Abstract
ASY1 is an Arabidopsis protein required for synapsis and crossover formation during meiosis. The chronology of meiotic recombination has been investigated in wild type and an asy1 mutant. We observe a delay between the appearance of chromatin-associated AtSPO11-1 foci and DNA double-strand break (DSB) formation, which occurs contemporaneously with chromosome axis formation and transition of ASY1 from chromatin-associated foci to a linear axis-associated signal. DSBs are formed independently of ASY1 in an AtSPO11-1-dependent manner. They are partially restored in Atspo11-1-3 using cisplatin, but their control appears abnormal. Axis morphogenesis is independent of ASY1, but axis structure may be compromised in asy1. Localization of the strand exchange proteins AtRAD51 and AtDMC1 to the chromatin occurs asynchronously shortly after DSB formation, with AtDMC1 localizing in advance of AtRAD51. In wild-type nuclei, both recombinases form numerous foci that persist for ∼12 h before gradually decreasing in number. In asy1, initial localization of AtDMC1 is normal, but declines abruptly such that interhomolog recombination is severely compromised. Limited ASY1-independent, DMC1-dependent interhomolog recombination remains, but appears restricted to subtelomeric sequences where the homologs are fortuitously in proximity. Thus, ASY1 plays a key role in coordinating the activity of the RecA homologs to create a bias in favor of interhomolog recombination.
Keywords: Arabidopsis, meiosis, ASY1, recombination, DNA double-strand breaks, strand invasion
The accurate reductional segregation of homologous chromosomes (homologs) at the first meiotic division is dependent on the formation of chiasmata during prophase I (Jones 1984). Chiasmata are cytological structures that occur at sites corresponding to the reciprocal exchange of DNA crossovers (COs) resulting from homologous recombination (HR). Chiasmata provide physical connections between nonsister chromatids that, together with bipolar attachment of the kinetochores, are required to ensure segregation of the recombined maternal and paternal homologs to opposite poles at the first division (Hawley 1988). HR is initiated at leptotene by the formation of a programmed set of DNA double-strand breaks (DSBs) catalyzed by SPO11, a topoisomerase-related enzyme (Keeney et al. 1997). In the majority of organisms the number of COs is low, typically one to three per chromosome pair, but the number of DSBs that are initially formed is far greater (10- to 40-fold) (Bishop 1994; Anderson et al. 2001; Moens et al. 2002). Thus, the majority of DSBs are destined to become non-CO products.
In many eukaryotes, the early stages of meiotic recombination are catalyzed by the concerted activity of two orthologs of the bacterial RecA protein, Rad51 and Dmc1 (Neale and Keeney 2006). Rad51 plays a crucial role in mitotic recombination, where it mediates strand invasion between sister chromatids, and in meiosis where it acts in conjunction with its meiosis-specific partner, Dmc1, to promote interhomolog recombination. Based on earlier studies, it was suggested that the switch from intersister recombination in vegetative cells to meiotic interhomolog recombination was dependent on Dmc1 (Bishop et al. 1992). However, subsequent investigations in budding yeast revealed that loss of the interhomolog recombination activity of Dmc1 could be overcome by overexpression of Rad51 or Rad54, which stimulates Rad51 activity, indicating that, in this species at least, the specificity of repair partner choice during meiosis does not entirely rest on Dmc1 (Bishop et al. 1999; Tsubouchi and Roeder 2003). This is supported by work that has revealed several proteins that are important regulators of this process in budding yeast. Studies indicate that a protein complex comprising Hop1, Red1, and Mek1 is instrumental in establishing interhomolog bias by virtue of its capacity to actively prevent Rad51-mediated intersister chromatid repair (Hollingsworth and Ponte 1997; Woltering et al. 2000; Niu et al. 2005). It has also been proposed that HIM-3, a protein that is related to Hop1 on the basis that it contains a HORMA domain and is a component of chromosome axes, may also prevent the use of sister chromatids for repair during meiosis in Caenorhabditis elegans (Zetka et al. 1999; Couteau et al. 2004).
ASY1 is an Arabidopsis HORMA domain protein that exhibits homology within this domain with Hop1 and HIM3 (Caryl et al. 2000). Immunolocalization studies have revealed that ASY1 is initially detected as numerous punctate foci in pollen mother cells (meiocytes) during G2. As G2 progresses, the foci assume a more linear nature such that by the onset of leptotene a linear signal running the full length of the homologous chromosomes is visible. Immunogold localization in conjunction with electron microscopy indicates that ASY1 is closely associated with the chromosome axes, but not the chromatin loops of the sister chromatids. The linear ASY1 signal persists until late pachytene before disappearing as the homologs desynapse (Armstrong et al. 2002). Mutants lacking ASY1 are asynaptic with a majority of chromosomes present as univalents at metaphase I, although some chiasmata are detected at a mean frequency of 1.39 per cell, which is ∼15% that of the wild-type plants (Ross et al. 1997; Sanchez-Moran et al. 2001).
Here, we have investigated the chronology of the early meiotic recombination pathway in Arabidopsis, focusing on the role of ASY1 and its interrelationship with the HR pathway. Time-course experiments reveal that formation of DSBs is synchronized with chromosome axis formation. We provide evidence that recombination is initiated normally in an asy1 mutant, but fails to progress as usual such that DSBs are repaired without the normal subset progressing to form COs. The data reveal that ASY1 acts at the interface between the developing chromosome axes and the recombination machinery, where it is required to ensure AtDMC1-mediated interhomolog recombination.
Results
Previously, we characterized an asy1 T-DNA mutant in a Wassilewskija (Ws) genomic background (Caryl et al. 2000). For this report, we have analyzed a T-DNA insertion line in a Columbia (Col-0) background. Both mutant lines showed identical asynaptic and chiasma frequency phenotypes (Supplementary Fig. 1).
In order to obtain an accurate chronology of the early recombination pathway in wild-type meiocytes and to investigate the role of ASY1 in these events, most of the immunolocalization of meiotic proteins was conducted in conjunction with prior BrdU pulse-labeling (2-h pulse) of the meiocytes during meiotic S phase as previously described (Fig. 1A; Armstrong et al. 2003).
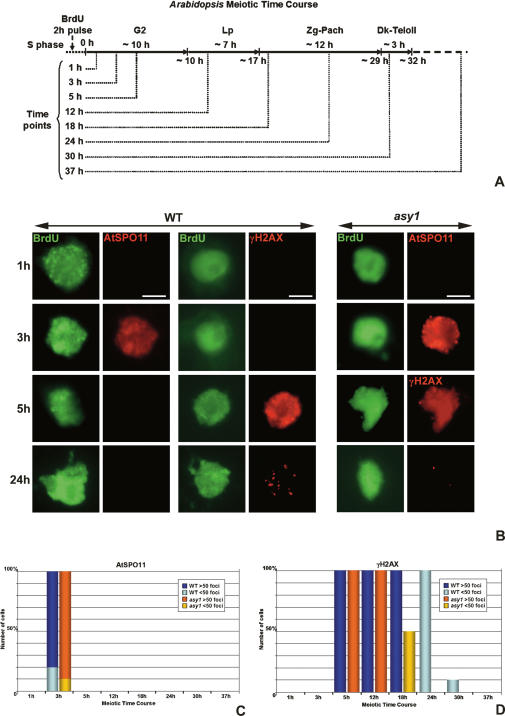
Figure 1.
Time course of DNA DSB formation. (A) Scheme of the Arabidopsis meiotic time course assessed by anti-BrdU pulse-chase labeling of nuclear DNA in meiotic S phase. Time points where immunolocalization of meiotic proteins was performed are indicated. (Lp) Leptotene, (Zg-Pach) zygotene–pachytene, (Dk-TeloII) diakinesis-telophase II. (B) Time course of AtSPO11-1 and γH2AX localization in squash preparations of meiocytes in Arabidopsis wild type (WT) and asy1 mutant. Note that for asy1, localization of AtSPO11 is shown at 1 h and 3 h and γH2AX at 5 h and 24 h. All the images are Z-stack projections. Bar, 5 μM. (C) Histogram showing AtSPO11-1 time course in wild type and asy1 mutant. (D) Histogram showing γH2AX time course in wild type and asy1 mutant. Percentage of nuclei with >50 or <50 foci are indicated.
Monitoring DNA DSB formation in Arabidopsis
In budding yeast, meiotic DSB formation is catalyzed by Spo11 (Keeney et al. 1997). It appears that DSBs are formed in the same manner in Arabidopsis by the homologous protein AtSPO11-1 in conjunction with AtSPO11-2 (Grelon et al. 2001; Stacey et al. 2006). To enable us to investigate the role of ASY1 in relation to the HR pathway, it was necessary to develop a procedure that would allow us to monitor DSB formation since this is the earliest step in the HR pathway. Two approaches were used. First, we immunolocalized AtSPO11-1 using an anti-AtSPO11-1 peptide antibody (Ab); and second, DSB formation was detected using an Ab that recognizes the phosphorylated form of the histone variant H2AX (γH2AX), which has previously been used for this purpose in other species (Paull et al. 2000; Mahadevaiah et al. 2001; Shroff et al. 2004).
