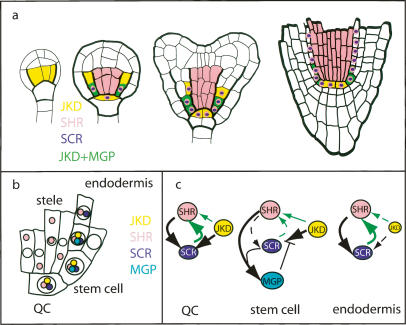
Abstract
In the Arabidopsis root, the SHORT-ROOT transcription factor moves outward to the ground tissue from its site of transcription in the stele and is required for the specification of the endodermis and the stem cell organizing quiescent center cells. In addition, SHORT-ROOT and the downstream transcription factor SCARECROW control an oriented cell division in ground tissue stem cell daughters. Here, we show that the JACKDAW and MAGPIE genes, which encode members of a plant-specific family of zinc finger proteins, act in a SHR-dependent feed-forward loop to regulate the range of action of SHORT-ROOT and SCARECROW. JACKDAW expression is initiated independent of SHORT-ROOT and regulates the SCARECROW expression domain outside the stele, while MAGPIE expression depends on SHORT-ROOT and SCARECROW. We provide evidence that JACKDAW and MAGPIE regulate tissue boundaries and asymmetric cell division and can control SHORT-ROOT and SCARECROW activity in a transcriptional and protein interaction network.
Keywords: Meristem, protein movement, stem cells, transcription factor, nuclear localization
The intercellular movement of transcription factors has emerged as a regulatory mechanism to direct higher plant development (Barton 2001; Wu et al. 2002). In the Arabidopsis root, radial patterning and growth depend upon the transcription factor SHORT-ROOT (SHR) that can move between cells (Helariutta et al. 2000; Nakajima et al. 2001). The root displays a radial organization in which concentric epidermal and ground tissue layers surround a central stele. In addition, a distal stem cell group that extends the tissues of the root is maintained by an organizing center, the quiescent center (QC). Together, the stem cells and the QC form a stem cell niche (Fig. 1a). SHR moves outward from its site of transcription within the stele, and it is required for the specification of QC and ground tissue identity (Helariutta et al. 2000; Nakajima et al. 2001; Sabatini et al. 2003). In shr mutants, the asymmetric division of the cortex/endodermis stem cell daughter (CED) does not occur, resulting in a single layer of ground tissue with cortex identity (Benfey et al. 1993). SHR promotes the expression of a related transcription factor, SCARECROW (SCR), which also patterns the QC and ground tissue (Di Laurenzio et al. 1996; Sabatini et al. 2003).
Figure 1.
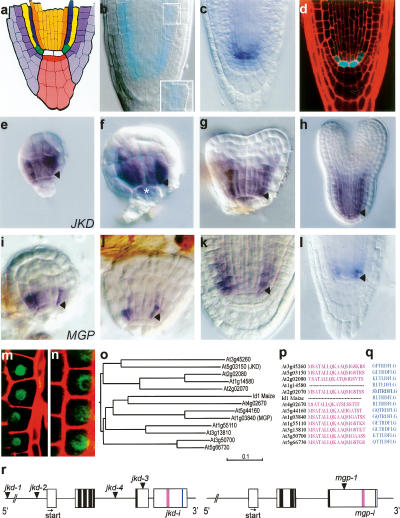
JKD encodes a C2H2 zinc finger expressed in the QC and ground tissue. (a) Median longitudinal view of the Arabidopsis root meristem (RM). (White) QC; (red) columella stem cells; (pink) columella root cap; (light purple) lateral root cap; (purple) epidermis; (green) cortical/endodermal stem cells; (yellow) cortex; (blue) endodermis; (orange) pericycle; (light orange) provascular cells. (b) GUS staining of the DWK15 promoter trap line (jkd-1) at 3-dpg; inset shows ectopic periclinal cortex division. (c) Whole-mount in situ hybridization with a JKD probe in 3-dpg wild-type Col-0 seedling. (d) Longitudinal confocal section of pJKD∷CFP. (e–h) Whole-mount in situ hybridization with JKD in wild-type 32-cell stage (e), globular stage (f), early heart stage (g), and torpedo stage (h) embryos. (i–l) MGP expression during embryogenesis (i–k) and in 3-dpg wild-type seedlings (l). Arrowheads mark cortex/endodermis stem cells. Asterix marks the QC cells. (m,n) Longitudinal confocal section of 35S∷JKD:GFP and 35S∷MGP:GFP in Arabdidopsis root epidermal cells. (o) Phylogenetic tree of JKD/ID1 protein family. (p,q) Amino acid sequence alignment of conserved C-terminal domains. (r) Schematic representation of JKD and MGP genes. Dark boxes indicate location of zinc finger domains, pink and blue boxes indicate conserved C-terminal domains. Arrowheads mark the site of T-DNA insertions, arrows mark the translation start.
Loss of SCR function produces roots with a single layer of ground tissue with mixed endodermal and cortical identity (Di Laurenzio et al. 1996). Cell-autonomous action of SCR in the QC maintains QC identity and the activity of the surrounding stem cells (Sabatini et al. 2003). Clonal activation and deletion of SCR suggests a second cell-autonomous requirement for SCR in the rotation of the cell division plane and asymmetry of division in the ground tissue and a third role in mature endodermis cells in restricting SHR expression beyond a single ground tissue cell file (Heidstra et al. 2004). Ectopic SHR expression also suggests that in the wild type, SHR movement must be restricted to a single cell layer surrounding the stele (Sena et al. 2004; Nakajima et al. 2001). Recent data reveal that SHR protein interacts with SCR for joint up-regulation of SCR expression, and reveal how SCR can restrict SHR in a feed-forward loop (Levesque et al. 2006; Cui et al. 2007).
The question arises as to which factors regulate the three spatially separate roles of SHR and SCR. Here, we describe the JACKDAW (JKD) and MAGPIE (MGP) genes, encoding members of a plant-specific family of C2H2 zinc finger proteins, expressed from early embryogenesis onward in a subset of the SHR/SCR expression domain. We show that JKD and MGP contribute to refining the range of SHR and SCR action. We provide evidence that JKD is required for radial patterning and stem cell maintenance and that its activity is counteracted by the SHR target MGP, and we demonstrate that JKD and MGP interact physically with SCR and SHR, indicating how the SHR/SCR feed-forward loop can be differentially regulated in the stem cell domain.
