Abstract
Mismatch repair (MMR) has important roles in meiotic and mitotic recombination, DNA damage signaling, and various aspects of DNA metabolism including class-switch recombination, somatic hypermutation, and triplet-repeat expansion. Defects in MMR are responsible for human cancers characterized by microsatellite instability. Intriguingly, MMR deficiency has been shown to rescue survival and proliferation of telomerase-deficient yeast strains. A putative role for MMR at mammalian telomeres that could have an impact on cancer and aging is, however, unknown. Here, we studied the role of MMR in response to dysfunctional telomeres by generating mice doubly deficient for telomerase and the PMS2 MMR gene (Terc−/−/PMS2−/− mice). PMS2 deficiency prolonged the mean lifespan and median survival of telomerase-deficient mice concomitant with rescue of degenerative pathologies. This rescue of survival was independent of changes in telomere length, in sister telomere recombination, and in microsatellite instability. Importantly, PMS2 deficiency rescued cell proliferation defects but not apoptotic defects in vivo, concomitant with a decreased p21 induction in response to short telomeres. The proliferative advantage conferred to telomerase-deficient cells by the ablation of PMS2 did not produce increased tumors. Indeed, Terc−/−/PMS2−/− mice showed reduced tumors compared with PMS2−/− mice, in agreement with a tumor suppressor role for short telomeres in the context of MMR deficiencies. These results highlight an unprecedented role for MMR in mediating the cellular response to dysfunctional telomeres in vivo by attenuating p21 induction.
Keywords: Telomeres, MMR, PMS2, telomerase knockout mice, aging, cancer
Mismatch repair (MMR) is a multifaceted DNA repair system with a crucial role in removing errors associated with DNA replication (for review, see Jiricny 2006). In addition, MMR inhibits recombination between nonidentical sequences and influences a number of other processes associated with DNA metabolism, including DNA-damage signaling, class-switch recombination, somatic hypermutation, and triplet-repeat expansion (for review, see Jiricny 2006). Loss of MMR results in the so-called “mutator phenotype,” which can lead to increased susceptibility to cancer (Fishel et al. 1993; Leach et al. 1993; Bronner et al. 1994; Nicolaides et al. 1994; Papadopoulos et al. 1994; Shibata et al. 1994). Indeed, a number of human tumors including cancers of the colon and the endometrium show a phenotype known as “microsatellite instability,” which is associated with defects in the MMR pathway (for review, see Modrich 1994). The mammalian MMR machinery is comprised of six homologs of the Escherichia coli MutS family of genes (MSH genes: MSH1 to MSH6) and four homologs of the MutL family of genes (MLH genes: MLH1, MLH3, PMS1, and PMS2). Mutations in both families of genes have been associated with human hereditary nonpolyposis colon cancer (Fishel et al. 1993; Leach et al. 1993; Bronner et al. 1994; Nicolaides et al. 1994; Papadopoulos et al. 1994). Mice deficient for different MMR genes show increased spontaneous mutation as well as an increased susceptibility to develop cancer, with MSH2 knockout mice showing the most severe phenotype (Baker et al. 1995; de Wind et al. 1995; Prolla et al. 1998). A role for MMR proteins in DNA damage signaling, which in turn could affect tumorigenesis, has also been proposed (Duckett et al. 1999; Peters et al. 2003; Luo et al. 2004). In particular, MMR proteins have been directly involved in signaling DNA damage through the p53 pathway (Duckett et al. 1999; Peters et al. 2003; Luo et al. 2004); in turn, DNA replication errors produced by MMR deficiency have been proposed to result in ATR/ATM-mediated DNA damage signaling (Brown et al. 2003).
Telomeres are protective structures at the ends of chromosomes that consist of tandem TTAGGG repeats and associated proteins (Chan and Blackburn 2002; de Lange 2005). Critical telomere shortening and loss of function of telomere-binding proteins results in loss of telomere protection, end-to-end chromosome fusions, and cell cycle arrest or apoptosis (van Steensel et al. 1998; Goytisolo and Blasco 2002; de Lange 2005). Telomere shortening is envisioned as a potent tumor suppressor mechanism (González-Suárez et al. 2000; Blasco and Hahn 2003; Blasco 2005). Telomerase activity, in turn, is able to elongate telomeres in those cells where it is highly expressed, such as in the vast majority of human cancers, and it is associated with cell immortalization (Shay and Wright 2006). Mice that lack telomerase activity show premature loss of organismal viability when their telomeres become critically short and are resistant to cancer (Lee et al. 1998; Greenberg et al. 1999; Herrera et al. 1999; González-Suárez et al. 2000), with the only exceptions being p53-deficient and TRF2-overexpressing genetic backgrounds (Chin et al. 1999; Artandi et al. 2000; Blanco et al. 2007).
