Abstract
Mutations in the RMRP gene lead to a wide spectrum of autosomal recessive skeletal dysplasias, ranging from the milder phenotypes metaphyseal dysplasia without hypotrichosis and cartilage hair hypoplasia (CHH) to the severe anauxetic dysplasia (AD). This clinical spectrum includes different degrees of short stature, hair hypoplasia, defective erythrogenesis, and immunodeficiency. The RMRP gene encodes the untranslated RNA component of the mitochondrial RNA–processing ribonuclease, RNase MRP. We recently demonstrated that mutations may affect both messenger RNA (mRNA) and ribosomal RNA (rRNA) cleavage and thus cell-cycle regulation and protein synthesis. To investigate the genotype-phenotype correlation, we analyzed the position and the functional effect of 13 mutations in patients with variable features of the CHH-AD spectrum. Those at the end of the spectrum include a novel patient with anauxetic dysplasia who was compound heterozygous for the null mutation g.254_263delCTCAGCGCGG and the mutation g.195C→T, which was previously described in patients with milder phenotypes. Mapping of nucleotide conservation to the two-dimensional structure of the RMRP gene revealed that disease-causing mutations either affect evolutionarily conserved nucleotides or are likely to alter secondary structure through mispairing in stem regions. In vitro testing of RNase MRP multiprotein-specific mRNA and rRNA cleavage of different mutations revealed a strong correlation between the decrease in rRNA cleavage in ribosomal assembly and the degree of bone dysplasia, whereas reduced mRNA cleavage, and thus cell-cycle impairment, predicts the presence of hair hypoplasia, immunodeficiency, and hematological abnormalities and thus increased cancer risk.
The regulation of cell growth and division is not only essential for normal stature but is also linked to further conditions promoted by impaired cell-growth regulation, such as cancer. Mutations in the RMRP gene lead to a wide spectrum of autosomal recessive skeletal dysplasias with different degrees of short stature, but also variably including hypotrichosis, hematological abnormalities, immunodeficiency, and joint laxity. Although short stature with metaphyseal dysplasia is a hallmark of the whole spectrum—ranging from the milder phenotypes metaphyseal dysplasia without hypotrichosis (MDWH [MIM 250460]) and cartilage hair hypoplasia (CHH [MIM 250250]) to the severe anauxetic dysplasia (AD [MIM 607095])1—Hirschsprung disease, immunodeficiency, hematological abnormalities, and malignancies have been reported only in patients with CHH. The clinical classification is based mainly on the degree of short stature and radiographic characteristics, consisting of mild metaphyseal dysplasia in MDWH, moderate metaphyseal dysplasia in CHH, and severe spondyloepimetaphyseal dysplasia in AD. In contrast to AD, the vertebral bodies in MDWH and CHH are mildly affected, the pelvic appearance is normal, and the epiphyses are usually only mildly affected (cone-shaped phalangeal epiphyses in CHH). The growth plate in AD is severely distorted, with few disseminated chondrocytes, almost no columnization, and highly irregular osteochondral ossification.2 The lack of correlation between the severity of skeletal manifestations and the occurrence of extraskeletal anomalies is a striking observation that presently is without explanation.3
The RMRP gene is transcribed by RNA polymerase III and encodes the untranslated RNA component of the mitochondrial RNA–processing ribonuclease RNase MRP, which is involved in ribosome assembly and cell-cycle regulation.1,4–6 Various mutations have been described to date. Apart from the founder mutation g.70A→G, which is present in 92% of Finnish and 48% of non-Finnish patients with CHH, a total of 25 insertions or duplications between the TATA box and the transcription start site and >62 other mutations within the RMRP gene have been identified in patients with phenotypes in the CHH-AD spectrum (table 1).1,3,7–16 We recently showed the first evidence of a genotype-phenotype correlation by demonstrating that the CHH founder mutation affects ribosome assembly and cell-cycle regulation intermediately, whereas AD mutations severely affect ribosomal assembly only.1 To further investigate genotype-phenotype correlations, we now analyze the functional effect of 13 mutations in patients with variable features in the CHH-AD spectrum, including a novel patient with AD.
Table 1. .
Overview of Known RMRP Mutations, SNPs, and Other Variants[Note]
| RMRP Alteration | Nationality |
| RMRP mutation: | |
| g.−26_−5dupTACTACTCTGTGAAGCTGAGAA7 | American |
| g.−25_−11dupACTACTCTGTGAAGC7 | American |
| g.−25_−11tripACTACTCTGTGAAGC3,8,9 | Swiss, English, Turkish |
| g.−25_−10tripACTACTCTGTGAAGCT3 | Italian |
