Abstract
Lethal congenital contractural syndrome (LCCS) is a severe form of arthrogryposis. To date, two autosomal recessive forms of the disease (LCCS and LCCS2) have been described and mapped to chromosomes 9q34 and 12q13, respectively. We now describe a third LCCS phenotype (LCCS3)—similar to LCCS2 yet without neurogenic bladder. Using 10K single-nucleotide–polymorphism arrays, we mapped the disease-associated gene to 8.8 Mb on chromosome 19p13. Further analysis using microsatallite markers narrowed the locus to a 3.4-Mb region harboring 120 genes. Of these genes, 30 candidates were sequenced, which identified a single homozygous mutation in PIP5K1C. PIP5K1C encodes phosphatidylinositol-4-phosphate 5-kinase, type I, gamma (PIPKIγ), an enzyme that phophorylates phosphatidylinositol 4-phosphate to generate phosphatidylinositol-4,5-bisphosphate (PIP2). We demonstrate that the mutation causes substitution of aspartic acid with asparagine at amino acid 253 (D253N), abrogating the kinase activity of PIPKIγ. Thus, a defect in the phosphatidylinositol pathway leading to a decrease in synthesis of PIP2, a molecule active in endocytosis of synaptic vesicle proteins, culminates in lethal congenital arthrogryposis.
Arthrogryposis multiplex congenita (AMC) is a heterogeneous group of disorders characterized by congenital nonprogressive joint contractures. It has a worldwide incidence of 1 in 3,000, but it is more common in isolated populations, such as Finland1 and the Bedouin community in Israel. To date, four genetic loci associated with autosomal recessive AMC syndromes have been described. AMC, neurogenic type (AMCN [MIM 208100], on chromosome 5q35), described in a large Israeli-Arab inbred kindred, is a nonprogressive multiple joint contracture disease that is not lethal.2,3 Arthrogryposis–renal dysfunction–cholestasis syndrome (ARC [MIM 208085], caused by VPS33B mutations, on chromosome 15q26.14) is a neurogenic AMC, with renal tubular dysfunction and neonatal cholestasis, that leads to death during the 1st year of life.5–9 Lethal congenital contractural syndrome (LCCS [MIM 253310], on chromosome 9q3410), reported in Finland,11 is characterized by early fetal hydrops and akinesia, the Pena-Shokeir phenotype, fractures, multiple pterygia, and a specific neuropathology in the spinal cord.12,13 LCCS2 (MIM 607598) is prevalent in a large inbred Israeli-Bedouin kindred.14 The phenotype, which is lethal during the neonatal period in most cases, is characterized by multiple joint contractures, micrognathia, anterior horn atrophy in the spinal cord, and a unique feature of a markedly distended urinary bladder. Individuals affected with LCCS2 lack the hydrops, pterygia, and multiple fractures that are seen in LCCS. The LCCS2 phenotype suggests a spinal cord neuropathic etiology.14 We have recently mapped the LCCS2 locus to chromosome 12q13.15
We now describe a novel autosomal recessive LCCS (type 3 [LCCS3]), similar to LCCS2 yet lacking the urogenic bladder defect. We mapped the genetic defect leading to this syndrome to 3.4 Mb on chromosome 19p13 and show that LCCS3 results from a mutation in PIP5K1C (GenBank accession number NM_012398).
Patients and Methods
Patient Samples
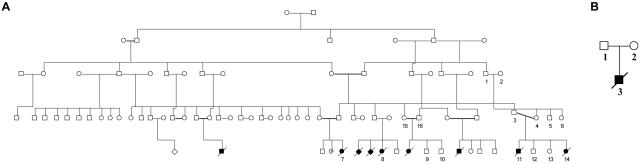
The pedigree of extended family A is shown in figure 1. Four affected and 23 unaffected individuals of the same consanguineous Bedouin tribe were subject to genetic analysis. Another unrelated Bedouin family (fig. 1B) presenting with an identical phenotype was also analyzed. Blood samples were obtained with informed consent after approval was received from the Soroka University Medical Center institutional review board. DNA was isolated from whole blood, tissue samples, or cultured cells, with use of the PUREGENE DNA Isolation Kit per the manufacturer’s instructions (Gentra Systems).
Figure 1. .
Israeli-Bedouin kindreds affected with LCCS3. Blackened and unblackened symbols represent affected and unaffected individuals, respectively. Numbers denote individuals whose DNA samples were analyzed. A, Pedigree of family A. B, Pedigree of family B.
Genetic Linkage Studies
Genomewide linkage analysis was undertaken on DNA samples of four affected individuals and two parents, with use of the Affymetrix GeneChip Human Mapping 10K Array Xba 142 2.0 according to the Affymetrix GeneChip Mapping Assay protocol at the biological services, Weizmann Institute of Science, Israel. GeneChip data analysis was done using the Genotyping Data Analysis Software to obtain SNP genotyping, genotype calls, marker orders, and map distance. An average of ∼11,500 SNPs for each person was genotyped, with an average SNP call rate of 99.1%. The results were organized in Excel software. Fine mapping was performed using microsatellite markers within the interval (listed in table 1) as follows: PCR products were separated on a 6% polyacrylamide gel and were visualized by silver staining.15 Haplotypes were manually constructed and analyzed. Microsatellite markers were selected from public genome databases (Marshfield, Généton, and deCODE Genetic maps). Novel microsatellite markers were planned using the Tandem Repeats Finder program and the University of California–Santa Cruz (UCSC) Genome Browser database and were designed using the Primer3 program. Two-point linkage analysis was performed using SUPERLINK software.16
Table 1. .
