Abstract
Autophagy is a major pathway used to degrade long-lived proteins and organelles. Autophagy is thought to promote both cell and organism survival by providing fundamental building blocks to maintain energy homeostasis during starvation. Under different conditions, however, autophagy may instead act to promote cell death through an autophagic cell death pathway distinct from apoptosis. Although several recent papers suggest that autophagy plays a role in cell death, it is not known whether autophagy can cause the death of an organism. Furthermore, why autophagy acts in some instances to promote survival but in others to promote death is poorly understood. Here we show that physiological levels of autophagy act to promote survival in Caenorhabditis elegans during starvation, whereas insufficient or excessive levels of autophagy contribute to death. We found that inhibition of autophagy decreases survival of wild-type worms during starvation, and that muscarinic signaling regulates starvation-induced autophagy, at least in part, through the death-associated protein kinase signaling pathway. Furthermore, we found that in gpb-2 mutants, in which muscarinic signaling cannot be down-regulated, starvation induces excessive autophagy in pharyngeal muscles, which in turn, causes damage that may contribute to death. Taken together, our results demonstrate that autophagy can have either prosurvival or prodeath functions in an organism, depending on its level of activation.
Keywords: Autophagy, muscarinic acetylcholine receptor signaling, starvation
Autophagy is a well-conserved lysosomal pathway used to degrade long-lived proteins and cytoplasmic organelles. It involves the formation of double-membrane vesicles called autophagosomes that sequester cytoplasmic material, and the subsequent fusion of an autophagosome with a lysosome that results in the degradation of cytoplasmic material (Levine and Klionsky 2004). Autophagy can be stimulated by both environmental stress (e.g., nutrient starvation, hypoxia, heat shock) and intracellular stress (e.g., damaged mitochondria, superfluous peroxisomes, protein aggregates, pathogens) (Levine and Klionsky 2004; Mizushima 2005). Once stimulated, autophagy helps alleviate the harmful effects of intracellular and extracellular stress.
For many years, it has been assumed that autophagy acts as a prosurvival response to starvation at the cell and organism levels. For example, when the supply of environmental nutrients is limited, autophagy can generate a source of metabolic substrates to maintain cellular ATP production, protein synthesis, and fatty acid synthesis, thereby sustaining cellular activity needed for the cell to stay alive (Lum et al. 2005). This autophagic response to starvation has been well studied in various organisms including yeast, worms, flies, and mice. Loss of autophagy results in death in nitrogen-starved yeast, defects in dauer formation of Caenorhabditis elegans, hypersensitivity to starvation in Drosophila, and early postnatal lethality in mice after termination of the placental nutrient supply (Tsukada and Ohsumi 1993; Melendez et al. 2003; Kuma et al. 2004; Scott et al. 2004). These lines of evidence support the view that autophagy is a survival mechanism for both cell and organism.
However, several recent studies indicate that autophagy also plays a prodeath role at the cellular level. Based on morphological evidence, it has been suggested that autophagy causes a nonapoptotic autophagic cell death during development and in response to toxins or stressful stimuli (Levine and Yuan 2005). In addition, it was shown that RNA interference (RNAi) knockdown of essential autophagy genes inhibits nonapoptotic cell death in a variety of cell types under different conditions (Shimizu et al. 2004; Yu et al. 2004; Reef et al. 2006). Together, these new findings strongly suggest that autophagy has a prodeath function at the cellular level. At the organismal level, excessive autophagy is associated with severe wasting of denervated myofibers in runx1 mutant mice (Wang et al. 2005), suggesting that autophagy could be harmful to the organism. It is not known, however, whether autophagy can lead to organismal death.
Despite intensive study, it remained elusive until recently how autophagy could perform these seemingly opposite roles as a prosurvival mechanism and a prodeath mechanism. An important clue to understand the mechanism by which autophagy performs dual roles with respect to cell survival came from studying the biological significance of the interaction between Bcl-2 and Beclin 1 (Pattingre et al. 2005). The researchers found that Bcl-2 proteins bind to Beclin-1 and inhibit Beclin 1-dependent autophagy in yeast and mammalian cells. Furthermore, they found that mutants of Beclin 1 that do not bind Bcl-2 demonstrate excessive levels of autophagy and promote autophagy gene-dependent cell death in MCF7 cells. Based on these new findings, a new model for understanding the roles of autophagy in cell survival and cell death presented itself: Physiological levels of autophagy are essential for normal cellular homeostasis and play a prosurvival role, whereas excessive levels of autophagy promote autophagic cell death and play a prodeath role at the cellular level (Pattingre et al. 2005). It is plausible to assume that the same mechanism might cause autophagy to have dual roles in the survival of a multicellular organism. For example, physiological levels of autophagy can act as a prosurvival role in an organism by providing energy during starvation (Kuma et al. 2004), whereas excessive autophagy might act as a prodeath role by causing damage in tissues essential for survival.
In this study, we use C. elegans as a model system to evaluate whether autophagy plays dual roles in the survival of a multicellular organism undergoing starvation. We find that insufficient levels of autophagy caused by knockdown of bec-1 promote the death of worms undergoing starvation. In contrast, gpb-2 mutants that are hypersensitive to starvation (You et al. 2006) have excessive levels of autophagy during starvation, and knockdown of bec-1 can rescue the starvation-induced death of gpb-2 mutants. Together, these findings suggest that autophagy can play both a prosurvival role and a prodeath role at the organismal level depending on its level of activation.