Intriguingly, previous work in budding yeast indicated that MMR abrogation rescued survival of telomerase-deficient strains (Rizki and Lundblad 2001). This rescue was attributed to the known role of MMR in repressing recombination between nonhomologous sequences, which in turn could favor survival in the absence of telomerase by sustaining recombination-based alternative elongation of telomeres, or ALT mechanisms. A putative role for MMR in mammalian telomere biology is unknown, however, except for the description of increased telomeric recombination in MMR-deficient human colon cancer cells (Bechter et al. 2004).
Here, we set out to address whether MMR-deficiency had any impact on survival and life span of telomerase-deficient mice by generating mice doubly deficient for telomerase and the MMR gene PMS2 (Terc−/−/PMS2−/− mice). PMS2-deficient mice have been previously shown to have a decreased MMR efficiency, increased microsatellite instability, as well as an increased susceptibility to develop sarcomas and lymphomas (Baker et al. 1995; Prolla et al. 1998). In addition, PMS2-deficient males are infertile due to abnormal chromosome synapsis in meiosis (Baker et al. 1995). Interestingly, PMS2 deficiency rescued median survival and lifespan of telomerase-deficient mice. This rescue in survival was concomitant with a decreased incidence of degenerative pathologies and, at the cellular level, with an increased proliferative potential. Interestingly, these effects were independent of telomere length and of the presence of severe telomere dysfunction as indicated by similar frequencies of end-to-end fusions, critically short telomeres, and γH2AX-positive cells. Similarly, the rescue of phenotypes associated with dysfunctional telomeres was not associated with significant differences in the frequency of homologous recombination events between sister telomeres (telomere sister chromatid exchanges, T-SCE), suggesting that PMS2 deficiency is not rescuing survival by favoring the activation of mammalian ALT pathways (Dunham et al. 2000; Muntoni and Reddel 2005). Similarly, we did not find significant differences in microsatellite instability between PMS2−/− and Terc−/−/PMS2−/− mice, which could explain the rescue in survival. Importantly, we found an attenuated p21 induction both in vivo and in vitro in mice and cells doubly deficient for telomerase and PMS2 compared with the single telomerase-deficient controls, which was concomitant with increased proliferation rates in vivo as well as in vitro. This decreased p21 response was paralleled by decreased p53 levels. All together, these results suggest that MMR deficiency rescues organismal survival and proliferation in telomerase-deficient mice by attenuating the anti-proliferative response associated with short telomeres.
Results
PMS2 deficiency rescues median survival and mean lifespan of mice with dysfunctional telomeres
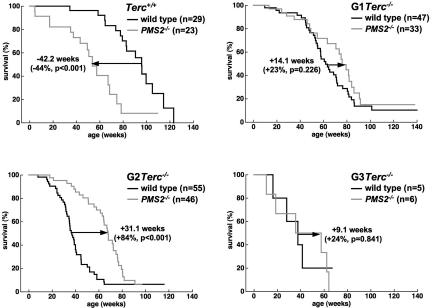
To study the impact of PMS2 deficiency in Terc−/− mouse survival, we generated increasing generations of double mutant Terc−/−/PMS2−/− mice as well as of the single Terc−/− controls (see Materials and Methods). In a C57BL6 genetic background, Terc−/− mice can be maintained for up to three generations (G1 to G3). First, we confirmed that PMS2-deficient mice have a decreased median survival compared with wild-type mice (Baker et al. 1995; P < 0.001; Fig. 1), and that increasing generations of telomerase-deficient mice show progressively decreased median survival compared with wild-type controls (Fig. 1; P < 0.005 for all comparisons between wild-type and G1–G3 Terc−/− mice; see “statistics” in Supplemental Material for comparisons between mouse generations; Herrera et al. 1999). PMS2 abrogation extended median survival of telomerase-deficient mice in G1 and G2 mouse generations, and this difference reached statistical significance when comparing G2 Terc−/−/PMS2−/− with single G2 Terc−/− mice (P < 0.001; Fig. 1). In addition, PMS2 deficiency reproducibly increased both median survival as well as mean lifespan of increasing generations of telomerase-deficient mice, and these differences reached statistical significance when comparing G2 Terc−/−/PMS2−/− with single G2 Terc−/− mice (P < 0.001; Supplemental Fig. 1A,B). The fact that the G3 Terc−/−/PMS2−/− to G3 Terc−/− comparisons did not reach statistical significance is probably due to the small numbers of mice obtained for this generation due to severe infertility of G2 Terc−/− mice (Herrera et al. 1999). Significantly, G1 and G2 Terc−/−/PMS2−/− mice showed increased median survival and mean lifespan compared with single PMS2−/− mice (P < 0.05 in both cases; Supplemental Fig. 1A,B), also suggesting that telomerase deficiency partially ameliorates PMS2-associated loss of organismal survival (see later). All together, these results suggest that PMS2 has a role in the organismal response to critically short and dysfunctional telomeres associated with telomerase deficiency. Of note, this is the second genetic alteration shown to rescue life span and survival of telomerase-deficient mice after a recent publication describing that p21 abrogation has a similar effect (Choudhury et al. 2007).