Genotyping of All Affected Individuals of Family A at the LCCS3 Chromosome 19 Locus[Note]
| Allele for Individual |
||||||||||
| Marker | Physical Map (kb) |
Marshfield Genetic Map (cM) |
7 | 8 | 11 | 14 | ||||
| D19S886 | 949,791 | .00 | 4 | 4 | 4 | 4 | 1 | 4 | 1 | 4 |
| D19S1166 | 3,026,947 | 9.84 | 2 | 4 | 4 | 4 | 4 | 4 | 1 | 4 |
| D19S247 | 3,091,202 | 9.84 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| D19S120 | 3,131,343 | 10.97 | … | … | … | … | 4 | 4 | 4 | 4 |
| D19S424 | 3,177,373 | … | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| rs2108389 | 3,542,590 | … | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Ch19_AAAT2 | 3,579,205 | … | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Ch19_GT | 3,669,061 | … | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Ch19_AAAG | 3,830,455 | … | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Ch19_CTTT | 4,025,911 | … | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D19S894 | 4,343,434 | 15.55 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| D19S216 | 4,900,376 | 20.01 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| D19S549 | 5,443,422 | 20.01 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| rs1384936 | 5,374,643 | … | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D19S177 | 5,468,626 | 20.75 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Ch19_ATTC | 5,747,132 | … | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| rs639251 | 5,913,530 | … | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| D19S1034 | 6,064,482 | 20.75 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D19S427 | 6,141,901 | 20.75 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Ch19_GAAT | 6,291,300 | … | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Ch19_CTAT | 6,431,574 | … | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 3 |
| rs2241393 | 6,636,304 | … | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| rs164022 | 6,779,040 | … | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Ch19_TTCC | 6,848,927 | … | 5 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| D19S905 | 7,576,090 | 25.17 | 4 | 2 | 6 | 1 | 2 | 3 | 2 | 3 |
| rs1862456 | 8,829,549 | … | 2 | 2 | 2 | 1 | 2 | 1 | 2 | 1 |
| D19S391 | 8,524,703 | 28.83 | 5 | 5 | 5 | 1 | 5 | 6 | 5 | 6 |
Note.— The shaded region indicates the area of the shared homozygous region. Microsatellite markers (ch19_AAAT2, ch19_GT, ch19_AAAG, ch19_CTTT, ch19_GAAT, ch19_CTAT, and ch19_TTCC) were designed using the Tandem Repeats Finder program and the UCSC Genome Browser database.
Sequence Analysis
Positional candidate genes were selected according to data in public databases (National Center for Biotechnology Information Map Viewer and UCSC Genome Browser). The coding sequences were amplified from genomic DNA or cDNA with PCR primers designed using the Primer3 program. RNA was extracted from cultured fibroblasts of affected individuals. PCR products were subject to agarose-gel electrophoresis and gel extraction (QIAGEN), followed by sequencing with either the forward or reverse primer on an ABI PRISM 377 DNA Sequencer (Applied Biosystems). Results were analyzed using ChromasPro software, and the obtained DNA sequences were compared with published sequences by use of BLAST. Primers used to amplify the coding sequence of PIP5K1C from cDNA of patients with LCCS3 are listed in table 2.
Table 2. .
Primers Used for Sequencing the cDNA of PIP5K1C
| Primer Sequence(5′→3′) |
||
| Primer | Forward | Reverse |
| PIP5K1C_CDS1 | GCCATGGAGCTGGAGGTA | CTTGAGGTGCATCTTGACCA |
| PIP5K1C_CDS2 | GCTGCTGCCCAAGTTCTATG | AGGTCGTACTGGGCCTCCT |
| PIP5K1C_CDS3 | TCATGAGCAACACGGTCTTT | GCCGGAGCAGAAGTGGAG |
Screening for the G757A Mutation in PIP5K1C
Genomic DNA flanking the G757A substitution was amplified (PIP5K1C-ex7F primer, 5′-GCA GAA CCT CAA CCA GAA CC-3′, and PIP5K1C-ex7R primer, 5′-GTC CTG CAT GAA GTC CAG GT-3′). The G757A mutation in the PIP5K1C gene abolishes a TaqI restriction site. Screening for the mutation was performed on the basis of differential restriction products.
Construction of Expression Plasmids
Full-length cDNA of normal and mutant PIP5K1C from cultured fibroblasts were PCR cloned into pGEX-4T-1 vector (Life Technologies), and the sequence was verified. Primers used for PCR amplification were 5′-AATTCATGGAGCTGGAGGTACCGG-3′ (forward) and 5′-TCGAGTTATGTGTCGCTCTCGCCGT-3′ (reverse).
Expression and Purification of the Recombinant GST-Fusion Protein
Proteins were expressed in Origami B (DE3) Escherichia coli (Novagen). In brief, overnight culture was diluted 1:100 and was grown to optical density OD600 ∼0.6. Protein expression was induced by the addition of 0.1 mM isopropyl β-d-1-thiogalactopyranoside at 15°C overnight. Cells were lysed (in 50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1 mg/ml lysozyme, 20 μg DNase I, and 0.1 M phenylmethylsulphonyl fluoride) at 4°C for 1 h and were sonicated. Proteins were purified with glutathione agarose beads (0.3 ml/liter of culture [Sigma]) for 1 h at 4°C and were eluted with 0.3 ml of 10-mM reduced glutathione (Sigma) in 50 mM Tris-HCl (pH 8.0).