Time-course studies using wild-type meiocytes pulse-labeled with BrdU revealed the accumulation of AtSPO11-1 foci in early G2 (Fig. 1B,C; Supplementary Fig. 2). At 1 h post-S phase, no foci were observed, but by 3 h a majority (80%) of meiocytes at this time point contained >50 foci. The average number of foci at 3 h was 88.4 (±8.7, n = 50). However, by 5 h, AtSPO11-1 foci were no longer detectable. This suggests that the protein undergoes a rapid cycle of accumulation and disappearance in meiocytes over a period of between 1 and 5 h post-S phase.
H2AX is a meiosis-specific isoform of histone H2A. Upon DSB formation, rapid accumulation of phosphorylated H2AX (γH2AX) occurs around the break site. Recent studies in yeast indicate that this modification extends over a region of 50 kb, although there appears to be little γH2AX in the 1–2 kb immediately flanking the DSB (Shroff et al. 2004). Nevertheless, H2AX phosphorylation is a useful cytological marker for DSB formation and has been applied in a range of species. Given the transient nature of AtSPO11-1, we investigated whether immunolocalization of γH2AX could provide an alternative approach to monitor DSB formation in Arabidopsis. As expected from the behavior of AtSPO11-1, γH2AX foci also accumulated in early G2. Immunolocalization studies in spread preparations of wild-type meiocytes at G2/early leptotene revealed the accumulation of numerous rather diffuse γH2AX foci throughout the chromatin (Fig. 1B,D). However, their accumulation was not contemporaneous with that of AtSPO11-1. At 3 h post-S when the number of AtSPO11-1 foci detected was maximal, no γH2AX foci were detected. During the 3- to 5-h window when AtSPO11-1 foci rapidly disappeared, there was an equally swift accumulation of γH2AX to a maximum of >50 diffuse foci. The level of γH2AX then remained constant for a further 13 h before undergoing a gradual decrease to 10–20 foci in the 18- to 24-h post-S period. By 30 h the foci had disappeared from the chromatin.
At pachytene (24 h), the mean number of γH2AX foci per nucleus was 10.0 (±0.98, n = 50). Since this approximates numerically the chiasma frequency in wild-type Arabidopsis, it suggests that these foci may represent CO sites. To investigate this, dual immunolocalization using anti-γH2AX Ab and an Ab against the MutL homolog AtMLH3, which in conjunction with its partner AtMLH1 localizes to the sites of COs, was conducted (Marcon and Moens 2003; Jackson et al. 2006). An average of 52% (n = 50) of the foci exhibited colocalization (Supplementary Fig. 3). This observation is consistent with a dynamic situation whereby AtMLH3 accumulates at CO sites at the time that γH2AX is disappearing. Similar patterns of turnover of recombination components, such as RPA and DMC1, have previously been reported (Moens et al. 2002).
Formation of γH2AX foci is AtSPO11-dependent
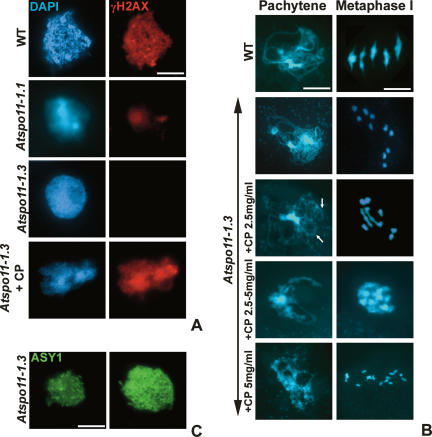
To confirm that the presence of γH2AX foci at G2/early leptotene was dependent on AtSPO11-1-induced DSBs, immunolocalization studies were conducted in two lines carrying different mutant alleles of AtSPO11-1. Line Atspo11-1-1 was previously reported to exhibit a substantial reduction in recombination such that the chiasma frequency at metaphase I was ∼7% that of wild type (Grelon et al. 2001). This would suggest that DSB formation is greatly reduced, but not entirely absent. It is reported on the basis of reduced seed-set that the Atspo11-1-3 allele is more severe than Atspo11-1-1 (Stacey et al. 2006). To confirm this was due a further reduction in chiasma formation, a sample of >300 Atspo11-1-3 meiocytes were analyzed at metaphase I. This revealed a total lack of chiasmata, suggesting that this allele is completely defective in SPO11-1-catalyzed DSB formation (data not shown). Immunolocalization of γH2AX in chromosome spreads of meiocytes at leptotene prepared from each mutant revealed an effect commensurate with the apparent reduction in AtSPO11-1 activity. In Atspo11-1-1 there was a substantial reduction in signal compared with wild type at the same meiotic stage, and in the case of Atspo11-1-3 there was a complete absence of γH2AX foci (Fig. 2A). These observations provide strong evidence that the formation of γH2AX foci directly reflects the activity of AtSPO11-1 and is thus a reliable method for monitoring DSB formation in Arabidopsis. Nevertheless, to substantiate this we investigated whether restoration of γH2AX foci occurred when artificial DSBs were introduced into Atspo11-1-3. Cis-platinum(II)diamine dichloride (cisplatin) is a chemotherapeutic agent that is capable of forming monoadducts (Kartalou and Essigmann 2001). Cisplatin reacts with DNA to form intra- and interstrand cross-links; the excision of DNA cross-links creates DSBs, which can stimulate recombinational repair and meiotic recombination (Wu and Lichten 1994; Hanneman et al. 1997). Chromosome spread preparations were prepared from meiocytes isolated from Atspo11-1-3 flower buds at different time points following a 2-h pulse of cisplatin applied at a range of concentrations. These were subjected to cytological and immunological analysis (Fig. 2B). Inspection of DAPI-stained prophase I chromosome spreads revealed that treatment with cisplatin at concentrations ≥1 mg/mL restored bivalent formation, indicating that DSB formation had occurred. Immunolocalization of the synaptonemal complex (SC) transverse filament protein ZYP1 (Higgins et al. 2005) also revealed the partial restoration of chromosome synapsis (Supplementary Fig. 4). The number of bivalents was broadly dose-dependent, with increasing numbers of chiasmata found at higher levels of cisplatin treatment, but with some differences compared with wild type. At a cisplatin concentration of 2.5 mg/mL, nuclei containing up to three rod bivalents were observed at metaphase I. However, a further increase in the level of cisplatin did not result in the formation of any cells with a normal complement of five bivalents. Chiasma frequency was increased, but inspection of a sample of 50 meiocytes revealed that this led to multivalent formation at 2.5–5 mg/mL cisplatin. At 5 mg/mL, extensive chromosome fragmentation was observed (Fig. 2B). Thus, it appears that while cisplatin does introduce DSBs, it does not directly compensate for the loss of SPO11-1 activity. Immunolocalization of γH2AX in cisplatin-treated meiocytes revealed the presence of numerous foci throughout the chromatin (Fig. 2A). The number of foci and intensity of γH2AX staining at G2/leptotene increased in a dose-dependent manner as the concentration of cisplatin was raised. As prophase I progressed, the foci decreased in number in the same manner as previously observed in wild-type meiocytes. We therefore conclude that immunolocalization of γH2AX provides a reliable method for monitoring meiotic DSB formation in Arabidopsis.
Figure 2.
γH2AX accumulation is dependent on DSB formation, whereas ASY1 localization is DSB independent. (A) γH2AX immunolocalization in meiocyte spread preparations of wild type, spo11-1-1, spo11-1-3, and spo11-1-3 treated with a cisplatin pulse. (B) Light micrographs of DAPI-stained pachytene and metaphase I nuclei of wild type, and spo11-1-3 following cisplatin (CP) treatment (0–5 mg/mL). Arrows indicate pairing and synapsis of some chromosome regions. (C) Immunolocalization of ASY1 in spo11-1-3 meiocytes at G2 (left) and leptotene (right). Bar, 5 μM.
ASY1 localization and DSB formation are independent
Mutation of ASY1 results in asynapsis and a severe reduction in chiasma formation at metaphase I, such that a majority of the chromosomes are present as univalents (Ross et al. 1997). We were therefore interested to determine if this observation reflected a reduction in DSB formation or a failure in recombination progression. Immunolocalization of both AtSPO11-1 and γH2AX was conducted in meiocytes from an asy1 mutant that had been pulse-labeled with BrdU during S phase. This revealed that the number of AtSPO11 foci and their chronology was identical to wild-type nuclei (Fig. 1B,C). This was also the case for the accumulation of foci corresponding to γH2AX at early time points (Fig. 1B,D). Together, these observations suggest that DSB formation is independent of ASY1. As prophase I progressed, one apparent difference did emerge in the γH2AX staining. Whereas in wild ty10–2 γH2AX foci remained at 18–24 h, only one to two foci remained at 18 h in the asy1 mutant (Fig. 1B,D). This is a substantial reduction relative to wild type, but does correspond to the number COs that remain in an asy1 mutant. Treatment of asy1 with cisplatin did not lead to detectable synapsis or an increase in chiasma frequency (data not shown). Together, these observations indicate that DSB formation occurs normally in asy1, but normal meiotic recombination progression fails at some critical early stage in the absence of the protein.
To examine the behavior of ASY1 in the absence of DSB formation, immunolocalization of ASY1 was carried out in chromosome spreads prepared from Atspo11-1-3. Distribution of the protein appeared identical to that in wild-type meiocytes, starting as numerous foci before forming linear signals extending the full length of the chromosome axes (Fig. 2C). Hence, the association of ASY1 with the chromosome axes and DSB formation occur as independent events. Nevertheless, it appears that in wild-type cells these events are temporally coordinated during G2/early prophase I.