Results
Autophagy is required for optimal survival of worms during starvation
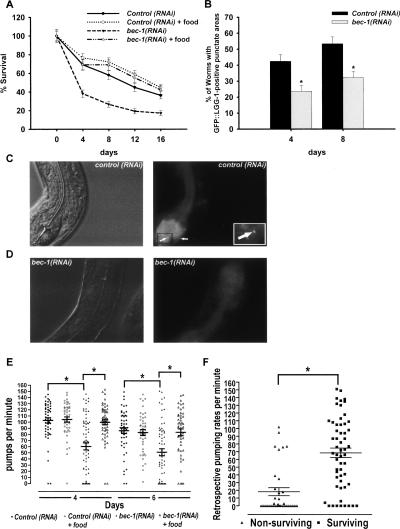
To test the role of autophagy in survival of C. elegans during starvation, we used RNAi to inhibit autophagy. Since animals in which autophagy is completely blocked by mutation are very unhealthy (unc-51 mutants) (data not shown) or die (bec-1 mutants) (Takacs-Vellai et al. 2005), we thought that partial inhibition by RNAi would be more appropriate to study the role of autophagy during starvation. We targeted bec-1 for RNAi. bec-1 is the C. elegans ortholog of yeast ATG6 and is essential for the autophagy process (Melendez et al. 2003). Depleting bec-1 function by RNAi decreased the survival of wild-type worms after starvation (Fig. 1A; see Materials and Methods for details of starvation survival analyses).
Figure 1.
Autophagy is required for optimal survival of C. elegans during starvation. Starvation survival analyses were performed as described in Materials and Methods. Each of the starvation survival curves presented in this and subsequent figures is the result of one representative trial. Additional trials are listed in Table 1. (A) Percent of worms surviving to adulthood on NGM plates with HB101 bacteria after incubation in M9 buffer in the absence of food for the indicated time. Error bars are standard errors estimated assuming a Poisson distribution. (B) Quantitation of autophagy in the pharyngeal muscles of control RNAi animals and bec-1(RNAi) animals in A. Error bars indicate standard error for proportions. (*) P < 0.001. (C) Representative images of control RNAi animals after 9 d of starvation. Differential interference contrast image of control RNAi animals (left) and green fluorescence image (right). The arrow shows representative GFP∷LGG-1-positive punctate structures that label preautophagosomal and autophagosomal structures. In the inset, the area marked by the box is magnified and contrast enhanced. (D) Representative images of bec-1(RNAi) animals after 9 d of starvation. Differential interference contrast image of bec-1(RNAi) animals (left) and green fluorescence image (right). (E) Pumping rates in control RNAi animals and bec-1(RNAi) animals after starvation. Each dot represents an individual worm. Horizontal lines represent the average (line) and SEM (error bars). (*) P < 0.0001 (Student’s t-test). (F) Retrospective pumping rates of surviving and nonsurviving bec-1(RNAi) animals after starvation. Each dot represents an individual worm. Horizontal lines represent the average (line) and SEM (error bars). (*) P < 0.0001 (Student’s t-test).
Because the feeding organ, the pharynx, is critical for recovery from starvation (You et al. 2006), we measured autophagy in pharyngeal muscle. We generated a strain carrying an integrated transgene that expressed a GFP-tagged version of lgg-1, which is a specific fluorescence marker for autophagy (Melendez et al. 2003; Mizushima 2004). When autophagy is induced, GFP∷LGG-1 changes from its diffuse cytoplasmic cellular localization to form punctate structures that label the preautophagosomal and autophagosomal membranes (Fig. 1C). We found that starvation induced autophagy in pharyngeal muscle, and that bec-1 RNAi treatment decreased the amount of autophagy induced by starvation (Fig. 1B–D). We also tested the effects on the autophagy process of RNAi of another C. elegans ortholog of a yeast autophagy gene. RNAi-mediated interference of the C. elegans ortholog of yeast ATG7 decreased survival of wild-type worms after starvation and also reduced autophagy in the pharyngeal muscle (Supplementary Fig. S2). This suggests that the effect of bec-1 RNAi on survival of wild-type worms during starvation is due to defects in autophagy rather than defects in other potential functions of bec-1. The survival of bec-1(RNAi) worms could be rescued by the addition of food (Fig. 1A), suggesting that a major cause of death in bec-1(RNAi) worms was a lack of nutrients during starvation, probably resulting from a decrease in autophagy.
Seeking to understand why starvation kills worms when autophagy is suppressed, we tested whether a decrease in autophagy affected the activity of the pharynx. We found that bec-1 RNAi treatment decreased pumping rates (Fig. 1E) and that pumping could be rescued by the addition of food, suggesting that autophagy is required to maintain basal activity of the pharynx during starvation. We next tested whether pumping could predict survival. To determine the importance of basal pumping rates for survival, we counted the pumping rates of individual worms that were still alive immediately after starvation, assessed their survival, then compared the average pumping rate of worms that eventually died to that of worms that survived (Fig. 1F). Worms that initially had high pumping rates survived better than those that had low pumping rates after starvation; pumping predicts survival with up to 80% accuracy. Our finding, combined with previous work showing that the pharynx is important for recovery from starvation (You et al. 2006), suggests that maintenance of basal pumping rates may be important for the survival of worms after starvation. Taken together, these results demonstrate that autophagy is required for the optimal survival of wild-type worms during starvation, perhaps providing an energy source or essential nutrients to maintain cellular activity.