PIP5K1C Kinase Activity Assay
The reaction was performed in a final volume of 50 μl kinase buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM ethylene glycol tetraacetic acid, 10 mM MgCl2, 80 μM phosphatidylinositol 4-phosphate (PI4P) (Sigma), 50 μM ATP, 10 μCi of [γ32P]ATP, and 0.4 μg of recombinant protein for 11 min at room temperature. The reaction was stopped by adding 100 μl of 1-M HCl. Lipids were extracted with 200 μl of chloroform/methanol (1:1) and were separated using thin-layer chromatography plates (Silica Gel 60 [Merck]) in chloroform/methanol/NH4OH/H2O (90:90:7:22). The labeled products were detected by autoradiography.17
Results
We describe a novel autosomal recessive LCCS, LCCS3, that is prevalent in a large, inbred Israeli-Bedouin kindred, with nine affected individuals identified to date (fig. 1A). The phenotype is similar to that of LCCS2 but lacks the urogenic bladder defect. The phenotype can be distinguished from the original LCCS by the absence of hydrops, fractures, and multiple pterygia.
Detailed Clinical Phenotype of LCCS3
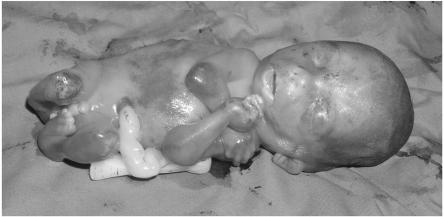
All affected newborns were small or borderline adequate for gestational age. The limbs showed severe multiple joint contractures with severe muscle wasting and atrophy, mainly in the legs. Prenatal sonographic examination and postnatal physical examination identified no additional abnormal findings in any of the affected subjects (fig. 2). Four of the nine gestations with affected embryos were terminated on the basis of ultrasonographic findings of fetal akinesia and fetal limb contractures. The other five subjects were born alive (two at 29 wk gestation and three at term) and died of respiratory insufficiency minutes to hours after birth. Karyotype, assayed for three of the subjects, was normal. None of the families agreed to postmortem examination.
Figure 2. .
LCCS3-affected fetus at 16 wk gestation
Homozygosity Mapping
Because the LCCS3 phenotype is similar to that of LCCS2, we first tested homozygosity of LCCS3-affected individuals at the LCCS2 locus. Linkage to the 12q13 LCCS2 locus was ruled out in the LCCS3-affected kindred—as was linkage to any of the three other loci associated with autosomal recessive arthrogryposis (AMCN at 5q35,2,3 ARC at 15q26.1,4 and LCCS at 9q3410) (data not shown).
To identify the locus harboring the disease-causing molecular defect, genomewide linkage analysis was performed. DNA samples of all four available affected individuals (7, 8, 11, and 14) of family A (fig. 1A) were tested using the Affymetrix 10K SNP arrays. We developed Excel-based software that allowed easy identification of regions of homozygosity; the results were organized according to the location of the markers on the chromosomes and were labeled according to their genotype (in table 3, italics indicate subjects homozygous for allele A, and bold type indicates those homozygous for allele B). A function in the Excel software was generated to calculate the length of the intervals of homozygosity and to score them according to the number of SNPs within each interval that shared homozygosity in all the affected individuals; the first homologous SNP got a score of 1, the second SNP got a score of 2, and so on (table 3). Regions that demonstrated a shared haplotype for at least two SNP markers in an interval of at least 1 Mb for all the affected individuals were further analyzed. Narrowing the number of the “suspected” regions was done by genotyping two of the parents (under the assumption that the disease-associated locus would show a shared homozygous haplotype in the affected individuals but not in their parents).
Table 3. .
Chromosome 19 Locus in LCCS3-Affected Family A—Linkage Analysis Results of the Affymetrix GeneChip SNP Arrays[Note]
| Haplotype |
||||||||||||
| Affected Individualsof Family A |
UnaffectedParents of Individuals11 and 14 |
Map Position |
||||||||||
| dbSNP | Positiona (bp) |
7 | 8 | 11 | 14 | 3 | 4 | Marshfield | Cytogenetic | No. of Homozygous SNPsb |
No. of SNPsc | Lengthd (kb) |
| rs2108389 | 3542590 | BB | BB | BB | BB | AB | BB | 12.15 | p13.3 | 1 | 1 | 0 |
| rs1384936 | 5374643 | AA | AA | AA | AA | AB | AB | 19.73 | p13.3 | 1 | 2 | 0 |
| rs639251 | 5913530 | BB | BB | BB | BB | BB | AB | 20.51 | p13.3 | 1 | 3 | 0 |
| rs2241393 | 6636304 | AA | AA | AA | AA | AA | AA | 21.99 | p13.3 | 1 | 4 | 0 |
| rs164022 | 6779040 | BB | BB | BB | BB | AB | BB | 22.34 | p13.3 | 1 | 5 | 8.83 |
| rs1862456 | 8829549 | BB | AB | AB | AB | AB | AA | 30.48 | p13.2 | 0 | 0 | 0 |
| rs2112527 | 9464751 | AB | AB | AA | AA | AA | AA | 32.66 | p13.2 | 0 | 0 | 0 |
| rs2009518 | 9660655 | BB | AB | AB | AB | BB | AB | 32.78 | p13.2 | 0 | 0 | 0 |
Note.— Alleles A and B are shown for each chromosome 19 SNP.
On the physical map.
SNPs that shared homozygosity in all the affected individuals are indicated as 1.
Score of the sequential homozygous SNPs (the number of SNPs in the homozygous block).
Length of the shared homozygosity interval.