ASY1 is required for normal meiotic recombination progression
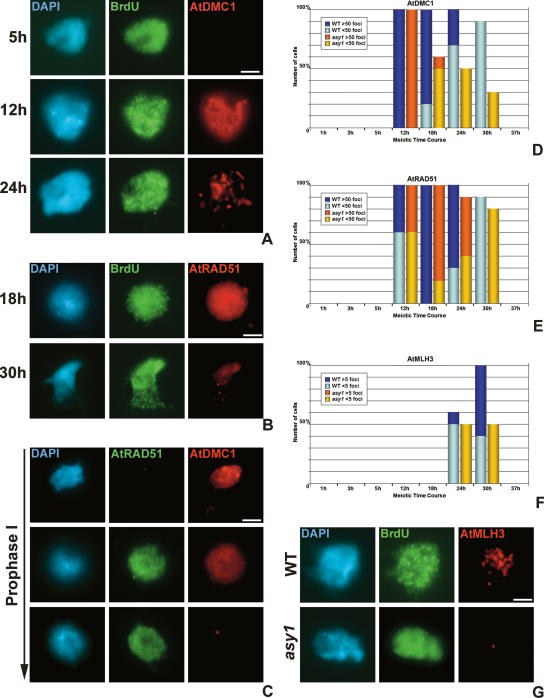
Our studies indicate that DSB formation is normal in an asy1 mutant, but recombination progression is perturbed such that the chiasma frequency at metaphase I is dramatically reduced. To investigate the basis of this observation, immunolocalization studies were conducted with Abs that recognize key recombination proteins. Localization of the Arabidopsis RecA homologs AtDMC1 and AtRAD51 (Klimyuk and Jones 1997; Li et al. 2004) was conducted in chromosome preparations from wild-type and asy1 meiocytes following BrdU pulse-labeling. In wild-type nuclei at 5 h post-S phase there was no evidence of AtDMC1 localization, but by 12 h there was significant accumulation of foci to >50 foci per meiocyte. By 18 h the majority (80%) of the meiocytes retained >50 foci, but in a few cases the number had reduced to <50. This reduction continued such that in the 24 h sample most nuclei contained <20 foci, and by 30 h only one or two foci remained. At 37 h no AtDMC1 foci were detectable (Fig. 3A,D). From the analysis of chromosome spread preparations, the peak number of AtDMC1 foci at mid-prophase I was 138 (±11.45, n = 50).
Figure 3.
ASY1 is required for normal progression of meiotic recombination. Time-course analysis of AtDMC1 (A) and AtRAD51 (B) localization. (C) Coimmunolocalization of AtDMC1 and AtRAD51 early to late prophase I. (D) Histogram of AtDMC1 time course in wild-type and asy1 cells. (E) Histogram of AtRAD51 time course. (F) Histogram of AtMLH3 time course. (G) Localization of AtMLH3 in wild-type and asy1 nuclei at 24 h (pachytene). Bar, 5 μM.
The general pattern of AtRAD51 localization mirrored that of AtDMC1, but with a difference in chronology. As with AtDMC1, no AtRAD51 foci were detected in the 5-h sample, but in 12-h post-S-phase nuclei, when >50 AtDMC1 foci were detectable in all nuclei, ∼55% still had <50 AtRAD51 foci. By 18 h, when a slight decrease in the number of AtDMC1 foci was apparent, all nuclei retained >50 AtRAD51 foci. At 24 h the proportion of nuclei with >50 AtRAD51 foci was still greater than those with <50, closely resembling the distribution of AtDMC1 foci at 18 h. There then followed a continual reduction of foci, such that 37 h none remained (Fig. 3B,E). The average number of AtRAD51 foci at mid-prophase I was 120 (±5.29, n = 50). These observations suggest that accumulation of AtDMC1 on the chromatin slightly precedes that of AtRAD51. Dual immunolocalization using anti-AtDMC1 and anti-AtRAD51 Abs reinforced this conclusion, since early prophase I nuclei with >50 AtDMC1 foci, but with little or no AtRAD51, were regularly observed, whereas nuclei stained with AtRAD51, but with little AtDMC1, were observed only at late stages of prophase I (Fig. 3C).
Initially, the localization of AtDMC1 in asy1 was identical to wild type. At 12 h, >50 AtDMC1 foci were detectable in all nuclei, again contrasting with AtRAD51, where a smaller proportion (∼60%) of nuclei at this time point had accumulated <50 foci. However, at 18 h, in marked contrast to wild-type cells, there was a dramatic reduction in the number of AtDMC1 foci such that only one or two remained per nucleus. At 24 h the mean number of foci per nucleus was 1.0 (n = 50), whereas the corresponding figure for wild type was 22.7 (n = 50). Occasional foci were detected in asy1 cells up to 30 h but had disappeared by 37 h (Fig. 3D). The pattern of AtRAD51 localization in asy1 was very similar to wild type save for the fact that reduction in the number of foci was slightly more rapid in the mutant. At 18 h all wild-type nuclei contained >50 AtRAD51 foci, whereas 20% of asy1 nuclei had <50. By 30 h, <50 foci remained in both wild-type and mutant cells, but the mean number of foci in the latter was 15.7 (n = 50), which was significantly lower than the wild type, which contained an average of 25.9 (n = 50) (t-test, P = 0.0049) (Fig. 3E). Overall, prophase I was completed 1–2 h faster in asy1 compared with wild type.
These observations suggest that in the absence of ASY1, AtDMCI fails to form a stable association with the majority of early recombination intermediates. Consistent with this, there was an effect on localization of proteins active at later stages in the recombination pathway. Immunolocalization of AtMLH3 revealed that the protein was initially detectable in wild-type cells at ∼24 h post-S phase. By 30 h, up to 11 foci per nucleus were observed, which is in accord with the number of chiasmata that are found in wild-type nuclei at metaphase I (Fig. 3F,G). Although there was no obvious effect on the chronology of AtMLH3 localization in the asy1 mutant, the number of foci was significantly reduced to approximately one per cell (t-test, P = 0.00024) (Fig. 3G). This corresponds to the number of chiasmata that remain in asy1.
Activity of the RecA homologs in an asy1 mutant
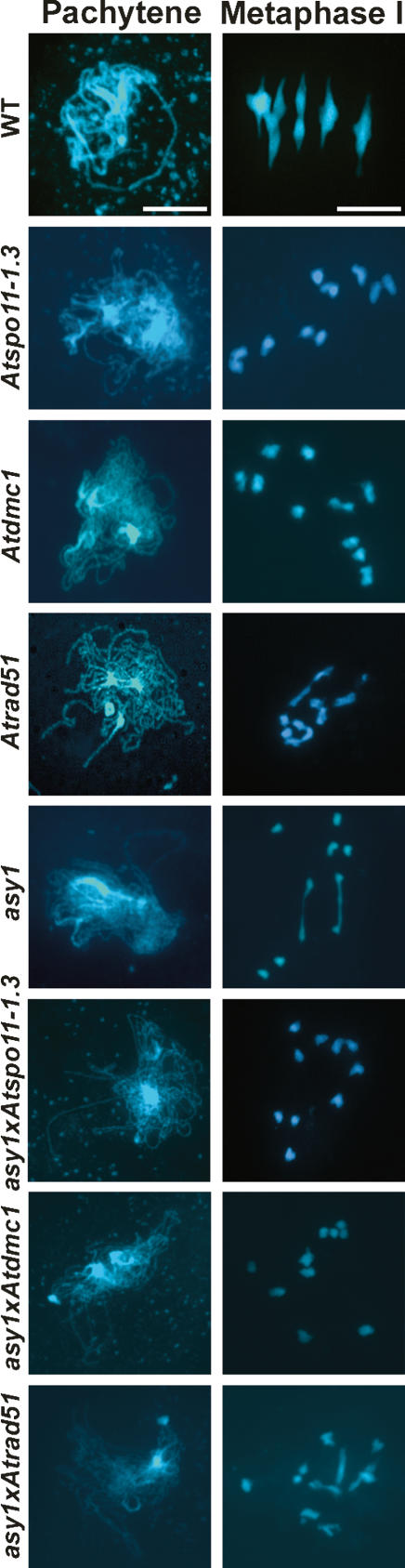
Overall, our observations indicate that in asy1 the subset of CO-designated DSBs do not progress as normal, but are nevertheless repaired since there is no evidence of chromosome fragmentation. This phenotype is reminiscent of that found in an Atdmc1 mutant (Couteau et al. 1999). To investigate this further, we constructed an asy1/Atrad51 double mutant. Cytological analysis of this mutant revealed extensive post-metaphase I chromosome fragmentation (Fig. 4). This result directly implicates AtRAD51 in the repair of DSBs in asy1 and is consistent with the observation that the localization of AtRAD51 at early prophase I is essentially normal.
Figure 4.
Light micrograph of DAPI-stained pachytene and metaphase I nuclei in wild type and single and double mutants (as indicated). Bar, 5 μM.