Overactivated muscarinic acetylcholine signaling induces excessive autophagy and causes death of worms during starvation
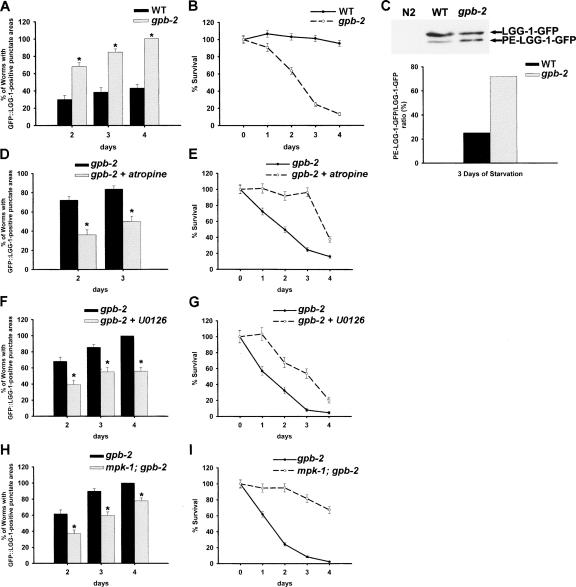
Previously, we showed that starvation activates MAPK (mitogen-activated protein kinase) in the pharyngeal muscle via a muscarinic acetylcholine receptor signaling pathway (muscarinic acetylcholine receptor → Gqα → PKC → MAPK) (You et al. 2006). In addition, this starvation signaling induces a change in the physiology of the pharyngeal muscle to increase its activity. However, when the starvation signal is overactivated, as in the case of gpb-2 (a G-protein β subunit involved in RGS-mediated inhibition of the Gqα pathway) mutants in which muscarinic acetylcholine receptor signaling cannot be down-regulated, starvation instead induces damage in the pharyngeal muscle that leads to aberrant activity and eventual death on recovery from starvation (You et al. 2006).
The environmental cue (starvation) that induces changes in the pharyngeal muscles of worms can also trigger autophagy (Levine and Klionsky 2004). Furthermore, it is known that overactivated autophagy causes damage at the cellular level (Codogno and Meijer 2005; Kroemer and Jaattela 2005; Levine and Yuan 2005); this is reminiscent of the way overactivated muscarinic signaling causes damage that leads to the malfunction of worm pharyngeal muscle. Based on these similarities, we hypothesized that overactivated muscarinic signaling in gpb-2 mutants induces unrestrained autophagy, which in turn, causes damage to the pharyngeal muscles and eventually contributes to death after starvation.
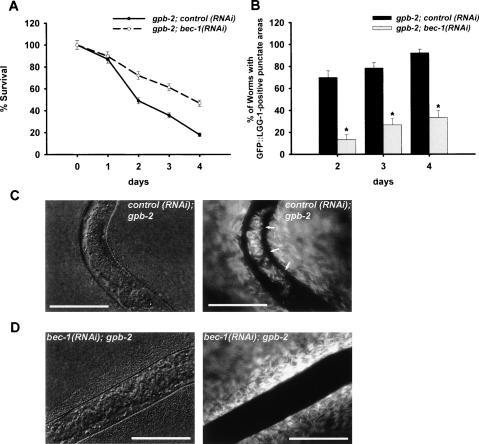
To test this, we examined the level of autophagy in the pharyngeal muscle of gpb-2 mutants during starvation. As reported previously (You et al. 2006), gpb-2 mutants were more sensitive to starvation than wild-type worms (Fig. 2B). Moreover, we found that autophagy is excessively induced following starvation in the pharyngeal muscle of gpb-2 mutants compared with that of wild-type worms as measured by GFP–LGG-1 punctate structures (Fig. 2A). It is unlikely that an increase of GFP–LGG-1 punctate structures in gpb-2 mutants results from defects in the turnover of autophagosomes because treatment with lysosomal inhibitor (NH4Cl) increases accumulation of GFP–LGG-1 punctate structures (Supplementary Fig. S3). We also confirmed induction of autophagy by detection of the modification of GFP∷LGG-1 using immunoblotting (Mizushima 2004). After starvation, the ratio of phosphatidylethanolamine-conjugated LGG-1 (PE-LGG-1∷GFP) to LGG-1 (LGG-1∷GFP) is higher in gpb-2 mutants than in wild-type worms (Fig. 2C), indicating that autophagy is excessively induced in gpb-2 mutants compared with wild-type worms following starvation. Death of starved gpb-2 mutants is likely caused by pharyngeal muscle malfunction, not cell death, because pharyngeal muscle cells of gpb-2 mutants show no obvious cell death, and they are still able to contract after starvation. Furthermore, mutation of ced-3 could not rescue death of gpb-2 mutants during starvation (data not shown), suggesting that apoptosis is not involved. To test whether muscarinic acetylcholine receptor signaling is involved in the regulation of autophagy, we used inhibitors of muscarinic signaling such as atropine, a muscarinic acetylcholine receptor antagonist, and U0126, which prevents MAPK activation by inhibiting the upstream kinase MEK (You et al. 2006). Treatment with either atropine or U0126 reduced autophagy in the pharyngeal muscle of gpb-2 mutants during starvation (Fig. 2D,F) and also partially rescued the starvation hypersensitivity of gpb-2 mutants (Fig. 2E,G). Furthermore, we found that mutation of mpk-1, which encodes the MAPK activated by starvation, also decreased autophagy in the pharyngeal muscle of gpb-2 mutants during starvation (Fig. 2H) and suppressed the starvation hypersensitivity phenotype (Fig. 2I). Taken together, these data indicate that under starvation conditions, overactivated muscarinic acetylcholine receptor signaling in gpb-2 mutants causes excessive autophagy. Furthermore, the fact that excessive autophagy is associated with the death of starved gpb-2 mutants strongly suggests that unrestrained autophagy may contribute to the death. It is important to note that mutation of mpk-1 rescues the death of gpb-2 mutants very efficiently, yet is less effective at reducing autophagy than atropine. It is possible that other unknown mechanisms also contribute to the death of starved gpb-2 mutants.
Figure 2.
Overactivated muscarinic signaling induces excessive levels of autophagy and causes death in gpb-2 mutants after starvation. (A,D,F,H) Quantitation of autophagy in the pharyngeal muscles of indicated genotype after incubation in M9 buffer in the absence of food with or without either atropine (10 mM) or U0126 (25 μM) for the indicated time. Error bars indicate standard error for proportions. (*) P < 0.0001 for all except 2 d in F and H, for which P < 0.005. See Supplementary Figure S1 for representative images. (B,E,G,I) Percent of animals surviving to adulthood in A, D, F, and H. Error bars are standard errors estimated assuming a Poisson distribution. (C) The relative levels of PE-conjugated LGG-1–GFP and LGG-1–GFP were determined by Western blotting. Starved L1 worms were prepared and analyzed by immunoblotting using an anti-GFP antibody. The arrows indicate LGG-1–GFP and PE-conjugated LGG-1–GFP. The Western blot is representative of what was seen in two independent experiments.