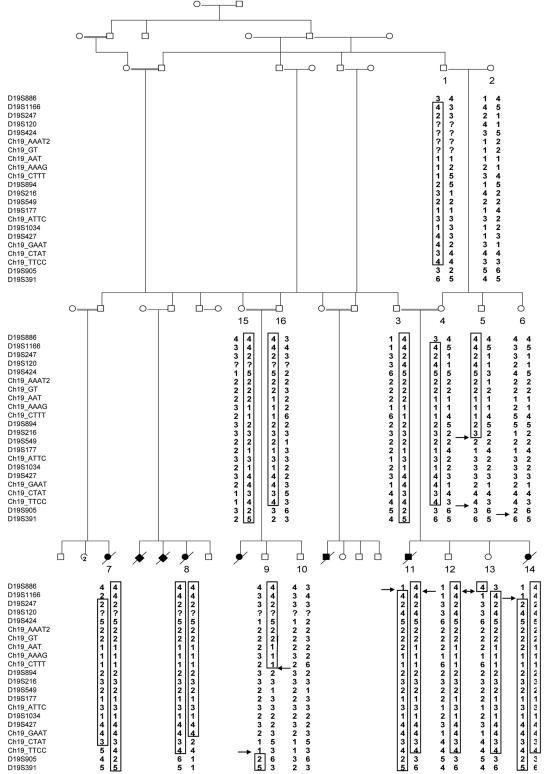
Approximately 260 regions of homozygosity (a minimum of two SNPs in a >1-Mb region) were found to be shared by all four affected individuals. Analysis of 10K SNP arrays of two obligate carrier parents (fig. 1, individuals 3 and 4 of family A, parents of individuals 11 and 14) reduced the number of “suspect” loci, demonstrating that only two of those regions were not homozygous in the obligate carriers (>2 SNPs in a >1-Mb region). One of these loci was excluded using microsatellite markers (data not shown). The second locus, on chromosome 19p13, contained five SNPs in an ∼8.8-Mb interval between the telomeric end and rs164022 (table 3). The interval of homozygosity was confirmed by testing DNA samples of all the affected individuals and 12 unaffected members of the extended family, with use of additional markers within the interval (D19S886, D19S1166, D19S247, D19S894, D19S549, D19S177, D19S427, and D19S905) (table 1). All the obligate carriers were shown to be heterozygous for the haplotype, whereas none of the unaffected family members was homozygous at this haplotype. Further detailed mapping of the LCCS3 locus was performed using 21 microsatellite markers. Thirteen microsatellite markers were selected from public genome databases (D19S886, D19S1166, D19S247, D19S120, D19S424, D19S894, D19S216, D19S549, D19S177, D19S1034, D19S427, D19S905, and D19S391) (table 1). Eight novel microsatellite markers were planned using the Tandem Repeats Finder program and the UCSC Genome Browser database (ch19_AAAT2, ch19_GT, ch19_AAAG, ch19_CTTT, ch19_ATTC, Ch19_GAAT, ch19_CTAT, and ch19_TTCC) (table 1). On the basis of this fine mapping, the LCCS3 locus was narrowed to 3.4 Mb on chromosome 19p13 between markers D19S1116 and ch19_CTAT in family A (fig. 3) and in a second, unrelated Bedouin family (family B). Two-point linkage analysis demonstrated a maximum LOD score of Zmax=3.9 at θ=0.0 (table 4).
Figure 3. .
Linkage results for chromosome 19 for a partial pedigree of family A. The haplotype showing the homozygosity region is boxed. Arrows indicate crossing-over events. The upper and lower boundaries were determined according to individuals 14 and 8, respectively.
Table 4. .
Two-Point LOD-Score Analysis of Family A for 12 Markers on Chromosome 19p13 with Use of SUPERLINK[Note]
| LOD at θ= |
||||||||
| Marker ID |
Marker Name | .00 | .01 | .05 | .10 | .20 | .30 | .40 |
| 1 | D19S886 | −5.185850 | −2.469052 | −1.816745 | −1.352973 | −.735673 | −.414711 | −.198239 |
| 2 | D19S1166 | −9.036289 | .117788 | 1.129059 | 1.282851 | 1.004234 | .558832 | .207434 |
| 3 | D19S247 | 3.155144 | 3.068566 | 2.724091 | 2.300226 | 1.495462 | .794720 | .273807 |
| 4 | D19S894 | 2.013806 | 1.973064 | 1.800441 | 1.570383 | 1.098154 | .658975 | .293541 |
| 5 | D19S216 | 1.727720 | 1.690233 | 1.538654 | 1.346473 | .962532 | .600143 | .279878 |
| 6 | D19S549 | .720431 | .695902 | .601303 | .491581 | .303699 | .160810 | .061703 |
| 7 | D19S177 | 3.122706 | 3.046224 | 2.738937 | 2.353873 | 1.596748 | .895484 | .325370 |
| 8 | D19S1034 | 3.303153 | 3.222378 | 2.896683 | 2.486550 | 1.677683 | .935451 | .350451 |
| 9 | D19S427 | 3.522624 | 3.436507 | 3.090021 | 2.654764 | 1.797075 | 1.008254 | .381538 |
| 10 | Ch19CTAT | 3.932675 | 3.840282 | 3.466661 | 2.992051 | 2.034242 | 1.128878 | .420811 |
| 11 | Ch19TTCC | −1.153815 | −.163561 | .373562 | .502131 | .455395 | .303296 | .144084 |
| 12 | D19S905 | −8.280259 | −2.263770 | −.386081 | .258759 | .556117 | .437770 | .194493 |
Note.— LOD scores >1.0 are shown in bold.