Although chromosome fragmentation is absent in both asy1 and Atdmc1, they are not phenotypically identical, as asy1 has a mean chiasma/CO frequency of 1.27 whereas Atdmc1 fails to form COs (Ross et al. 1997; Couteau et al. 1999). To investigate the origin of the remaining COs in asy1, we constructed an asy1/Atdmc1 double mutant. Cytological analysis of this line revealed a complete absence of COs (n = 50) (Fig. 4). Thus, it appears that the residual COs in asy1 are AtDMC1-dependent. This is consistent with the observation that despite a dramatic decrease in the number of AtDMC1 foci I in asy1 compared with wild type at mid-prophase I, one or two foci persist in the mutant.
Chromosome axis organization in asy1
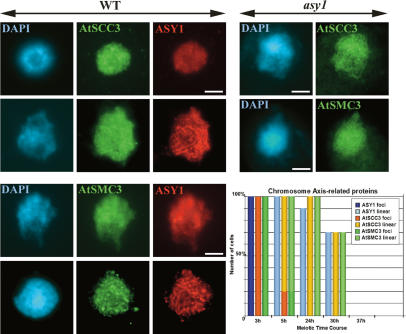
Previous studies have indicated that ASY1 localizes to the chromatin that is associated with the chromosome axes (Armstrong et al. 2002). We were therefore interested to determine the chronology of axis formation in relation to the juxtaposition of ASY1 foci to form a linear axis-associated signal. The chronology of ASY1 localization together with the cohesins AtSCC3 and AtSMC3, which are components of the complex responsible for sister chromatid cohesion (Nasmyth 2001; Chelysheva et al. 2005; Lam et al. 2005), was investigated in meiocytes pulse-labeled with BrdU. All three proteins were initially detected as numerous diffuse foci in all nuclei 3 h post-S phase. As the chromosome axes were elaborated, the foci disappeared and the proteins became detectable as linear signals that extended fully along each homolog. By 5 h all the ASY1 foci and ∼90% of the AtSCC3 and AtSMC3 foci had been replaced by linear signals. At 24 h all three proteins remained detectable as signals running the full length of each homolog. These signals persisted in a majority (∼75%) of nuclei until 30 h, but by 37 h the signal was no longer apparent in any of the nuclei observed (Fig. 5). These studies reveal that the transition of ASY1 foci to a form a linear axis-associated signal is concomitant with axis formation as a cytologically identifiable entity. Moreover, this transition, although independent of DSB formation, occurs at the time that γH2AX foci first appear.
Figure 5.
Chromosome axis formation in wild type (WT) and asy1 mutant. Immunolocalization of ASY1, AtSCC3, and AtSMC3 in wild-type and asy1 nuclei. Bar, 5 μM. Histogram of the time course of localization of axis-associated proteins in wild type.
Given the close temporal coordination of localization of the axis-associated proteins, we next explored if loss of ASY1 had a direct impact on chromosome axis formation. Previous inspection of DAPI-stained chromosome spread preparations from male and female meiocytes suggested chromosome axis formation is essentially normal in an asy1 mutant (Armstrong et al. 2002). Immunolocalization of AtSCC3 and AtSMC3 in asy1 revealed that they were present as normal along the chromosome axes, thereby lending support to this conclusion (Fig. 5).
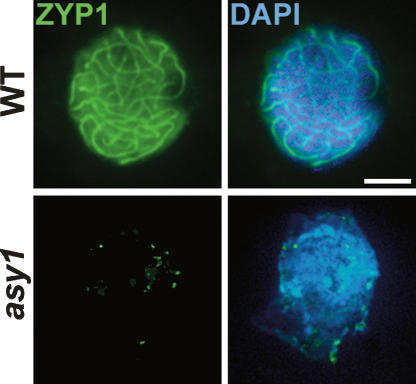
The asy1 mutant was originally characterized as asynaptic. Loss of the SC protein ZYP1 also results in the expected failure to form SC, but has an additional effect on recombination. Since recombination is initiated in asy1, it was of interest to investigate the behavior of ZYP1 in the mutant. Immunolocalization using anti-ZYP1 Ab revealed the formation of numerous foci in asy1 at early leptotene, but no subsequent polymerization to form linear SC (Fig. 6).
Figure 6.
Immunolocalization of the SC transverse element protein ZYP1 in wild-type and asy1 nuclei. Bar, 5 μM.
Discussion
The role of AtSPO11-1 in programmed DSB formation
The Arabidopsis genome contains three paralogs of the budding yeast gene Spo11, which encodes a topoisomerase II-like protein that is responsible for meiotic DSB formation (Hartung and Puchta 2000; Grelon et al. 2001). Currently, it is suggested that two of these, AtSPO11-1 and AtSPO11-2, work in combination, and this might involve a direct interaction to form a functional complex or, if they share an overlapping function, that both are required to ensure sufficient “dosage” to achieve wild-type levels of DSBs (Stacey et al. 2006). Our analysis of Atspo11-1-3 failed to detect any evidence of DSB formation. This indicates that AtSPO11-2 cannot catalyze DSB formation in the absence of AtSPO11-1, implying that the two proteins may function as a heterodimer rather than functioning additively.
Immunolocalization of AtSPO11-1 conducted in conjunction with a BrdU time course revealed that the protein localizes as discrete foci within 1–3 h of S phase. The maximum number of foci is achieved some 3 h after S before rapidly disappearing over a further 2-h period. In this study, up to ∼150 AtDMC1 foci were detected in the Col-0 wild type at mid-prophase I. This is somewhat less than the figure reported in a recent study using the Arabidopsis ecotype Ws, where >200 AtDMC1 were recorded (Chelysheva et al. 2007). Nevertheless, in both cases the figures are considerably greater than the peak number of AtSPO11-1 foci (∼90) that we observed. Most probably this discrepancy reflects the rapid turnover of AtSPO11-1 foci. A similar situation has been reported in budding yeast (Prieler et al. 2005). Intriguingly, it appears that there is a significant delay in the detection of γH2AX foci following the appearance of AtSPO11-1 foci. Since it is reported that phosphorylation of H2AX occurs within minutes of DSB formation, this suggests that following the association of AtSPO11-1 with the DNA there is a delay before DSBs occur (Rogakou et al. 1998; Shroff et al. 2004; Friesner et al. 2005). Studies in budding yeast have indicated that in itself association of Spo11 with the chromatin is not sufficient for DSB formation (Prieler et al. 2005; Robine et al. 2007). This implies that further criteria must be met before DSB formation proceeds. It has been suggested that the nascent recombination complex assembles within a chromatin loop, but becomes axis-associated before DSB formation occurs within a so-called tethered-loop/axis complex (Blat et al. 2002). However, the alternative possibilities that DSBs are transported to the chromosome axes after formation or occur concomitantly with axis formation have been postulated (van Heemst and Heyting 2000; Kleckner 2006; Lorenz et al. 2006). Our finding that the appearance of γH2AX is coincident with the transition of the axis-associated proteins ASY1, SMC3, and SCC3 to linear signals signifying axis development is consistent with the proposal that DSBs and axis formation are concomitant, spatially coordinated events.
In both wild-type and asy1 nuclei, a subset of γH2AX foci was found to persist until later stages of prophase I. Numerically, these corresponded to the number of COs, and a proportion of them colocalized with AtMLH3. Following DNA break repair, γH2AX foci are removed from the chromatin by dephosphorylation and/or histone exchange (Chowdhury et al. 2005). This suggests that the DSBs that do not progress to form COs are repaired more quickly than their CO-designated counterparts. This observation is consistent with studies in budding yeast that indicate that non-CO products are formed earlier than COs (Allers and Lichten 2001).
In the filamentous fungus Sordaria macrospora, ionizing radiation can partially restore chiasma formation and synapsis in a spo11 mutant (Storlazzi et al. 2003). However, this was accompanied by the presence of fragmented chromosomes and complex associations of nonhomologous chromosomes. These findings are very similar to those obtained in this study. A 2-h pulse of cisplatin at increasing concentrations was found to restore bivalent formation in Atspo11-1-3 meiocytes. In wild-type meiosis, the distribution of COs is subject to stringent control. Although the underlying mechanisms remain elusive, this control ensures that each pair of chromosomes undergoes a minimum of at least one CO, referred to as the obligate CO (Jones 1984; Jones and Franklin 2006). Also, subsequent COs are subject to interference, which reduces the probability that two COs occur within adjacent chromosome regions. When cisplatin was used to induce DSBs, chiasma formation initially appeared dose-dependent and it was possible to identify meiocytes with up to three bivalents, each with a single chiasma. However, when the cisplatin dose was increased beyond a critical level, rather than observing five bivalents with the normal set of one to three chiasmata per bivalent, only nuclei with extensive multivalent formation were found, together with evidence of chromosome fragmentation. This suggests that cisplatin-induced DSBs are not subject to the controls that govern their formation and/or fate of those induced by AtSPO11-1.
DSB formation is independent of ASY1
Mutation of the budding yeast HOP1 gene results in a reduction of CO frequency to between 10% and 20% of wild-type level (Hollingsworth and Johnson 1993). DSB formation is compromised in hop1 mutants, and in some genetic backgrounds the effect is severe (Mao-Draayer et al. 1996; Woltering et al. 2000). Extrapolating from the chiasma frequency at metaphase I, it appears that mutation of ASY1 has a similar overall effect on recombination. In contrast to hop1 mutants, DSB formation in asy1 meiocytes appears to occur at or near wild-type levels. It is possible to account for a high level of γH2AX signal in asy1 by a delayed turnover of fewer DSBs. However, the chromosome fragmentation phenotype of the asy1/Atrad51 double mutant is indistinguishable from that of the Atrad51 mutant (Fig. 4). Thus, the substantial reduction in CO formation in asy1 is probably not a result of any major defect in DSB formation. This suggests that although ASY1 and HOP1 exhibit homology, they may show some functional divergence.