Figure 1.
PMS2 deficiency rescues organismal survival of mice with dysfunctional telomeres. Kaplan–Meyer survival curve of mice of the indicated genotype. (n) Number of mice of each genotype. Statistical comparisons between genotypes using the log rank test are also shown. For statistical comparisons between mouse generations, see “statistics” in the Supplemental Material.
PMS2 deficiency rescues degenerative pathologies in mice with dysfunctional telomeres without accelerating carcinogenesis
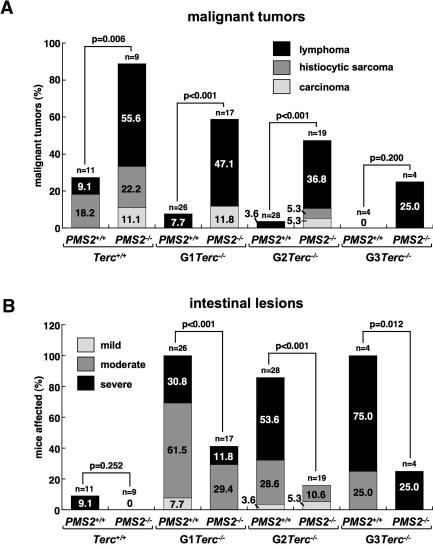
Next, we performed a full histopathological analysis to determine the cause of death of the different mouse cohorts. As previously described, single PMS2-deficient mice showed an increased cancer susceptibility, with a high incidence of lymphomas (55.6%), followed by histiocytic sarcomas (22%; liver, lung, kidney) and colon carcinomas (11.1%) (Fig. 2A; Baker et al. 1995). Similarly, PMS2 abrogation also increased the incidence of these tumors in the context of Terc-deficient mice (P < 0.001 for all comparisons except G3; Fig. 2A). However, increasing generations of Terc−/−/PMS2−/− mice showed a progressively reduced incidence of tumors compared with the single PMS2−/− mice. In particular, lymphomas were reduced from 55% in the single PMS2−/− mice to only 25% in the G3 Terc−/−/PMS2−/− (Fig. 2A). Similarly, histiocytic sarcomas and carcinomas were also decreased with increasing Terc−/−/PMS2−/− mouse generations compared with the PMS2−/− controls (Fig. 2A). These differences were statistically significant when comparing all malignancies in G2–G3 Terc−/−/PMS2−/− mice with the single PMS2−/− controls (P < 0.05 for all comparisons; see “statistics” in Supplemental Material). Single Terc-deficient mice also showed decreased tumors with increasing mouse generations (Fig. 2A; P = 0.028 when comparing wild-type and G2 Terc−/− mice; see “statistics” in Supplemental Material). These results are in agreement with short telomeres acting as potent tumor suppressors in the context of telomerase deficiency (González-Suárez et al. 2000; Blasco and Hahn 2003). Finally, these results indicate that telomerase deficiency and short telomeres suppress PMS2-induced tumorigenesis, which in turn could contribute to the increased lifespan of G1 and G2 Terc−/−/PMS2−/− mice compared with single PMS2-mutant mice, as well as suggest that telomerase inhibitors would be effective in ceasing the growth of PMS2-mutant tumors.
Figure 2.