Selection of Candidate Genes in the LCCS3 Interval
Of ∼120 positional candidate genes in the 3.4-Mb LCCS3 locus, 30 were fully sequenced. Genes selected to be sequenced (table 5) were those previously known to be expressed in the spinal cord and to participate in neurogenesis or known to encode proteins that function in the neural system (such as SEMA6B, PTPRS, and NRTN). Another group of candidates were genes known to be associated with vesicle transport, such as SH3GL1, M6PRBP1, and PIP5K1C. No mutation was found in any of those genes, except PIP5K1C.
Table 5. .
Candidate Genes in the LCCS3 Interval[Note]
| Position (bp) |
Gene | Type of DNA Sequenceda |
| 3581182 | PIP5K1C | C |
| 3701771 | APBA3 | C |
| 3831672 | ATCAY | C |
| 3996217 | ZBTB7 | C |
| 4041322 | MAP2K2 | C |
| 4125106 | SIRT6 | C |
| 4198087 | Q7LE05 | C |
| 4229598 | NP_064594.2 | C |
| 4255668 | FSD1 | C |
| 4275041 | STAP2 | C |
| 4294551 | Q9Y2P1 | C |
| 4311368 | SH3GL1 | C |
| 4396009 | UBXD1 | C |
| 4493600 | SEMA6B | C |
| 4590530 | NP_689575.1 | C |
| 4608559 | CS010 | C |
| 4789354 | M6PRBP1 | C |
| 5157379 | PTPRS | C |
| 5406442 | ZNRF4 | C |
| 5509185 | PLAC2 | C |
| 5538010 | SAFB2 | C |
| 5629433 | NP_991330.1 | C |
| 5774818 | NRTN | C |
| 5781798 | FUT6 | G |
| 5793902 | FUT3 | G |
| 5817612 | FUT5 | G |
| 5865219 | CAPS | C |
| 5867158 | RANBP3 | C |
| 6163966 | MLLT1 | C |
| 6257725 | ASAH3 | C |
Note.— The candidate genes in the interval were arranged in the table by their location on the chromosome, according to the Map Viewer, build 36, and the UCSC Genome Browser.
G indicates that genomic DNA was sequenced. C indicates that cDNA was sequenced.
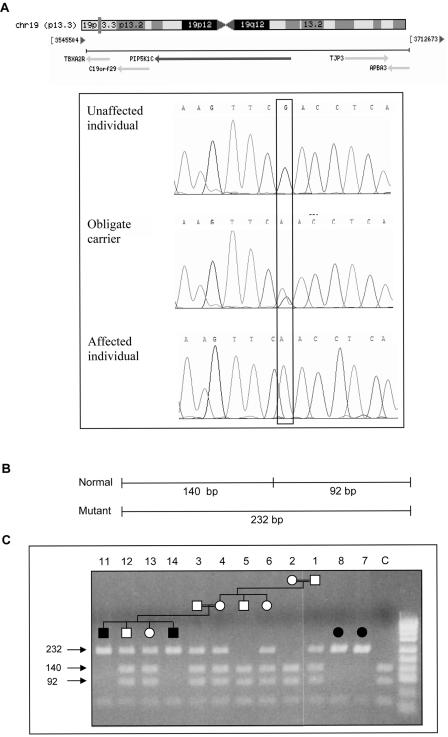
Identification of the PIP5K1C Mutation
A G757A missense mutation (fig. 4A) was identified in exon 7 of PIP5K1C, encoding phosphatidylinositol-4-phosphate 5-kinase, type I, gamma (PIPKIγ). PIPKIγ is an enzyme that phophorylates PI4P to generate phosphatidylinositol-4,5-bisphosphate (PIP2).18,19
Figure 4. .
The G757A mutation in exon 7 of PIP5K1C. A, Sequence analysis of the A→G substitution in an unaffected individual, obligate carrier, and affected individual. chr = Chromosome. B, PIP5K1C exon 7 mutation scanned with TaqI restriction enzyme, with schematic diagram illustrating the Taq1 fragments. C, Mutation analysis of family A members. PCR amplification products were digested with Taq1 restriction enzyme and were separated on 2% agarose gel. C = unrelated healthy control. Individuals 7, 8, 11, and 14 were homozygous for the mutated allele. Individuals 1, 3, 4, 6, 12, and 13 were heterozygous for the wild-type and mutated alleles. Individuals 2 and 5 were homozygous for the wild-type allele.
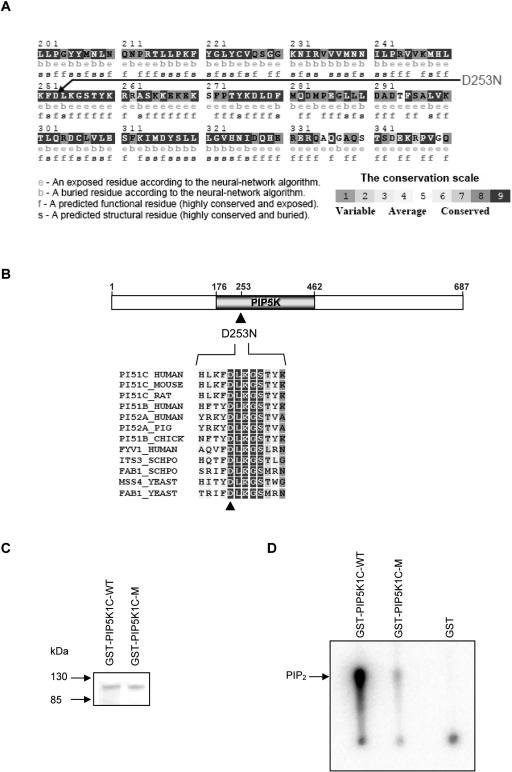
The G→A mutation in PIP5K1C abolishes a TaqI restriction site. Restriction-analysis screening for the mutation was performed for all available family members. The mutation segregated with the disease in 16 affected and unaffected individuals in extended families A and B (fig. 4B) and was not detected in 340 alleles of unrelated Bedouin individuals (data not shown). The mutation causes an aspartic acid→asparagine substitution at amino acid 253 (D253N). The aspartic acid at position 253 is conserved among other PIPK proteins and among species, from human to Saccharomyces cerevisiae, as can be seen in the ConSeq results (fig. 5A). The aspartic acid in position 253 is located in the PIPK domain within a conserved DLKGS motif (fig. 5B).
