodomain proteins acts as a recognition helix both for DNA and protein interactions. Nucleic Acids Res 33:2661–2675 10.1093/nar/gki562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koizumi K, Lintas C, Nirenberg M, Maeng JS, Ju JH, Mack JW, Gruschus JM, Odenwald WF, Ferretti JA (2003) Mutations that affect the ability of the vnd/NK-2 homeoprotein to regulate gene expression: transgenic alterations and tertiary structure. Proc Natl Acad Sci USA 100:3119–3124 10.1073/pnas.0438043100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ju JH, Maeng JS, Zemedkun M, Ahronovitz N, Mack JW, Ferretti JA, Gelmann EP, Gruschus JM (2006) Physical and functional interactions between the prostate suppressor homeoprotein NKX3.1 and serum response factor. J Mol Biol 360:989–999 10.1016/j.jmb.2006.05.064 [DOI] [PubMed] [Google Scholar]
- 30.Zhao XX, Suzumori N, Yamaguchi M, Suzumori K (2005) Mutational analysis of the homeobox region of the human NOBOX gene in Japanese women who exhibit premature ovarian failure. Fertil Steril 83:1843–1844 10.1016/j.fertnstert.2004.12.031 [DOI] [PubMed] [Google Scholar]
- 31.Suzumori N, Pangas SA, Rajkovic A (2007) Candidate genes for premature ovarian failure. Curr Med Chem 14:353–357 10.2174/092986707779941087 [DOI] [PubMed] [Google Scholar]