| g.−25_−6dupACTACTCTGTGAAGCTGAGA7 | Belgium |
| g.−25_−5dupACTACTCTGTGAAGCTGAGGA3 | German |
| g.−24_−15dupCTACTCTGTG7 | American |
| g.−23_−15dupTACTCTGTG3 | French |
| g.−23_−14dupTACTCTGTGA3,8,9 | Finnish, French |
| g.−23_−4dupTACTCTGTGAAGCTGAGGAC7 | German |
| g.−22_−10dupACTCTGTGAAGCT3 | French |
| g.−21_−1dupCTCTGTGAAGCTGAGGACGTG7 | American |
| g.−20_−14dupTCTGTGA3,7,8 | American |
| g.−20_−4dupTCTGTGAAGCTGAGGAC3,9–11 | English, Japan, Swiss |
| g.−16_−7dupTGAAGCTGAG8 | French |
| g.−16_−1insCTCTGTGAAGCTGAGG10 | Japan |
| g.−15_2dupGAAGCTGAGGACGTGGT10,11 | Japan |
| g.−14_−7dupAAGCTGAG8 | Mexican |
| g.−14_−3dupAAGCTGAGGACG12 | Swiss/Danish |
| g.−14_−1dupAAGCTGAGGACGTG8 | American |
| g.−13_−1dupAGCTGAGGACGTG8 | American |
| g.−8_−1dupAGGACGTG3 | Canadian |
| g.−7_3dupGGACGTGGTT8 | Brazilian |
| g.−7_−1insCCTGAG9 | Finnish, German |
| g.−4_6insGGACGTGGTT8 | American |
| g.4C→T3,8,13 | English, German, Spanish |
| g.9T→C7 | German |
| g.14G→T7 | American |
| g.14G→A1 | German |
| g.35C→T3 | French |
| g.40G→A3 | Dutch |
| g.45_53dupTGTTCCTCC3 | Dutch |
| g.57_64insTTCCGCCT8 | French |
| g.63C→T3,8,14 | Australian, Dutch |
| g.64T→C3 | Italian |
| g.64T→A15 | Chinese |
| g.70A→G3,7–9,12 | Variousa |
| g.79G→A8 | American |
| g.79G→T15 | Chinese |
| g.80G→A7 | American |
| g.89C→G7 | American |
| g.90_91AG→GC1 | German |
| g.91G→A7 | Belgium |
| g.93G→C3 | Dutch |
| g.92_93insA3 | Turkish |
| g.94_95delAG8,16 | Amish, English |
| g.97G→A3,7 | American |
| g.96_97dupTG3,8,9 | Canadian, Turkish |
| g.101C→T7 | Belgium |
| g.111_112insACTGTAGACATTCCT1 | Jordanian |
| g.116A→G7 | American |
| g.118A→G8 | German |
| g.124C→T7 | American |
| g.126C→T3,8 | Arabian, Italian |
| g.127G→A3 | Italian, Canadian |
| g.146G→A3,8 | Chinese, French |
| g.146G→C3 | Italian |
| g.152A→G8 | Canadian |
| g.154G→T8 | Finnish |
| g.168G→A10 | Japan |
| g.180G→A7,8 | Mexican, American |
| g.182G→C8 | Dutch, English |
| g.182G→A10,11 | Japan |
| g.182G→T3,7 | German |
| g.193G→A3,8,9 | English, Canadian |
| g.195C→T3,7,8,12 (present article) | Variousb |
| g.194_195insT7,14 | American |
| g.211C→G7,8 | Variousc |
| g.213C→G3,7 | German |
| g.214A→T8 | German |
| g.217C→T10 | Japan |
| g.218A→G10,11 | Japan |
| g.220T→C3 | German, Italian |
| g.230C→T8 | Polish |
| g.236A→G8 | American |
| g.238C→T3,8,12 | Variousd |
| g.242A→G3,8 | Canadian, Brazilian |
| g.243C→T8 | Canadian |
| g.244G→A3 | German |
| g.248C→T3 | Canadian |
| g.254C→G1 | German |
| g.254_263delCTCAGCGCGG (present article) | Spanish/Mexican |
| g.261C→T3,8 | Israeli, Trinidad |
| g.262G→C7 | German |
| g.262G→T8,9 | Finnish |
| g.264C→A8 | Austrian |
| RMRP polymorphism: | |
| g.−58T→C3,11,12 | … |
| g.−56A→G1,3,11,12 | … |
| g.−48C→A1,3,11,12 | … |
| g.−6G→A1,3,11,12 | … |
| g.127G→C3,7 | … |
| g.156G→C1,3,11,12 | … |
| g.177C→T1,3,11,12 | … |
| g.*7 (272)T→C7,12 | … |
| g.*9 (274)T→C1,3,11,12 | … |
| RMRP variant: | |
| g.−24C→G12 | … |
| g.36T→G11 | … |
| g.55_56InsC11 | … |
| g.57_58InsA12 | … |
| g.162C→T11 | … |
| g.172C→T11 | … |
| g.250C→T12 | … |
Note.— Mutations analyzed in the functional assays in this study are in bold font. Mutations observed in patients with AD are in italics.
Finnish, Swiss/Danish, American, Canadian, Dutch, Turkish, English, Amish, Australian, French, German, Brazilian.
Swiss/Danish, Israeli, Brazilian, Spanish/French.
Finnish, American, Polish, German.
Austrian, Israeli, American, Australian.
Material and Methods
This study was approved by the ethical review board of the medical faculty of the Friedrich-Alexander University of Erlangen-Nuremberg, Germany.
Novel Patient with AD
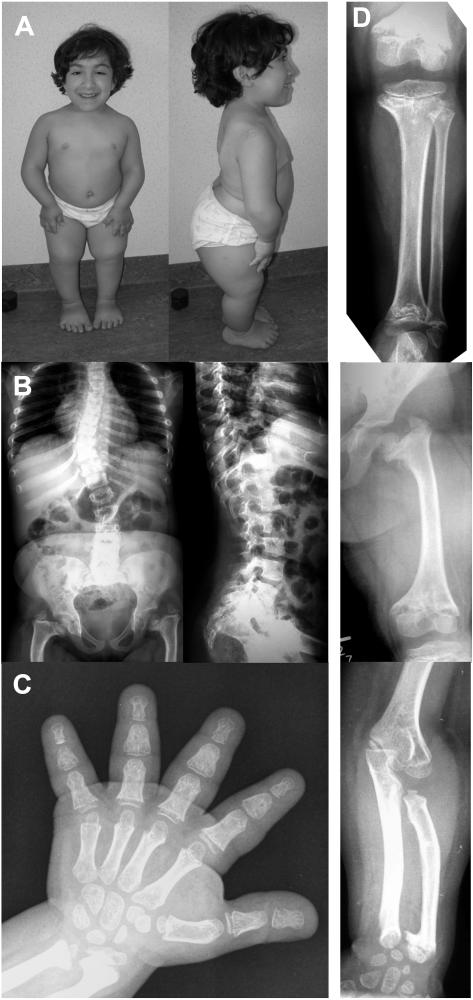
This 11-year-old girl (fig. 1a), the first child of nonconsanguineous, healthy parents, was born at 40.5 wk with a body length of 39 cm (<−5 SD), a weight of 3,000 g (<−1 SD), and a normal head circumference (mean for gestational age). Short limbs were noted on prenatal ultrasound at ∼17 wk of gestation. The postnatal course was uneventful. She had a normal psychomotor development. She underwent an adenoidectomy at age 7 years and, 1 year later, had a supernumerary tooth removed. Her major medical problem was related to her short stature and severe growth failure. She progressively developed scoliosis and joint pain in the lower limbs. At age 9 years, her height was 83 cm (−8 SD), weight was 16.6 kg (−4 SD), and head circumference was 52 cm (mean for age). Clinical evaluation at age 10 years revealed a normal face, normal hair, and disproportionate short stature, with short limbs and severe thoracolumbar scoliosis with right-sided gibbus. She was not able to fully extend her elbows but did show pronounced ligamentous laxity in the fingers and toes and also in her knees. Her hands and feet were very short.
Figure 1. .