Rescue of malignant tumors and degenerative pathologies in Terc−/−/PMS2−/− mice. (A) Percentage of mice of the indicated genotype showing malignant tumors (lymphoma, histiocytic sarcoma, carcinoma) at the time of death. Statistical comparisons using the χ2 test are also shown. For statistical comparisons between mouse generations, see “statistics” in the Supplemental Material. (B) Percentage of mice of the indicated genotype showing mild, moderate, and severe intestinal atrophy at the time of death (see Materials and Methods for description of the lesions). Statistical comparisons using the χ2 test are also shown. For statistical comparisons between mouse generations, see “statistics” in the Supplemental Material.
In agreement with previous reports, single Terc-deficient mice showed an increased incidence of various degenerative pathologies in the different mouse generations that was not observed in PMS2−/− mice, intestinal atrophies being the most abundant lesions (Supplemental Fig. 2; Lee et al. 1998; Herrera et al. 1999). In particular, the incidence of severe intestinal lesions (see Materials and Methods for detailed description of these lesions) was increased from 30.8% in G1 Terc−/− to 75% in G3 Terc−/− mice (Fig. 2B; P < 0.001 when comparing wild-type mice with each Terc−/− generation; see “statistics” in Supplemental Material). Remarkably, PMS2 deficiency rescued degenerative pathologies in every Terc−/−/PMS2−/− mouse generation compared with the single Terc−/− mice (P < 0.05 for all comparisons; Fig. 2B). These results suggest that PMS2 abrogation ameliorates degenerative pathologies associated with short telomeres, in agreement with the fact that PMS2 deficiency rescues both median life span and survival of telomerase-deficient mice (Fig. 1).
PMS2 deficiency rescues proliferation defects but not apoptosis in telomerase-deficient mice with short telomeres
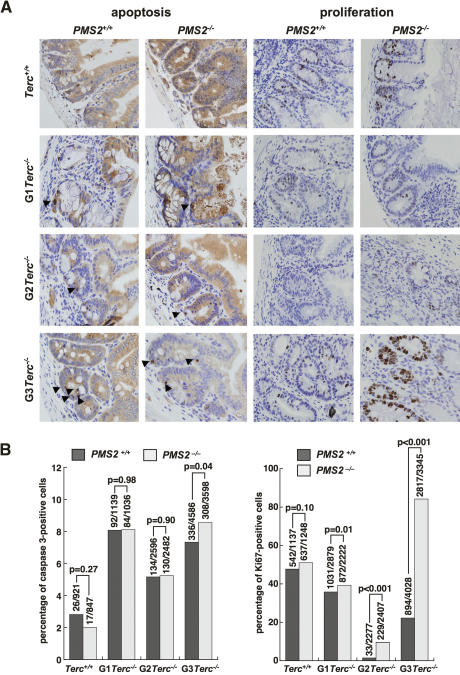
Decreased survival of late-generation Terc-deficient mice has been previously shown to be the consequence of severe proliferative defects and increased apoptosis in tissues characterized by high regeneration rates, such as the gastrointestinal (GI) tract (Lee et al. 1998; Herrera et al. 1999). Indeed, according to histopathological analysis of Terc-deficient moribund mice (Supplemental Fig. 2), the GI tract was the most frequently affected organ. For this reason we focused our analysis at the cellular level on the intestine. First, we confirmed that G1 to G3 Terc-deficient mice showed increased apoptosis compared with wild-type controls in the GI tract (Fig. 3A,B; P < 0.005 for all comparisons between generations in “statistics” in Supplemental Material). This increased apoptosis, however, was not significantly rescued by PMS2 deficiency with increasing generations of Terc−/−/PMS2−/− mice compared with the single Terc−/− controls (Fig. 3A,B), indicating that PMS2 abrogation does not rescue degenerative pathologies and mouse survival by decreasing apoptotic rates in Terc−/−/PMS2−/− mice. Indeed, apoptosis was significantly increased in late generation G3 Terc−/−/PMS2−/− mice compared with the single G3 Terc−/− controls (P = 0.04; Fig. 3B). In marked contrast, we observed a significant rescue of proliferative defects in the GI tract of different generation Terc−/−/PMS2−/− mice compared with the single Terc-deficient controls (P ≤ 0.01 for all comparisons; Fig. 3A,B), suggesting that PMS2 deficiency specifically rescues proliferation defects but not apoptosis associated with dysfunctional telomeres. This rescue of proliferation defects may explain the rescue of degenerative pathologies and survival in Terc−/−/PMS2−/− mice. This is analogous to what has been recently described for Terc−/−/p21−/− mice, in which p21 deficiency rescued survival and proliferative defects but not apoptotic defects associated with ablation of telomerase (Choudhury et al. 2007). Finally, several mechanisms can be envisioned by which PMS2 deficiency may be rescuing proliferative defects and organismal survival associated with dysfunctional telomeres. On one hand, PMS2 deficiency may be rescuing telomere length or telomere capping defects associated with Terc deficiency. In this regard, PMS2 abrogation may be favoring telomere recombination events and therefore activation of ALT pathways. This would be in agreement with its known role in inhibiting mitotic recombination (Modrich 1994; Jiricny 2006), similarly to that previously proposed for simultaneous telomerase and MMR deficiency in yeast (Rizki and Lundblad 2001). Alternatively, PMS2 may have a prominent role in the cellular response to short dysfunctional telomeres, resulting in a rescue of proliferative defects, similarly to that recently described for p21 deficiency (Choudhury et al. 2007).