Results
JACKDAW and MAGPIE gene expression in the stem cell region and ground tissue depends on SHR and SCR
The moving transcription factor SHR specifies the QC and the endodermis as a single cell layer and regulates stem cell daughter divisions. To identify new factors involved in this process, we screened ∼15,000 T-DNA insertion lines harboring a promoterless β-glucuronidase (GUS) gene adjacent to the right border of the T-DNA (Bechtold et al. 1993) for expression patterns similar to SCR and mutations affecting asymmetric division in the ground tissue. We identified one line, DWK15, with strong GUS staining in the ground tissue and QC (Fig. 1b). DWK15 homozygotes reveal ectopic periclinal divisions in the ground tissue (Fig. 1b and see below). Based on the contrasting phenotype in the ground tissue to scr mutants, we designated this mutation as jackdaw-1 (jkd-1). The flanking sequence of the insertion in jkd-1 identified a T-DNA insertion residing in the promoter region of the At5g03150 gene, 3.5 kb upstream of the predicted coding region (Fig. 1r).
JKD mRNA accumulates in the QC, the ground tissue stem cells, and to a lesser extent in mature cortex and endodermis cells (Fig. 1c). This pattern resembles the GUS staining pattern of the jkd-1 promoter trap line, and 2.8 kb of the JKD promoter fused to CFP essentially recapitulates the expression pattern observed by in situ hybridization (Fig. 1d). JKD transcript first accumulates during the 16- to 32-cell stage of embryogenesis, where it is strongest in the lower tier ground tissue or its precursor cells (Fig. 1e). At the globular stage, JKD transcript expands into the lens-shaped upper derivative of the hypophysis (Fig. 1f), and by heart stage (Fig. 1g) it takes on the QC and ground tissue expression found also in later-stage embryos (Fig. 1h). A translational JKD:GFP fusion driven by the constitutive CaMV 35S promoter localizes to the nucleus of Arabidopsis root cells (Fig. 1m).
The predicted JKD protein is a member of a large family of zinc finger proteins (Fig. 1o), defined by the Maize protein INDETERMINATE (ID1), a regulator of flowering time (Colasanti et al. 1998; Kozaki et al. 2004). The protein family is defined by the N-terminal ID domain, a highly conserved amino acid sequence consisting of a putative nuclear localization signal (NLS) followed by two C2H2 and two C2HC zinc finger motifs (Fig. 1r; Kozaki et al. 2004). We additionally identified two C-terminal domains of unknown function conserved in many members of this protein family (Fig. 1p,q). The two middle zinc fingers (Z2 and Z3) of ID1 are required for binding to a specific 11-base-pair (bp) DNA sequence (Kozaki et al. 2004), indicating that one function of ID-like proteins is transcriptional regulation.
We searched the AREX data set (Birnbaum et al. 2003) for ground tissue-expressed JKD homologs and identified At1g03840, which we designated MAGPIE (MGP), a gene independently discovered as a SHR target (Levesque et al. 2006). Like JKD, MGP:GFP protein localizes to the nucleus (Fig. 1n). MGP transcript accumulates in ground tissue and stele cells of embryos and 2-d post-germination (dpg) wild-type roots but, in contrast to JKD, mRNA is absent from the QC (Fig. 1i–l).
The initiation of JKD expression is not affected in scr and shr mutant embryos (Fig. 2a–f). However, post-embryonic QC and ground tissue stem cell expression of JKD is markedly reduced in shr-1 and shr-2 roots (Fig. 2h; data not shown) and less reduced in scr-4 mutants (Fig. 2i; Supplementary Fig. 1g–i).
Figure 2.
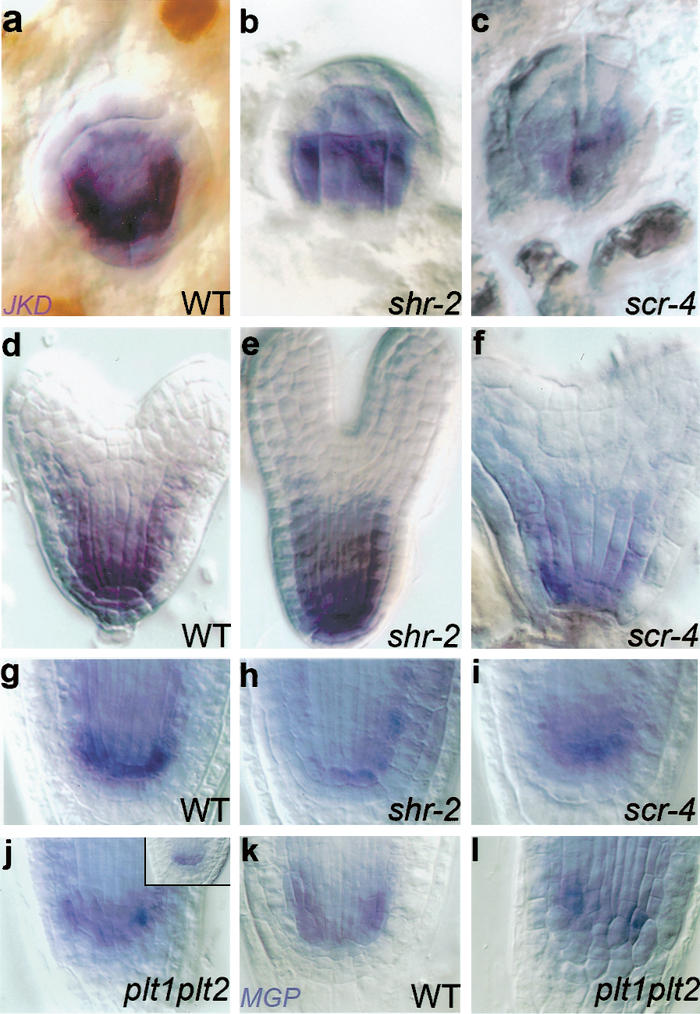
Regulation of JKD and MGP expression in the ground tissue. Whole-mount in situ hybridization of JKD (a–j) and MGP (k,l) in embryos of wild type (a,d), shr-2 (b,e), and scr-4 (c,f), and in 2-dpg seedlings of wild type (g, inset of j,k), shr-2 (h), scr-4 (i), and plt1,plt2 (j,l).
In contrast to our results with JKD, MGP transcript in a shr-2 mutant background is barely detectable by in situ hybridization at any stage (Supplementary Fig. 1b,e), whereas expression is strongly reduced in scr-4 (Supplementary Fig. 1c,f). RT–PCR analysis confirmed that SHR is required for MGP expression (Supplementary Fig. 2g,i).
Because both JKD and MGP transcription are closely associated with the stem cell niche, we assessed whether their expression is controlled by the PLT1 and PLT2 genes, which redundantly control QC specification and stem cell activity in a SHR- and SCR-independent way (Aida et al. 2004). In plt1,plt2 double mutants, neither JKD nor MGP transcript accumulation was affected in seedlings (Fig. 2j–l) or embryos (data not shown).