Clinical and radiographic features of the novel patient with the AD phenotype. A, Female patient at age 10 years, showing severe disproportionate short stature. B and D, Radiographs of the axial and appendicular skeleton at age 8 years. The spine is abnormal, with thoracolumbar scoliosis and foreshortened vertebral bodies (on lateral film) showing anterior and posterior scalloping at the lumbar level. The end plates are slightly convex. The pelvis shows coxa vara with dysplastic femoral heads and severe shortening of the femoral necks. The femur, tibia, and fibula are short, with severe metaphyseal changes. The knee epiphyses are small and flattened. There is mesomelic shortening in the upper limbs. C, Hand radiograph taken at age 9 years. Metaphyseal changes are apparent in the wrists, with shortening of the distal ulna and ulnar kinking of the distal radius. The phalanges and metacarpals are short and broad (bullet-shaped middle phalanges), with small epiphyses already attached to the metaphyses.
Evaluation of the radiographs taken at age ∼8–9 years revealed a spondyloepimetaphyseal involvement. All tubular bones were shortened, with widened and irregular metaphyses and rather small epiphyses (fig. 1b). For most of these bones, the epiphyses were already attached to the corresponding metaphyses, indicating a premature fusion of the growth plate. The hands showed brachydactyly with short and broad phalanges and metacarpals (fig. 1c). The spine did not show platyspondyly but rather foreshortened vertebral bodies with posterior scalloping. The pelvis revealed severe coxa vara with short femoral necks and dysplastic capital femoral epiphyses.
After receiving informed consent, we obtained peripheral blood samples from her and her parents. Sequencing of the whole known promoter and transcript region of the RMRP gene was performed as described elsewhere1 and revealed two heterozygous mutations, g.195C→T and g.254_263delCTCAGCGCGG. The different parental origin of these mutations confirmed compound heterozygosity. Both mutations affect evolutionarily highly conserved regions and were absent in 378 control chromosomes. In addition, we identified the three known frequent SNPs: g.−58T→C and g.−48C→A, which were both heterozygous, and g.*9T→C, which was homozygous.
Phenotype Scoring
Since most mutations have been described in a compound heterozygous state with other mutations, we scored the phenotype of patients whose case was published with sufficient clinical data and who harbored at least 1 of the 13 mutations analyzed with respect to degree of bone dysplasia, hair hypoplasia, and additional features, such as immune deficiency and hematological abnormalities, and we calculated an average score for each trait in each mutation (table 2). Mild bone dysplasia was identified by the shortening of bones and recognizable metaphyseal changes; intermediate bone dysplasia was identified by shortened long bones associated with severe, pronounced metaphyseal changes and phalangeal epiphyseal (cone-shaped) changes. Additional involvement of the spine and severe epiphyseal changes were the hallmarks of severe bone dysplasia. Hair hypoplasia with thin, often lightly colored hair was scored as present or not. The classification of immunodeficiency and hematological abnormalities was scored as none when no signs of immunodeficiency (e.g., recurrent infections) were present and hematological parameters—for example, red and white blood cell counts—were within the normal age-related range, was scored mild when the patients showed recurrent infections or mild anemia and/or leukopenia that did not need treatment, was scored intermediate when severe immunodeficiency or severe anemia and/or leukopenia were present, and was scored severe when patients had severe immunodeficiency with opportunistic infections and severe anemia and/or leukopenia leading to bone marrow transplantation.
Table 2. .
Phenotype Scoring in Mutations Selected for Functional Studies[Note]
| Selected Mutation and Second Allele |
Bone Dysplasia | Hair Hypoplasia | Immunodeficiency and Hematological Abnormalities | Reference |
| g.195C→T: | ||||
| g.−14_−3dupAAGCTGAGGACG | 1 | 0 | 1 | Bonafe et al.12 |
| g.146G→C | 2 | 0 | 0 | Bonafe et al.3 |
| g.220T→C | 2 | 1 | 3 | Bonafe et al.3 |
| g.−14_−3dupAAGCTGAGGACG | 2 | 0 | 0 | Bonafe et al.3 |
| g.70A→G | 1 | 0 | 0 | Bonafe et al.3 |
| g.254_263delCTCAGGCGCGG | 3 | 0 | 0 | Present article |
| Mean score | 1.83 | .17 | .67 | |
| g.63C→T: | ||||
| g.70A→G | 2 | 0 | 2 | Ridanpaa et al.8 |
| g.195insT | 2 | 1 | 3 | Kuijpers et al.14 |
| g.4C→T | 1 | 1 | 0 | Bonafe et al.3 |
| g.70A→G | 2 | 0 | 0 | Bonafe et al.3 |
| g.−23_−14dupTACTCTGTGA | 1 | 0 | 1 | Bonafe et al.3 |
| g.70A→G | 1 | 0 | 1 | Bonafe et al.3 |
| Mean score | 1.5 | .33 | 1.17 | |
| g.70A→G: | ||||
| g.−23_−15dupTACTCTGTG | 2 | 0 | 1 | Bonafe et al.3 |
| g.−8_−1dupAGGACGTG | 1 | 1 | 2 | Bonafe et al.3 |