Figure 5. .
The D253N PIP5K1C mutation in LCCS3, which abolishes the PIP5K1C kinase activity. A, Prediction of the structural and functional importance of the aspartic acid residue at position 253, done using the ConSeq server. The ConSeq conservation grade was 9 (highly conserved). B, The aspartic acid at position 253, which is within a conserved DLKGS motif in the PIPK domain of the PIP5K1C protein. C, Purified wild-type (WT) and mutant (M) recombinant GST-PIP5K1C fusion proteins (SDS-PAGE). D, Kinase activity of wild-type (WT) and mutant (M) PIP5K1C recombinant proteins, tested in a reaction mixture containing PI4P (80 μM) as substrate and [γ32P]ATP. The labeled PIP2 product was separated using thin-layer chromatography and was quantified by phosphor-imaging analysis. PIP2 (mean±SD of three independent experiments) synthesized by mutant recombinant PIP5K1C was 1.7%±0.56% of PIP2 synthesized by equal amount of wild-type PIP5K1C. GST = glutathione S-transferase (negative control).
Kinase Activity of the Recombinant PIP5K1C Protein
To test whether the mutation had a functional effect on the kinase activity of the PIPKIγ protein, recombinant wild-type and mutant proteins were generated. Mutant and native full-length cDNA samples of PIP5K1C were RT-PCR amplified from fibroblasts of affected individuals and healthy controls, respectively. The cDNA samples were cloned into pGEX plasmids, and frame and sequence were verified. Recombinant proteins were generated and affinity purified. Kinase-activity assay was performed using PI4P as a substrate and [γ32P]ATP. The labeled PIP2 product was separated using thin-layer chromatography. Analysis of kinase activity of normal and mutant recombinant PIP5K1C proteins showed that the D253N mutation reduced the PIP5K1C kinase activity to barely detectable levels (fig. 5C). The mutation abrogates the kinase activity, regardless of varying substrate concentrations—in tests of the enzymatic activity of recombinant PIP5K1C at substrate (PI4P) concentrations of 20 μM versus 30, 50, 100, and 200 μM, the activity of the native enzyme was significantly augmented at higher substrate concentrations (nearly 1,000-fold at the highest PI4P concentration), whereas the enzymatic activity of the mutant PIP5K1C was unaltered and remained nearly undetectable (data not shown).
Discussion
We describe a novel arthrogryposis phenotype, LCCS3, that is identical to LCCS2 except for the absence of a neurogenic bladder. Genomewide linkage analysis of four affected individuals from three branches of a single inbred consanguineous kindred yielded ∼260 candidate loci. Interestingly, analysis of two obligate carrier parents of affected individuals was extremely effective in narrowing the number of candidate loci to two.
A homozygous PIP5K1C missense mutation (G757A) was found in LCCS3-affected individuals. PIP5K1C is a member of the type I PIPKIγ family of enzymes that phosphorylates the fifth hydroxyl of PI4P to generate PIP2. Although both PIP5K types I and II enzymes synthesize PIP2, type I PIP kinases account for the majority of PIP2 synthesis. The type I PIP5K family includes three catalytically active homologues: PIP5Kα, PIP5Kβ, and PIP5Kγ (also called “PIP5K1A,” “PIP5K1B,” and “PIP5K1C,” respectively). Of these three homologues, PIP5Kγ is thought to be the main enzyme responsible for PIP2 synthesis in the brain.20,21 The aspartic acid at position 253 of PIP5Kγ is located in the catalytic core domain (PIPK domain) within a DLKGS motif. Both the DLKGS motif and the aspartic acid in this motif are well conserved among other PIPK proteins and among species (from human to S. cerevisiae). Mutation of Asp216 in the DLKGS domain of PIPKIIβ was shown to abolish its activity.22 Similarly, we show that the Asp253Asn mutation found in the DLKGS domain of PIP5Kγ in LCCS3-affected patients significantly reduced the kinase activity of PIP5Kγ to barely detectable levels, regardless of substrate concentration. The human disease caused by a homozygous defect in PIP5K1C is reminiscent of the phenotype of the null mutants of its mouse orthologue; PIPKγ−/− mice die within the 1st day of life. Although no major developmental defects are observed in these mice, their impaired mobility and inability to feed after birth strongly suggest the occurrence of major neurological defects.20 Low PIP2 concentrations have been demonstrated in neurons of Pipkγ−/− mice,20 in line with the defective PIP2 synthesis we demonstrate in human LCCS3.