JKD is required for QC specification and stem cell activity and maintains SCR expression
To study the function of the JKD gene, we identified three independent jkd insertion lines and constructed a line with reduced JKD levels by RNA interference (RNAi) referred to as jkd-i (see Materials and Methods). Severely reduced levels of JKD were detected in jkd-4 homozygotes, while no signal was detected in jkd-2 homozygotes (data not shown), indicating that jkd-2 is a null mutant.
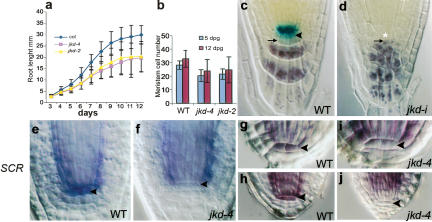
jkd mutants display a slight reduction in root length compared with wild-type (Fig. 3a), and ∼15% of jkd-4 and jkd-2 seedlings show an early initiation of lateral roots (data not shown). To determine if root meristem size is also affected in jkd mutants, we measured meristem cell number in wild-type, jkd-4, and jkd-2 seedlings. At both 5 and 12 dpg, the meristem cell number of jkd-4 and jkd-2 roots is reduced compared with that of wild type (Fig. 3b).
Figure 3.
JKD is required for QC identity and stem cell maintenance. Root length (a) and meristem cell number (b) are reduced in jkd-4 and jkd-2 compared with that of wild-type (Col-0). QC25 expression in 10-dpg wild-type (c) and jkd-i (d) roots. Lack of lugol staining marks columella stem cells in wild-type roots (arrowhead). Lugol-staining cells (arrow) designate differentiated columella cells, which neighbor defective QC cells (asterisk) in jkd-i. SCR expression in 2-dpg wild-type (e), and jkd-4 (f) roots, and in wild-type (g,h) and jkd-4 (i,j) embryos at heart stage (g,i) and torpedo stage (h,j). SCR is absent from jkd mutant QC cells (arrowhead) from heart stage onward.
The reduction in meristem size and root growth suggested a role for JKD in QC/stem cell specification and/or maintenance. To investigate JKD function in the QC, we examined several independent QC markers in jkd mutants. We were unable to detect the QC-specific promoter trap QC46 (Sabatini et al. 1999) in jkd mutants (data not shown), while QC25 expression (Sabatini et al. 1999) is normal from germination to 7 dpg but disappears at 8–9 dpg, with a concomitant disorganization of the QC and columella (Fig. 3c,d). Cells at the position of the QC in jkd mutants of that age often neighbor columella cells containing starch granules, indicating the absence of columella stem cells (Fig. 3d). Expression of QC25 and QC46 requires cell-autonomous SCR activity (Sabatini et al. 2003). We could not detect SCR expression in the QC of jkd-4 mutants (Fig. 3e,f) and detected no QC expression of pSCR∷YFPER in jkd mutant roots (Fig. 4b). To determine at what developmental time point JKD is required for QC expression of SCR, we analyzed SCR expression in jkd-4 mutant embryos. From early heart stage onward, QC expression of SCR in jkd-4 mutant embryos is absent (Fig. 3g–i), indicating a requirement for JKD in the regulation of SCR expression.
Figure 4.
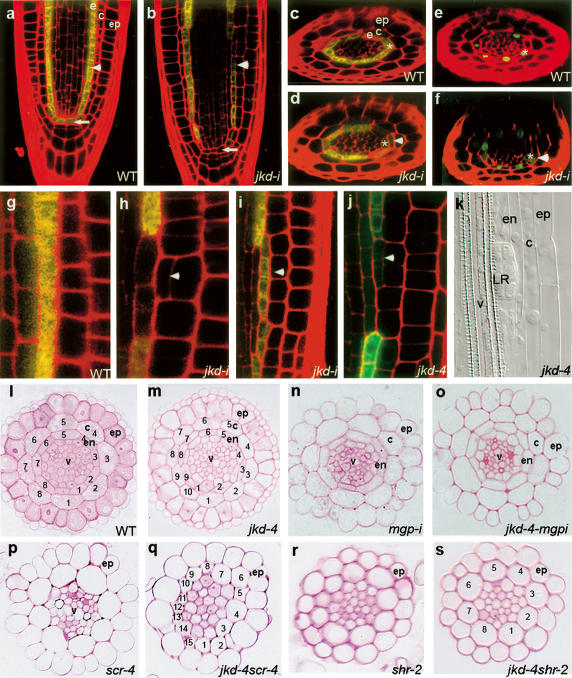
Mutations in jkd cause ectopic divisions and misexpression of SCR in the ground tissue. Longitudinal (a,b) and radial (c–f) confocal sections of 3-dpg wild-type root carrying pSCR∷YFP (c) or pSCR∷SCR:GFP (e) with two layers of ground tissue and jkd-i (d,f) roots with ectopic periclinal divisions in cortex cells (arrowhead). Arrow marks QC cells and asterix mark the endodermal cells. (g–j) Longitudinal confocal sections of pSCR∷FPER in 3-dpg wild-type (g), jkd-i (h,i), and jkd-4 roots (j). SCR is expressed in endodermal cells and is absent from cortex cells in wild type. Ectopic periclinal cortex divisions (arrowhead) in jkd mutants coincide with a reduction of SCR promoter activity in adjacent endodermis cells and induction of SCR promoter activity in the middle ground tissue layer. (k) Lateral root primordium emerging from the pericycle in jkd-4. (l–s) Root tissue sections from 10-d-old wild type (l) ([ep] epidermis; [c] cortex; [e] endodermis; [v] vascular bundle); jkd-4 (m); mgpi (n); jkd-4mgpi (o) (note that cell number in the ground tissue layers is restored to wild type); scr-4 (p); jkd-4scr-4 (q); shr-2 (r); and jkd-4shr-2 (s).
In jkd-4 scr-4 and jkd-i scr-3 double homozygotes, cells in the root meristem differentiate earlier and root lengths are shorter than in scr single mutants (Supplementary Fig. 2a,c), indicating that JKD regulates additional factors required for QC and stem cell maintenance. One candidate is the SHR protein, which regulates SCR-independent factors in the QC (Sabatini et al. 2003). To test whether JKD could act independently of SHR, we constructed jkd-4 shr-2 and jkd-i shr-2 double mutants. We observed only a slight reduction in root length compared with shr-2 single mutants (Supplementary Fig. 1b,d; t-test, P = 0.005), indicating that JKD largely but not exclusively acts through SHR in the QC.