| g.96_97dupTG | 2 | 0 | 0 | Bonafe et al.3 |
| g.−20_−14dupTCTGTGA | 2 | 0 | 2 | Bonafe et al.3 |
| g.63C→T | 2 | 0 | 2 | Ridanpaa et al.8 |
| g.63C→T | 2 | 0 | 0 | Bonafe et al.3 |
| g.70A→G | 2 | 1 | 1 | Bonafe et al.3 |
| g.70A→G | 2 | 1 | 0 | Bonafe et al.3 |
| g.63C→T | 1 | 0 | 1 | Bonafe et al.3 |
| g.238C→T | 1 | 0 | 0 | Bonafe et al.3 |
| g.70A→G | 2 | 0 | 0 | Bonafe et al.3 |
| g.70A→G | 1 | 1 | 0 | Bonafe et al.3 |
| g.35C→T | 1 | 1 | 2 | Bonafe et al.3 |
| g.195C→T | 1 | 0 | 0 | Bonafe et al.3 |
| g.70A→G | 1 | 0 | 0 | Bonafe et al.3 |
| g.238C→T | 1 | 0 | 0 | Bonafe et al.3 |
| g.−20_−14dupTCTGTGA | 2 | 1 | 1 | Hermanns et al.7 |
| g.70A→G | 2 | 1 | 0 | Hermanns et al.7 |
| g.70A→G | 2 | 1 | 1 | Hermanns et al.7 |
| g.70A→G | 2 | 0 | 0 | Hermanns et al.7 |
| g.70A→G | 2 | 1 | 2 | Hermanns et al.7 |
| Mean score | 1.62 | .43 | .71 | |
| g.96_97dupTG: | ||||
| g.70A→G | 2 | 0 | 0 | Bonafe et al.3 |
| g.−25_−11tripACTACTCT | 2 | 0 | 2 | Bonafe et al.3 |
| Mean score | 2 | 0 | 1 | |
| g.126C→T: | ||||
| g.64T→C | 2 | 0 | 0 | Bonafe et al.3 |
| g.92_93insA | 1 | 0 | 0 | Bonafe et al.3 |
| g.126C→T | 2 | 0 | 0 | Ridanpaa et al.8 |
| Mean score | 1.67 | 0 | 0 | |
| g.146G→A: | ||||
| g.195C→T | 2 | 0 | 0 | Bonafe et al.3 |
| g.−22_−10dupACTCTGTGAAGCT | 1 | 1 | 2 | Bonafe et al.3 |
| g.146G→A | 1 | 0 | 0 | Ridanpaa et al.8 |
| Mean score | 1.33 | .33 | .67 | |
| g.4C→T: | ||||
| g.−25_−10tripACTACTCTGTGAAGCT | 1 | 0 | 1 | Bonafe et al.3 |
| g.−20_−4dupTCTGTGAAGCTGAGGAC | 1 | 0 | 3 | Bonafe et al.3 |
| g.220T→C | 2 | 0 | 1 | Bonafe et al.3 |
| g.63C→T | 1 | 1 | 0 | Bonafe et al.3 |
| g.238C→T | 1 | 0 | 2 | Bonafe et al.3 |
| g.−25_−11dupACTACTCTGTGAAGCTGAGAA | 1 | 0 | 2 | Munoz-Robles et al.13 |
| Mean score | 1.17 | .17 | 1.5 | |
| g.220T→C: | ||||
| g.195C→T | 2 | 1 | 3 | Bonafe et al.3 |
| g.4C→T | 2 | 0 | 1 | Bonafe et al.3 |
| Mean score | 2 | .5 | 2 | |
| g.248C→T: | ||||
| g.127G→A | 1 | 1 | 3 | Bonafe et al.3 |
| Score | 1 | 1 | 3 | |
| g.261C→T: | ||||
| g.261C→T | 2 | 0 | 0 | Bonafe et al.3 |
| Score | 2 | 0 | 0 |
Note.— For bone dysplasia, 1 = mild; 2 = intermediate; 3 = severe. For hair hypoplasia, 0 = absent; 1 = present. For immunodeficiency and hematological abnormalities, 0 = none; 1 = mild; 2 = intermediate; 3 = severe.
Multiple Sequence Alignment of the RMRP Gene Region
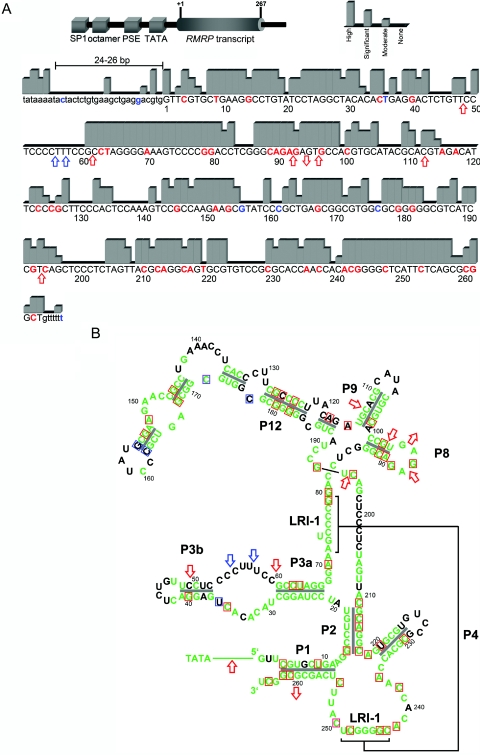
We analyzed evolutionary conservation by comparing the human RMRP gene against the orthologous sequences of mouse (Mus musculus), rat (Rattus norvegicus), rabbit (Oryctolagus cuniculus), dog (Canis familiaris), armadillo (Dasypus novemcinctus), elephant (Loxodonta africana), opossum (Monodelphis domestica), and pipid frog (Xenopus tropicalis) from the National Center for Biotechnology Information (NCBI) and the University of California–Santa Cruz (UCSC Genome Browser) databases, with use of the ClustalW Multiple Sequence Alignment algorithm (Baylor College of Medicine) and AlignX of the Vector NTI Suite 6.0 (InforMax). Evolutionary conservation was considered high when all species showed the same nucleotide, was significant when at least eight of nine species had the same nucleotide, and was moderate when more than six species had a conserved nucleotide at the position (fig. 2a). The degree of conservation and the position of sequence alterations were then transferred to the two-dimensional model of the RMRP transcript (fig. 2b).
Figure 2. .
A, Schematic drawing of RMRP (top) and degree of RMRP evolutionary conservation (bottom), categorized into four levels by sequence alignment of nine species. The length between the TATA box and the transcription start site is conserved at 24–26 bp. This distance is known to be important for proper binding of the RNA polymerase III transcription factor complex (TFIIIB) and is altered by insertions, duplications, and triplications, leading to reduced transcription.9,17,18 SP1 = SP1 binding site; octamer = octamer element; PSE = proximal sequence element. Nucleotides for which disease-related mutations have been published are in red; polymorphic sites are in blue. B, Mapping of evolutionary conservation of RMRP nucleotides to its two-dimensional structure.4 Green indicates moderately to highly conserved nucleotides. Nucleotides for which disease-related mutations have been published are boxed in red; polymorphic sites are boxed in blue. Red arrows indicate insertion or deletion mutations; blue arrows indicate insertion or deletion polymorphisms. Note that disease-related mutations, which do not affect conserved nucleotides, lead to mispairing in stem structures.
RNA Analysis
The ratio of the mRNA levels of the mutations g.254_263delCTCAGCGCGG and g.195C→T, detected in our novel patient, was analyzed by sequencing of the RT-PCR products from cDNA synthesized using Superscript II Reverse Transcriptase Kit with random hexamer primers (Invitrogen) from RNA extracted from a lymphoblastoid cell line of the patient.
