Abstract
Transcriptional mechanisms that govern cellular differentiation typically include sequence-specific DNA-binding proteins and chromatin-modifying activities. These regulatory factors are assumed necessary and sufficient to drive both divergent programs of proliferation and terminal differentiation. By contrast, potential contributions of the basal transcriptional apparatus to orchestrate cell-specific gene expression have been poorly explored. In order to probe alternative mechanisms that control differentiation, we have assessed the fate of the core promoter recognition complex, TFIID, during skeletal myogenesis. Here we report that differentiation of myoblast to myotubes involves the disruption of the canonical holo-TFIID and replacement by a novel TRF3/TAF3 (TBP-related factor 3/TATA-binding protein-associated factor 3) complex. This required switching of core promoter complexes provides organisms a simple yet effective means to selectively turn on one transcriptional program while silencing many others. Although this drastic but parsimonious transcriptional switch had previously escaped our attention, it may represent a more general mechanism for regulating cell type-specific terminal differentiation.
Keywords: TFIID, TAF3, TRF3, differentiation, myogenesis
Spatial and temporal patterns of transcription are exquisitely controlled during cellular differentiation as specific lineages become established throughout embryonic development of metazoan organisms. Within differentiated tissues, specialization and maintenance of function is achieved through expression of cell type-specific genes. The responsibility of directing cell- and gene-selective transcriptional programs has generally been attributed to the well-documented role of sequence-specific DNA-binding transcription factors. By contrast, the core promoter recognition apparatus, commonly referred to as the “basal machinery,” has largely been relegated to a supporting role that did not call for differential gene or cell type-selective functions. Indeed, many studies carried out in the past 20 years have found that the core machinery (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, and RNA polymerase II [Pol II]) remains largely invariant in different cell types (Thomas and Chiang 2006). This relatively myopic view of the core apparatus may result in part because most studies have used either unicellular organisms or only a handful of metazoan cell types (i.e., HeLa and Drosophila S2 cells). However, recent advances in both biochemical and molecular genetic tools have provided new opportunities to challenge this reasonable but unverified assumption. At the same time, the discovery of cell type-specific subunits and orthologs of core promoter recognition components suggests that the potential role of the core apparatus to drive cell type-specific programs of transcription may have been underappreciated (Thomas and Chiang 2006).
An early clue pointing to the differential function of the core machinery came from studies that identified cell type-specific TAF (TATA-binding protein-associated factor) subunits of the TFIID complex. The first of these cell type-specific subunits was TAFII105 (renamed TAF4b), the key component of an alternative TFIID complex essential in development of mouse ovaries and in spermatogenesis (Freiman et al. 2001; Falender et al. 2005). Similarly, in mouse testes, a TAF7 paralog (TAF7L) was reported to have specific roles in spermatid differentiation (Pointud et al. 2003). In Drosophila melanogaster, several testes-specific TAFs have been found to play a critical role during terminal differentiation of male germ cells (Hiller et al. 2004). In addition to cell type-specific TAFs, other TAFs have important roles in tissue development (Indra et al. 2005). TAF3 is a subunit of TFIID that was initially identified through its interaction with a developmentally regulated transcription factor Bric a brac 1, which is implicated in pattern formation and tissue morphogenesis in Drosophila (Pointud et al. 2001; Couderc et al. 2002). Although a TAF3 homolog has been identified in mouse and human (Gangloff et al. 2001), its involvement in mediating transcriptional programs during development remained to be elucidated.
In line with the discovery of cell type-specific TAFs, studies have also revealed the existence of TBP-related factors (TRFs) adding another piece of the puzzle to the metazoan core promoter recognition machinery. The original TRF (TRF1) in Drosophila has been implicated to function in both Pol II and Pol III transcription (Holmes and Tjian 2000; Takada et al. 2000). Another more distant relative of TBP (TRF2) was identified in a number of organisms (Moore et al. 1999; Ohbayashi et al. 1999a, b; Rabenstein et al. 1999; Teichmann et al. 1999). More recently, TRF3, the closest relative of TBP, was identified in zebrafish, frogs, and mammals, but not found in urochordates and lower eukaryotes such as flies and worms (Persengiev et al. 2003). Therefore, TRF3 appears to be a vertebrate-specific TRF that is broadly expressed from fish to human. Unlike TRF2, TRF3 has retained at least some ability to recognize and bind TATA elements (Bartfai et al. 2004; Jallow et al. 2004). Although there are striking similarities in protein sequence and structure between TBP and TRF3, analyses of various stages in zebrafish development showed complementary expression patterns during embryogenesis, suggesting potential functional divergence between these two related factors. Likewise, the expression of TRF3 in frog embryos depleted of TBP resulted in only a partial rescue providing further support for nonredundant functions of TBP and TRF3 (Bartfai et al. 2004; Jallow et al. 2004). Despite these early studies of TRF3, well-defined target genes and functional analyses of its potential role in development remained to be unveiled. In particular, functionally relevant TRF3-associated factors have not been identified. Throughout these previous studies, potential cell type-specific TAFs and TRFs were invariably found to coexist with the canonical TFIID complex, thus adding specificity but not replacing the core machinery.
In an attempt to gain more insight into the role of TAFs and TRFs during cellular differentiation, we have begun to dissect their potential functional specificity during myogenesis. In mice, the multistage muscle differentiation process involves the induction of proliferating myoblasts (determined muscle precursor cells) to terminally differentiate into myotubes that subsequently form the contractile myofibers of skeletal muscle (Pownall et al. 2002). This terminal differentiation event is characterized by an irreversible loss of proliferative capacity coupled with both selective gene silencing and gene activation events. The myoblast to myotube transformation requires a transcriptional program mediated by a cascade of cell type-specific basic helix–loop–helix (bHLH) activators (Myf5, MyoD, Myogenin, and Mrf4). These myogenic transcription factors, in combination with other transcriptional regulators, collectively establish the myogenic identity of cells and maintain the differentiated state critical to skeletal muscle formation (Berkes and Tapscott 2005; Sartorelli and Caretti 2005; Palacios and Puri 2006).
Here we have used a combination of model cell lines and bona fide mouse skeletal muscle cells to probe the potential differential requirements for TAF3 and the prototypic TFIID complex during myogenesis. In addition, through biochemistry as well as molecular genetic loss-of-function studies, we have begun to elucidate a mechanism that involves the apparent destruction of TFIID and replacement with a novel core promoter recognition apparatus containing both TAF3 and TRF3 that directs cell type-specific gene expression during muscle differentiation. Thus, vertebrates have apparently evolved the use of alternative core promoter complexes to establish cell-type specialization of skeletal muscle. Although unprecedented in animal cells, this mechanism is reminiscent of σ-factor switching in bacteria wherein alternative RNA polymerase holoenzymes are used to direct different programs of transcription during sporulation, heat shock, and phage infection (Gross et al. 1998).
Results
Differential expression of TAF3 and TFIID subunits in myotubes and skeletal muscle
To investigate the role of TFIID (including TAF3) during skeletal myogenesis, we utilized the myoblast cell line C2C12. This cell line has been determined to display an expression profile closely resembling that of primary myoblasts (Blais et al. 2005). In addition, this myoblast model system has also been used as a differentiation control for various TAFs (Perletti et al. 2001; Guermah et al. 2003). However, to date, none of these studies have measured the levels of TAF3 and importantly, all previous studies have used a mixed cell population consisting of myotubes and “undifferentiated” reserve cells. These reserve cells retain Myf5 expression even under differentiation conditions and have remained serum-responsive to re-enter the cell cycle as myoblasts (Kitzmann et al. 1998). In order to circumvent this potential complication of mixed cell types, we have taken specific precautions to isolate nearly homogenous (∼95%) populations of myotubes and reserve cells derived from differentiated C2C12 cells by taking advantage of the selective capacity of reserve cells to remain attached to plates upon trypsinization. By contrast, myotubes are largely nonadherent to cell culture plates, and therefore can be readily separated from reserve cells (Fig. 1A).
Figure 1.
TAF3 and TFIID subunits are differentially expressed in myotubes. (A) Phase-contrast images and DAPI/Phalloidin staining of myoblast, myotubes, and reserve cells from C2C12 cells. (B) Immunoblot analyses of whole-cell extracts from highly purified myoblasts, myotubes, and reserve cells for the expression of TAF3, canonical TFIID subunits, other general transcription factors, and muscle-specific transcription factors and proteins. Ponceau S staining was used to control for equal protein loading per sample. (C–E) Myoblasts, myotubes, and reserve cells were analyzed by immunofluorescence for TAF3, TAF4, TBP, and Pol II localization. Bar, 10 μm.