JKD and MGP oppositely control SHR–SCR action in the ground tissue
Wild-type roots contain ground tissue with an inner endodermis and a single outer cortex layer (Figs. 1a, 4a,c). In contrast, we observed ectopic periclinal divisions in the cortex of all jkd mutants generating ground tissue with three layers (Fig. 4b,d,f). Close to the root tip, patches of ectopic cells revealed that these derive from divisions of cortex cells based on the length of the adjacent cells relative to one another and the position of subsequent divisions (Fig. 4b, h–j). These cell clones join to form a continuous extra layer higher up in the meristem (Supplementary Fig. 2h–k).
To determine whether these ectopic divisions were asymmetric, we analyzed the expression of the SCR promoter fused to ER-targeted YFP or GFP, together referred to as pSCR∷FPER. In wild-type roots, pSCR∷FPER is found in the QC, ground tissue stem cells, and endodermis (Fig. 4a,c). In both jkd4 and jkd-i, small clones of cortex cells (one to three cells) containing periclinal divisions do not show pSCR∷FPER expression. However, endodermal cells adjacent to these cells display a reduced level of pSCR∷FPER expression compared with endodermal cells above and below, and this reduction can be observed prior to the periclinal divisions (Fig. 4d, h–j). The inner cells of larger clones (more than three cells) with ectopic cortical periclinal divisions show expression of pSCR∷FPER (Fig. 4i,j), demonstrating that the ectopic divisions are asymmetric and that the inner cells take on endodermis characteristics. These new cells express also a SCR protein fusion (Fig. 4f; Supplementary Fig. 2h,i). Interestingly, in many cases, former endodermal cells adjacent to large clones no longer express pSCR∷FPER (Fig. 4i,j) and eventually acquire pericycle fate based on their capacity to generate lateral root primordia (Fig. 4k). These data, together with the reduction of pSCR∷FPER in cells adjacent to smaller clones, demonstrate a requirement for JKD in the restriction of SCR transcription in the ground tissue region.
We did not observe the ectopic periclinal divisions found in cortex cells of jkd mutants in jkd-4 scr-4 and jkd-4 shr-2 double mutants (Fig. 4p–s), indicating that these divisions require SHR and SCR and that JKD acts in the ground tissue by modifying the activity of SHR and SCR.
It is of note that the jkd mutant showed increased cortex and endodermis cell numbers within layers (Fig. 4l-m; Supplementary Table 1), revealing its requirement for restricting cell number in the ground tissue. This increase in numbers is suppressed in jkd-4 shr-2 double mutants but not in jkd-4 scr-4 mutants (Fig. 4p–s), suggesting that JKD controls cell numbers in the circumference of the ground tissue through SHR but independent of SCR.
To assess whether MGP acts in a similar process as JKD, we analyzed MGP RNAi (mgp-i) lines in wild-type and in jkd-4 background. While mgp-i lines reveal no phenotype on their own (Fig. 4n; Supplementary Fig. 2e), combination with jkd-4 homozygotes largely complements the jkd-4 ground tissue phenotype and restores pSCR∷SCR:GFP expression (Fig. 4o; Supplementary Fig. 2g,l,m).
JKD affects intracellular SHR localization in the QC and SHR expression range in the ground tissue
To determine how JKD could affect SHR activity in the QC, we analyzed pSHR∷GFP:SHR expression in the QC of jkd-4 and jkd-i roots. In contrast to the nuclear-localized GFP in wild-type roots (Fig. 5a,d), we observed cytoplasmic GFP:SHR in the majority of jkd-4 and jkd-i mutant QC cells (Fig. 5b,e; data not shown). Cui et al. (2007) demonstrated a role for SCR in sequestering SHR within the endodermis. To determine whether loss of SCR function in the QC could affect GFP:SHR localization, we examined pSHR∷GFP:SHR expression in scr-4 mutants. Despite the low expression of pSHR∷GFP:SHR in scr-4 roots, GFP:SHR mostly localizes to both nucleus and cytoplasm in cells at the position of the QC (Fig. 5c,f); however, occasionally we did observe nuclear localization of SHR (Fig. 5c, inset), as in jkd-4 (Fig. 5b, inset). Together, these results suggest that JKD controls the nuclear localization of SHR in the QC mostly through its effect on the maintenance of SCR expression.
Figure 5.
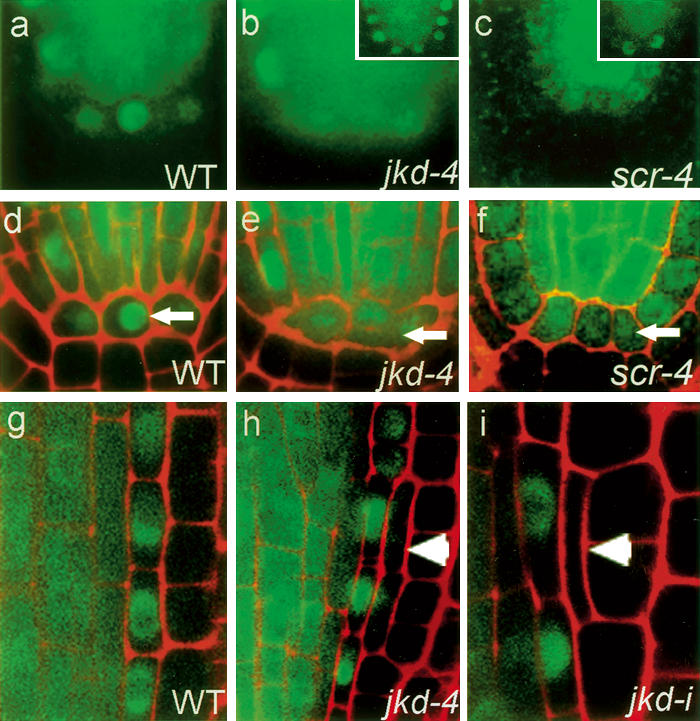
Subcellular localization of SHR in the QC and the ground tissue layers of jkd mutants. Longitudinal confocal sections of pSHR∷GFP:SHR in wild type (a,d,g), jkd-4 (b,e,h), and scr-4 (c,f). (a–c) Green channel. (d–f) Overlay. Arrows mark GFP:SHR localization in the QC cells. SHR protein is nuclear in wild type and cytoplasmic in jkd-4 and scr-4. Insets show nuclear SHR localization occasionally observed in both jkd-4 and scr-4. (g) In wild-type roots, GFP:SHR localizes to the cytoplasm and nucleus in the stele and to the nucleus in the endodermis, and is absent from cortex cells. (h,i) Ectopic periclinal cortex divisions (arrowhead) in jkd mutants cause SHR:GFP expression in two of the three ground tissue layers.