Figure 3.
Rescue of proliferation defects but not apoptosis in Terc−/−/PMS2−/− mice. (A) Representative examples of sections of the small intestine from mice of the indicated genotypes stained for caspase 3 as a marker for apoptosis (left) and Ki67 as marker for proliferating cells (right). Note similar numbers of caspase 3-positive cells in single G3 Terc−/− and double mutant G3Terc−/−/PMS2−/− mice. (Arrows) Caspase 3-positive cells. Note the increase of Ki67-positive cells (brown staining) in double mutant G3 Terc−/−/PMS2−/− mice compared with the single G3 Terc−/− controls. (B) Quantification of percentage of cells showing caspase 3- and Ki67-positive staining at the GI tract in mice of the indicated genotypes. The total number of cells scored for the analysis is also indicated on top of each bar. Statistical comparisons using the χ2 test are also shown.
PMS2 deficiency promotes cellular immortalization independently of telomere length
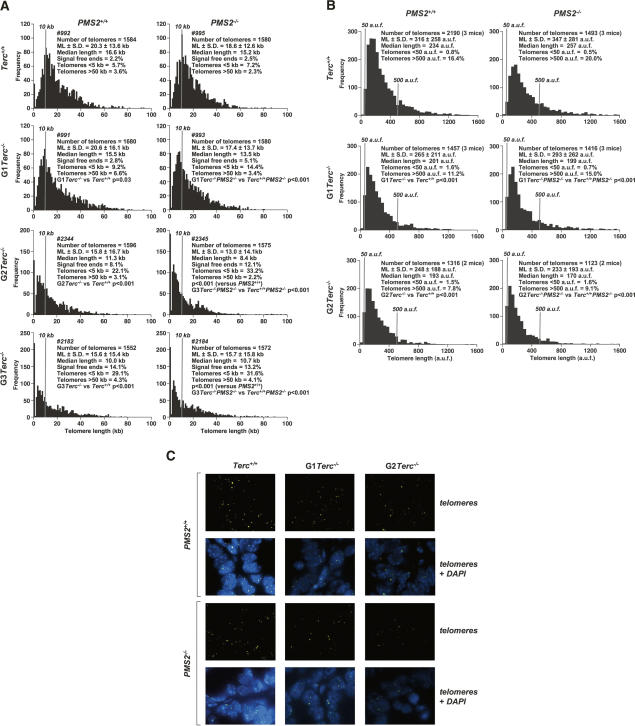
To address whether the improved survival of Terc−/−/PMS2−/− mice compared with single Terc−/− mice was associated with changes in telomere length caused by PMS2 deficiency, we first measured telomere length using quantitative fluorescence in situ hybridization, Q-FISH, on metaphase spreads from primary (passage 2) littermate mouse embryonic fibroblasts (MEF) of the different genotypes performed in parallel (see Materials and Methods; numbers in Fig. 4A refer to individual littermate MEF). We found a similar rate of telomere loss with increasing generations (G1 to G3) of single Terc−/− or double Terc−/−/PMS2−/− MEF (P < 0.05 for all comparisons between G1–G3 Terc−/− or double Terc−/−/PMS2−/− littermate mice and the corresponding Terc+/+ controls; Fig. 4A), indicating that PMS2 deficiency does not impact telomere length regulation. Furthermore, in agreement with telomere shortening along increasing generations of telomerase deficiency, we detected increased frequencies of very short telomeres (<5 kb) in single G1–G3 Terc−/− MEF (P < 0.001 for all comparisons between generations; Supplemental Fig. 3), which were further increased in the corresponding double mutant Terc−/−/PMS2−/− MEF (P < 0.001 for comparisons between G1 and G2 Terc−/− MEF with the corresponding double Terc−/−/PMS2−/− MEF; Supplemental Fig. 3), again ruling out that the increased survival of Terc−/−/PMS2−/− mice is due to a rescue of critically short telomeres in the absence of PMS2. The fact that the frequency of very short telomeres (<5 kb) was increased in double mutant Terc−/−/PMS2−/− MEF compared with the single Terc−/− MEF may be the consequence of increased proliferation rates in the absence of PMS2 (see Fig. 3A,B), which in turn may result in increased telomere erosion. Finally, these telomere length results obtained with littermate MEF were confirmed by increasing the number of MEF per genotype (Supplemental Fig. 4). Furthermore, they were also confirmed by using Southern-based “terminal restriction fragment” (TRF) analysis (Supplemental Fig. 5; Blasco et al. 1997).