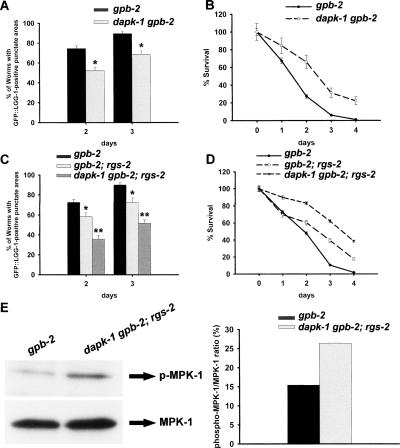
To identify downstream target genes of muscarinic acetylcholine receptor signaling responsible for the regulation of autophagy, we used RNAi to reduce the expression of several candidate genes. One of them is dapk-1, the C. elegans ortholog of death-associated protein kinase (DAP kinase). Previous cell culture studies that showed that DAP kinase mediates autophagy (Inbal et al. 2002), together with the fact that ERK can phosphorylate and activate DAP kinase in cell culture (Chen et al. 2005), led us to hypothesize that muscarinic signaling regulates autophagy through DAP kinase. We found that dapk-1 RNAi reduces starvation-induced autophagy and partially rescues starvation hypersensitivity of gpb-2 mutants (Table 1). To verify the RNAi result, we made dapk-1 gpb-2 double mutants and found that mutation of dapk-1 also decreases starvation-induced autophagy (Fig. 3A) and partially rescues death of gpb-2 mutants (Fig. 3B). It is unlikely that mutation of dapk-1 rescues death of gpb-2 mutants during starvation by the inhibition of apoptosis because mutation of ced-3 did not rescue death of gpb-2 mutants (data not shown). Since mutation of dapk-1 only partially rescued gpb-2 mutants, it is likely that other unknown signaling molecules also function downstream from muscarinic signaling to regulate autophagy. In fact, we found that mutation of rgs-2, a regulator of G protein signaling, further decreases autophagy in dapk-1 gpb-2 mutants and rescues death of dapk-1 gpb-2 during starvation (Fig. 3C,D). It is possible, however, that RGS-2 negatively regulates muscarinic signaling to decrease autophagy through GOA-1, which is a target of RGS-2, and is known to antagonize muscarinic signaling (Miller et al. 1999). To test this possibility, we examined the activation of MPK-1 in dapk-1 gpb-2; rgs-2 mutants. We found that MPK-1 is activated in dapk-1 gpb-2; rgs-2 mutants as it is in gpb-2 mutants (Fig. 3E), suggesting that RGS-2 is unlikely to negatively regulate muscarinic signaling via a feedback loop. A previous study showing that GAIP, the mammalian ortholog of rgs-2, functions downstream from ERK to regulate autophagy in cell culture (Ogier-Denis et al. 2000) further supports our hypothesis that RGS-2 functions downstream from muscarinic signaling to regulate autophagy. Thus, these results suggest that DAPK-1 acts, at least in part, downstream from or in parallel to muscarinic signaling in the regulation of autophagy, possibly with RGS-2.
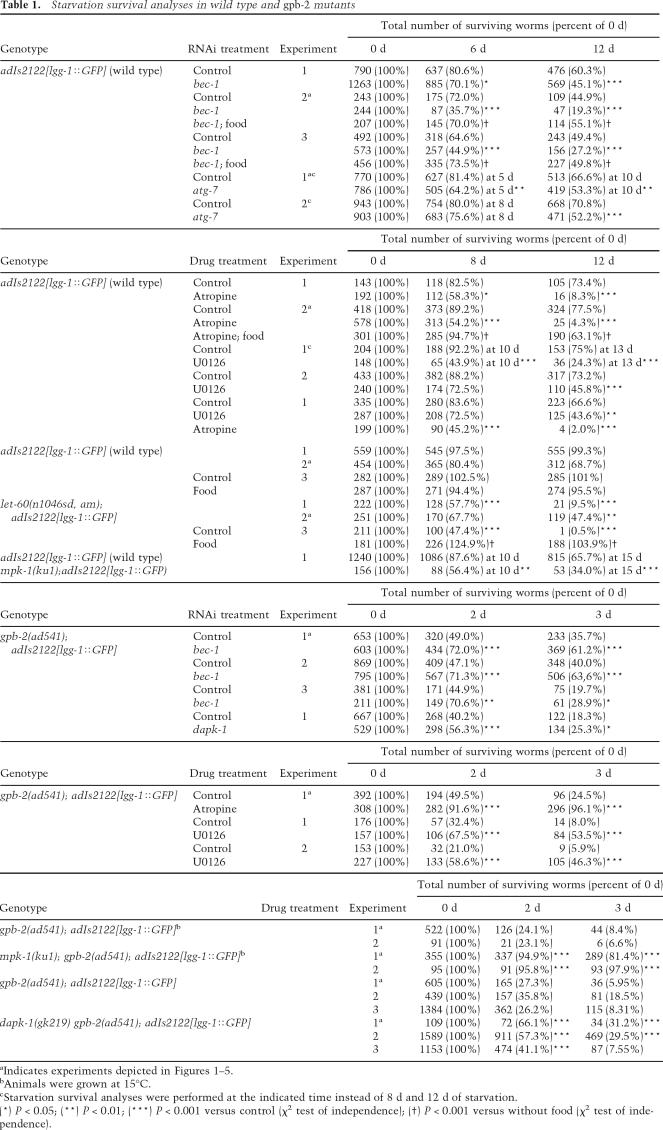
Table 1.