A key role for ASY1 at the chromosome axis-recombination machinery interface
Immunofluorescence studies reveal that ASY1 protein is initially distributed as numerous foci throughout the chromatin. During early G2, the foci are juxtaposed to the nascent chromosome axes to form a continuous axis-associated signal. The immunolocalization studies conducted in conjunction with a BrdU time course provide some new insights into these early meiotic events. They reveal that DSB formation and localization of ASY1 to the chromosome axes can occur as independent events. Nevertheless, in wild-type meiocytes they occur as temporally coordinated events occurring within a window of 2 h. The association of ASY1 with the chromosome axes is concurrent with axis morphogenesis rather than associating with a preformed axis.
Previous immunogold studies have suggested that ASY1 is axis associated rather than an integral axis component (Armstrong et al. 2002). In the absence of ASY1, localization of AtSCC3 and AtSMC3 appears normal and there are no obvious defects at the level of light microscopy. A recent ultrastructural analysis of the chromosome axes in asy1 also revealed clearly defined chromosome axes. However, in contrast to wild type, some small breaks/discontinuities were apparent in the axes (Pradillo et al. 2007). Thus, in the absence of ASY1, the axes may not mature to form continuous linear entities, or possibly they do form intact structures, but these are more fragile than normal and hence susceptible to the introduction of breaks during preparation of the samples for electron microscopy. Overall, these observations do suggest that loss of ASY1 may have a minor effect on normal chromosome axis organization. We propose that ASY1 associates at sites along the axes, providing an interface between the axes and the chromatin. This may define a chromatin loop–axis organization/environment that favors AtDMC1-mediated interhomolog repair. How axis morphogenesis and recombination initiation are coordinated remains to be resolved. One potential candidate is the SWITCH1 (SWI1) protein (Mercier et al. 2001, 2003). Mutants lacking SWI1 fail to initiate recombination, do not form axial elements, and ASY1 localization does not progress beyond the stage of chromatin-associated foci, implying that these may be interdependent processes.
Normal levels of interhomolog recombination require ASY1
Despite no apparent defect in DSB formation in asy1, CO formation is severely compromised. Immunolocalization of recombination pathway proteins indicates that the fate of the majority of DSBs is different in asy1 compared with wild-type meiocytes. Most significant is the differential effect that loss of ASY1 has on the localization of the RecA homologs AtRAD51 and AtDMC1. Whereas AtRAD51 is not substantially affected by the loss of ASY1, localization of AtDMC1 is severely compromised.
Our results suggest that at least one role of ASY1 is to stabilize loading of AtDMC1 onto DSBs. Without this, there is a dramatic reduction in CO formation, and the presynaptic alignment of homologous chromosomes fails to progress beyond initial nucleolus-associated telomere pairing (Armstrong et al. 2001). This implies that the interhomolog interactions that lead to presynaptic alignment are AtDMC1-dependent and is consistent with the finding that COs are not formed in an Atdmc1 mutant (Couteau et al. 1999). In the case of the Atdmc1 study, it is suggested that the DSBs are repaired via AtRAD51-mediated recombination with the sister chromatid. This is a reasonable suggestion, and in light of the studies on Hop1, discussed below, a likely explanation of the results obtained in this study. Also, previous evidence indicates that in an asy1 mutant any interaction between homologs, other than at subtelomeric regions, is likely to be at best transient (Armstrong et al. 2001). Nevertheless, the possibility that interhomolog repair occurs cannot be excluded.
Studies have demonstrated that the RecA homologs function in a coordinated manner to promote early steps in the meiotic recombination pathway (Neale and Keeney 2006; Sheridan and Bishop 2006), a key feature of which is a preferential bias that ensures that recombination occurs between homologous nonsister chromosomes (Schwacha and Kleckner 1997). In budding yeast, it is proposed that an axis-associated complex of Hop1, Red1, and Mek1 actively suppress DMC1-independent strand invasion during meiosis (Niu et al. 2005). The bias in favor of interhomolog DSB repair is due to the establishment of a barrier to sister chromatid repair that arises through Hop1-promoted recruitment and dimerization of Mek1. This protein kinase is proposed to phosphorylate a substrate in the vicinity of the DSB to block intersister recombination and thus promote an interhomolog interaction. In addition, a recent study has identified Hed1, a meiosis-specific protein that is also important for coordinating the activity of Rad51 and Dmc1 by suppressing the activity of Rad51 when Dmc1 is absent (Tsubouchi and Roeder 2006). Thus, in budding yeast it appears that at least two independent pathways influence the activity of the recombinases to favor interhomolog recombination. Nevertheless, efficient repair of all the meiotic DSBs does appear to require Rad51 activity. Hence, it has been proposed that the barrier to intersister repair is eventually lifted, such that Rad51-mediated repair can occur (Sheridan and Bishop 2006). Similarly, in Arabidopsis, AtRAD51 is essential to ensure that the repair of meiotic DSBs is efficient, as DNA fragmentation is observed in an Atrad51 mutant (Li et al. 2004).
Our data in asy1 are compatible with the operation of an analogous mechanism (or mechanisms) in Arabidopsis. Initially, it appears that AtDMC1 associates with the chromatin as normal in the absence of ASY1. However, in contrast to wild-type cells, the protein does not persist. This observation indicates that ASY1 is not required for AtDMC1 loading, but when absent a barrier to AtRAD51 is not established. This allows it to displace AtDMC1 from the recombination intermediates, thereby disfavoring interhomolog interactions.
A further observation in asy1 consistent with the removal of a barrier to AtRAD51-dependent DSB repair was the small, yet noticeable effect on the chronology of the protein. The abundance and kinetics of appearance of AtRAD51 foci on the chromatin was indistinguishable from wild-type nuclei. However, it did appear that the reduction in AtRAD51 foci that occurs as meiosis proceeds was quicker in asy1. This could be indicative of the removal of a barrier that would allow the protein to initiate and presumably complete DSB repair at an earlier time point than in wild-type cells. An additional consideration is that in budding yeast there is evidence to suggest that the interhomolog bias is established before or during DSB formation and is subsequently maintained until joint molecule recombination intermediates are established between homologs (Schwacha and Kleckner 1997). The timing of ASY1 localization is such that it may participate in the establishment and/or maintenance of the interhomolog bias. However, further studies will be needed to confirm if the mechanism that ensures the interhomolog bias during meiotic recombination in Arabidopsis is the same as that in budding yeast, particularly as there appear to be no obvious homologs of either Red1 or Hed1 in higher eukaryotes. It remains possible that ASY1 operates via a different type of mechanism. For example, it may play a role in capturing the homologous partner axis by the axis-associated recombination complex, as proposed from studies in Sordaria (Tesse et al. 2003). If so, in the absence of ASY1 this process would break down, leading to use of the sister chromatid as the default for repair of the DSB.
Chromatin loading of AtDMC1 and AtRAD51 is asynchronous
Studies in budding yeast have led to the proposal that there may be inherent asymmetry in the way that Dmc1 and Rad51 interact with the two ends of the meiotic DSBs (Shinohara et al. 2000; Hunter and Kleckner 2001). Consistent with this are immunological observations that reveal a side-by-side distribution of the two recombinases (Tarsounas et al. 1999; Shinohara et al. 2000). In addition, biochemical studies indicate that the two ends of the resected DSB behave differently, with one, proposed to be Dmc1-coated, responsible for the initial strand invasion of the homolog, while the second is captured at a later stage in the recombination pathway (Hunter and Kleckner 2001). It has been speculated that evolutionary divergence of Rad51 and Dmc1 may have resulted in the latter forming more rigid nucleoprotein structures that favor interhomolog interactions (Sheridan and Bishop 2006). This differentiation of the two ends of the DSB is further emphasized in recent studies that have revealed that the endonucleolytic processing of DSBs that precedes resection is asymmetric (Neale et al. 2005). Our observation in wild-type and asy1 cells that accumulation of AtDMC1 slightly precedes AtRAD51 is another reflection of the asymmetry associated with the early stages of meiotic recombination. Asynchrony in the loading of the two recombinases in this way could provide an additional mechanism to promote interhomolog recombination.