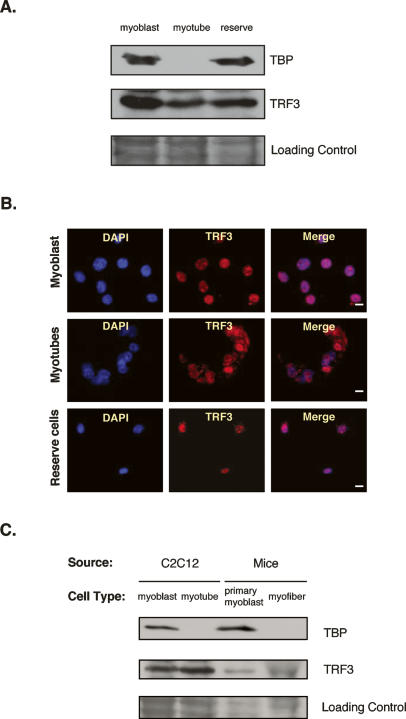
We have used these highly purified myotubes as well as myoblasts and reserve cells to analyze the expression levels of TAF3 and the other TFIID subunits by immunoblotting. As expected for dividing cells, myoblasts contain prototypic TFIID subunits as well as TAF3 (Fig. 1B). Surprisingly, we found that myotubes contain dramatically reduced transcript and protein levels of TAF1, TAF4, and TBP, three canonical subunits of TFIID (Fig. 1B; Supplementary Fig. 1). Throughout our studies, these three subunits were routinely tracked because TAF1 and TBP are TFIID-specific components while TAF4 is a core subunit essential for the stability of the complex (Wright et al. 2006). We also observed that TAF9 and TAF10 (subunits of TFIID as well as TFTC and STAGA), are dramatically reduced in myotubes (data not shown). Remarkably, we establish that myotubes may have essentially shed all of its TFIID. By contrast, TAF3 protein levels in differentiated myotubes remained high (Fig. 1B). The levels of other general transcription factors and RNA Pol II subunits remained unchanged during myoblast differentiation into myotubes (Fig. 1B). As a control, we monitored various myogenic transcription factors and other muscle-specific markers to gauge the efficiency of our myotube differentiation and isolation. Consistent with previous muscle differentiation patterns, we observed Myf5 expression in proliferating myoblast cells and reserve cells, while Myogenin and Myosin were both expressed in isolated myotubes but not myoblasts or reserve cells, confirming that we indeed had efficiently isolated myotubes (Fig. 1B).
Next, we carried out immunofluorescence studies to confirm and survey the differential levels of TAF3 and other TFIID subunits in intact myoblasts, myotubes, and reserve cells (Fig. 1C–E). Antibody staining of these cells revealed a barely detectable diffuse cellular background staining for TFIID subunits, TAF4, and TBP in myotubes, closely reflecting the loss of TFIID observed in immunoblot analyses. In contrast, TAF3 and Pol II showed a strong and distinct nuclear stain in myotubes (Fig. 1D). As expected, all these factors (TAF3, TAF4, TBP, and Pol II) strongly stained the nucleus of undifferentiated myoblasts and reserve cells (Fig. 1C,E). As a control, we confirmed the nuclear stain of Myf5 in myoblast and reserve cells while Myogenin stained myotubes consistent with their well-documented and differential expression patterns (Supplementary Fig. 1A). Thus, in contrast to previous studies that utilized the C2C12 cell line (Guermah et al. 2003), the isolation of highly pure populations of myotubes in combination with cell imaging, has uncovered a dramatic and possibly complete loss of key TFIID subunits (including TAF1, TAF4, and TBP) during differentiation of muscle cells with a concomitant and striking retention of TAF3 in differentiated myotubes (Fig. 1B,D). Taken together, these observations suggest that the very basic composition of the basal transcription machinery may be critically altered, with perhaps various core components (i.e., TFIID) even discarded as the cells transition into a post-mitotic state of terminal differentiation. Furthermore, these findings suggest that TAF3 may emerge to play a regulatory role involved in directing transcription in differentiated myotubes by possibly replacing the canonical TFIID complex.
Although C2C12 cells represent a useful model cell line to study certain stages of muscle development, there are limitations and differences relative to in vivo muscle differentiation (Pownall et al. 2002). In order to investigate the relevance of the observed differential expression of TAF3 versus TFIID made in C2C12 cells we analyzed various cell types within bona fide skeletal muscle. First, we isolated primary myoblast cells from newborn mice by a combination of enzymatic digestion and manual disaggregation of isolated limb muscle (Rando and Blau 1994). The primary myoblast cells were then analyzed for myogenic transcription factors by immunofluorescence. This analysis revealed that the expression pattern of various myogenic transcription factors in primary myoblast cells are strikingly similar to our observations of C2C12 reserve cells and myoblasts wherein both Myf5 and MyoD are expressed but not Myogenin (Supplementary Fig. 1B). These primary myoblast cells were also analyzed for TAF3, TAF4, TBP, and Pol II expression by immunofluorescence and found to express all of these factors in a nuclear staining pattern indistinguishable from those we observed in both C2C12 myoblasts and reserve cells (Fig. 2A).
Figure 2.
TAF3 and TFIID subunits are differentially expressed in skeletal muscle. (A,B) Primary myoblast cells and myofibers were analyzed by immunofluorescence for TAF3, TAF4, TBP, and Pol II staining patterns. Also included are the phase-contrast image and DAPI/Phalloidin staining of the isolated myofibers. Bar, 10 μm. (C) Immunoblot analyses of TAF3, TAF4, TBP, and RNA Pol II protein levels in primary myoblast cells and myofibers. As controls, the expression of these factors was compared with their expression profiles in C2C12 myoblast and myotube cells. Myf5 and Myosin expression in primary myoblast cells or myofibers were also analyzed as additional controls. Ponceau S staining was used as loading control.
Next, individual myofibers from adult mice were isolated through a series of enzymatic digestion and manual trituration steps (Rosenblatt et al. 1995). As a control, we found that myofibers express the appropriate myogenic factor Mrf4, a characteristic late-differentiation marker (Supplementary Fig. 2C). In addition, to eliminate the possibility that satellite cells were coisolated with the myofibers, we analyzed for Pax7 expression (Seale et al. 2000; Pownall et al. 2002) by immunofluorescence and found no Pax7-positive cells (data not shown). In contrast to primary myoblast cells, antibody staining of individually isolated myofibers revealed the virtually complete absence of TAF4 and TBP in the nucleus. As expected, a strong and distinct nuclear staining for TAF3 and Pol II was observed in these myofibers (Fig. 2B). Thus, the pattern of TAF3 levels in bona fide myofibers closely matches our findings in cell culture myotubes.
Protein levels determined by immunoblot of TAF3, TAF4, TBP, and Pol II in myofibers and primary myoblast cells were also compared with the levels found in C2C12 derived myoblasts and myotubes (Fig. 2C). This study confirmed that myofibers lack the conventional TFIID subunits while expressing high levels of TAF3 in a pattern indistinguishable from C2C12 derived myotubes and distinctly different from myoblasts, reserve cells, and primary myoblast cells. These results strongly suggest that loss of TFIID subunits is likely linked to post-mitotic events specific to terminally differentiated cell types such as myotubes/myofibers. Furthermore, in cell types that are actively dividing or have the capacity to re-enter the cell cycle (i.e., myoblasts and reserve cells), the canonical TFIID complex remains intact and presumably active. This suggests that loss of the holo-TFIID complex may be part of an important and dramatic regulatory switch in the transcription mode as myoblast cells undergo terminal differentiation.