Analysis of Mutant RMRP Constructs and Real-Time PCR Assays
To identify the influence of different mutations on ribosomal RNA (rRNA) and mRNA processing and the role of the involved RMRP domains, we selected 13 mutations distributed over the different base-pairing stem regions and RNA-protein–binding domains for which detailed clinical information was available. This included three AD mutations (g.111_112insACTGTAGACATTCCT, g.90_91AG→GC, and g.254C→G), nine mutations associated with milder phenotypes (g.63C→T, g.70A→G, g.96_97dupTG, g.126C→T, g.146G→A, g.4C→T, g.220T→C, g.248C→T, and g.261C→T), and the g.195C→T mutation, which occurred in our novel patient with AD but was previously described in association with CHH. After PCR amplification, the RMRP wild type was cloned into the pcDNA3.1 vector with the Directional TOPO Expression Kit vector (Invitrogen). Each mutation was inserted in a different RMRP wild-type clone by use of the Quik-Change site-directed mutagenesis kit (Strategene), in accordance with the manufacturer’s instructions.
Transient transfection of normal human fibroblasts with each of the different RMRP constructs was performed as described elsewhere.1 After RNA and DNA extraction with the TRIzol Reagent (Invitrogen), RMRP, CCNB2, 5.8S rRNA, and ITS-1–bound 5.8S rRNA expression levels were quantified by real-time PCR on a 7900HT (Applied Biosystems) with the QuantiTect Probe RT-PCR Kit (Qiagen). Results were normalized against the mean of four endogeneous controls (β-2-microglobulin, β-actin, hypoxanthine phosphoribosyltransferase 1, and RNA polymerase II). Relative cell count was measured by targeting albumin genomic DNA levels. The assay for each sample was performed in 384-well plates with a final volume of 20 μl each. Expression levels were calculated using the ΔΔCt method and were normalized to the expression levels in cells transfected with wild-type RMRP, which were set to 1. mRNA cleavage activity was calculated as the inverse relative increase of CCNB2 mRNA level. rRNA cleavage activity was expressed as the ratio of levels of cleaved to uncleaved 5.8S rRNA.
Results
Conservation of the RMRP Gene Region and Mutational Position
In most parts, conservation of the transcribed region was organized in blocks of high-to-significant conservation, predicting sites of base-pairing stems or RNA-DNA interaction (fig. 2a). Interestingly, two blocks of continuous high conservation comprise the regions 61–81 and 241–258, which provide a major tertiary interaction to form the so-called LRI-1 element (fig. 2a), the central core of the functional domain 1.19 The distance between the TATA box and the transcription start site, which is important for maintaining transcription levels, showed a conserved length of 24–26 bp. Of the 46 known putative pathogenetic single-nucleotide mutations, 33 were located in highly conserved regions, 8 within significantly conserved regions, and 2 in moderately conserved regions. In contrast, five putative polymorphisms within the RMRP transcript resided in not conserved or moderately conserved regions. Mapping of the conservation information to the two-dimensional structure of the RMRP transcript showed highly conserved stem and loop structures within the P1, P2, P3, P8, P9, and P12 domains (fig. 2b). Conservation is exceptionally high within regions of base-pairing stems and sites of proposed RNA-protein interaction; hence, mutations predominantly affect these regions. Four mutations (g.116A→G, g.118A→G, g.126C→T, and g.220T→C) were situated in evolutionarily nonconserved regions. Three of these mutations (g.118A→G, g.126C→T, and g.220T→C) have an impact on interaction with the corresponding nucleotide within conserved stem structures. The mutation at position 116, which also affects a nonconserved nucleotide, seems to not be part of a stem structure, but this position might be necessary for RNA-protein interaction.
Sequencing of the RMRP RT-PCR product of our novel patient showed absence of detectable levels of the mutation allele g.254_263delCTCAGCGCGG, whereas the g.195C→T allele was present (data not shown). The first of these mutations and the g.14G→A mutation, identified as compound heterozygous in another patient with AD,1 are likely to result in an unstable RNA and therefore represent null alleles. Hence, the mutations g.254_263delCTCAGCGCGG and g.14G→A were not analyzed in the functional assays. Mutations increasing the distance between the TATA box and the transcription start site, which are presumed to decrease the RMRP transcription level, were observed only as compound heterozygous in patients with CHH or MDWH.
Genotype-Phenotype Correlation
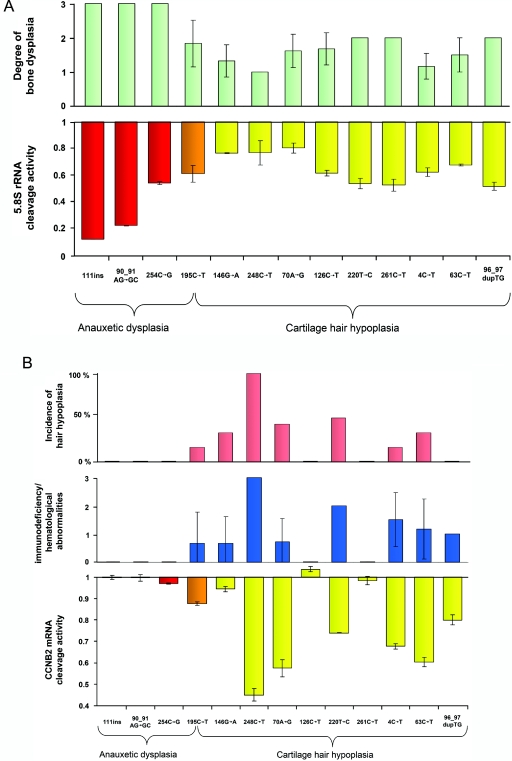
Comparison of phenotype scores and rRNA and mRNA cleavage activities revealed significant negative correlations between the degree of bone dysplasia and rRNA cleavage activity (correlation coefficient R=-0.8346; P=.0008), between the degree of immunodeficiency or hematological abnormalities and mRNA cleavage activity (R=-0.8429; P=.0007), and between the incidence of hair hypoplasia and mRNA cleavage activity (R=-0.8115; P=.001). Compound heterozygosity of mutations g.111_112insACTGTAGACATTCCT, g.90_91AG→GC, and g.254C→G results in AD that presents with the highest degree of bone dysplasia and without any additional obvious features, such as hair hypoplasia, immunodeficiency, or hematological abnormalities, in the affected patients (table 2 and fig. 3a and 3b). Mutations g.126C→T and g.261C→T were also associated, in most cases, with bone dysplasia only. No hair hypoplasia but bone dysplasia and different degrees of additional features were present in patients compound heterozygous for mutations g.4C→T, g.96_97dupTG, and g.146G→A. Mutations g.63C→T, g.70A→G, g.220T→C, and g.248C→T were identified as compound heterozygous in patients with bone dysplasia, hair hypoplasia, and different degrees of additional features. The mutation g.195C→T, identified as compound heterozygous with a null mutation in our novel patient with AD, was described as compound heterozygous with the mutations g.−20_−14dupTCTGTGA, g.−16_−7dupTGAAGCTGAG, g.−20_−4dupTCTGTGAAGCTGAGGAC, g.64T→C, g.146G→C, g.220T→C, and g.242A→G in patients with an intermediate level of bone dysplasia, mild hair hypoplasia, and mild additional features or who were given the classification of CHH without further details.3,7,8,12
Figure 3. .