AtDMC1-mediated interhomolog recombination occurs in the absence of ASY1 in some chromosome regions
The failure of AtDMC1 to persist on the chromatin in the absence of ASY1 has a profound effect on chromosome alignment and synapsis as well as recombination. Nevertheless, some residual chiasmata, ∼15% that of wild type, remain, indicating that interhomolog recombination is not completely abolished in an asy1 mutant. Moreover, our studies indicate that these residual chiasmata remain AtDMC1 dependent since they do not occur in an Atdmc1/asy1 double mutant. Also, despite the disruption of AtDMC1 localization when ASY1 is not present, occasional AtDMC1 persist in some nuclei. Numerically, these correspond to both the residual chiasmata and the number of AtMLH3 foci that mark CO sites. This would imply that in a few cases that AtDMC1 mediated, interhomolog recombination prevails over intersister repair. However, this does not appear to be a simply stochastic event because the distribution of the chiasmata in asy1 is markedly different from wild type in that they are predominantly subterminal (Sanchez-Moran et al. 2001). Our previous studies have demonstrated that clustering and pairing of homologous telomeres occurs in late G2 as a prelude to full presynaptic alignment and synapsis (Armstrong et al. 2001). These early pairing events are not dependent on ASY1. Hence, it appears that homolog juxtaposition in some chromosomal regions is to a limited extent independent of recombination-dependent alignment and that in these regions AtDMC1 is able to mediate interhomolog recombination in the absence of ASY1. It is conceivable that in such regions the chromosome axes would develop in the context of homologous chromatin that is already in close proximity. Consequently, some recombination intermediates would be fortuitously localized at sites on the aligned homologous axes where they could progress form to a CO. In this instance, ASY1 does not seem to be essential for AtDMC1-mediated interhomolog recombination. Thus, in telomeric regions where the homologous chromosomes have a degree of prealignment, this combined with the earlier appearance of AtDMC1 relative to AtRAD51 would in some instances enable interhomolog strand invasion to initiate and proceed to a point where it cannot be (re)directed to the sister chromatid despite the removal of any barrier. Similarly, if, as proposed above, ASY1 has a role in “reeling in” the homologous partner, then this activity may also be dispensable in these aligned subtelomeric regions. However, since there is no general prealignment of chromosomal regions in Arabidopsis, ASY1 would be essential to facilitate AtDMC1-mediated interactions at most sites.
Materials and methods
The Arabidopsis thaliana ecotype Columbia (Col-0) was used in this study for wild-type analysis. The T-DNA insertion lines SALK_144182 (asy1), SALK_146172 (Atspo11-1-3), SALK_056177 (Atdmc1), SAIL_873_C08 (Atrad51), SALK_067823 (Atmre11), SALK_084967 (Atrad50), and SALK_061706 (Atcom1), were obtained from the Salk Institute via NASC for mutant analysis (Alonso et al. 2003). The T-DNA insertion site was mapped with specific primers designed by the T-DNA primer design tool (http://signal.salk.edu/tdnaprimers.2.html) from the Salk Institute Genomic Analysis Laboratory.
Atspo11-1-1 seeds were kindly donated by Mathilde Grelon (INRA, Versailles, France). Plant growth, material collection, and nucleic acid extractions were performed as described by Higgins et al. (2004).
Double mutants were generated by crossing heterozygotes of appropriate mutant lines. Double mutants were identified by PCR of the F2 population derived from selfing F1 plants heterozygous for both alleles.
Nucleic acid sequencing
Automated nucleotide sequencing was carried out by the Genomics Laboratory, Biosciences, University of Birmingham.
Antibodies used in the investigation
The following Abs were used in the study: anti-ASY1 (rabbit/rat, dilution 1 in 500), anti-AtSPO11 (rabbit, dilution 1 in 100), anti-γH2AX (ser 139, catalog no. 07-164, Upstate Biotechnology; rabbit, dilution 1 in 200), anti-AtRAD51 (rabbit, dilution 1 in 200), anti-AtDMC1 (rabbit, dilution 1 in 200), anti-MLH3 (rat, dilution 1 in 200), anti-ZYP1 (rabbit/rat, dilution 1 in 500), anti-AtSMC3 (rat, dilution 1 in 500), and anti-AtSCC3 (rat, dilution 1 in 500) (Armstrong et al. 2002; Mercier et al. 2003; Higgins et al. 2004, 2005; Sanchez-Moran et al. 2004; Jackson et al. 2006).
Anti-AtSPO11-1 Ab was raised against a multiple antigenic peptide (MAP) comprising amino acid residues 189–206 of the protein (AltaBioscience, University of Birmingham). A rabbit polyclonal antiserum was produced against the peptide (ISL). This peptide is not found in AtSPO11-2, which is also expressed during meiosis. To confirm that the Ab specifically identified AtSPO11-1, comparative immunolocalization was conducted on meiotic chromosomes prepared from wild-type and Atspo11-1-3 meiocytes at early G2 (3 h post-S phase). This confirmed that the AtSPO11-1 signal found in wild type was absent in the mutant line (Supplementary Fig. 2).
Anti-DMC1 Ab was produced in the same way as anti-SPO11-1 using a MAP based on amino acid residues 17–35 of AtDMC1 that are specific to the protein. To verify that the anti-AtDMC1 Ab did not cross-react with AtRAD51, and that likewise the anti-RAD51 Ab did not cross-react with AtDMC1, immunolocalization was carried out on chromosome spreads from wild type plus the Atdmc1 and Atrad51 mutants. As expected, the anti-AtDMC1 Ab did not give a signal in Atdmc1, but did in both wild type and Atrad51, whereas the anti-RAD51 Ab did not give a signal in Atrad51, but numerous foci were observed in both the wild type and Atdmc1 (Supplementary Fig. 5). These data confirm the specificity of the two Abs.
Cytological procedures and data analysis
Meiotic time-course experiments (n = 4) were conducted using BrdU pulse-labeling combined with cytological analyses as previously described (Armstrong et al. 2003; Sanchez-Moran et al. 2004), with the modification that a meiocyte squash technique was used in addition to chromosome spreading (Page et al. 1998). Following the immunolocalization procedures, slides were analyzed by fluorescence microscopy using a Nikon Eclipse T300 microscope. Capture and image analysis was conducted using Analysis (Olympus). Individual images were acquired as Z-stacks of 10 multicolor fluorescent sections (∼1 μm each) (Supplementary Fig. 6). Images are presented as mean intensity projections. To quantify the distribution of each protein, a minimum of 10 nuclei from each time point were analyzed. Where appropriate, additional nuclei were analyzed as indicated in the text. Numerical data were analyzed using standard statistical methods as indicated in the text.
Cisplatin treatment
The inflorescence stems of Atspo11-1-3 and asy1 mutants were cut under water and placed in different concentrations of cisplatin (2.5, 3, 3.5, 4, 4.5, and 5 mg/mL) as previously described for aminopeptidase inhibitor assays (Sanchez-Moran et al. 2004).
Acknowledgments
We thank Steve Price and Karen Staples for technical assistance and Nancy Hollingsworth for comments on our data. We thank the anonymous referees for their valuable comments. F.C.H.F. and G.H.J. are grateful to the Biotechnology and Biotechnology Research Council for financial support. J.L.S. is supported by the Ministero de Educación y Ciencia de España (Grant: esBFU2005-02431).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.439007
References
- Allers T., Lichten M., Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Zimmerman J., Barajas P., Cheuk R., Barajas P., Cheuk R., Cheuk R., et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Anderson L.K., Hooker K.D., Stack S.M., Hooker K.D., Stack S.M., Stack S.M. The distribution of early recombination nodules on zygotene bivalents from plants. Genetics. 2001;159:1259–1269. doi: 10.1093/genetics/159.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong S.J., Franklin F.C.H., Jones G.H., Franklin F.C.H., Jones G.H., Jones G.H. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J. Cell Sci. 2001;114:4207–4217. doi: 10.1242/jcs.114.23.4207. [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Caryl A.P., Jones G.H., Franklin F.C.H., Caryl A.P., Jones G.H., Franklin F.C.H., Jones G.H., Franklin F.C.H., Franklin F.C.H. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 2002;115:3645–3655. doi: 10.1242/jcs.00048. [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Franklin F.C.H., Jones G.H., Franklin F.C.H., Jones G.H., Jones G.H. A meiotic time-course for Arabidopsis thaliana. Sex. Plant Reprod. 2003;16:141–149. [Google Scholar]