Expression of a TRF in myotubes and myofibers
With the apparent loss of conventional TFIID in differentiated myotubes, TAF3 may switch into playing a primary role in the transcriptional regulation of muscle-specific gene expression. In order to potentiate this regulatory function, it is expected that TAF3 will need to be recruited to the core promoters of these genes. Because all of our attempts to detect any sequence-specific DNA-binding activity attributed to TAF3 have failed, we hypothesize that TAF3 alone likely has no intrinsic sequence-specific DNA-binding capacity or core promoter recognition properties, characteristic of some TAFs (Albright and Tjian 2000). On the other hand, in line with other prototypic TBP/TAF-containing complexes, TAF3 may work in conjunction with a partner or partners to carry out core promoter recognition of target genes. One possibility is that in the absence of TBP, as previously documented during embryonic stages in several model systems such as frogs and zebrafish, cell type-specific transcription may instead enlist the function of TRFs (Bartfai et al. 2004; Jallow et al. 2004). Thus, we set out to determine whether a TRF may help recruit TAF3 to muscle-specific genes. As an initial approach to test this idea, we focused our efforts on the expression profile of known TRFs. We and others have identified and characterized several TRFs, and these include TRF1, TRF2, and TRF3 (Crowley et al. 1993; Hansen et al. 1997; Ohbayashi et al. 1999a, b; Rabenstein et al. 1999; Persengiev et al. 2003). When compared with either TBP or TRF levels by quantitative PCR (Q-PCR) and immunoblot analyses of C2C12 cells we observed that TRF3 expression is strikingly similar to the expression profile of TAF3 in myotubes (Figs. 1B, 3A; data not shown).
Figure 3.
TRF3 Expression in terminally differentiated myotubes and myofibers. (A) Immunoblot analyses for TRF3 expression in myoblasts, myotubes, and reserve cell populations. (B) Representative cell immunofluorescence analyses of TRF3 localization in various C2C12 cell types. Bar, 10 μm. (C) Immunoblot analyses of TRF3 expression in isolated myofibers. Ponceau S was used to assess the total protein loaded per sample for immunoblot analyses.
To further confirm these observations, the localization of TRF3 was analyzed by immunofluorescence in C2C12 cells (Fig. 3B). This approach showed that TRF3, like TAF3, exhibits a strong and distinct nuclear stain in myotubes while TBP and other TFIID subunits have been lost in nuclei of myotube cells (Figs. 1D, 3B). Likewise, TRF3 was found in myofibers while no TBP and TAF4 subunits were detected (Figs. 2C, 3C). Thus, both TRF3 and TAF3 protein levels remain high in differentiated cell types such as myotubes and myofibers concomitant with a dramatic loss of TBP and TFIID. These observations suggest that in the absence of TBP and other conventional TFIID subunits, TAF3 together with TRF3, may functionally substitute for essential core promoter recognition activity in differentiated myotubes/myofibers to direct muscle-specific gene transcription.
TAF3 and TRF3 interaction in myotubes
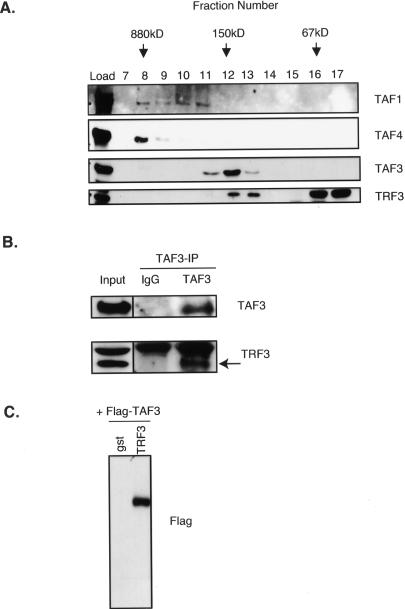
With the loss of conventional TFIID subunits in myotubes, it seemed possible that TAF3, perhaps in conjunction with TRF3, would functionally replace the canonical TFIID to help direct muscle gene transcription. To date, specific TRF3-associated factors have not been documented. Thus, in order to address the potential regulatory roles of TAF3 and TRF3 in myotubes, we first determined whether TAF3 might associate with TRF3 during C2C12 differentiation. To investigate this possibility we performed size exclusion chromatography analyses and found that the majority of TAF3 in differentiated cells elutes with a native molecular weight of ∼150–200 kDa, which is distinct from TFIID (∼800 kDa) (Fig. 4A). In these experiments, we also detected some level of TAF1 and TAF4, since we used a mixed population of differentiated C2C12 cells that contain both myotubes and reserve cells. These observations suggest that the atypical TAF3 subunit in muscle cells associates with other factors forming a significantly smaller complex than TFIID. Next, we determine whether TRF3 may fractionate with TAF3 by size exclusion chromatography. The majority of TRF3 elutes in fractions smaller than 67 kDa, but a significant portion of TRF3 coelutes with TAF3 at ∼150–200 kDa (Fig. 4A). The cofractionation of TAF3 and TRF3 suggests that these proteins may form a complex in myotubes. Indeed, the observed molecular weight of TAF3 and TRF3 is ∼180 kDa, consistent with the calculated molecular weight of a heterodimer between these two proteins. We also performed size exclusion chromatography with extracts derived from proliferating myoblasts and found a similar TAF3 and TRF3 elution profile to that observed in differentiated cells (data not shown). Next, we tested the possibility that TAF3 is in a complex with TRF3 in myotubes. We found that an antibody directed against TAF3 efficiently immunoprecipitates TRF3 from myotube extracts (Fig. 4B). A similar analysis using myoblasts extracts confirmed that TAF3 coimmunoprecipitates with TRF3 (data not shown). In addition, our in vitro binding reactions using recombinant proteins indicate a direct interaction between TAF3 and TRF3 (Fig. 4C). Taken together, these studies suggest that TAF3 and TRF3 can interact directly with each other to form a novel complex that is present in myotubes when the conventional TBP/TFIID complex becomes depleted. We therefore propose that a TAF3/TRF3 complex may play a significant functional role in the regulation of cell type-specific genes during muscle differentiation.
Figure 4.
TAF3 and TRF3 form a complex in differentiated myotubes. (A) Size exclusion chromatography of nuclear extracts prepared from differentiated C2C12 cells that contain both myotubes and reserve cell populations. Fractions were analyzed by immunoblot for TAF3 and TRF3. The representative molecular weights of protein standards are indicated in kilodaltons. (B) TAF3-specific antibody was used to coimmunoprecipitate associated proteins from myotube nuclear extracts and was subsequently analyzed by immunoblot. (C) In vitro binding assay using Glutathione Sepharose-bound gst or gst-TRF3- and Flag-TAF3-transfected SF9 protein extracts. Samples were analyzed by immunoblot.
Targeted depletion of TAF3 or TRF3 expression inhibits myoblast differentiation
To investigate the functional relevance of the TAF3 and TRF3 complex during myotube formation, we generated stable RNA interferance (RNAi) lines of both TAF3 and TRF3 in C2C12 cells. For specificity, we used a scrambled sequence to generate a stable RNAi control line in C2C12. Using these RNAi cell lines to knockdown TAF3 and TRF3, we analyzed several parameters including cell morphology and the expression patterns of myogenic transcription factors during myoblast to myotube differentiation to determine the potential effects of depleting these two factors.
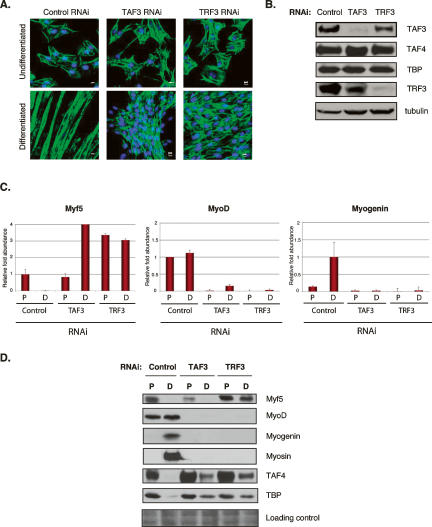
First, we observed that by knocking down TAF3 expression C2C12 cells exhibit a differentiation defective phenotype wherein cells partially align and fuse, but no normal myotubes are formed (Fig. 5A). Likewise, depletion of TRF3 by RNAi also blocks the ability of C2C12 myoblasts to form normal myotubes. Indeed, no obvious cell alignment or cell fusion events were observed in the TRF3 knockdown (Fig. 5A). The apparent differentiation defect resulting from the depletion of either TAF3 or TRF3 in the RNAi cells is likely not due to a failure to exit the cell cycle, since both cell lines exhibit the prototypic loss of cell cycle markers such as Cyclin B1 expression when induced to differentiate (data not shown). Also, these cell lines proliferate at a similar rate when compared with control RNAi cells and wild-type C2C12 cells (data not shown).
Figure 5.