Figure 4.
PMS2 deficiency does not rescue telomere shortening in telomerase-deficient mice. (A) Telomere fluorescence as determined by Q-FISH in primary MEF (passage 2) of the indicated genotypes. Numbers identify individual MEF cultures. MEFs #991, 992, 993, and 995; #2344 and 2345; as well as #2182 and 2184 were derived from littermate embryos. For each genotype, >1500 telomeres were analyzed by Q-FISH. Mean telomere length in kilobases and standard error are shown. Statistical significance using the Wilcoxon–Mann–Whitney rank sum test is indicated. It is important to note that telomere length values shown in this experiment can be compared between genotypes, as Q-FISH was performed on the same day and in parallel. These values, however, cannot be directly compared with those shown in Supplemental Fig. 6A, as they correspond to an independent Q-FISH experiment with immortalized (passage 35) MEF. (B) Q-FISH analysis of small intestine sections in mice of indicated genotypes. Between 60 and 110 nuclei from two to three mice of each genotype were analyzed. Statistical significance using the Wilcoxon–Mann–Whitney rank sum test is indicated. (C) Representative images of telomere fluorescence (telomeres), and combined telomere fluorescence and DAPI staining on small intestine sections as determined by Q-FISH.
These results with cultured MEF were also recapitulated when measuring telomere length using Q-FISH directly on small intestine sections (see Materials and Methods; González-Suárez et al. 2000; Muñoz et al. 2005). Again, we found a similar rate of telomere loss with increasing generations (G1 to G2) of single Terc−/− or double Terc−/−/PMS2−/− MEF (P < 0.0001 for all comparisons between G1–G2 Terc−/− or double Terc−/−/PMS2−/− mice and the corresponding Terc+/+ controls; Fig. 4B for quantification and Fig. 4C for representative images), indicating that PMS2 deficiency does not rescue telomere length in the in vivo setting of the Terc−/− small intestine.
We next studied whether PMS2 deficiency could rescue in vitro proliferative defects in late-generation Terc−/− MEF. In particular, we have previously shown that late-generation primary (passage 2) Terc−/− MEF showed decreased immortalization ability in culture compared with wild-type controls (Espejel and Blasco 2002). To this end, we studied the spontaneous immortalization ability of increasing generation (G1 to G3) single Terc−/− as well as double mutant Terc−/−/PMS2−/− primary MEF using the 3T3 cell passage protocol (see Materials and Methods; Todaro and Green 1963). G3 Terc−/− MEF showed a decreased immortalization ability compared with wild-type MEF as indicated by the higher number of passages required to escape the so-called “premature senescence” arrest or “culture shock” and to achieve exponential growth in vitro (Supplemental Fig. 6A,B). In contrast, double mutant G2 and G3 Terc−/−/PMS2−/− MEF were able to immortalize at lower passages compared with the corresponding single mutant Terc-deficient controls, reflecting on their higher proliferative capacity (Supplemental Fig. 6B). Importantly, the higher immortalization ability of Terc−/−/PMS2−/− MEF compared with the single Terc−/− MEF was independent of telomere length, which was comparable in immortalized (passage 35) single and double mutant MEF (Supplemental Fig. 6A; histograms reflect individual MEF cultures).