Normal levels of SCR transcript in the endodermis require functional SHR (Helariutta et al. 2000). To determine if the alterations in SCR transcription in jkd mutants were due to changes in SHR expression, we analyzed pSHR∷GFP:SHR expression in jkd-4 and jkd-i roots. In wild type, GFP:SHR localizes to both cytoplasm and nuclei in stele cells, its site of transcription, and exclusively in the nuclei of endodermis cells (Fig. 5g; Nakajima et al. 2001). In large clones of cortex cells that had divided periclinally, we observed GFP:SHR in the inner cells (Fig. 5h), while smaller clones showed no GFP: SHR (Fig. 5i). All former endodermis cells still express nuclear-localized GFP:SHR (Fig. 5 h,i) at a stage when SCR promoter activity is already reduced (cf. Fig. 4h,i). The expansion of pSHR∷GFP:SHR expression in jkd mutants is completely suppressed in the mgpi background (Supplementary Fig. 2n,o). These data show that the expression of new SCR in jkd associates with an expanded SHR domain, but lack of maintenance of SCR expression in the inner ground tissue layer is not caused by down-regulation of SHR.
JKD and MGP physically interact with SCR and SHR
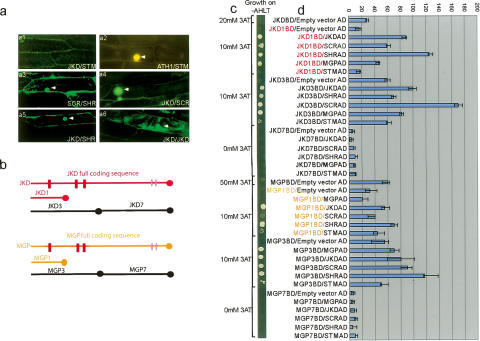
SHR and SCR proteins interact with each other, and a SHR–SCR complex binds promoters of SCR and MGP (Cui et al. 2007). To assess whether JKD and MGP could influence SHR and SCR activity at least in part through protein interaction in the stem cell area where they are jointly expressed, we performed in planta Bimolecular Fluorescence Complementation (BiFC) assays using JKD, MGP, SCR, and SHR cDNAs. We observed no fluorescence following coexpression of free nonfused YFP subfragments and negative control proteins (Supplementary Table 2). Consistent with reported interactions, fluorescence accumulated in nuclei of cells expressing SCR and SHR, and ATH1 and STM (Cole et al. 2006), but notably also in most of the combinations with JKD and MGP, suggesting multiple interactions (Fig. 6; Supplementary Table 2). These interactions were confirmed using the yeast two-hybrid system. We did not observe growth when fusion proteins (BD or AD) were tested with control empty vectors and when JKD and SCR were tested with a negative control protein. Next, we confirmed that SHR interacted strongly with SCR (Fig. 6c,d; Supplementary Fig. 2; see also Cui et al. 2007). Autoactivation domains in JKD and MGP were identified by comparing full-length cDNAs with fragments and eliminated from the analysis by selecting fragments with low autoactivation potential for subsequent experiments (Supplementary Figs. 2, 4).
Figure 6.
JKD and MGP protein interaction studies. (a) In planta analysis using BiFC. Confocal laser scanning microscopy images of bombarded onion epidermal cells coexpressing JKD with SHOOT MERISTEMLESS (STM) (panel a1), SCR with SHR (panel a3), JKD with SCR (panel a4), JKD with SHR (panel a5), and JKD with itself (panel a6). (Panel a2) A. THALIANA HOMEOBOX GENE1 (ATH1) was used as positive control for STM. (b–d) Yeast two-hybrid analysis. (b) Deletions used for mapping JKD and MGP interacting domains. (c) Growth rates on selective medium Ade–His–Leu-–Trp–(AHLT) supplemented with different 3amino triazol (3AT) concentrations; (d) Protein interactions quantified by measuring lacZ activity depicted as units of β-galactosidase. CPRG was used as the substrate. Error bars show standard deviation of the mean of triplicate assays.
Next, we showed that all interactions involving JKD and MGP required their first zinc finger domain (located between 107and 215 for JKD and 95 and 202 for MGP, designated as JKD1 and MGP1) (Fig. 6b). In pairwise combinations we found that SHR interacted with JKD1 and MGP1 (Fig. 6; Supplementary Fig. 2); JKD1 could interact with itself and with both SCR and MGP1. We detected weak interaction between SCR and MGP1 but no self-interaction for MGP and SCR. Taken together, our data strongly indicate that JKD and MGP can be part of nuclear SHR–SCR complexes.
Discussion
Here, we demonstrate that JKD and MGP, two members of a large plant C2H2 zinc finger protein family, regulate the range of SHR action. JKD transcription is initiated early in the ground tissue by a SHR- and SCR-independent mechanism, whereas MGP is activated later and depends on SHR and SCR activity. Notably, SHR and SCR can directly bind to the MGP promoter, corroborating that MGP is a direct target (Levesque et al. 2006; Cui et al. 2007). Later on, maintenance of JKD also becomes dependent on SHR and SCR. Together, these transcriptional dependencies create an intriguing pattern of gene expression where the QC and ground tissue layers are marked by JKD expression, but MGP is transcribed only in those cells that perform asymmetric cell divisions (Fig. 7a). Protein–protein interactions among SHR, SCR, MGP, and JKD indicated by independent methods suggest that the proteins form nuclear complexes in cells where they are coexpressed (Fig. 7b). Our phenotypic analyses show that JKD promotes SCR transcription and SHR nuclear localization in the QC, and that it counteracts the occurrence of supernumerary SHR–SCR-mediated asymmetric cell divisions to regulate cell type specification and stable boundary formation. MGP function is harder to assess because single mutants reveal no defects. Even double mutants, where the coregulated and phylogenetically related NUTCRACKER gene activity (Levesque et al. 2006) also has been reduced, reveal no phenotype, suggesting extensive redundancy (data not shown). However, MGP reduction in the jkd background reveals a specific phenotype: In the ground tissue, JKD restricts the range of SHR action through counteracting a MGP-dependent cell division-promoting activity that is also dependent on SHR and SCR. Given the abundant protein interactions, a plausible scenario is that MGP redundantly facilitates the asymmetric cell division-promoting activity of the SHR–SCR complex while being incorporated in it. To explain the epistasis of mgp over jkd mutants, JKD would have to inhibit this activity either by competing for binding in the same complex or by interactions within a complex that includes MGP. Future analysis on competitive binding of MGP and JKD on the SHR–SCR complex should distinguish between these possibilities.
Figure 7.