TAF3 and TRF3 are required factors for myoblast differentiation. (A) Representative images of DAPI/Phalloidin staining using control, TAF3, and TRF3 RNAi cell lines grown under proliferation and differentiation conditions. (B) TAF3 and TRF3 RNAi myoblast cells were analyzed by immunoblot for TAF3, TAF4, TBP, and TRF3 protein levels. Anti-tubulin antibody was used as loading control. (C,D) Control, TAF3, and TRF3 RNAi cells grown under either proliferation (P) or differentiation (D) conditions, were analyzed by Q-PCR and immunoblot for Myf5, MyoD, and Myogenin levels. Each sample was normalized to the abundance of U6 RNA and presented as means ± SD from four independent reactions. In addition, the protein levels of TAF4 and TBP were also analyzed. Ponceau S stain was used to control for equal protein loading.
Next, we analyzed the protein levels of canonical TFIID subunits and found that the levels of TAF4 and TBP remained unchanged in myoblasts when either TAF3 or TRF3 were targeted for depletion (Fig. 5B). We did, however, observe a slight decrease in TRF3 levels in TAF3-depleted cells. Likewise, TRF3 depletion also resulted in a slight decrease in TAF3 levels, further supporting a functional and structural association between these factors. These observations suggest that TFIID subunit expression in these stable myoblast RNAi cell lines remain largely unaffected, and the complex presumably remains functional. Our findings reveal that reducing the levels of TAF3 and/or TRF3 in C2C12 cells results in a disruption of myoblast differentiation into myotubes in support of a critical functional role for these two transcription factors.
Since the depletion of TAF3 and TRF3 disrupted the process of myoblast differentiation into myotubes, we next determined the stage at which this differentiation block may have occurred by analyzing the expression patterns of key myogenic transcription factors. We observed that Myf5 is expressed in control and RNAi knockdown cells. On the other hand, MyoD, Myogenin and Myosin failed to be expressed in RNAi-treated cells (Fig. 5C,D). Consistent with previous studies (Rudnicki et al. 1992, 1993) we also observed that these cells show an increased level of Myf5 (Fig. 5C). Importantly, cell lines lacking TAF3 and TRF3 appear unable to express certain key regulators of myogenic differentiation, such as Myogenin, a transcription factor that specifies the differentiated state.
Interestingly, a significant and aberrant level of residual TAF4 and TBP proteins were detected even after treatment to induce differentiation in TAF3 and TRF3 RNAi knockdown cells (Fig. 5D). Although, the levels of TBP and TAF4 were reduced by comparison with the levels in proliferating stable TAF3 and TRF3 RNAi cells, it was intruiging to find that the loss of TAF3/TRF3 in cells grown under differentiation conditions can concomitantly lead to the persistent retention of TBP and TFIID that ordinarily would have been largely devoid of these canonical subunits. This observation suggests that the ectopic presence of TBP and TFIID in myotubes is no substitute for TAF3/TRF3 required to regulate the expression of genes driving muscle differentiation (i.e., Myogenin), providing further support that TAF3 and TRF3 likely play primary regulatory roles in muscle gene expression and are not redundant with TBP and TFIID.
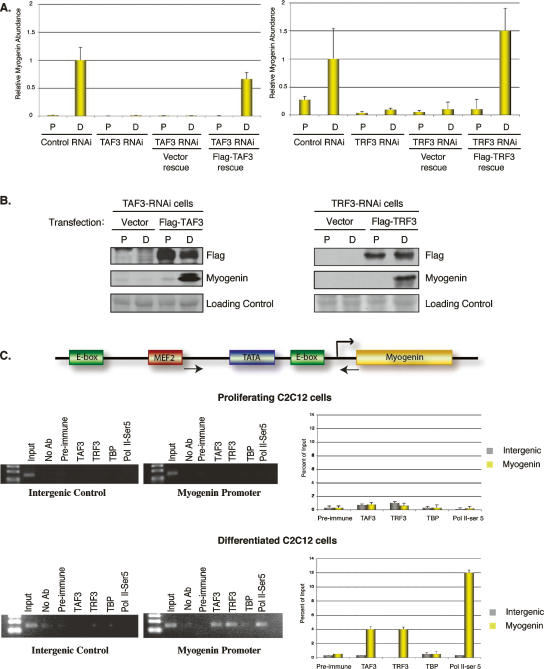
As a control, we confirmed that the differentiation block observed in both TAF3 and TRF3 RNAi cell lines is a direct and specific consequence of the targeted depletion of these two factors and are not due to off-target effects. We carried out rescue experiments by re-expressing TAF3 or TRF3 in the differentiation-deficient TAF3 or TRF3 RNAi cell lines. We found that the TAF3- and TRF3-rescued RNAi cells regained the ability to form myotubes (Supplementary Fig. 2A). In addition, we also observed that Myogenin expression is fully restored in both TAF3-transfected TAF3-RNAi cells and TRF3-transfected TRF3-RNAi cells (Fig. 6A,B).
Figure 6.
TAF3 and TRF3 are enriched on the myogenin promoter in differentiated C2C12 cells. (A) The rescue phenotype was analyzed by Q-PCR for the recovery of Myogenin expression in Flag-TAF3- or Flag-TRF3-rescued TAF3-RNAi or TRF3-RNAi cell lines. Samples were normalized to U6 RNA levels, and are presented as mean ± SD from four independent reactions. (B) The rescued RNAi cell lines grown in either proliferation (P) or differentiation (D) conditions were analyzed for endogenous levels of Myogenin by immunoblot. The expression levels of the tagged versions of TAF3 and TRF3 used in the rescue experiment were also analyzed. Ponceau S stain was used as loading control. (C) The mouse Myogenin promoter consists of a TATA box and is flanked by activator sites (MEF2 and E-box). Arrowheads correspond to the region spanning the Myogenin primer sequences used to analyze the ChIP. Intergenic primers were used to amplify a region ∼10 kb upstream of the Myogenin transcription start site. ChIP from either proliferating or differentiated C2C12 cells (after 3 d of induction) using the indicated antibodies. ChIP samples were also analyzed by Q-PCR using primers directed at either intergenic regions or the Myogenin core promoter region. Values were normalized to input and presented as percent of input. The data reflect the mean ± SD from four independent reactions.
Based on observations in wild-type cells and loss-of-function experiments, the TAF3/TRF3 complex appears to represent a key replacement factor complex for the prototypic TFIID that directs cell type-specific transcription of Myogenin during C2C12 differentiation. Thus, the Myogenin core promoter could be a direct regulatory target of both TAF3 and TRF3 during differentiation. To address this possibility, we analyzed the recruitment of these two transcription factors to the endogenous Myogenin promoter by chromatin immunoprecipitation (ChIP) using specific antibodies to TAF3 and TRF3. By surveying both intergenic regions and Myogenin-specific core promoter sequences combined with Q-PCR quantitation, we performed ChIPs from undifferentiated, early differentiated (after 24 h of induction) (data not shown) and late differentiated (after 72 h of induction) (Fig. 6C) C2C12 cells. These experiments revealed that TAF3 and TRF3 are both highly enriched at the Myogenin promoter only in differentiated samples but not at intergenic control sites or in undifferentiated cells (Fig. 6C). We observed some recruitment of TAF3 and TRF3 to the Myogenin promoter was observed as early as 24 h after inducing differentiation (data not shown). As a control, we also included Pol II Ser-5 and TBP ChIP in our analyses, and found that, as expected, Pol II Ser5 is efficiently recruited to the Myogenin promoter along with TAF3 and TRF3 in differentiated samples, but not in proliferating C2C12 cells (Fig. 6C). In contrast, no TBP enrichment was observed at the Myogenin promoter. Taken together, our loss-of-function, rescue experiments, and ChIP analyses all strongly point to an important and direct role of TAF3 and TRF3 during muscle differentiation.