Effects of PMS2 deficiency on telomere dysfunction in late-generation Terc-deficient mice
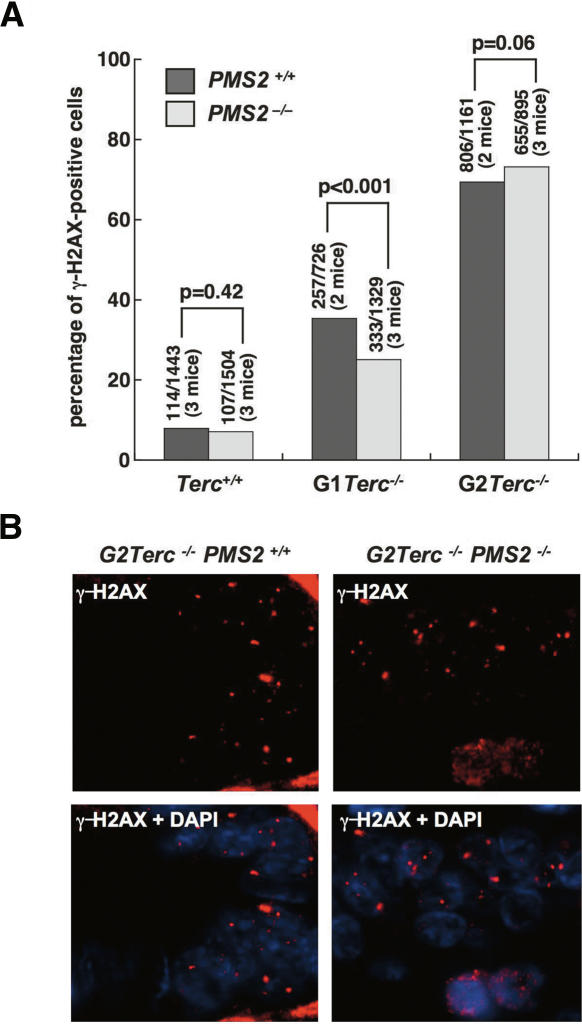
To address the possible involvement of PMS2 on telomere dysfunction, we first quantified the frequency of cells showing γ-H2AX foci in small intestine sections from late-generation (G2) single Terc−/− and double Terc−/−/PMS2−/− mice. γ-H2AX foci have been previously described to mark double-strand breaks as well as critically short and dysfunctional telomeres (Modesti and Kanaar 2002; d’Adda di Fagagna et al. 2003). As shown in Figure 5, A and B, increasing generations of Terc−/− mice showed progressively higher numbers of small intestine cells with γ-H2AX foci compared with wild-type controls, in agreement with progressively shorter telomeres in these cells. Importantly, the percentage of cells with γ-H2AX foci was not significantly rescued by PMS2 deficiency in late-generation Terc−/−/PMS2−/− mice (Fig. 5A,B), suggesting that mutant Terc−/−/PMS2−/− cells bear a similarly high load of DNA damage and telomere dysfunction to that of the single mutant Terc−/− controls.
Figure 5.
Similar numbers of γH2AX-positive cells in Terc−/−/PMS2+/+ and Terc−/−/PMS2−/− small intestine sections. (A) Quantification of the percentage of nuclei containing γH2AX foci in small intestine sections from mice of the indicated genotypes. Between 700 and 1500 nuclei were examined in a total of two to three mice per genotype. Statistical calculations using the χ2 test are shown. (B) Representative images of γH2AX-positive cells in small intestine sections from G2 Terc−/−/PMS2+/+ and G2 Terc−/−/PMS2−/− mice are shown.