Starvation survival analyses in wild type and gpb-2 mutants
aIndicates experiments depicted in Figures 1–5.
bAnimals were grown at 15°C.
cStarvation survival analyses were performed at the indicated time instead of 8 d and 12 d of starvation.
(*) P < 0.05; (**) P < 0.01; (***) P < 0.001 versus control (χ2 test of independence); (†) P < 0.001 versus without food (χ2 test of independence).
Figure 3.
DAPK-1 acts downstream from or in parallel to muscarinic signaling in the regulation of autophagy, probably with RGS-2. (A,C) Quantitation of autophagy in the pharyngeal muscles of the indicated genotype after incubation in M9 buffer in the absence of food for the indicated time. Error bars indicate standard error for proportions. (*) P < 0.005; (**) P < 0.0001. (B,D) Percent of worms surviving to adulthood in A and C. Error bars are standard errors estimated assuming a Poisson distribution. (E) The relative levels of phospho-MPK-1 and MPK-1 were determined by Western blotting. Starved L1 worms were prepared 32.5 h after egg preparation and were analyzed by immunoblotting using an anti-phospho-MPK-1 antibody and an anti-MPK-1 antibody. The Western blot is representative of what was seen in two independent experiments.
Excessive autophagy causes defects in the pharyngeal muscles of gpb-2 mutants and contributes to death
To directly examine the role of excessive autophagy in the death of starved gpb-2 mutants, we inhibited autophagy in these mutants by bec-1 RNAi. bec-1 RNAi treatment decreased autophagy in the pharyngeal muscle of starved gpb-2 mutants (Fig. 4B). It also partially rescued the death of starved gpb-2 mutants (Fig. 4A), suggesting that excessive autophagy contributes to the death of the gpb-2 mutants during starvation conditions. We also verified these results by atg-7 RNAi (data not shown).
Figure 4.
Unrestrained autophagy causes defects in the pharyngeal muscles of gpb-2 mutants and contributes to death after starvation. (A) Percent of worms surviving to adulthood on NGM plates with HB101 bacteria after incubation in M9 buffer in the absence of food for the indicated time. Error bars are standard errors estimated assuming a Poisson distribution. (B) Quantitation of autophagy in A. bec-1(RNAi) reduced excessive levels of autophagy in the pharyngeal muscles of gpb-2 during starvation. Error bars indicate standard error for proportions. (*) P < 0.0001. See Supplementary Figure S1 for representative images. (C,D) gpb-2 control RNAi animals (C) and gpb-2; bec-1(RNAi) animals (D) after 2 d of starvation followed by growth on GFP-expressing E. coli. (Left) Differential interference contrast image. (Right) Green fluorescence image. Arrows in C indicate unground bacteria in the intestine. Bars, 20 μm.
Starvation induces malfunction (hypercontraction) of pharyngeal muscle in gpb-2 mutants after starvation, so that the worms cannot grind up their bacterial food (You et al. 2006). To investigate the effects of excessive autophagy in gpb-2 mutants on the ability of pharyngeal muscle to grind bacteria, we used GFP-expressing Escherichia coli (You et al. 2006). If worms grind bacteria efficiently, green fluorescence cannot be observed in their intestines. If not, however, the intestines are fluorescent with unground bacteria. As previously shown (You et al. 2006), gpb-2 mutants cannot grind bacteria after starvation (Fig. 4C). In contrast gpb-2 mutants in which bec-1 expression was reduced by RNAi can grind bacteria after starvation (Fig. 4D). Together, these results suggest that excessive autophagy causes defects in the pharyngeal muscles of gpb-2 mutant and eventually contributes to death after starvation. It is possible, however, that excessive autophagy in other tissues contributes to death of gpb-2 mutants.
Muscarinic signaling plays a role in regulating autophagy during starvation
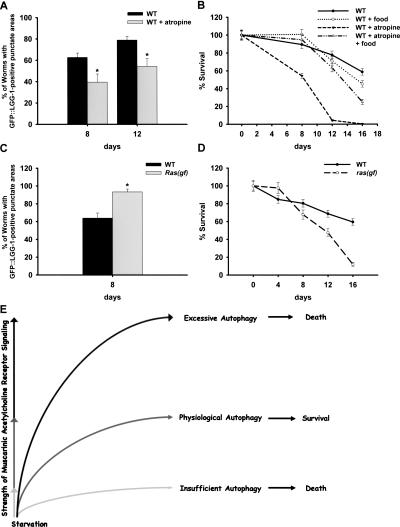
Our data in gpb-2 mutants indicate that overactivated muscarinic signaling induces excessive autophagy during starvation. To evaluate whether muscarinic acetylcholine receptor signaling can also regulate autophagy in wild-type worms during starvation, we inhibited muscarinic signaling by treatment with atropine. Treatment with atropine decreased pharyngeal muscle autophagy below basal levels as compared with controls (Fig. 5A), indicating that blocking muscarinic signaling decreases the levels of autophagy in wild-type worms during starvation. Treatment with atropine also decreased survival of wild-type worms after starvation (Fig. 5B), confirming that physiological levels of autophagy are required for optimal survival of wild-type worms under starvation. The survival of wild-type worms treated with atropine could be rescued partially by the addition of food (Fig. 5A), suggesting that the effect of atropine on survival of wild-type worms during starvation may be due to decreases in autophagy rather than drug toxicity only. We also found that treatment with U0126 reduced pharyngeal muscle autophagy (data not shown) and decreased survival of wild-type worms after starvation to a lesser extent than treatment with atropine (Table 1). In contrast, a Ras gain-of-function mutation (which has a Gly-to-Glu substitution at codon 13 and mimics the effect of overactivated muscarinic signaling on MAPK) induced excessive autophagy in the pharyngeal muscles during starvation (Fig. 5C). It also decreased survival of worms after starvation (Fig. 5D). Thus, overactivated muscarinic signaling increases the levels of autophagy above physiological levels during starvation. Taken together, these data suggest that muscarinic acetylcholine receptor signaling positively regulates autophagy in wild-type worms under starvation and is important for the optimal survival of starved wild-type worms.