JKD and MGP action in QC and ground tissue. (a) Summary of onset of expression of JKD, SHR, SCR, and MGP genes. Only SHR and SCR are drawn in the nucleus for simplicity. (b) Model incorporating the partially cytoplasmic localization of SHR in the stele and the presumed nuclear localization and protein interactions of JKD and MGP. Note the existence of three separate zones in QC and ground tissue with different protein composition. (c) Current view on the interaction network between SHR, SCR, JKD, and MGP revealing differences in QC, stem cells, and mature endodermis. (Black arrows) Transcriptional regulation, (green arrows) regulation of nuclear localization/protein movement. Arrow thickness represents regulation strength.
In the QC, SCR is required for maintaining its own expression and nuclear localization of SHR (Heidstra et al. 2004; Cui et al. 2007; this study), and in the ground tissue, lower levels of SCR promote extra asymmetric divisions (Cui et al. 2007). Thus, the effects of jkd mutants resemble either elimination of SCR (in the QC) or reduction of SCR (in the ground tissue). Therefore, the major function of JKD might be to promote rapid up-regulation of SCR by SHR. The delay of this up-regulation by the presence of MGP could specifically poise the stem cell area to perform asymmetric divisions. The roles of JKD in nuclear sequestration of SHR in the QC and in restricting its range of action in the ground tissue might also be explained by restriction of SHR by JKD-regulated SCR, although we cannot exclude that JKD binding to SHR directly affects these processes. It is intriguing that MGP itself is a SHR target and that we could not establish a role in MGP transcriptional regulation for the second root stem cell regulatory system through the PLT genes (Aida et al. 2004). In this context, it is noteworthy that the complex transcriptional regulation and the potential to form different protein complexes indicate different and superimposed dynamic feedbacks that might establish different networks and different activities of SCR in different cells (Fig. 7c). Before all redundant partners in this network have been identified, it seems premature to speculate on the precise dynamics of this system. It is nevertheless gratifying that the presence of different networks in QC, stem cells, and endodermal cells correlates with the different activities in the ground tissue layer and with the different functions of SCR as deduced from clonal analysis.
In conclusion, SHR might be sequestrated to the nucleus in different complexes by its downstream target SCR and MGP and by JKD, whose expression is set up independently.
jkd and mgp mutants have subtle phenotypes, which could argue against a pivotal role for JKD and MGP in control of SHR activity. However, we observed similar genetic interactions between jkd and a mgp homolog (I. Blilou, H. Hassan, and B. Scheres, unpubl.). Therefore, we suspect extensive redundancy within the JKD clade. Taken together, all available data hint at an elaborate mechanism to restrict the activity of SHR action. Although such a dynamic restriction of the activity range of moving transcription factors seems to be novel, the principle of controlling the movement of instructive factors other than transcription factors is broadly used. In the Drosophila wing disc, for example, the Hedgehog (Hh) protein, initially expressed in the posterior compartment, moves one cell layer into the anterior compartment where it binds to and up-regulates through a signal transduction cascade the expression of the transmembrane receptor Patched (Ptc). Binding to Ptc serves to limit the movement of Hh beyond this single cell layer (Chen and Struhl 1996; Nybakken and Perrimon 2002). It will be interesting to further investigate whether plants have evolved different restriction mechanisms directly at the level of instructive transcription factors.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana ecotypes Wassilewskija (WS) and Columbia (Col) were used. Origins and ecotypes of mutant and transgenic lines are as follows: jkd-1 (WS), INRA T-DNA lines (Bechtold et al. 1993); jkd-3, jkd-4, and jkd-2 (Col), the SALK t-DNA collection (http://signal.salk.edu); scr-3 (Col) and scr-4 (WS) (Fukaki et al. 1998); shr-1 (WS) (Benfey et al. 1993); QC25 and QC46 (WS) (Sabatini et al. 2003); shr-2 and pSHR∷GFP:SHR (Col) (Nakajima et al. 2001); pSCR∷YFP (Col) (Aida et al. 2004). Growth conditions of seedlings and embryos are described in Sabatini et al. (1999) and Scheres et al. (1995), respectively.
Insertion sites of jkd mutants
For determination of the insertion site of jkd-1, the sequence flanking the left border (LB) of the T-DNA was amplified by Vectorette-PCR (Morrison and Markham 1995). The T-DNA insertion sites in jkd-3, jkd-4, and jkd-2 are described in the Salk Institute Web site (http://signal.salk.edu) and were confirmed by PCR-based genotyping: jkd-1, −3450 bp from the predicted translation start point; jkd-3, 1940 bp from the predicted translation start point; jkd-4, 1381 bp from the predicted translation start point; and jkd-2, −206 bp from the predicted translation start point.
Transgenic plants
The pGreenII vector set (Hellens et al. 2000; http://www.pgreen.ac.uk) was used for plant transformation. For the 35S∷JKD:GFP and 35S∷MGP:GFP translational fusion, xGFP was amplified by PCR from pGIIxGFP using the primers xGFP-L (5′-GCGAGCT CACTAGTAAAGGAGAAGAACTTTTCA-3′) and xGFP-R (5′-CGGAATTCTCATTTGTATAGTTCATCCATGCCATGT-3′) and was cloned into a cassette containing the 35S promoter. The coding sequence of JKD was amplified by PCR using the primers JKD-HA-0C-L (5′-GCGGATCCATGCAGATGATTCCAGGA GATCCAT-3′) and JKD-HA-1N-R (5′-GCGAATTCTCAAC CCAATGGAGCAAACC-3′) from root cDNA and fused in-frame before xGFP. 35S∷JKD:GFP was transferred into pGreen carrying the methotrexate resistance cassette (Heidstra et al. 2004). For pJKD:CFP, a 3.5-kb fragment of the JKD promoter was PCR-amplified using the primers K15prom-dn-L (5′-GGC GCGCTGTTCGATATCACATTTTGAC-3′) and K15prom-dn-R-mk2 (5′-GCGCGTGCTTGACTCTTTGGTTATGCC-3′) and placed before ER-CFP and the NOS terminator and transferred into pGreenII carrying the nos-basta resistance cassette. For the JKD-i and MGP-i constructs, 500-bp fragments of the JKD and MGP coding sequence were amplified with the primers K15ZFRNAi-L (5′-ATTCTAGACTCGAGCATCATCATCCCT CCCTGAT-3′) and K15ZFRNAi-R (5′-ATATCGATGGTAC CAACCTTGCGAGTTCTTGAGG-3′) for JKD and MGPi FW primer (5′-CTCGAGGGATCCGACGCTTTAGCAGAAGAAA CCGC-3′) and MGPi REV primer (5′-GGTACCATCGATATT GGTCGGTAGTAATCGTCGTC-3′) for MGP, and cloned into the pKANNIBAL vector (Wesley et al. 2001). JKD-i and MGP-i were transferred into pGreenII carrying the norflurazon resistance or methotrexate resistance cassette, respectively (Heidstra et al. 2004).