Correlation of phenotype scores for mutations analyzed, with reference to degree of bone dysplasia, hair hypoplasia, and immunodeficiency/hematological abnormalities. A, Results of 5.8S rRNA cleavage activity normalized to wild-type activity as a function of ribosomal assembly in 13 mutations, compared with the respective skeletal phenotype score, revealing a significant negative correlation between degree of bone dysplasia and rRNA cleavage activity (R=-0.8346; P=.0008). rRNA cleavage was most affected by the AD mutations g.111_112insACGTAGACATTCCT and g.90_91AG→GC. An intermediate effect was observed for the AD mutation g.254C→G; mutation g.195C→T, identified in the new patient with AD as well as in patients with CHH and MDWH; and mutations g.126C→T, g.220T→C, g.261C→T, g.4C→T, g.63C→T, and g.96_97dupTG, leading to the milder phenotypes. The least effect was seen for g.146G→A, g.248C→T, and g.70A→G. Red columns represent AD mutations, yellow columns represent CHH and MDWH mutations, and orange columns represent AD, CHH, and MDWH mutations. B, Results of mRNA cleavage activity normalized to wild-type activity as a function of cell-cycle progression in 13 mutations, compared with the respective immunological/hematological phenotype score and the incidence of hair hypoplasia. Notably, the presence of impaired mRNA cleavage activity strongly correlates with the immunological/hematological phenotype and the likelihood of hair hypoplasia (R=-0.8429 [P=.0007] and R=-0.8115 [P=.001], respectively).
Cleavage Activity in Mutant RMRP Constructs
In accordance with our previous findings,1 human fibroblast cultures transiently transfected with the RMRP wild-type construct showed an increase in endonucleolytic cleavage activity at the ITS-1–5.8S rRNA junction site, whereas cells transfected with the mutant constructs presented with variably impaired cleavage activity represented by the decrease in the ratio of cleaved to uncleaved 5.8S rRNA when normalized to the wild-type transfected cells (fig. 3a). The decrease in cleavage activity was most pronounced in cells transfected with the AD mutations g.111_112insACGTAGACATTCCT and g.90_91AG→GC. Mutations g.254C→G and g.195C→T, which occurred as compound heterozygotes with g.90_91AG→GC and a null allele in patients with the severe skeletal dysplasia phenotype AD, led to a decrease in cleavage activity in the intermediate range. Only mild impairment was observed for the mutations g.148G→A, g.248C→T, and g.70A→G.
We also showed elsewhere that cells overexpressing RMRP wild-type constructs presented with a decrease in cyclin B2 mRNA correlating with an increase in B2 mRNA cleavage activity.1 The most significant impairment of cyclin B2 mRNA cleavage was seen for the mutations g.248C→T, g.70A→G, and g.63C→T (fig. 3b). The AD mutations g.111_112insACGTAGACATTCCT, g.90_91AG→GC, and g.254C→G, as well as the mutations g.126C→T and g.261C→T seen in milder phenotypes, showed no significant alteration compared with the wild type. The mutation g.195C→T, associated with AD in our novel patient but also with milder phenotypes in compound heterozygosity with other mutations, showed a mild degree of cyclin B2 mRNA cleavage activity.
Discussion
Mutations of the RMRP gene can be primarily classified into alterations within the distance between the TATA box and the transcription start site (insertions, duplications, and triplications) and mutations within the RMRP transcript. Although the first group reduces the RMRP transcription level and should therefore be considered hypomorphic alleles,9 the second group previously showed no obvious genotype-phenotype correlation. Because the RMRP gene is not translated into protein, even the distinction between rare variants and mutations has been difficult, especially since there was no strict correlation of affected nucleotides with evolutionary conservation among mammals.3 According to our alignment data in nine phylogenetically diverse species, all but four known pathogenic mutations are located within highly or moderately conserved regions. In contrast, the known common polymorphisms g.156G→C and g.177C→T are positioned within nonconserved areas. The same holds true for rare, probably not disease-related variants g.36T→G, g.55_56insC, g.57_58insA, and g.162C→T. The impact of the other rare variants, g.172C→T and g.250C→T—which affect a moderately and highly conserved nucleotide close to an important interaction site between 72–79 bp and 243–249 bp, respectively—remains unknown.
To gain further insight into the genotype-phenotype correlation, we mapped nucleotide conservation to the recently confirmed two-dimensional structure of the RMRP transcript.4,19 It became obvious that highly conserved regions are part of stem and loop regions assumed to be important for the two-dimensional structure and RNA-protein binding, respectively. g.118A→G, g.126C→T, and g.220T→C, three of the four mutations not affecting conserved nucleotides by sequence alignment, were part of stem regions with high conservation of the type of interacting bases that allow for base pairing in the secondary structure. Accordingly, 20 of 23 disease-causing mutations within these stem structures are G/C→A/T, or vice versa, substitutions. The transitions (C/G→G/C or A/T→T/A) g.89C→G, g.213C→G, and g.214A→T, for which no SNPs have been described yet, are part of RNA-protein–binding domains and they might therefore alter protein-binding capacities. The mutations g.14G→A1 and g.254_263delCTCAGCGCGG (the present article), both of which are not detectable by RT-PCR and thus apparently lead to mRNA instability, are located in the highly conserved P1 and P2 domains, which might also be important for the formation of the LRI-1 central core domain. The strikingly different phenotype for mutations in the P8 domain can be explained by the fact that, in contrast to the g.96_97dupTG mutation, the g.90_90AG→GC mutation not only changes the base-pair composition of the stem region but also affects the G of the GNRA tetraloop, the RNA-protein–binding site of the P8 domain.1 The mutation 96_97dupTG on the other side changes neither the stem region nor the GNRA tetraloop but might also influence the steric conformation of this loop, with only a milder effect on the phenotype. In contrast, the SNPs are located within nonconserved loop regions, affect stem regions in which both interacting nucleotides are nonconserved (g.156G→C and g.162C→T), or represent transitions within stem regions (g.127G→C, g.156G→C, and g.162C→T). Notably, transitions at positions 127, 182, and 262 represent SNPs, whereas substitutions G/C→A/T, or vice versa, have been associated with a disease phenotype.