PIP2 plays an essential role in clathrin-dependent endocytosis of synaptic vesicle proteins: impairment of PIP2 hydrolysis results in the accumulation of clathrin-coated vesicles, suggesting that the membrane concentration of PIP2 controls the stability of endocytic clathrin coats.23 In yeast, PI3K-induced PIP2 depletion blocks endocytosis23; ex vivo, neurons of Pipkγ−/− mice demonstrate defects in synaptic transmission, endocytic delay, and changes suggestive of exocytic defects.20 Thus, it is plausible that the LCCS3 arthrogryposis phenotype caused by inactivation of PIP5K1C leading to low PIP2 levels results in defective synaptic vesicle trafficking, which incurs neuronal malfunction. It should also be noted that both PIP2 and syntaxin are essential for exocytosis and that efficient soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE)–dependent fusion requires the generation of PIP2.24,25 This is in line with the fact that ARC, another arthrogryposis syndrome, is caused by mutations in the VPS33B gene,4 which encodes a Sec1/Munc-18 protein belonging to a subfamily of class C vacuolar sorting proteins (VPS).4,26 VPS proteins are essential for intracellular membrane-fusion reactions and are associated with severe vacuolar protein–sorting and –morphology defects.27
We have shown28(in this issue) that LCCS2 is caused by a mutation in ERBB3. ERBB3, known to heterodimerize with ERBB2, is involved in activating the PI3K pathway, which leads to phosphorylation of PIP2 and to generation of phosphatidylinositol-3,4,5-triphosphate (PIP3).29–31 Since the LCCS2 and LCCS3 phenotypes are very similar, it is plausible that at least part of the defect seen in LCCS3 is caused by diminished synthesis of PIP3, as a result of reduced synthesis of its precursor, PIP2.
Acknowledgments
We sincerely thank the Kahn Family Foundation for Humanitarian Support for making this study possible.
Web Resources
The accession number and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/blast/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for PIP5K1C [accession number NM_012398])
- Map Viewer, http://www.ncbi.nlm.nih.gov/mapview/ (for build 36)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AMCN, ARC, LCCS, and LCCS2)
- Primer3, http://www.genome.wi.mit.edu/cgi-bin/primer/primer3.cgi (for primer design)
- SUPERLINK, http://bioinfo.cs.technion.ac.il/superlink/ (for two-point linkage analysis)
- UCSC Genome Browser, http://www.genome.ucsc.edu/
References
- 1.Pakkasjarvi N, Ritvanen A, Herva R, Peltonen L, Kestila M, Ignatius J (2006) Lethal congenital contracture syndrome (LCCS) and other lethal arthrogryposes in Finland—an epidemiological study. Am J Med Genet A 140:1834–1839 [DOI] [PubMed] [Google Scholar]
- 2.Shohat M, Lotan R, Magal N, Shohat T, Fischel-Ghodsian N, Rotter JI, Jaber L (1997) A gene for arthrogryposis multiplex congenita neuropathic type is linked to D5S394 on chromosome 5qter. Am J Hum Genet 61:1139–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanamy MG, Magal N, Halpern GJ, Jaber L, Shohat M (2001) Fine mapping places the gene for arthrogryposis multiplex congenita neuropathic type between D5S394 and D5S2069 on chromosome 5qter. Am J Med Genet 104:152–156 10.1002/ajmg.10030 [DOI] [PubMed] [Google Scholar]
- 4.Gissen P, Johnson CA, Morgan NV, Stapelbroek JM, Forshew T, Cooper WN, McKiernan PJ, Klomp LW, Morris AA, Wraith JE, et al (2004) Mutations in VPS33B, encoding a regulator of SNARE-dependent membrane fusion, cause arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome. Nat Genet 36:400–404 10.1038/ng1325 [DOI] [PubMed] [Google Scholar]
- 5.Denecke J, Zimmer KP, Kleta R, Koch HG, Rabe H, August C, Harms E (2000) [Arthrogryposis, renal tubular dysfunction, cholestasis (ARC) syndrome: case report and review of the literature]. Klin Padiatr 212:77–80 10.1055/s-2000-9656 [DOI] [PubMed] [Google Scholar]
- 6.Di Rocco M, Callea F, Pollice B, Faraci M, Campiani F, Borrone C (1995) Arthrogryposis, renal dysfunction and cholestasis syndrome: report of five patients from three Italian families. Eur J Pediatr 154:835–839 10.1007/BF01959793 [DOI] [PubMed] [Google Scholar]
- 7.Eastham KM, McKiernan PJ, Milford DV, Ramani P, Wyllie J, van’t Hoff W, Lynch SA, Morris AA (2001) ARC syndrome: an expanding range of phenotypes. Arch Dis Child 85:415–420 10.1136/adc.85.5.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horslen SP, Quarrell OW, Tanner MS (1994) Liver histology in the arthrogryposis multiplex congenita, renal dysfunction, and cholestasis (ARC) syndrome: report of three new cases and review. J Med Genet 31:62–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdullah MA, Al-Hasnan Z, Okamoto E, Abomelha AM (2000) Arthrogryposis, renal dysfunction and cholestasis syndrome. Saudi Med J 21:297–299 [PubMed] [Google Scholar]