Figure 5.
Muscarinic signaling positively regulates autophagy during starvation. (A) Quantitation of autophagy in the pharyngeal muscles of wild-type animals treated with atropine (10 mM) after incubation in M9 buffer in the absence of food for the indicated time. (C) Quantitation of autophagy in the pharyngeal muscles of ras(gf) animals. Error bars indicate standard error for proportions. (*) P < 0.01. (B,D) Percent of worms surviving to adulthood in A and C. Error bars are standard errors estimated assuming a Poisson distribution. See Supplementary Figure S1 for representative images. (E) Model of dual roles of autophagy in the survival of C. elegans.
Discussion
Dual roles of autophagy in the survival of a multicellular organism during starvation
Our results demonstrate that physiological levels of autophagy are required for optimal survival of C. elegans during starvation, and that muscarinic acetylcholine receptor signaling is important for the induction of physiological levels of autophagy. In contrast, insufficient or excessive levels of autophagy render C. elegans hypersensitive to starvation. These results lead us to conclude that autophagy can play opposing roles promoting survival or death at the organismal level during starvation, depending on its level (Fig. 5E). The mechanism by which autophagy plays dual roles with respect to survival was not well understood until recently. As described in the Introduction, previous studies suggested the intriguing possibility that it is the level of autophagy that is critical in deciding between a prosurvival or a prodeath role (Pattingre et al. 2005). Our results provide in vivo evidence that a similar model may apply at the whole organism level: Physiological levels of autophagy are prosurvival, whereas insufficient or excessive levels of autophagy are prodeath.
The muscarinic acetylcholine pathway as an autophagy-inducing signaling pathway
During the past decade, molecular mechanisms involved in the regulation of autophagy have been discovered (Levine and Klionsky 2004; Codogno and Meijer 2005; Yorimitsu and Klionsky 2005). Despite this significant progress, the signaling pathway involved in the regulation of autophagy at the organismal level has been extensively characterized only in the context of autophagy-inhibiting signals. It is well known that the type I PI3K–mTOR signaling pathway negatively regulates autophagy at both the cellular and organismal levels (Scott et al. 2004; Shintani and Klionsky 2004; Codogno and Meijer 2005). Although several pathways such as eIF2α kinase, ERK1/2, and DAP kinase pathways have been implicated as autophagy-inducing signaling pathways at the cellular level (Inbal et al. 2002; Talloczy et al. 2002; Pattingre et al. 2003), little is known about autophagy-inducing signaling pathways in the multicellular organism. Our results demonstrate that the muscarinic acetylcholine pathway functions as an autophagy-inducing signaling pathway in the pharyngeal muscles of C. elegans during starvation. We found that during starvation, overactivation of muscarinic signaling in gpb-2 mutants causes excessive autophagy in the pharyngeal muscles, whereas inhibition of muscarinic signaling either by treatment with inhibitors or by mutation decreases autophagy in the pharyngeal muscles. Furthermore, we found that either mutation of dapk-1 or rgs-2 reduces starvation-induced autophagy and partially rescues the death of gpb-2 during starvation, suggesting that DAPK-1 and perhaps RGS-2 may act, at least in part, downstream from or in parallel to muscarinic signaling in the regulation of autophagy. Our results combined with the previous study showing that starvation activates muscarinic acetylcholine signaling in the pharyngeal muscles of C. elegans (You et al. 2006) suggest that the muscarinic acetylcholine signaling pathway positively regulates autophagy in response to starvation, probably through DAP kinase and RGS-2.
Previous studies showed that the muscarinic acetylcholine signaling pathway increases pumping rates to enhance feeding response by altering pharyngeal muscle physiology as a response to starvation (You et al. 2006). This change may render worms more competent to digest food when food becomes available once again. Because it is obvious that this change is an energy-demanding process, the question remains how energy is provided to increase pumping rates during starvation. Since it is known that autophagy can provide energy to maintain energy homeostasis (Kuma et al. 2004; Lum et al. 2005), one intriguing possibility is that autophagy in the pharyngeal muscles induced by the muscarinic acetylcholine signaling pathway may provide the energy needed to increase pumping rates. In fact, our result that shows reduced autophagy by bec-1 RNAi decreases pumping rates during starvation supports this possibility.
Several lines of evidence indicate that autophagy may play important roles in tumor development (Cuervo 2004; Kondo et al. 2005; Levine and Yuan 2005; Levine 2006). Autophagy can act to suppress cancer development by removing damaged organelles and promoting autophagic cell death. On the other hand, autophagy can contribute to cancer development by providing an energy source to cancer cells located far from the blood supply, where nutrients are extremely limited. Our observation that overactivation of the Ras signaling pathway induced autophagy, combined with previous studies showing that Ras signaling is frequently overactivated in various cancers, leads to the intriguing hypothesis that autophagy activated by overactivated Ras signaling may contribute to the survival of cancer cells under nutrient-limited conditions. However, this hypothesis must be viewed with caution because Ras signaling is known to inhibit autophagy in other systems and autophagy can function as a tumor suppressor depending on the circumstances (Kondo et al. 2005; Levine and Yuan 2005; Levine 2006). Nevertheless, it would be interesting to test if inhibition of autophagy affects the survival of cancer cells in the central area of the large tumor masses, where the nutrient supply is limited.