- Bishop D.K. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear-complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Park D., Xu L.Z., Kleckner N., Park D., Xu L.Z., Kleckner N., Xu L.Z., Kleckner N., Kleckner N. Dmc1—A meiosis-specific yeast homolog of Escherichia coli RecA required for recombination, synaptonemal complex formation, and cell-cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Nikolski Y., Oshiro J., Chon J., Shinohara M., Chen X., Nikolski Y., Oshiro J., Chon J., Shinohara M., Chen X., Oshiro J., Chon J., Shinohara M., Chen X., Chon J., Shinohara M., Chen X., Shinohara M., Chen X., Chen X. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells. 1999;4:425–443. doi: 10.1046/j.1365-2443.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- Blat Y., Protacio R.U., Hunter N., Kleckner N., Protacio R.U., Hunter N., Kleckner N., Hunter N., Kleckner N., Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Caryl A.P., Armstrong S.J., Jones G.H., Franklin F.C.H., Armstrong S.J., Jones G.H., Franklin F.C.H., Jones G.H., Franklin F.C.H., Franklin F.C.H. A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma. 2000;109:62–71. doi: 10.1007/s004120050413. [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Diallo S., Vezon D., Gendrot G., Vrielynck N., Belcram K., Rocques N., Marquez-Lema A., Bhatt A.M., Horlow C., Diallo S., Vezon D., Gendrot G., Vrielynck N., Belcram K., Rocques N., Marquez-Lema A., Bhatt A.M., Horlow C., Vezon D., Gendrot G., Vrielynck N., Belcram K., Rocques N., Marquez-Lema A., Bhatt A.M., Horlow C., Gendrot G., Vrielynck N., Belcram K., Rocques N., Marquez-Lema A., Bhatt A.M., Horlow C., Vrielynck N., Belcram K., Rocques N., Marquez-Lema A., Bhatt A.M., Horlow C., Belcram K., Rocques N., Marquez-Lema A., Bhatt A.M., Horlow C., Rocques N., Marquez-Lema A., Bhatt A.M., Horlow C., Marquez-Lema A., Bhatt A.M., Horlow C., Bhatt A.M., Horlow C., Horlow C., et al. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J. Cell Sci. 2005;118:4621–4632. doi: 10.1242/jcs.02583. [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Gendrot G., Vezon D., Doutriaux M.P., Mercier R., Grelon M., Gendrot G., Vezon D., Doutriaux M.P., Mercier R., Grelon M., Vezon D., Doutriaux M.P., Mercier R., Grelon M., Doutriaux M.P., Mercier R., Grelon M., Mercier R., Grelon M., Grelon M. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 2007;3:e83. doi: 10.1371/journal.pgen.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D., Keogh M.C., Ishii H., Peterson C.L., Buratowski S., Lieberman J., Keogh M.C., Ishii H., Peterson C.L., Buratowski S., Lieberman J., Ishii H., Peterson C.L., Buratowski S., Lieberman J., Peterson C.L., Buratowski S., Lieberman J., Buratowski S., Lieberman J., Lieberman J. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Couteau F., Belzile F., Horlow C., Grandjean O., Vezon D., Doutriaux M.P., Belzile F., Horlow C., Grandjean O., Vezon D., Doutriaux M.P., Horlow C., Grandjean O., Vezon D., Doutriaux M.P., Grandjean O., Vezon D., Doutriaux M.P., Vezon D., Doutriaux M.P., Doutriaux M.P. Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell. 1999;11:1623–1634. doi: 10.1105/tpc.11.9.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Nabeshima K., Villeneuve A., Zetka M., Nabeshima K., Villeneuve A., Zetka M., Villeneuve A., Zetka M., Zetka M. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 2004;14:585–592. doi: 10.1016/j.cub.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Friesner J.D., Liu B., Culligan K., Britt A.B., Liu B., Culligan K., Britt A.B., Culligan K., Britt A.B., Britt A.B. Ionizing radiation-dependent γ-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and rad3-related. Mol. Biol. Cell. 2005;16:2566–2576. doi: 10.1091/mbc.E04-10-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M., Vezon D., Gendrot G., Pelletier G., Vezon D., Gendrot G., Pelletier G., Gendrot G., Pelletier G., Pelletier G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001;20:589–600. doi: 10.1093/emboj/20.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanneman W.H., Legare M.E., Sweeney S., Schimenti J.C., Legare M.E., Sweeney S., Schimenti J.C., Sweeney S., Schimenti J.C., Schimenti J.C. Cisplatin increases meiotic crossing-over in mice. Proc. Natl. Acad. Sci. 1997;94:8681–8685. doi: 10.1073/pnas.94.16.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Puchta H., Puchta H. Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res. 2000;28:1548–1554. doi: 10.1093/nar/28.7.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley R.S. Exchange and chromosome segregation in eukaryotes. In: Kucherlapati R., Smith G.R., Smith G.R., editors. Genetic recombination. American Society for Microbiology; Washington, DC: 1988. pp. 497–528. [Google Scholar]
- Higgins J.D., Armstrong S.J., Franklin F.C.H., Jones G.H., Armstrong S.J., Franklin F.C.H., Jones G.H., Franklin F.C.H., Jones G.H., Jones G.H. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes & Dev. 2004;18:2557–2570. doi: 10.1101/gad.317504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Sanchez-Moran E., Armstrong S.J., Jones G.H., Franklin F.C.H., Sanchez-Moran E., Armstrong S.J., Jones G.H., Franklin F.C.H., Armstrong S.J., Jones G.H., Franklin F.C.H., Jones G.H., Franklin F.C.H., Franklin F.C.H. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes & Dev. 2005;19:2488–2500. doi: 10.1101/gad.354705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N.M., Johnson A.D., Johnson A.D. A conditional allele of the Saccharomyces cerevisiae Hop1 gene is suppressed by overexpression of two other meiosis-specific genes—Red1 and Rec104. Genetics. 1993;133:785–797. doi: 10.1093/genetics/133.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N.M., Ponte L., Ponte L. Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997;147:33–42. doi: 10.1093/genetics/147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Kleckner N., Kleckner N. The single-end invasion: An asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Jackson N., Sanchez-Moran E., Buckling E., Armstrong S.J., Jones G.H., Franklin F.C.H., Sanchez-Moran E., Buckling E., Armstrong S.J., Jones G.H., Franklin F.C.H., Buckling E., Armstrong S.J., Jones G.H., Franklin F.C.H., Armstrong S.J., Jones G.H., Franklin F.C.H., Jones G.H., Franklin F.C.H., Franklin F.C.H. Reduced meiotic crossovers and delayed prophase I progression AtMLH3-deficient Arabidopsis. EMBO J. 2006;25:1315–1323. doi: 10.1038/sj.emboj.7600992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G.H. The control of chiasma distribution. Symp. Soc. Exp. Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- Jones G.H., Franklin F.C.H., Franklin F.C.H. Meiotic crossing-over: Obligation and interference. Cell. 2006;126:246–248. doi: 10.1016/j.cell.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Kartalou M., Essigmann J.M., Essigmann J.M. Mechanisms of resistance to cisplatin. Mutat. Res. 2001;478:23–43. doi: 10.1016/s0027-5107(01)00141-5. [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C.N., Kleckner N., Giroux C.N., Kleckner N., Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–194. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- Klimyuk V.I., Jones J.D.G., Jones J.D.G. AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: Characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J. 1997;11:1–14. doi: 10.1046/j.1365-313x.1997.11010001.x. [DOI] [PubMed] [Google Scholar]
- Lam W.S., Yang X.H., Makaroff C.A., Yang X.H., Makaroff C.A., Makaroff C.A. Characterization of Arabidopsis thaliana SMC1 and SMC3: Evidence that AtSMC3 may function beyond chromosome cohesion. J. Cell Sci. 2005;118:3037–3048. doi: 10.1242/jcs.02443. [DOI] [PubMed] [Google Scholar]
- Li W.X., Chen C.B., Markmann-Mulisch U., Timofejeva L., Schmelzer E., Ma H., Reiss B., Chen C.B., Markmann-Mulisch U., Timofejeva L., Schmelzer E., Ma H., Reiss B., Markmann-Mulisch U., Timofejeva L., Schmelzer E., Ma H., Reiss B., Timofejeva L., Schmelzer E., Ma H., Reiss B., Schmelzer E., Ma H., Reiss B., Ma H., Reiss B., Reiss B. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. 2004;101:10596–10601. doi: 10.1073/pnas.0404110101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A., Estreicher A., Kohli J., Loidl J., Estreicher A., Kohli J., Loidl J., Kohli J., Loidl J., Loidl J. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma. 2006;115:330–340. doi: 10.1007/s00412-006-0053-9. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah S.K., Turner J.M.A., Baudat F., Rogakou E.P., de Boer P., Blanco-Rodriguez J., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S., Turner J.M.A., Baudat F., Rogakou E.P., de Boer P., Blanco-Rodriguez J., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S., Baudat F., Rogakou E.P., de Boer P., Blanco-Rodriguez J., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S., Rogakou E.P., de Boer P., Blanco-Rodriguez J., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S., de Boer P., Blanco-Rodriguez J., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S., Blanco-Rodriguez J., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S., Keeney S., Bonner W.M., Burgoyne P.S., Bonner W.M., Burgoyne P.S., Burgoyne P.S. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 2001;27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- Mao-Draayer Y., Galbraith A.M., Pittman D.L., Cool M., Malone R.E., Galbraith A.M., Pittman D.L., Cool M., Malone R.E., Pittman D.L., Cool M., Malone R.E., Cool M., Malone R.E., Malone R.E. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics. 1996;144:71–86. doi: 10.1093/genetics/144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon E., Moens P., Moens P. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics. 2003;165:2283–2287. doi: 10.1093/genetics/165.4.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R., Vezon D., Bullier E., Motamayor J.C., Sellier A., Lefevre F., Pelletier G., Horlow C., Vezon D., Bullier E., Motamayor J.C., Sellier A., Lefevre F., Pelletier G., Horlow C., Bullier E., Motamayor J.C., Sellier A., Lefevre F., Pelletier G., Horlow C., Motamayor J.C., Sellier A., Lefevre F., Pelletier G., Horlow C., Sellier A., Lefevre F., Pelletier G., Horlow C., Lefevre F., Pelletier G., Horlow C., Pelletier G., Horlow C., Horlow C. SWITCH1 (SWI1): A novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes & Dev. 2001;15:1859–1871. doi: 10.1101/gad.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R., Armstrong S.J., Horlow C., Jackson N.P., Makaroff C.A., Vezon D., Pelletier G., Jones G.H., Franklin F.C.H., Armstrong S.J., Horlow C., Jackson N.P., Makaroff C.A., Vezon D., Pelletier G., Jones G.H., Franklin F.C.H., Horlow C., Jackson N.P., Makaroff C.A., Vezon D., Pelletier G., Jones G.H., Franklin F.C.H., Jackson N.P., Makaroff C.A., Vezon D., Pelletier G., Jones G.H., Franklin F.C.H., Makaroff C.A., Vezon D., Pelletier G., Jones G.H., Franklin F.C.H., Vezon D., Pelletier G., Jones G.H., Franklin F.C.H., Pelletier G., Jones G.H., Franklin F.C.H., Jones G.H., Franklin F.C.H., Franklin F.C.H. The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development. 2003;130:3309–3318. doi: 10.1242/dev.00550. [DOI] [PubMed] [Google Scholar]