To address the question of genotype-phenotype correlation with respect to functional impairment of the ribosomal assembly as a prerequisite for protein synthesis and cell-cycle progression, we analyzed the effect of 13 different mutations on the phenotypic expression in correlation with rRNA and mRNA processing. Whereas mutations causing the severe AD skeletal phenotype were not associated with nonskeletal features, mutations described in patients with the milder skeletal phenotypes showed variable additional features, such as hair hypoplasia, immunodeficiency, and hematological abnormalities (table 2 and fig. 3a and 3b).
Because our first results indicated a function of the RNAse MRP complex in cleavage of 5.8S rRNA, as well as cleavage of cyclin B2 mRNA, we measured the degree of impairment of specific rRNA and mRNA cleavage activity caused by the 13 chosen mutations. Our results showed diminished rRNA cleavage for all mutations, which was most pronounced in the AD mutations and was only mildly to intermediately affected by the CHH and MDWH mutations. The degree of decrease in rRNA cleavage thus strongly correlated with the degree of bone dysplasia, as ascertained by our phenotype scoring (fig. 3a). We therefore conclude that the impairment of rRNA cleavage by RMRP mutations is the leading cause of bone dysplasia in patients with features in the CHH-AD spectrum.
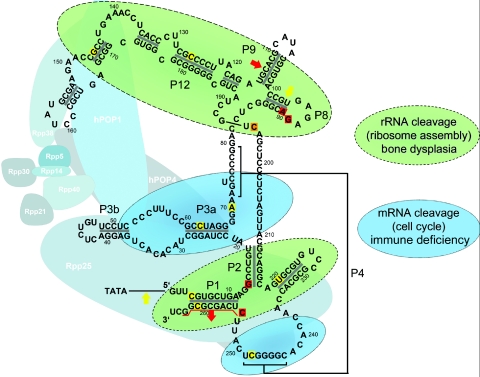
In comparisons of the cyclin B2 mRNA cleavage activity for the different mutations, a significant decrease in mRNA cleavage activity was observed only for mutations associated with the milder skeletal phenotypes. Because overexpression of cyclin B2 leads to accumulation of cells in late mitosis and contributes to chromosomal instability,20–22 we assumed that diminished mRNA cleavage was associated with susceptibility to cancer and proliferative bone marrow dysfunctions, such as immunodeficiency and anemia, as well as hair hypoplasia. Comparison of the mRNA cleavage activity with these features revealed that a decrease in mRNA cleavage activity indeed predicts the presence of immunodeficiency and hematological abnormalities and increases the likelihood of hair hypoplasia (fig. 3b). Our results also indicate a predominant participation of the domains P1, P2, and P19, as well as P8, P9, and P12, in the rRNA cleavage function, whereas parts of the P3 and P4 domains seem to be involved mainly in mRNA cleavage (fig. 4). This might suggest respective functions of the associated proteins of the RNAse MRP complex. On the basis of the observed role of the domains P1, P2, P19, P8, P9, and P12 in rRNA cleavage, the interacting proteins hPOP1 and Rpp25 could also be involved in rRNA cleavage, whereas the protein hPOP4 might be involved in mRNA cleavage. Hence, these proteins could be candidates for mutational screening, in as-yet-unexplained disorders of body and hair growth and bone marrow function, but further experimental support for this hypothesis is needed.
Figure 4. .
Two-dimensional structure of the RNase MRP complex. The RMRP gene encodes the untranslated RNA component of the mitochondrial RNA–processing ribonuclease. Nucleotides mutated in milder phenotypes are shaded yellow, those mutated in the severe skeletal phenotype are shaded red, and the mutation observed in both milder and severe skeletal phenotypes is shaded orange. Recently, it was shown that the proteins Rpp20, Rpp21, Rpp25, Rpp30, Rpp38, hPop1, and hPop4 interact directly with the RNA frame formed by the RMRP transcript, whereas hPop5 and Rpp14 do not.4,23,24 Our results also indicate a predominant participation of the P1, P2, and P19 domains, as well as the P8, P9, and P12 domains, in the rRNA cleavage function, whereas parts of the P3 and P4 domains seem to be involved mainly in mRNA cleavage.
However, the actual phenotype of patients with compound heterozygous mutations might be quite variable, depending on the functional impairment resulting from the respective combination of mutations. Accordingly, the novel patient with the severe AD skeletal phenotype was compound heterozygous for the null mutation g.254_263delCTCAGCGCGG and the moderately impairing mutation g.195C→T, which was previously described in patients with milder phenotypes. Those patients were compound heterozygous with insertions and/or duplications in the promoter region (g.−20_−14dupTCTGTGA, g.−16_−7dupTGAAGCTGAG, and g.−20_−4dupTCTGTGAAGCTGAGGAC) or single-nucleotide changes within the RMRP transcript (g.64T→C, g.146G→C, g.220T→C, and g.242A→G), leading to mild-to-moderate functional alterations only.3,12,7,8 Nevertheless, as in virtually every known genetic disorder, even the same RMRP genotype may be associated with different degrees of phenotypic manifestations in a subset of patients, probably depending on nonallelic modifiers.
In conclusion, we showed that the milder skeletal phenotypes are caused either by two mutations leading to mild-to-intermediate functional alteration of mRNA and rRNA cleavage or by compound heterozygosity with one of the mutations leading to mild-to-intermediate functional alteration and one allele reducing the transcription level by alterations within the distance between the TATA box and the transcription start site. In contrast, the more severe skeletal phenotype of AD is caused either by two mutations leading to intermediate-to-severe functional alteration of rRNA cleavage or by compound heterozygosity of the mutations leading to intermediate-to-severe functional alteration, with an allele leading to absence of the respective mRNA. We also found a strong correlation between the decrease in rRNA cleavage and the degree of bone dysplasia, whereas reduced mRNA cleavage and thus cell-cycle impairment predicts the presence of immunodeficiency, hematological abnormalities, and, thus, increased cancer risk, as well as increased likelihood of hair hypoplasia.
Acknowledgments
We thank the family for their kind cooperation. This work was supported by the Bundesministerium für Bildung und Forschung network grant “SKELNET” GFGM01141901 (to A. Rauch and A. Reis).