Discussion
A combination of cell biology and genetic studies previously identified key myogenic bHLH DNA-binding transcription factors responsible for regulating the different stages of skeletal myogenesis. These myogenic transcription factors have, in turn, been implicated in the recruitment of chromatin-modifying complexes to target genes in the multistep process of orchestrating gene expression cascades during myogenesis (Berkes and Tapscott 2005; de la Serna et al. 2005). Importantly, myogenesis and other terminal differentiation programs will also likely require batteries of genes to be kept off, perhaps permanently. Thus, although sequence-specific activators/repressors, chromatin modifiers, and other epigenetic markings assuredly contribute to directing myoblast differentiation, it remained an open question whether other key control mechanisms may be required to govern new networks of transcription during cellular differentiation. Here, we report that an unexpected critical component for directing cell type specification involves dramatic changes to the composition of the core promoter-binding complex. Indeed, previous studies had pointed to the likely importance of cell type-specific subunits appended within prototypic multisubunit ensembles such as TFIID and BAF chromatin remodeling complexes (Freiman et al. 2001; Olave et al. 2002; Pointud et al. 2003; Hiller et al. 2004). However, there was little to suggest that initiation and maintenance of transcription programs during myoblast differentiation would be accompanied by shedding the prototypic holo-TFIID and wholesale replacement by an alternative core promoter recognition complex. This surprising observation may, however, help explain a previous report that in C2C12 cells, TAF4, and TBP levels are reduced as a result of targeted proteolytic degradation during myoblast differentiation (Perletti et al. 2001). Since we recently established that TAF4 is a linchpin subunit of TFIID required for stability of the complex (Wright et al. 2006), we hypothesize that the destruction of one or a few cornerstone subunits could be sufficient to severely compromise the stability of the entire TFIID complex and thus trigger the elimination of the holo complex that we observed. Our studies suggest that this unprecedented removal of functional TFIID may be a prerequisite step during differentiation of myoblasts into myotubes. Consistent with the notion that TFIID plays a role during cellular proliferation and cell cycle regulation (Wang and Tjian 1994; Martin et al. 1999; Metzger et al. 1999; Voss et al. 2000; Martianov et al. 2002; Mohan et al. 2003), we speculate that removing key general transcription factors such as TFIID could provide a direct and efficient mechanism for cells to irreversibly exit the cell cycle and enter into a terminally differentiated state. In short, the replacement of TFIID with an alternative core complex would provide a relatively parsimonious means to ensure stably shutting off many competing transcription programs (i.e., proliferation genes) while turning on a new battery of genes responsible for terminal differentiation (Fig. 7).
Figure 7.
Proposed model for transcription initiation in myoblast and myotube cells. The novel core transcription initiation complex composed of TAF3 and TRF3 functionally replaces the canonical TFIID complex during the differentiation of myoblasts into myotubes. This mechanism of switching the core transcription machinery may regulate the unique transcriptional program during cell type-specific terminal differentiation.
Importantly, we have discovered that the loss of TFIID and its replacement by the novel TAF3/TRF3 core promoter complex in myotubes is likely a multistep process that occurs as myoblast cells transition into a differentiated state. Intriguingly, TAF3/TRF3 complex coexists with the prototypic TFIID complex in myoblast cells. As differentiation ensues, the levels of TFIID diminish and virtually disappear altogether while TAF3/TRF3 complex persists and becomes dominant. Consistent with some requirement for TAF3/TRF3 in myoblasts, we observed compromised expression of key myogenic marker (i.e., MyoD) in both proliferating and early differentiated myoblasts upon depletion of TAF3 or TRF3. Furthermore, consistent with a mutually dependent switching of core promoter recognition complexes, we observed an aberrant retention of TAF4 and TBP levels in differentiation-defective muscle precursor cells depleted of TAF3 and TRF3. The recovery of myotube formation and Myogenin expression observed in TAF3 and TRF3 rescue experiments further strengthened the correlation between the loss of TFIID and its replacement by the TAF3/TRF3 complex during myotube formation.
In light of our current findings, it would be of interest in future studies to determine whether a similar switch in the core transcription machinery also occurs during terminal differentiation of other cell types. A hint that such switching of the core machinery may be more broadly linked to differentiation can be deduced from a recent study that reported a decrease in TAF4 protein levels when cortical neuronal stem cells were induced to differentiate (Brunkhorst et al. 2005). Another equally interesting question is whether other “prototypic” components of the preinitiation complex (i.e., CRSP/Mediator, SAGA, etc.) might also be switched or discarded during differentiation. In addition, since TBP also functions in RNA Pol I- and III-mediated transcription as part of SL1 and TFIIIB complexes, respectively (Kassavetis and Geiduschek 2006; Russell and Zomerdijk 2006), it may be revealing to determine the potential role of TRF3 and other TRFs in these complexes in differentiated myotubes. The observed colocalization of RNA Pol I and TRF3 within the nucleolus is intriguing, and hints at the possibility of TRF3 replacing TBP in other classes of promoter recognition assemblies (Yang et al. 2006). Furthermore, the relevance of an altered core promoter recognition complex in myotubes may also reveal interesting insights into how genes that must be expressed in both myoblasts and myotubes (i.e., housekeeping genes) are controlled. If the initiation and maintenance of a specific terminally differentiated state is most economically achieved by a complete overhaul of certain basal transcription apparatus, such a switching mechanism must also accommodate the necessary transcription of essential housekeeping genes, perhaps by using tandem core promoters. Thus, the diversification of both sequence-specific activators/repressors as well as core promoter recognition assemblies would significantly enhance the ability of metazoan organisms to accommodate tissue-specific patterns of gene expression by essentially adapting a more elaborate eukaryotic version of the well-established bacterial “σ-factor switching” model for reprogramming transcription during development and differentiation (Gross et al. 1998).
Materials and methods
Plasmids, cell culture, stable RNAi cell lines, and rescue cell lines
The full-length mouse TAF3 was cloned from NIH3T3 cDNA. The mouse TAF3 fragment was cloned into mammalian expression vector p3XFlag-CMV (Sigma). The full-length mouse TRF3 construct was obtained from Open Biosystems and subcloned into p3XFlag-CMV vector. The C2C12 cells were purchased from and maintained in recommended cell culture conditions from American Type Culture Collection. To generate the stable TAF3 cell line in C2C12 cells, oligonucleotide sequences encoding for short hairpin RNA to target TAF3 depletion were cloned into pSHAG-MAGIC2 vector (Paddison et al. 2004). The pSHAG-MAGIC2 constructs used to deplete TRF3 were purchased from Open Biosystems. The RNAi cell lines were selected and maintained in the same cell culture media containing puromycin (Sigma). TAF3 and TRF3 rescue cell lines were generated by re-expression of p3XFlag-CMV TAF3 or TRF3 into TAF3 RNAi cell lines and TRF3 RNAi cell lines, respectively. The transfected cells were then maintained in both puromycin and G418 drug selections. All the transfections in C2C12 cells were done using Lipofectamine transfection reagent (Invitrogen) following the manufacturer’s instructions.
Immunofluorescence and antibodies
Immunofluorescence analyses were performed following standard formaldehyde fixation conditions and antibody incubations. Samples used in immunofluorescence analyses were fixed in 3.7% formaldehyde solution. Samples were then permeabilized in 0.1% TX-100, 1× PBS solution. The blocked samples were incubated in primary antibody and Alexa 594-conjugated secondary antibodies (Molecular Probes). The samples were also stained with Alexa 488 phalloidin (Molecular Probes) to visualize cell morphology. The DNA was stained using DAPI (Molecular Probe). Samples were mounted and visualized in a 510 Meta Confocal Microscope (Zeiss).
TAF3 polyclonal antibodies for immunoblot and immunofluorescence experiments were prepared against amino acid residues 102–175 or 801–871 of the mouse TAF3 protein. TRF3 polyclonal antibody was generated against amino acid residues 24–122 of the mouse TRF3. Both TAF3 and TRF3 antibodies used for immunofluorescence were affinity-purified using MBP-tagged versions of the respective antigens. In addition, the following antibodies were used in this study: α-TAF4, α-MyoD, α-Myogenin (BD Bioscience Pharmingen); α-TBP (Biodesign International); α-TAF1, α-TFIIA, α-TFIIB, α-TFIIE, α-TFIIF, α-TFIIH, α-Myf5, α-Mrf4 (Santa Cruz Biotechnology); α-CD34 (Abcam); α-RNA Polymerase II Ser5 (Novus); α-Myogenin, α-tubulin (Developmental Studies Hybridoma Bank); α-Myosin Heavy Chain (Upstate Biotechnology Cell Signaling Solutions); and α-Flag (Sigma).
Nuclear extract preparations and whole-cell extract preparation
Nuclear extracts were prepared from roller bottles of C2C12 cells grown in proliferating conditions (with 10% FBS; Hyclone) or induced to differentiate with 10% horse sera (Invitrogen). Cells were harvested and washed, and nuclei were isolated. Nuclear extract was prepared using Buffer C (20 mM HEPES, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.5 M EDTA, 1 mM DTT, 1 mM sodium metabisulfite, protease inhibitor tablets [Roche]). Cells were homogenized and spun for 30 min at 15,000 rpm. Nuclear extract from homogenously isolated myotubes was prepared in a similar manner.