- 10.Mäkelä-Bengs P, Järvinen N, Vuopala K, Suomalainen A, Ignatius J, Sipilä M, Herva R, Palotie A, Peltonen L (1998) Assignment of the disease locus for lethal congenital contracture syndrome to a restricted region of chromosome 9q34, by genome scan using five affected individuals. Am J Hum Genet 63:506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herva R, Leisti J, Kirkinen P, Seppanen U (1985) A lethal autosomal recessive syndrome of multiple congenital contractures. Am J Med Genet 20:431–439 10.1002/ajmg.1320200303 [DOI] [PubMed] [Google Scholar]
- 12.Vuopala K, Herva R (1994) Lethal congenital contracture syndrome: further delineation and genetic aspects. J Med Genet 31:521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vuopala K, Leisti J, Herva R (1994) Lethal arthrogryposis in Finland—a clinico-pathological study of 83 cases during thirteen years. Neuropediatrics 25:308–315 [DOI] [PubMed] [Google Scholar]
- 14.Landau D, Mishori-Dery A, Hershkovitz R, Narkis G, Elbedour K, Carmi R (2003) A new autosomal recessive congenital contractural syndrome in an Israeli Bedouin kindred. Am J Med Genet A 117:37–40 10.1002/ajmg.a.10894 [DOI] [PubMed] [Google Scholar]
- 15.Narkis G, Landau D, Manor E, Elbedour K, Tzemach A, Fishelson M, Geiger D, Ofir R, Carmi R, Birk OS (2004) Homozygosity mapping of lethal congenital contractural syndrome type 2 (LCCS2) to a 6 cM interval on chromosome 12q13. Am J Med Genet A 130:272–276 10.1002/ajmg.a.30266 [DOI] [PubMed] [Google Scholar]
- 16.Fishelson M, Geiger D (2002) Exact genetic linkage computations for general pedigrees. Bioinformatics Suppl 18:S189–S198 [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Li G, Sugita S (2005) A central kinase domain of type I phosphatidylinositol phosphate kinases is sufficient to prime exocytosis: isoform specificity and its underlying mechanism. J Biol Chem 280:16522–16527 10.1074/jbc.M413263200 [DOI] [PubMed] [Google Scholar]
- 18.Boronenkov IV, Anderson RA (1995) The sequence of phosphatidylinositol-4-phosphate 5-kinase defines a novel family of lipid kinases. J Biol Chem 270:2881–2884 10.1074/jbc.270.7.2881 [DOI] [PubMed] [Google Scholar]
- 19.Loijens JC, Anderson RA (1996) Type I phosphatidylinositol-4-phosphate 5-kinases are distinct members of this novel lipid kinase family. J Biol Chem 271:32937–32943 10.1074/jbc.271.51.32937 [DOI] [PubMed] [Google Scholar]
- 20.Di Paolo G, Moskowitz HS, Gipson K, Wenk MR, Voronov S, Obayashi M, Flavell R, Fitzsimonds RM, Ryan TA, De Camilli P (2004) Impaired PtdIns(4,5)P2 synthesis in nerve terminals produces defects in synaptic vesicle trafficking. Nature 431:415–422 10.1038/nature02896 [DOI] [PubMed] [Google Scholar]
- 21.Wenk MR, De Camilli P (2004) Protein-lipid interactions and phosphoinositide metabolism in membrane traffic: insights from vesicle recycling in nerve terminals. Proc Natl Acad Sci USA 101:8262–8269 10.1073/pnas.0401874101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson RA, Boronenkov IV, Doughman SD, Kunz J, Loijens JC (1999) Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J Biol Chem 274:9907–9910 10.1074/jbc.274.15.9907 [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez-Escudero I, Roelants FM, Thorner J, Nombela C, Molina M, Cid VJ (2005) Reconstitution of the mammalian PI3K/PTEN/Akt pathway in yeast. Biochem J 390:613–623 10.1042/BJ20050574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoyagi K, Sugaya T, Umeda M, Yamamoto S, Terakawa S, Takahashi M (2005) The activation of exocytotic sites by the formation of phosphatidylinositol 4,5-bisphosphate microdomains at syntaxin clusters. J Biol Chem 280:17346–17352 10.1074/jbc.M413307200 [DOI] [PubMed] [Google Scholar]
- 25.Vicogne J, Vollenweider D, Smith JR, Huang P, Frohman MA, Pessin JE (2006) Asymmetric phospholipid distribution drives in vitro reconstituted SNARE-dependent membrane fusion. Proc Natl Acad Sci USA 103:14761–14766 10.1073/pnas.0606881103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo B, Li L, Gissen P, Christensen H, McKiernan PJ, Ye C, Abdelhaleem M, Hayes JA, Williams MD, Chitayat D, et al (2005) Requirement of VPS33B, a member of the Sec1/Munc18 protein family, in megakaryocyte and platelet α-granule biogenesis. Blood 106:4159–4166 10.1182/blood-2005-04-1356 [DOI] [PubMed] [Google Scholar]
- 27.Carim L, Sumoy L, Andreu N, Estivill X, Escarceller M (2000) Cloning, mapping and expression analysis of VPS33B, the human orthologue of rat Vps33b. Cytogenet Cell Genet 89:92–95 10.1159/000015571 [DOI] [PubMed] [Google Scholar]
- 28.Narkis G, Ofir R, Manor E, Landau D, Elbedour K, Birk OS (2007) Lethal congenital contractural syndrome type 2 (LCCS2) is caused by a mutation in ERBB3 (Her3), a modulator of the PI3K/Akt pathway. Am J Hum Genet 81:589–595 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD (2001) Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem 70:535–602 10.1146/annurev.biochem.70.1.535 [DOI] [PubMed] [Google Scholar]
- 30.Halstead JR, Jalink K, Divecha N (2005) An emerging role for PtdIns(4,5)P2-mediated signalling in human disease. Trends Pharmacol Sci 26:654–660 10.1016/j.tips.2005.10.004 [DOI] [PubMed] [Google Scholar]
- 31.Hellyer NJ, Cheng K, Koland JG (1998) ErbB3 (HER3) interaction with the p85 regulatory subunit of phosphoinositide 3-kinase. Biochem J 333:757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]