Materials and methods
Strains
Strains were maintained as described (Brenner 1974) at 19°C. All worms were maintained and grown on E. coli HB101 bacteria. adEx2066[rol-6(d) lgg-1∷GFP], an extrachromosomal array that expresses an N-terminal GFP∷LGG-1, was a gift from K. Jia and B. Levine (Melendez et al. 2003). adEx2066 was integrated by γ-irradiation, followed by outcrossing seven times to N2, to generate an integration line, adIs2122[lgg-1∷GFP rol-6(df)]. adIs2122 was used as a wild-type control for all experiments. The following strains were generated using standard genetic procedures: gpb-2(ad541) I; adIs2122, mpk-1(ku1) III; adIs2122, gpb-2(ad541) I; mpk-1(ku1) III; adIs2122, let-60(n1046sd,am) IV; adIs2122, gpb-2(ad541) I; ced-3 (n717) IV; adIs2122, dapk-1 (gk219) I; adIs2122, gpb-2(ad541) dapk-1(gk219) I; adIs2122, gpb-2(ad541) I; rgs-2(vs22) X; adIs2122, gpb-2(ad541) dapk-1(gk219) I; rgs-2(vs22) X; adIs2122.
RNAi
RNAi by feeding was performed as described (Kamath et al. 2003) except for a few modifications. Briefly, RNAi clones were inoculated overnight at 37°C in LB plus tetracycline at 10 μg/mL and ampicillin at 75 μg/mL. After 2 h induction with 0.4 mM IPTG, RNAi clones were seeded onto NG–ampicillin–IPTG plates. L1 animals were transferred to RNAi plates containing bacteria induced to express an RNAi clone or carrying an empty RNAi feeding vector as a control and were allowed to grow to adulthood. After they had laid eggs, the adult worms were removed from the RNAi plates. The plates were then incubated for either 21 h (in wild type) or 17 h (in gpb-2 mutants) at 19°C. L1 worms were collected from RNAi plates and washed three times with M9 solution. The collected L1 worms were used for starvation survival analyses. The length of the incubation on RNAi plates allowed us to adjust the strength of the RNAi treatment. Since autophagy is necessary for optimal survival of wild type during starvation, we anticipated that too effective bec-1(RNAi) would also decrease starvation survival of gpb-2 worms; a prosurvival effect was expected only from a partial knockdown of bec-1 expression. Indeed, in preliminary exploratory experiments, bec-1(RNAi) for longer than 17 h actually decreased starvation survival of gpb-2 worms.
Starvation survival analyses
Starvation survival analyses were performed as described (You et al. 2006) with few modifications. After collection of L1 worms from RNAi plates or synchronization by egg preparation, we incubated them in 3 mL of sterilized M9 buffer for the time indicated in the figures or Table 1 at 19°C. At each time point, an aliquot from each sample tube was placed on a plate seeded with E. coli HB101. The number of worms surviving to L4 or adulthood was determined after 3 d further growth at 19°C. The number from day 0 of starvation was used as control and as the denominator to calculate the percentage of worms recovering after starvation.
Bacteria preparation for liquid cultivation
E. coli HB101 was prepared as described (You et al. 2006) with few modifications. An E. coli HB101 culture in LB broth of optical density 1.0∼1.2 (600 nm) was washed three times with M9 buffer and resuspended in the same volume of M9 buffer. This E. coli HB101 suspension was added to the worm liquid culture at a dilution between 1:4 and 1:6 in order to provide food during starvation survival analyses.
C. elegans autophagy analysis
Autophagy analysis was performed as described (Melendez et al. 2003) with few modifications. For light microscopic analysis of autophagy, starved L1 animals carrying an integrated transgene that expresses a GFP∷LGG-1 fusion were collected at each time point. GFP-positive punctate regions were visualized in the pharyngeal muscles of L1 animals using a Zeiss Axioplan 2 compound microscope (Zeiss Corporation). The percentage of animals with GFP-positive punctate regions was determined. For Western blot analysis of autophagy, starved L1 animals carrying an integrated transgene that expresses a GFP∷LGG-1 fusion were prepared at each time point and analyzed by immunoblotting using an anti-GFP antibody. The relative levels of PE-conjugated LGG-1–GFP and LGG-1–GFP were calculated by densitometry using ImageJ (version 1.36b, National Institutes of Health).
Drug treatment during starvation survival analyses
Drug treatment was performed as described (You et al. 2006).
Western blot analysis
Western blot analysis was performed as described (You et al. 2006).
Statistical analysis
Statistical differences between groups were analyzed using the χ2 test of independence unless otherwise indicated.
Acknowledgments
We thank B. Levine and S. Cameron for helpful discussions, B. Levine and K. Jia for providing strain and RNAi constructs, C. Glynn for critical reading, the C. elegans Genetics Center and the C. elegans Reverse Genetics Core Facility at UBC for providing strains, and M.S. Kim for unfailing support and encouragement. This work was supported by research grant HL46154 from the U.S. Public Health Service.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1573107
References
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.H., Wang W.J., Kuo J.C., Tsai H.C., Lin J.R., Chang Z.F., Chen R.H., Wang W.J., Kuo J.C., Tsai H.C., Lin J.R., Chang Z.F., Chen R.H., Kuo J.C., Tsai H.C., Lin J.R., Chang Z.F., Chen R.H., Tsai H.C., Lin J.R., Chang Z.F., Chen R.H., Lin J.R., Chang Z.F., Chen R.H., Chang Z.F., Chen R.H., Chen R.H. Bidirectional signals transduced by DAPK–ERK interaction promote the apoptotic effect of DAPK. EMBO J. 2005;24:294–304. doi: 10.1038/sj.emboj.7600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codogno P., Meijer A.J., Meijer A.J. Autophagy and signaling: Their role in cell survival and cell death. Cell Death Differ. 2005;12 (Suppl. 