Web Resources
The URLs for data presented herein are as follows:
- ClustalW, http://www.ebi.ac.uk/clustalw/
- NCBI, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MDWH, CHH, and AD)
- UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway
References
- 1.Thiel CT, Horn D, Zabel B, Ekici AB, Salinas K, Gebhart E, Ruschendorf F, Sticht H, Spranger J, Muller D, et al (2005) Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am J Hum Genet 77:795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menger H, Mundlos S, Becker K, Spranger J, Zabel B (1996) An unknown spondylo-meta-epiphyseal dysplasia in sibs with extreme short stature. Am J Med Genet 63:80–83 [DOI] [PubMed] [Google Scholar]
- 3.Bonafe L, Dermitzakis ET, Unger S, Greenberg CR, Campos-Xavier BA, Zankl A, Ucla C, Antonarakis SE, Superti-Furga A, Reymond A (2005) Evolutionary comparison provides evidence for pathogenicity of RMRP mutations. PLoS Genet 1:e47 10.1371/journal.pgen.0010047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welting TJ, van Venrooij WJ, Pruijn GJ (2004) Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res 32:2138–2146 10.1093/nar/gkh539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hermanns P, Bertuch AA, Bertin TK, Dawson B, Schmitt ME, Shaw C, Zabel B, Lee B (2005) Consequences of mutations in the non-coding RMRP RNA in cartilage-hair hypoplasia. Hum Mol Genet 14:3723–3740 10.1093/hmg/ddi403 [DOI] [PubMed] [Google Scholar]
- 6.Martin AN, Li Y (2007) RNase MRP RNA and human genetic diseases. Cell Res 17:219–226 [DOI] [PubMed] [Google Scholar]
- 7.Hermanns P, Tran A, Munivez E, Carter S, Zabel B, Lee B, Leroy JG (2006) RMRP mutations in cartilage-hair hypoplasia. Am J Med Genet A 140:2121–2130 [DOI] [PubMed] [Google Scholar]
- 8.Ridanpaa M, Sistonen P, Rockas S, Rimoin DL, Makitie O, Kaitila I (2002) Worldwide mutation spectrum in cartilage-hair hypoplasia: ancient founder origin of the major70A→G mutation of the untranslated RMRP. Eur J Hum Genet 10:439–447 10.1038/sj.ejhg.5200824 [DOI] [PubMed] [Google Scholar]
- 9.Ridanpaa M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, et al (2001) Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 104:195–203 10.1016/S0092-8674(01)00205-7 [DOI] [PubMed] [Google Scholar]
- 10.Hirose Y, Nakashima E, Ohashi H, Mochizuki H, Bando Y, Ogata T, Adachi M, Toba E, Nishimura G, Ikegawa S (2006) Identification of novel RMRP mutations and specific founder haplotypes in Japanese patients with cartilage-hair hypoplasia. J Hum Genet 51:706–710 10.1007/s10038-006-0015-3 [DOI] [PubMed] [Google Scholar]
- 11.Nakashima E, Mabuchi A, Kashimada K, Onishi T, Zhang J, Ohashi H, Nishimura G, Ikegawa S (2003) RMRP mutations in Japanese patients with cartilage-hair hypoplasia. Am J Med Genet A 123:253–256 10.1002/ajmg.a.20281 [DOI] [PubMed] [Google Scholar]
- 12.Bonafe L, Schmitt K, Eich G, Giedion A, Superti-Furga A (2002) RMRP gene sequence analysis confirms a cartilage-hair hypoplasia variant with only skeletal manifestations and reveals a high density of single-nucleotide polymorphisms. Clin Genet 61:146–151 10.1034/j.1399-0004.2002.610210.x [DOI] [PubMed] [Google Scholar]
- 13.Munoz-Robles J, Allende LM, Clemente J, Calleja S, Varela P, Gonzalez L, de Pablos P, Paz E, Morales P (2006) A novel RMRP mutation in a Spanish patient with cartilage-hair hypoplasia. Immunobiology 211:753–757 10.1016/j.imbio.2006.05.001 [DOI] [PubMed] [Google Scholar]
- 14.Kuijpers TW, Ridanpaa M, Peters M, de Boer I, Vossen JM, Pals ST, Kaitila I, Hennekam RC (2003) Short-limbed dwarfism with bowing, combined immune deficiency, and late onset aplastic anaemia caused by novel mutations in the RMPR gene. J Med Genet 40:761–766 10.1136/jmg.40.10.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam AC, Chan DH, Tong TM, Tang MH, Lo SY, Lo IF, Lam ST (2006) Metaphyseal chondrodysplasia McKusick type in a Chinese fetus, caused by novel compound heterozygosity 64T>A and 79G>T in RMRP gene. Prenat Diagn 26:1018–1020 10.1002/pd.1547 [DOI] [PubMed] [Google Scholar]
- 16.McKusick VA, Eldridge R, Hostetler JA, Ruangwit U, Egeland JA (1965) Dwarfism in the Amish. II. Cartilage-hair hypoplasia. Bull Johns Hopkins Hosp 116:285–326 [PubMed] [Google Scholar]
- 17.Gunnery S, Mathews MB (1995) Functional mRNA can be generated by RNA polymerase III. Mol Cell Biol 15:3597–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schramm L, Hernandez N (2002) Recruitment of RNA polymerase III to its target promoters. Genes Dev 16:2593–2620 10.1101/gad.1018902 [DOI] [PubMed] [Google Scholar]
- 19.Schmitt ME (1999) Molecular modeling of the three-dimensional architecture of the RNA component of yeast RNase MRP. J Mol Biol 292:827–836 10.1006/jmbi.1999.3116 [DOI] [PubMed] [Google Scholar]
- 20.Cai T, Reilly TR, Cerio M, Schmitt ME (1999) Mutagenesis of SNM1, which encodes a protein component of the yeast RNase MRP, reveals a role for this ribonucleoprotein endoribonuclease in plasmid segregation. Mol Cell Biol 19:7857–7869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarafan-Vasseur N, Lamy A, Bourguignon J, Pessot FL, Hieter P, Sesboue R, Bastard C, Frebourg T, Flaman JM (2002) Overexpression of B-type cyclins alters chromosomal segregation. Oncogene 21:2051–2057 10.1038/sj.onc.1205257 [DOI] [PubMed] [Google Scholar]
- 22.Cai T, Aulds J, Gill T, Cerio M, Schmitt ME (2002) The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics 161:1029–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein DJ, Schmeing TM, Moore PB, Steitz TA (2001) The kink-turn: a new RNA secondary structure motif. EMBO J 20:4214–4221 10.1093/emboj/20.15.4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pluk H, van Eenennaam H, Rutjes SA, Pruijn GJ, van Venrooij WJ (1999) RNA-protein interactions in the human RNase MRP ribonucleoprotein complex. RNA 5:512–524 10.1017/S1355838299982079 [DOI] [PMC free article] [PubMed] [Google Scholar]