- Moens P.B., Kolas N.K., Tarsounas M., Marcon E., Cohen P.E., Spyropoulos B., Kolas N.K., Tarsounas M., Marcon E., Cohen P.E., Spyropoulos B., Tarsounas M., Marcon E., Cohen P.E., Spyropoulos B., Marcon E., Cohen P.E., Spyropoulos B., Cohen P.E., Spyropoulos B., Spyropoulos B. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA–DNA interactions without reciprocal recombination. J. Cell Sci. 2002;115:1611–1622. doi: 10.1242/jcs.115.8.1611. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Neale M.J., Keeney S., Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M.J., Pan J., Keeney S., Pan J., Keeney S., Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H.Y., Wan L., Baumgartner B., Schaefer D., Loidl J., Hollingsworth N.M., Wan L., Baumgartner B., Schaefer D., Loidl J., Hollingsworth N.M., Baumgartner B., Schaefer D., Loidl J., Hollingsworth N.M., Schaefer D., Loidl J., Hollingsworth N.M., Loidl J., Hollingsworth N.M., Hollingsworth N.M. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page J., Suja J.A., Santos J.L., Rufas J.S., Suja J.A., Santos J.L., Rufas J.S., Santos J.L., Rufas J.S., Rufas J.S. Squash procedure for protein immunolocalization in meiotic cells. Chromosome Res. 1998;6:639–642. doi: 10.1023/a:1009209628300. [DOI] [PubMed] [Google Scholar]
- Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M., Kirchgessner C.U., Gellert M., Bonner W.M., Gellert M., Bonner W.M., Bonner W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Pradillo M., Lopez E., Romero C., Sanchez-Moran E., Cunado N., Santos J.L., Lopez E., Romero C., Sanchez-Moran E., Cunado N., Santos J.L., Romero C., Sanchez-Moran E., Cunado N., Santos J.L., Sanchez-Moran E., Cunado N., Santos J.L., Cunado N., Santos J.L., Santos J.L. An analysis of univalent segregation in meiotic mutants of Arabidopsis thaliana: A possible role for synaptonemal complex. Genetics. 2007;175:505–511. doi: 10.1534/genetics.106.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieler S., Penkner A., Borde V., Klein F., Penkner A., Borde V., Klein F., Borde V., Klein F., Klein F. The control of Spo11’s interaction with meiotic recombination hotspots. Genes & Dev. 2005;19:255–269. doi: 10.1101/gad.321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N., Uematsu N., Amiot F., Gidrol X., Barillot E., Nicolas A., Borde V., Uematsu N., Amiot F., Gidrol X., Barillot E., Nicolas A., Borde V., Amiot F., Gidrol X., Barillot E., Nicolas A., Borde V., Gidrol X., Barillot E., Nicolas A., Borde V., Barillot E., Nicolas A., Borde V., Nicolas A., Borde V., Borde V. Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:1868–1880. doi: 10.1128/MCB.02063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M., Orr A.H., Ivanova V.S., Bonner W.M., Ivanova V.S., Bonner W.M., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Ross K.J., Fransz P., Armstrong S.J., Vizir I., Mulligan B., Franklin F.C.H., Jones G.H., Fransz P., Armstrong S.J., Vizir I., Mulligan B., Franklin F.C.H., Jones G.H., Armstrong S.J., Vizir I., Mulligan B., Franklin F.C.H., Jones G.H., Vizir I., Mulligan B., Franklin F.C.H., Jones G.H., Mulligan B., Franklin F.C.H., Jones G.H., Franklin F.C.H., Jones G.H., Jones G.H. Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Res. 1997;5:551–559. doi: 10.1023/a:1018497804129. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E., Armstrong S.J., Santos J.L., Franklin F.C.H., Jones G.H., Armstrong S.J., Santos J.L., Franklin F.C.H., Jones G.H., Santos J.L., Franklin F.C.H., Jones G.H., Franklin F.C.H., Jones G.H., Jones G.H. Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res. 2001;9:121–128. doi: 10.1023/a:1009278902994. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E., Jones G.H., Franklin F.C.H., Santos J.L., Jones G.H., Franklin F.C.H., Santos J.L., Franklin F.C.H., Santos J.L., Santos J.L. A puromycin-sensitive aminopeptidase is essential for meiosis in Arabidopsis thaliana. Plant Cell. 2004;16:2895–2909. doi: 10.1105/tpc.104.024992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N., Kleckner N. Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Sheridan S., Bishop D.K., Bishop D.K. Red-Hed regulation: Recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes & Dev. 2006;20:1685–1691. doi: 10.1101/gad.1447606. [DOI] [PubMed] [Google Scholar]
- Shinohara M., Gasior S.L., Bishop D.K., Shinohara A., Gasior S.L., Bishop D.K., Shinohara A., Bishop D.K., Shinohara A., Shinohara A. Tid1/Rdh54 promotes colocalization of Rad51 and Dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. 2000;97:10814–10819. doi: 10.1073/pnas.97.20.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R., Arbel-Eden A., Pilch D., Ira G., Bonner W.M., Petrini J.H., Haber J.E., Lichten M., Arbel-Eden A., Pilch D., Ira G., Bonner W.M., Petrini J.H., Haber J.E., Lichten M., Pilch D., Ira G., Bonner W.M., Petrini J.H., Haber J.E., Lichten M., Ira G., Bonner W.M., Petrini J.H., Haber J.E., Lichten M., Bonner W.M., Petrini J.H., Haber J.E., Lichten M., Petrini J.H., Haber J.E., Lichten M., Haber J.E., Lichten M., Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey N.J., Kuromori T., Azumi Y., Roberts G., Breuer C., Wada T., Maxwell A., Roberts K., Sugimoto-Shirasu K., Kuromori T., Azumi Y., Roberts G., Breuer C., Wada T., Maxwell A., Roberts K., Sugimoto-Shirasu K., Azumi Y., Roberts G., Breuer C., Wada T., Maxwell A., Roberts K., Sugimoto-Shirasu K., Roberts G., Breuer C., Wada T., Maxwell A., Roberts K., Sugimoto-Shirasu K., Breuer C., Wada T., Maxwell A., Roberts K., Sugimoto-Shirasu K., Wada T., Maxwell A., Roberts K., Sugimoto-Shirasu K., Maxwell A., Roberts K., Sugimoto-Shirasu K., Roberts K., Sugimoto-Shirasu K., Sugimoto-Shirasu K. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 2006;48:206–216. doi: 10.1111/j.1365-313X.2006.02867.x. [DOI] [PubMed] [Google Scholar]
- Storlazzi A., Tesse S., Gargano S., James F., Kleckner N., Zickler D., Tesse S., Gargano S., James F., Kleckner N., Zickler D., Gargano S., James F., Kleckner N., Zickler D., James F., Kleckner N., Zickler D., Kleckner N., Zickler D., Zickler D. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes & Dev. 2003;17:2675–2687. doi: 10.1101/gad.275203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M., Morita T., Pearlman R.E., Moens P.B., Morita T., Pearlman R.E., Moens P.B., Pearlman R.E., Moens P.B., Moens P.B. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J. Cell Biol. 1999;147:207–219. doi: 10.1083/jcb.147.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesse S., Storlazzi A., Kleckner N., Gargano S., Zickler D., Storlazzi A., Kleckner N., Gargano S., Zickler D., Kleckner N., Gargano S., Zickler D., Gargano S., Zickler D., Zickler D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc. Natl. Acad. Sci. 2003;100:12865–12870. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H., Roeder G.S., Roeder G.S. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H., Roeder G.S., Roeder G.S. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes & Dev. 2006;20:1766–1775. doi: 10.1101/gad.1422506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heemst D., Heyting C., Heyting C. Sister chromatid cohesion and recombination in meiosis. Chromosoma. 2000;109:10–26. doi: 10.1007/s004120050408. [DOI] [PubMed] [Google Scholar]
- Woltering D., Baumgartner B., Bagchi S., Larkin B., Loidl J., de los Santos T., Hollingsworth N.M., Baumgartner B., Bagchi S., Larkin B., Loidl J., de los Santos T., Hollingsworth N.M., Bagchi S., Larkin B., Loidl J., de los Santos T., Hollingsworth N.M., Larkin B., Loidl J., de los Santos T., Hollingsworth N.M., Loidl J., de los Santos T., Hollingsworth N.M., de los Santos T., Hollingsworth N.M., Hollingsworth N.M. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol. Cell. Biol. 2000;20:6646–6658. doi: 10.1128/mcb.20.18.6646-6658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.C., Lichten M., Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- Zetka M.C., Kawasaki I., Strome S., Muller F., Kawasaki I., Strome S., Muller F., Strome S., Muller F., Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes & Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]