Nuclear extracts were used in size exclusion analyses (as mentioned in the text).
Whole-cell extracts were prepared in high-salt buffer (250 mM NaCl) with 0.5% NP-40.
Immunoprecipitation
Briefly, immunoprecipitation studies were performed using ∼200 μg of nuclear extracts and TAF3 antibody or preimmune sera that has been immobilized on Protein A Sepharose beads (Amersham). Samples were mixed overnight at 4°C. Samples were washed twice in NETN-240 = 20 mM HEPES 7.6, 240 mM NaCl, 0.5% TX-100, 0.1 mM EDTA, 1 mM DTT, and protease inhibitor cocktail, then washed twice in NETN with 500 mM NaCl and once in NETN-240.
In vitro binding assay
Recombinant gst and gst-TRF3 proteins were expressed and purified from bacteria and subsequently bound on Glutathione Sepharose 4B beads (Amersham). Bound proteins were washed in gst wash buffer 1 (100 mM Tris 8.0, 120 mM NaCl, 0.1% TX-100) and gst wash buffer 2 (gst wash buffer 1 containing 500 mM NaCl). Flag-TAF3 was expressed in SF9 cells following Gene Juice (Novagen) transfection conditions. Transfected SF9 cells were lysed in 50 mM Tris 7.4, 150 mM NaCl, 10% glycerol, and 0.5% Tween 20. To set up the binding reaction, 30 μL of bound gst and gst-TRF3 beads were incubated with 200 μL of Flag-TAF3 transfected SF9 lysate. The samples were incubated overnight at 4°C. Samples were washed using the following wash buffers: wash buffer 1 (50 mM Tris 7.4, 500 mM NaCl, 10% glycerol, 0.5% Tween 20) and wash buffer 2 (wash buffer 1 containing 1 M NaCl).
ChIP
Chromatin samples were isolated from both undifferentiated and differentiated C2C12 cells. Briefly, cells were cross-linked using a final concentration of 1% formaldehyde for 10 min. The cross-linking reaction was stopped by the addition of 1 M glycine. Cross-linked cells were washed in 1× PBS and collected. Cells were resuspended in cell lysis buffer (5 mM PIPES at pH 8, 85 mM KCl, 0.5% NP-40, protease inhibitor cocktail). Nuclei were then lysed in 50 mM Tris-HCl (pH 8.1), 10 mM EDTA, 1% SDS, and protease inhibitor cocktail (Roche). Isolated chromatin was then sonicated to an ∼500-bp fragment. Precleared chromatin was then used for ChIP using specific antibodies and blocked magnetic Protein A beads (Dynal). Samples were washed in buffer 1 (0.01% SDS, 1.1% TX-100, 1.2 mM EDTA, 16.7 mM Tris-HCl at pH 8.1, 167 mM NaCl) followed by another wash using buffer 2 (buffer 1 and 500 mM NaCl) for 1 h at 4°C. Following this, the samples were washed with buffer 3 (10 mM Tris-HCl at pH 8.0), 0.25 mM LiCl, 0.5% NP40, 0.5% deoxycholate, and 1 mM EDTA) and then 1× TE (pH 8.0). Precipitated chromatin complex was then treated with 50 mM Tris-HCl (pH 8.0), 25 mM EDTA, 1.25% SDS, 0.2 mg/mL RNase A, and 0.1 mg/mL proteinase K. Samples were incubated for 2 h at 45°C and temperature was increased to 67°C to reverse the cross-link overnight. The supernatant was collected and DNA was purified using the manufacturer’s instruction for QIAquick spin columns (Qiagen).
Q-PCR analyses
Total RNA was isolated from various cell lines mentioned in this study. Reverse transcription reaction was performed following the outlined instructions from a Cells-to-cDNA kit from Ambion. Reverse-transcribed samples were then subjected to real-time PCR using SYBR Green PCR master mix (Bio-Rad) and DNA Opticon 2 (MJ Research).
Acknowledgments
We thank M. Haggart and S. Zheng for assistance. We thank Si Wu for assistance in image processing. We also thank K. Wright, M. Marr, P. Hu, D. Rio, and R. Losick for discussions and review of the manuscript. We thank the Tjian laboratory members for valuable suggestions. M.D. is a predoctoral fellow of the California Institute for Regenerative Medicine (CIRM). R.T. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1583407
References
- Albright S.R., Tjian R., Tjian R. TAFs revisited: More data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- Bartfai R., Balduf C., Hilton T., Rathmann Y., Hadzhiev Y., Tora L., Orban L., Muller F., Balduf C., Hilton T., Rathmann Y., Hadzhiev Y., Tora L., Orban L., Muller F., Hilton T., Rathmann Y., Hadzhiev Y., Tora L., Orban L., Muller F., Rathmann Y., Hadzhiev Y., Tora L., Orban L., Muller F., Hadzhiev Y., Tora L., Orban L., Muller F., Tora L., Orban L., Muller F., Orban L., Muller F., Muller F. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr. Biol. 2004;14:593–598. doi: 10.1016/j.cub.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Berkes C.A., Tapscott S.J., Tapscott S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Blais A., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B.D., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B.D., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B.D., Sharan R., Kluger Y., Dynlacht B.D., Kluger Y., Dynlacht B.D., Dynlacht B.D. An initial blueprint for myogenic differentiation. Genes & Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkhorst A., Karlen M., Shi J., Mikolajczyk M., Nelson M.A., Metsis M., Hermanson O., Karlen M., Shi J., Mikolajczyk M., Nelson M.A., Metsis M., Hermanson O., Shi J., Mikolajczyk M., Nelson M.A., Metsis M., Hermanson O., Mikolajczyk M., Nelson M.A., Metsis M., Hermanson O., Nelson M.A., Metsis M., Hermanson O., Metsis M., Hermanson O., Hermanson O. A specific role for the TFIID subunit TAF4 and RanBPM in neural progenitor differentiation. Mol. Cell. Neurosci. 2005;29:250–258. doi: 10.1016/j.mcn.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Couderc J.L., Godt D., Zollman S., Chen J., Li M., Tiong S., Cramton S.E., Sahut-Barnola I., Laski F.A., Godt D., Zollman S., Chen J., Li M., Tiong S., Cramton S.E., Sahut-Barnola I., Laski F.A., Zollman S., Chen J., Li M., Tiong S., Cramton S.E., Sahut-Barnola I., Laski F.A., Chen J., Li M., Tiong S., Cramton S.E., Sahut-Barnola I., Laski F.A., Li M., Tiong S., Cramton S.E., Sahut-Barnola I., Laski F.A., Tiong S., Cramton S.E., Sahut-Barnola I., Laski F.A., Cramton S.E., Sahut-Barnola I., Laski F.A., Sahut-Barnola I., Laski F.A., Laski F.A. The bric a brac locus consists of two paralogous genes encoding BTB/POZ domain proteins and acts as a homeotic and morphogenetic regulator of imaginal development in Drosophila. Development. 2002;129:2419–2433. doi: 10.1242/dev.129.10.2419. [DOI] [PubMed] [Google Scholar]
- Crowley T.E., Hoey T., Liu J.K., Jan Y.N., Jan L.Y., Tjian R., Hoey T., Liu J.K., Jan Y.N., Jan L.Y., Tjian R., Liu J.K., Jan Y.N., Jan L.Y., Tjian R., Jan Y.N., Jan L.Y., Tjian R., Jan L.Y., Tjian R., Tjian R. A new factor related to TATA-binding protein has highly restricted expression patterns in Drosophila. Nature. 1993;361:557–561. doi: 10.1038/361557a0. [DOI] [PubMed] [Google Scholar]