2):1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- Cuervo A.M. Autophagy: In sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Inbal B., Bialik S., Sabanay I., Shani G., Kimchi A., Bialik S., Sabanay I., Shani G., Kimchi A., Sabanay I., Shani G., Kimchi A., Shani G., Kimchi A., Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J. Cell Biol. 2002;157:455–468. doi: 10.1083/jcb.200109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Le Bot N., Moreno S., Sohrmann M., Moreno S., Sohrmann M., Sohrmann M., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kondo Y., Kanzawa T., Sawaya R., Kondo S., Kanzawa T., Sawaya R., Kondo S., Sawaya R., Kondo S., Kondo S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- Kroemer G., Jaattela M., Jaattela M. Lysosomes and autophagy in cell death control. Nat. Rev. Cancer. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Ohsumi Y., Tokuhisa T., Mizushima N., Tokuhisa T., Mizushima N., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Levine B. Unraveling the role of autophagy in cancer. Autophagy. 2006;2:65–66. doi: 10.4161/auto.2.2.2457. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D.J., Klionsky D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Levine B., Yuan J., Yuan J. Autophagy in cell death: An innocent convict? J. Clin. Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum J.J., DeBerardinis R.J., Thompson C.B., DeBerardinis R.J., Thompson C.B., Thompson C.B. Autophagy in metazoans: Cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- Melendez A., Talloczy Z., Seaman M., Eskelinen E.L., Hall D.H., Levine B., Talloczy Z., Seaman M., Eskelinen E.L., Hall D.H., Levine B., Seaman M., Eskelinen E.L., Hall D.H., Levine B., Eskelinen E.L., Hall D.H., Levine B., Hall D.H., Levine B., Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Miller K.G., Emerson M.D., Rand J.B., Emerson M.D., Rand J.B., Rand J.B. Goα and diacylglycerol kinase negatively regulate the Gqα pathway in C. elegans. Neuron. 1999;24:323–333. doi: 10.1016/s0896-6273(00)80847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The pleiotropic role of autophagy: From protein metabolism to bactericide. Cell Death Differ. 2005;12 (Suppl. 2):1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- Ogier-Denis E., Pattingre S., El Benna J., Codogno P., Pattingre S., El Benna J., Codogno P., El Benna J., Codogno P., Codogno P. Erk1/2-dependent phosphorylation of Gα-interacting protein stimulates its GTPase accelerating activity and autophagy in human colon cancer cells. J. Biol. Chem. 2000;275:39090–39095. doi: 10.1074/jbc.M006198200. [DOI] [PubMed] [Google Scholar]
- Pattingre S., Bauvy C., Codogno P., Bauvy C., Codogno P., Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J. Biol. Chem. 2003;278:16667–16674. doi: 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B., Mizushima N., Packer M., Schneider M.D., Levine B., Packer M., Schneider M.D., Levine B., Schneider M.D., Levine B., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Reef S., Zalckvar E., Shifman O., Bialik S., Sabanay H., Oren M., Kimchi A., Zalckvar E., Shifman O., Bialik S., Sabanay H., Oren M., Kimchi A., Shifman O., Bialik S., Sabanay H., Oren M., Kimchi A., Bialik S., Sabanay H., Oren M., Kimchi A., Sabanay H., Oren M., Kimchi A., Oren M., Kimchi A., Kimchi A. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol. Cell. 2006;22:463–475. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Scott R.C., Schuldiner O., Neufeld T.P., Schuldiner O., Neufeld T.P., Neufeld T.P. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Shimizu S., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y., Mizuta T., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y., Thompson C.B., Tsujimoto Y., Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Shintani T., Klionsky D.J., Klionsky D.J. Autophagy in health and disease: A double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs-Vellai K., Vellai T., Puoti A., Passannante M., Wicky C., Streit A., Kovacs A.L., Muller F., Vellai T., Puoti A., Passannante M., Wicky C., Streit A., Kovacs A.L., Muller F., Puoti A., Passannante M., Wicky C., Streit A., Kovacs A.L., Muller F., Passannante M., Wicky C., Streit A., Kovacs A.L., Muller F., Wicky C., Streit A., Kovacs A.L., Muller F., Streit A., Kovacs A.L., Muller F., Kovacs A.L., Muller F., Muller F. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr. Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- Talloczy Z., Jiang W., Virgin H.W., Leib D.A., Scheuner D., Kaufman R.J., Eskelinen E.L., Levine B., Jiang W., Virgin H.W., Leib D.A., Scheuner D., Kaufman R.J., Eskelinen E.L., Levine B., Virgin H.W., Leib D.A., Scheuner D., Kaufman R.J., Eskelinen E.L., Levine B., Leib D.A., Scheuner D., Kaufman R.J., Eskelinen E.L., Levine B., Scheuner D., Kaufman R.J., Eskelinen E.L., Levine B., Kaufman R.J., Eskelinen E.L., Levine B., Eskelinen E.L., Levine B., Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc. Natl. Acad. Sci. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Wang X., Blagden C., Fan J., Nowak S.J., Taniuchi I., Littman D.R., Burden S.J., Blagden C., Fan J., Nowak S.J., Taniuchi I., Littman D.R., Burden S.J., Fan J., Nowak S.J., Taniuchi I., Littman D.R., Burden S.J., Nowak S.J., Taniuchi I., Littman D.R., Burden S.J., Taniuchi I., Littman D.R., Burden S.J., Littman D.R., Burden S.J., Burden S.J. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes & Dev. 2005;19:1715–1722. doi: 10.1101/gad.1318305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D.J., Klionsky D.J. Autophagy: Molecular machinery for self-eating. Cell Death Differ. 2005;12 (Suppl. 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y.J., Kim J., Cobb M., Avery L., Kim J., Cobb M., Avery L., Cobb M., Avery L., Avery L. Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 2006;3:237–245. doi: 10.1016/j.cmet.2006.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Alva A., Su H., Dutt P., Freundt E., Welsh S., Baehrecke E.H., Lenardo M.J., Alva A., Su H., Dutt P., Freundt E., Welsh S., Baehrecke E.H., Lenardo M.J., Su H., Dutt P., Freundt E., Welsh S., Baehrecke E.H., Lenardo M.J., Dutt P., Freundt E., Welsh S., Baehrecke E.H., Lenardo M.J., Freundt E., Welsh S., Baehrecke E.H., Lenardo M.J., Welsh S., Baehrecke E.H., Lenardo M.J., Baehrecke E.H., Lenardo M.J., Lenardo M.J. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]