Microscopy and mRNA detection
For DIC optics, seedlings and embryos were cleared and mounted according to Willemsen et al. (1998). Starch granules and β-glucuronidase activity were visualized as described (Willemsen et al. 1998). Root length was measured as described (Willemsen et al. 1998). The number of root meristematic cells was obtained by counting cortex cells showing no signs of rapid elongation. For confocal microscopy, roots were mounted in 10 μM propidium iodide. In situ hybridization on whole-mount tissues was performed manually as described (Hejátko et al. 2006). Root tissue sections were performed as described (Willemsen et al. 1998).
Yeast two-hybrid assay
Yeast two-hybrid interactions were studied using the ProQuest Two Hybrid System (Invitrogen Life Technologies). The coding sequences of JKD, MGP, SCR, and SHR were amplified and fused to both the pDEST32 BD and pDEST22 AD vectors. Primers used to amplify full-length cDNAs and deletion fragments are described in Supplementary Table 3.
Autoactivation of yeast containing bait was tested using selective medium −His −leu and supplied with 2, 5, 10, 25, 50, 75, and 100 mM 3AT (3-Amino-1,2,4-Triazol, Fluka).
The bait and prey constructs were transformed into the yeast strains Pj694α and Pj694a, respectively. Mating and selection for interactions were performed as described in James et al. (1996). β-Gal activity was measured using CPRG as substrate according to the manufacturer’s protocol (Clontech).
Bimolecular fluorescence complementation assay
For “Split-YFP” analysis (Walter et al. 2004), the coding sequence of Enhanced Yellow Fluorescence Protein (EYFP, Clontech Laboratories) was fragmented into an N-terminal domain of 154 amino acids (YFP/N) and a C-terminal domain of 84 amino acids (YFP/C). Two sets of vectors were generated: one for C-terminal fusions of the two YFP fragments to proteins of interest (pARC235 and pARC236), and one for N-terminal fusions (pARC233 and pARC234). The YFP/N- and YFP/C-encoding fragments were generated by PCR and cloned as XbaI– BamHI fragments into pGD120, which drives expression from the CaMV35S promoter and is suitable for transient assays (Nougalli-Tonaco et al. 2006). An Ochre stop codon was introduced after codon 154 of YFP/N for the pARC233 C-terminal fusion construct. For the pARC236 N-terminal fusion construct, the stop codon of YFP/C was left out. The four pGD120-derived vectors were made Gateway compatible by introducing the RFB Gateway cassette (Invitrogen) into the BamHI-digested and -blunted vector (N-terminal fusion constructs) or XbaI-digested and -blunted vectors (C-terminal fusion constructs). This resulted in pARC233–pARC236. JKD, MGP, SCR, and SHR cDNAs were inserted to generate N-terminal YFP fusions (pARC233 and pARC234) and C-terminal fusions (pARC235 and pARC236). YFP fluorescence after bombardment of onion epidermal cells was recorded using a Leica SP2 CLSM.
Acknowledgments
We are grateful to the Benfey laboratory for sharing materials and results prior to publication, and to Viola Willemsen for screening and first identifying the DWK15 promoter trap. This work was sponsored by an NWO-PIONIER grant to B.S. and an NWO-VIDI grant to I.B.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.440307
References
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.-S., Amasino R., Scheres B., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.-S., Amasino R., Scheres B., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.-S., Amasino R., Scheres B., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.-S., Amasino R., Scheres B., Blilou I., Galinha C., Nussaume L., Noh Y.-S., Amasino R., Scheres B., Galinha C., Nussaume L., Noh Y.-S., Amasino R., Scheres B., Nussaume L., Noh Y.-S., Amasino R., Scheres B., Noh Y.-S., Amasino R., Scheres B., Amasino R., Scheres B., Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Barton M.K. Giving meaning to movement. Cell. 2001;107:129–132. doi: 10.1016/s0092-8674(01)00532-3. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Ellis J., Pelletier G., Ellis J., Pelletier G., Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Compt. Rendus Acad. Sci. III Sci. Vie. 1993;316:1194–1199. [Google Scholar]
- Benfey P.N., Linstead P.J., Roberts K., Schiefelbein J.W., Hauser M.T., Aeschbacher R.A., Linstead P.J., Roberts K., Schiefelbein J.W., Hauser M.T., Aeschbacher R.A., Roberts K., Schiefelbein J.W., Hauser M.T., Aeschbacher R.A., Schiefelbein J.W., Hauser M.T., Aeschbacher R.A., Hauser M.T., Aeschbacher R.A., Aeschbacher R.A. Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development. 1993;119:57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N., Lambert G.M., Galbraith D.W., Benfey P.N., Galbraith D.W., Benfey P.N., Benfey P.N. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Chen Y., Struhl G.I., Struhl G.I. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- Colasanti J., Yuan Z., Sundaresan V., Yuan Z., Sundaresan V., Sundaresan V. The indeterminate gene encodes a zinc finger protein and regulates a leaf generated signal required for the transition to flowering in maize. Cell. 1998;93:593–603. doi: 10.1016/s0092-8674(00)81188-5. [DOI] [PubMed] [Google Scholar]
- Cole M., Nolte C., Werr W., Nolte C., Werr W., Werr W. Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 2006;34:1281–1292. doi: 10.1093/nar/gkl016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K., Wang J.Y., Blilou I., Scheres B., Benfey P.N., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K., Wang J.Y., Blilou I., Scheres B., Benfey P.N., Vernoux T., Jung J.W., Paquette A.J., Gallagher K., Wang J.Y., Blilou I., Scheres B., Benfey P.N., Jung J.W., Paquette A.J., Gallagher K., Wang J.Y., Blilou I., Scheres B., Benfey P.N., Paquette A.J., Gallagher K., Wang J.Y., Blilou I., Scheres B., Benfey P.N., Gallagher K., Wang J.Y., Blilou I., Scheres B., Benfey P.N., Wang J.Y., Blilou I., Scheres B., Benfey P.N., Blilou I., Scheres B., Benfey P.N., Scheres B., Benfey P.N., Benfey P.N. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N., Hahn M.G., Feldmann K.A., Benfey P.N., Feldmann K.A., Benfey P.N., Benfey P.N. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Fukaki H., Wysocka-Diller J., Kato T., Fujisawa H., Benfey P.N., Tasaka M., Wysocka-Diller J., Kato T., Fujisawa H., Benfey P.N., Tasaka M., Kato T., Fujisawa H., Benfey P.N., Tasaka M., Fujisawa H., Benfey P.N., Tasaka M., Benfey P.N., Tasaka M., Tasaka M. Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 1998;14:425–430. doi: 10.1046/j.1365-313x.1998.00137.x. [DOI] [PubMed] [Google Scholar]