- de la Serna I.L., Ohkawa Y., Berkes C.A., Bergstrom D.A., Dacwag C.S., Tapscott S.J., Imbalzano A.N., Ohkawa Y., Berkes C.A., Bergstrom D.A., Dacwag C.S., Tapscott S.J., Imbalzano A.N., Berkes C.A., Bergstrom D.A., Dacwag C.S., Tapscott S.J., Imbalzano A.N., Bergstrom D.A., Dacwag C.S., Tapscott S.J., Imbalzano A.N., Dacwag C.S., Tapscott S.J., Imbalzano A.N., Tapscott S.J., Imbalzano A.N., Imbalzano A.N. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falender A.E., Freiman R.N., Geles K.G., Lo K.C., Hwang K., Lamb D.J., Morris P.L., Tjian R., Richards J.S., Freiman R.N., Geles K.G., Lo K.C., Hwang K., Lamb D.J., Morris P.L., Tjian R., Richards J.S., Geles K.G., Lo K.C., Hwang K., Lamb D.J., Morris P.L., Tjian R., Richards J.S., Lo K.C., Hwang K., Lamb D.J., Morris P.L., Tjian R., Richards J.S., Hwang K., Lamb D.J., Morris P.L., Tjian R., Richards J.S., Lamb D.J., Morris P.L., Tjian R., Richards J.S., Morris P.L., Tjian R., Richards J.S., Tjian R., Richards J.S., Richards J.S. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes & Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman R.N., Albright S.R., Zheng S., Sha W.C., Hammer R.E., Tjian R., Albright S.R., Zheng S., Sha W.C., Hammer R.E., Tjian R., Zheng S., Sha W.C., Hammer R.E., Tjian R., Sha W.C., Hammer R.E., Tjian R., Hammer R.E., Tjian R., Tjian R. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- Gangloff Y.G., Pointud J.C., Thuault S., Carre L., Romier C., Muratoglu S., Brand M., Tora L., Couderc J.L., Davidson I., Pointud J.C., Thuault S., Carre L., Romier C., Muratoglu S., Brand M., Tora L., Couderc J.L., Davidson I., Thuault S., Carre L., Romier C., Muratoglu S., Brand M., Tora L., Couderc J.L., Davidson I., Carre L., Romier C., Muratoglu S., Brand M., Tora L., Couderc J.L., Davidson I., Romier C., Muratoglu S., Brand M., Tora L., Couderc J.L., Davidson I., Muratoglu S., Brand M., Tora L., Couderc J.L., Davidson I., Brand M., Tora L., Couderc J.L., Davidson I., Tora L., Couderc J.L., Davidson I., Couderc J.L., Davidson I., Davidson I. The TFIID components human TAFII140 and Drosophila BIP2 (TAFII155) are novel metazoan homologues of yeast TAFII47 containing a histone fold and a PHD finger. Mol. Cell. Biol. 2001;21:5109–5121. doi: 10.1128/MCB.21.15.5109-5121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C.A., Chan C., Dombroski A., Gruber T., Sharp M., Tupy J., Young B., Chan C., Dombroski A., Gruber T., Sharp M., Tupy J., Young B., Dombroski A., Gruber T., Sharp M., Tupy J., Young B., Gruber T., Sharp M., Tupy J., Young B., Sharp M., Tupy J., Young B., Tupy J., Young B., Young B. The functional and regulatory roles of σ factors in transcription. Cold Spring Harb. Symp. Quant. Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- Guermah M., Ge K., Chiang C.M., Roeder R.G., Ge K., Chiang C.M., Roeder R.G., Chiang C.M., Roeder R.G., Roeder R.G. The TBN protein, which is essential for early embryonic mouse development, is an inducible TAFII implicated in adipogenesis. Mol. Cell. 2003;12:991–1001. doi: 10.1016/s1097-2765(03)00396-4. [DOI] [PubMed] [Google Scholar]
- Hansen S.K., Takada S., Jacobson R.H., Lis J.T., Tjian R., Takada S., Jacobson R.H., Lis J.T., Tjian R., Jacobson R.H., Lis J.T., Tjian R., Lis J.T., Tjian R., Tjian R. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell. 1997;91:71–83. doi: 10.1016/s0092-8674(01)80010-6. [DOI] [PubMed] [Google Scholar]
- Hiller M., Chen X., Pringle M.J., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T.Y., Marino S., Fuller M.T., Chen X., Pringle M.J., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T.Y., Marino S., Fuller M.T., Pringle M.J., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T.Y., Marino S., Fuller M.T., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T.Y., Marino S., Fuller M.T., Sancak Y., Viswanathan S., Bolival B., Lin T.Y., Marino S., Fuller M.T., Viswanathan S., Bolival B., Lin T.Y., Marino S., Fuller M.T., Bolival B., Lin T.Y., Marino S., Fuller M.T., Lin T.Y., Marino S., Fuller M.T., Marino S., Fuller M.T., Fuller M.T. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- Holmes M.C., Tjian R., Tjian R. Promoter-selective properties of the TBP-related factor TRF1. Science. 2000;288:867–870. doi: 10.1126/science.288.5467.867. [DOI] [PubMed] [Google Scholar]
- Indra A.K., Mohan W.S., Frontini M., Scheer E., Messaddeq N., Metzger D., Tora L., Mohan W.S., Frontini M., Scheer E., Messaddeq N., Metzger D., Tora L., Frontini M., Scheer E., Messaddeq N., Metzger D., Tora L., Scheer E., Messaddeq N., Metzger D., Tora L., Messaddeq N., Metzger D., Tora L., Metzger D., Tora L., Tora L. TAF10 is required for the establishment of skin barrier function in foetal, but not in adult mouse epidermis. Dev. Biol. 2005;285:28–37. doi: 10.1016/j.ydbio.2005.05.043. [DOI] [PubMed] [Google Scholar]
- Jallow Z., Jacobi U.G., Weeks D.L., Dawid I.B., Veenstra G.J., Jacobi U.G., Weeks D.L., Dawid I.B., Veenstra G.J., Weeks D.L., Dawid I.B., Veenstra G.J., Dawid I.B., Veenstra G.J., Veenstra G.J. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc. Natl. Acad. Sci. 2004;101:13525–13530. doi: 10.1073/pnas.0405536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G.A., Geiduschek E.P., Geiduschek E.P. Transcription factor TFIIIB and transcription by RNA polymerase III. Biochem. Soc. Trans. 2006;34:1082–1087. doi: 10.1042/BST0341082. [DOI] [PubMed] [Google Scholar]
- Kitzmann M., Carnac G., Vandromme M., Primig M., Lamb N.J., Fernandez A., Carnac G., Vandromme M., Primig M., Lamb N.J., Fernandez A., Vandromme M., Primig M., Lamb N.J., Fernandez A., Primig M., Lamb N.J., Fernandez A., Lamb N.J., Fernandez A., Fernandez A. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J. Cell Biol. 1998;142:1447–1459. doi: 10.1083/jcb.142.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I., Viville S., Davidson I., Viville S., Davidson I., Davidson I. RNA polymerase II transcription in murine cells lacking the TATA binding protein. Science. 2002;298:1036–1039. doi: 10.1126/science.1076327. [DOI] [PubMed] [Google Scholar]
- Martin J., Halenbeck R., Kaufmann J., Halenbeck R., Kaufmann J., Kaufmann J. Human transcription factor hTAFII150 (CIF150) is involved in transcriptional regulation of cell cycle progression. Mol. Cell. Biol. 1999;19:5548–5556. doi: 10.1128/mcb.19.8.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D., Scheer E., Soldatov A., Tora L., Scheer E., Soldatov A., Tora L., Soldatov A., Tora L., Tora L. Mammalian TAFII30 is required for cell cycle progression and specific cellular differentiation programmes. EMBO J. 1999;18:4823–4834. doi: 10.1093/emboj/18.17.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan W.S., Scheer E., Wendling O., Metzger D., Tora L., Scheer E., Wendling O., Metzger D., Tora L., Wendling O., Metzger D., Tora L., Metzger D., Tora L., Tora L. TAF10 (TAFII30) is necessary for TFIID stability and early embryogenesis in mice. Mol. Cell. Biol. 2003;23:4307–4318. doi: 10.1128/MCB.23.12.4307-4318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.A., Ozer J., Salunek M., Jan G., Zerby D., Campbell S., Lieberman P.M., Ozer J., Salunek M., Jan G., Zerby D., Campbell S., Lieberman P.M., Salunek M., Jan G., Zerby D., Campbell S., Lieberman P.M., Jan G., Zerby D., Campbell S., Lieberman P.M., Zerby D., Campbell S., Lieberman P.M., Campbell S., Lieberman P.M., Lieberman P.M. A human TATA binding protein-related protein with altered DNA binding specificity inhibits transcription from multiple promoters and activators. Mol. Cell. Biol. 1999;19:7610–7620. doi: 10.1128/mcb.19.11.7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi T., Kishimoto T., Makino Y., Shimada M., Nakadai T., Aoki T., Kawata T., Niwa S., Tamura T., Kishimoto T., Makino Y., Shimada M., Nakadai T., Aoki T., Kawata T., Niwa S., Tamura T., Makino Y., Shimada M., Nakadai T., Aoki T., Kawata T., Niwa S., Tamura T., Shimada M., Nakadai T., Aoki T., Kawata T., Niwa S., Tamura T., Nakadai T., Aoki T., Kawata T., Niwa S., Tamura T., Aoki T., Kawata T., Niwa S., Tamura T., Kawata T., Niwa S., Tamura T., Niwa S., Tamura T., Tamura T. Isolation of cDNA, chromosome mapping, and expression of the human TBP-like protein. Biochem. Biophys. Res. Commun. 1999a;255:137–142. doi: 10.1006/bbrc.1999.0159. [DOI] [PubMed] [Google Scholar]
- Ohbayashi T., Makino Y., Tamura T.A., Makino Y., Tamura T.A., Tamura T.A. Identification of a mouse TBP-like protein (TLP) distantly related to the Drosophila TBP-related factor. Nucleic Acids Res. 1999b;27:750–755. doi: 10.1093/nar/27.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave I., Wang W., Xue Y., Kuo A., Crabtree G.R., Wang W., Xue Y., Kuo A., Crabtree G.R., Xue Y., Kuo A., Crabtree G.R., Kuo A., Crabtree G.R., Crabtree G.R. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes & Dev. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison P.J., Cleary M., Silva J.M., Chang K., Sheth N., Sachidanandam R., Hannon G.J., Cleary M., Silva J.M., Chang K., Sheth N., Sachidanandam R., Hannon G.J., Silva J.M., Chang K., Sheth N., Sachidanandam R., Hannon G.J., Chang K., Sheth N., Sachidanandam R., Hannon G.J., Sheth N., Sachidanandam R., Hannon G.J., Sachidanandam R., Hannon G.J., Hannon G.J. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat. Methods. 2004;1:163–167. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- Palacios D., Puri P.L., Puri P.L. The epigenetic network regulating muscle development and regeneration. J. Cell. Physiol. 2006;207:1–11. doi: 10.1002/jcp.20489. [DOI] [PubMed] [Google Scholar]
- Perletti L., Kopf E., Carre L., Davidson I., Kopf E., Carre L., Davidson I., Carre L., Davidson I., Davidson I. Coordinate regulation of RARγ2, TBP, and TAFII135 by targeted proteolysis during retinoic acid-induced differentiation of F9 embryonal carcinoma cells. BMC Mol. Biol. 2001;2:4. doi: 10.1186/1471-2199-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persengiev S.P., Zhu X., Dixit B.L., Maston G.A., Kittler E.L., Green M.R., Zhu X., Dixit B.L., Maston G.A., Kittler E.L., Green M.R., Dixit B.L., Maston G.A., Kittler E.L., Green M.R., Maston G.A., Kittler E.L., Green M.R., Kittler E.L., Green M.R., Green M.R. TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc. Natl. Acad. Sci. 2003;100:14887–14891. doi: 10.1073/pnas.2036440100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointud J.C., Larsson J., Dastugue B., Couderc J.L., Larsson J., Dastugue B., Couderc J.L., Dastugue B., Couderc J.L., Couderc J.L. The BTB/POZ domain of the regulatory proteins Bric a brac 1 (BAB1) and Bric a brac 2 (BAB2) interacts with the novel Drosophila TAFII factor BIP2/dTAFII155. Dev. Biol. 2001;237:368–380. doi: 10.1006/dbio.2001.0358. [DOI] [PubMed] [Google Scholar]
- Pointud J.C., Mengus G., Brancorsini S., Monaco L., Parvinen M., Sassone-Corsi P., Davidson I., Mengus G., Brancorsini S., Monaco L., Parvinen M., Sassone-Corsi P., Davidson I., Brancorsini S., Monaco L., Parvinen M., Sassone-Corsi P., Davidson I., Monaco L., Parvinen M., Sassone-Corsi P., Davidson I., Parvinen M., Sassone-Corsi P., Davidson I., Sassone-Corsi P., Davidson I., Davidson I. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J. Cell Sci. 2003;116:1847–1858. doi: 10.1242/jcs.00391. [DOI] [PubMed] [Google Scholar]
- Pownall M.E., Gustafsson M.K., Emerson C.P., Gustafsson M.K., Emerson C.P., Emerson C.P. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Rabenstein M.D., Zhou S., Lis J.T., Tjian R., Zhou S., Lis J.T., Tjian R., Lis J.T., Tjian R., Tjian R. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc. Natl. Acad. Sci. 1999;96:4791–4796. doi: 10.1073/pnas.96.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando T.A., Blau H.M., Blau H.M. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 1994;125:1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J.D., Lunt A.I., Parry D.J., Partridge T.A., Lunt A.I., Parry D.J., Partridge T.A., Parry D.J., Partridge T.A., Partridge T.A. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell. Dev. Biol. Anim. 1995;31:773–779. doi: 10.1007/BF02634119. [DOI] [PubMed] [Google Scholar]
- Rudnicki M.A., Braun T., Hinuma S., Jaenisch R., Braun T., Hinuma S., Jaenisch R., Hinuma S., Jaenisch R., Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Rudnicki M.A., Schnegelsberg P.N., Stead R.H., Braun T., Arnold H.H., Jaenisch R., Schnegelsberg P.N., Stead R.H., Braun T., Arnold H.H., Jaenisch R., Stead R.H., Braun T., Arnold H.H., Jaenisch R., Braun T., Arnold H.H., Jaenisch R., Arnold H.H., Jaenisch R., Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Russell J., Zomerdijk J.C., Zomerdijk J.C. The RNA polymerase I transcription machinery. Biochem. Soc. Symp. 2006;73:203–216. doi: 10.1042/bss0730203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V., Caretti G., Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr. Opin. Genet. Dev. 2005;15:528–535. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A., Mansouri A., Gruss P., Rudnicki M.A., Gruss P., Rudnicki M.A., Rudnicki M.A. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Takada S., Lis J.T., Zhou S., Tjian R., Lis J.T., Zhou S., Tjian R., Zhou S., Tjian R., Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- Teichmann M., Wang Z., Martinez E., Tjernberg A., Zhang D., Vollmer F., Chait B.T., Roeder R.G., Wang Z., Martinez E., Tjernberg A., Zhang D., Vollmer F., Chait B.T., Roeder R.G., Martinez E., Tjernberg A., Zhang D., Vollmer F., Chait B.T., Roeder R.G., Tjernberg A., Zhang D., Vollmer F., Chait B.T., Roeder R.G., Zhang D., Vollmer F., Chait B.T., Roeder R.G., Vollmer F., Chait B.T., Roeder R.G., Chait B.T., Roeder R.G., Roeder R.G. Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc. Natl. Acad. Sci. 1999;96:13720–13725. doi: 10.1073/pnas.96.24.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.C., Chiang C.M., Chiang C.M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Voss A.K., Thomas T., Petrou P., Anastassiadis K., Scholer H., Gruss P., Thomas T., Petrou P., Anastassiadis K., Scholer H., Gruss P., Petrou P., Anastassiadis K., Scholer H., Gruss P., Anastassiadis K., Scholer H., Gruss P., Scholer H., Gruss P., Gruss P. Taube nuss is a novel gene essential for the survival of pluripotent cells of early mouse embryos. Development. 2000;127:5449–5461. doi: 10.1242/dev.127.24.5449. [DOI] [PubMed] [Google Scholar]
- Wang E.H., Tjian R., Tjian R. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- Wright K.J., Marr M.T., Tjian R., Marr M.T., Tjian R., Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc. Natl. Acad. Sci. 2006;103:12347–12352. doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Cao J., Huang L., Fang H.Y., Sheng H.Z., Cao J., Huang L., Fang H.Y., Sheng H.Z., Huang L., Fang H.Y., Sheng H.Z., Fang H.Y., Sheng H.Z., Sheng H.Z. Regulated expression of TATA-binding protein-related factor 3 (TRF3) during early embryogenesis. Cell Res. 2006;16:610–621. doi: 10.1038/sj.cr.7310064. [DOI] [PubMed] [Google Scholar]