- Heidstra R., Welch D., Scheres B., Welch D., Scheres B., Scheres B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes & Dev. 2004;18:1964–1969. doi: 10.1101/gad.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N., Jung J., Sena G., Hauser M.T., Benfey P.N., Sena G., Hauser M.T., Benfey P.N., Hauser M.T., Benfey P.N., Benfey P.N. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M., Leyland N.R., Bean S., Mullineaux P.M., Bean S., Mullineaux P.M., Mullineaux P.M. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Hejátko J., Blilou I., Brewer P., Friml J., Scheres B., Benková E., Blilou I., Brewer P., Friml J., Scheres B., Benková E., Brewer P., Friml J., Scheres B., Benková E., Friml J., Scheres B., Benková E., Scheres B., Benková E., Benková E. In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat. Protoc. 2006;1:1939–1946. doi: 10.1038/nprot.2006.333. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A., Halladay J., Craig E.A., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki A., Hake S., Colasanti J., Hake S., Colasanti J., Colasanti J. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 2004;32:1710–1720. doi: 10.1093/nar/gkh337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque M., Vernoux T., Cui H., Wang J.Y., Blilou I., Hassan H., Nakajima K., Matsumoto N., Scheres B., Benfey P., Vernoux T., Cui H., Wang J.Y., Blilou I., Hassan H., Nakajima K., Matsumoto N., Scheres B., Benfey P., Cui H., Wang J.Y., Blilou I., Hassan H., Nakajima K., Matsumoto N., Scheres B., Benfey P., Wang J.Y., Blilou I., Hassan H., Nakajima K., Matsumoto N., Scheres B., Benfey P., Blilou I., Hassan H., Nakajima K., Matsumoto N., Scheres B., Benfey P., Hassan H., Nakajima K., Matsumoto N., Scheres B., Benfey P., Nakajima K., Matsumoto N., Scheres B., Benfey P., Matsumoto N., Scheres B., Benfey P., Scheres B., Benfey P., Benfey P. A whole-genome meta-analysis predicts the SHORT-ROOT developmental subnetwork in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison J.F., Markham A.F., Markham A.F. PCR-based approaches to human genome mapping. In: McPherson M.J., et al., editors. PCR2— A practical approach. Oxford University Press; Oxford, UK: 1995. pp. 165–196. [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N., Sena G., Nawy T., Benfey P.N., Nawy T., Benfey P.N., Benfey P.N. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Nougalli Tonaco I.A., Borst J.W., de Vries S.C., Angenent G.C., Immink R.G.H., Borst J.W., de Vries S.C., Angenent G.C., Immink R.G.H., de Vries S.C., Angenent G.C., Immink R.G.H., Angenent G.C., Immink R.G.H., Immink R.G.H. In-vivo imaging of MADS box transcription factor interactions. J. Exp. Bot. 2006;57:33–42. doi: 10.1093/jxb/erj011. [DOI] [PubMed] [Google Scholar]
- Nybakken K., Perrimon N., Perrimon N. Hedgehog signal transduction: Recent findings. Curr. Opin. Genet. Dev. 2002;12:503–511. doi: 10.1016/s0959-437x(02)00333-7. [DOI] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Benfey P., Leyser O., Bechtold N., Weisbeek P., Leyser O., Bechtold N., Weisbeek P., Bechtold N., Weisbeek P., Weisbeek P., et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B., Heidstra R., Wildwater M., Scheres B., Wildwater M., Scheres B., Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes & Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B., Dilaurenzio L., Willemsen V., Hauser M.T., Janmaat K., Weisbeek P., Benfey P.N., Dilaurenzio L., Willemsen V., Hauser M.T., Janmaat K., Weisbeek P., Benfey P.N., Willemsen V., Hauser M.T., Janmaat K., Weisbeek P., Benfey P.N., Hauser M.T., Janmaat K., Weisbeek P., Benfey P.N., Janmaat K., Weisbeek P., Benfey P.N., Weisbeek P., Benfey P.N., Benfey P.N. Mutations affecting the radial organization of the Arabidopsis root display specific defects throughout the embryonic axis. Development. 1995;121:53–62. [Google Scholar]
- Sena G., Jung J.W., Benfey P.N., Jung J.W., Benfey P.N., Benfey P.N. A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development. 2004;131:2817–2826. doi: 10.1242/dev.01144. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Blazevic D., Grefen C., Schumacher K., Oecking C., Grefen C., Schumacher K., Oecking C., Schumacher K., Oecking C., Oecking C., et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- Wesley S.V., Helliwell C.A., Smith N.A., Wang M.B., Rouse D.T., Liu Q., Gooding P.S., Singh S.P., Abbott D., Stoutjesdijk P.A., Helliwell C.A., Smith N.A., Wang M.B., Rouse D.T., Liu Q., Gooding P.S., Singh S.P., Abbott D., Stoutjesdijk P.A., Smith N.A., Wang M.B., Rouse D.T., Liu Q., Gooding P.S., Singh S.P., Abbott D., Stoutjesdijk P.A., Wang M.B., Rouse D.T., Liu Q., Gooding P.S., Singh S.P., Abbott D., Stoutjesdijk P.A., Rouse D.T., Liu Q., Gooding P.S., Singh S.P., Abbott D., Stoutjesdijk P.A., Liu Q., Gooding P.S., Singh S.P., Abbott D., Stoutjesdijk P.A., Gooding P.S., Singh S.P., Abbott D., Stoutjesdijk P.A., Singh S.P., Abbott D., Stoutjesdijk P.A., Abbott D., Stoutjesdijk P.A., Stoutjesdijk P.A., et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Willemsen V., Wolkenfelt H., de Vrieze G., Weisbeek P., Scheres B., Wolkenfelt H., de Vrieze G., Weisbeek P., Scheres B., de Vrieze G., Weisbeek P., Scheres B., Weisbeek P., Scheres B., Scheres B. The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development. 1998;125:521–531. doi: 10.1242/dev.125.3.521. [DOI] [PubMed] [Google Scholar]
- Wu X., Weigel D., Wigge P.A., Weigel D., Wigge P.A., Wigge P.A. Signaling in plants by intercellular RNA and protein movement. Genes & Dev. 2002;16:151–158. doi: 10.1101/gad.952002. [DOI] [PubMed] [Google Scholar]