To further study the possible involvement of PMS2 in the generation of chromosomal instability triggered by dysfunctional telomeres, we performed cytogenetic analysis of primary (passage 3) MEF derived from G1 to G3 single Terc−/− and double mutant Terc−/−/PMS2−/− mice using telomere Q-FISH on metaphase spreads (see Materials and Methods). Again, in agreement with progressive telomere shortening associated with telomerase deficiency, we detected increased frequencies of chromosome ends lacking the TTAGGG signal (“signal-free” ends) in G1–G3 Terc−/− MEF (P < 0.001 for all comparisons; Supplemental Fig. 7A), which were further increased in the corresponding double mutant Terc−/−/PMS2−/− MEF (P < 0.001 when comparing G2–G3 Terc−/− MEF to G2–G3 Terc−/−/PMS2−/− MEF; Supplemental Fig. 7A). Accordingly, end-to-end fusions lacking TTAGGG signals at the fusion point were increased both in late-generation G3 Terc−/− MEF and G3 Terc−/−/PMS2−/− MEF (P < 0.001; Supplemental Fig. 7A). Of note, abrogation of only PMS2 also resulted in slightly increased end-to-end fusions (P = 0.013; Supplemental Fig. 7A); however, these fusions are unlikely to result from telomere shortening, as PMS2-deficient cells show normal telomere length (Fig. 4A,B). PMS2-deficient cells also showed increased frequencies of breaks and fragments (Supplemental Fig. 7A), as well as of a number of complex aberrations including bivalent recombination figures and chromatid cross-links, complex chromosome fusions, and minichromosomes (Supplemental Fig. 7B), probably reflecting on the known roles of PMS2 in MMR and mitotic recombination (Jiricny 2006). Some of these aberrations, such as breaks and fragments, bivalent recombination figures, and complex aberrations, were also increased in late-generation G3 Terc−/− mice (P < 0.05 for all comparisons; Supplemental Fig. 7A,B), suggesting that they may be also triggered by short dysfunctional telomeres. Interestingly, the increased bivalent recombination figures and chromatid cross-links detected in late-generation G3 Terc−/− mice were rescued in the absence of PMS2, suggesting a role for MMR in the origin of these lesions (Supplemental Fig. 7B). In turn, the increased number of complex chromosome aberrations and fusions, as well as increased minichromosomes found in single PMS2-deficient cells, were partially rescued by Terc-deficiency and short telomeres (Supplemental Fig. 7B). This partial rescue of complex chromosomal aberrations cannot be explained by a lower proliferative potential of double mutant Terc−/−/PMS2−/− MEF compared with the single PMS2−/− controls, as Terc−/−/PMS2−/− MEF have increased proliferative rates compared with the single controls (Supplemental Fig. 6B). Finally, the partial rescue of PMS2-associated complex chromosomal aberrations and minichromosomes in double mutant Terc−/−/PMS2−/− mice may be related, at least in part, to the decreased tumorigenesis shown by these mice compared with the single PMS2-deficient controls (see Fig. 2A).
All together, these results indicate that mice and cells doubly deficient for PMS2 and telomerase show severe telomere dysfunction as indicated by similar frequencies of γ-H2AX foci, as well as by similar frequencies of end-to-end fusions and signal-free ends compared with single Terc-deficient controls, suggesting that PMS2 deficiency does not rescue survival or proliferation of Terc-deficient mice by lowering DNA damage associated with telomere dysfunction.
Sister telomere recombination frequencies in double mutant Terc−/−/PMS2−/− mice
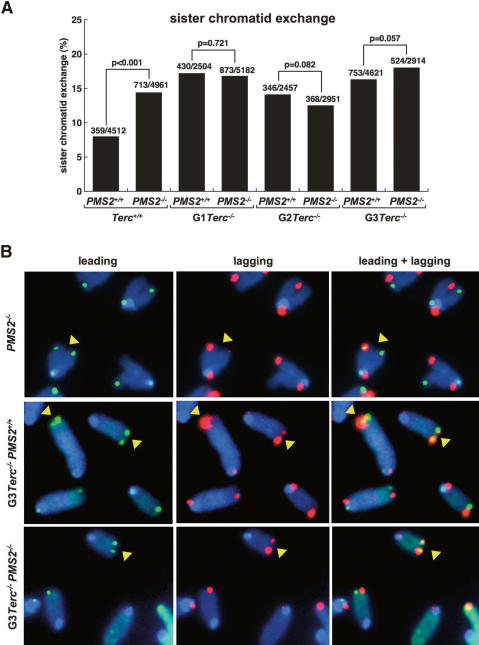
The fact that MMR deficiency was able to rescue proliferation defects in telomerase-deficient yeast strains was interpreted according to the known role of MMR in inhibiting recombination between imperfect repeats (Modrich and Lahue 1996), which in turn could be favoring activation of ALT pathways for telomere maintenance (Rizki and Lundblad 2001). Similarly, the fact that double mutant Terc−/−/PMS2−/− mice show an improved survival compared with single telomerase-deficient mice may be suggesting a similar role for mammalian PMS2 in repressing telomere recombination. In order to directly test this, we determined the frequency of telomeric sister chromatid exchange events (T-SCE) in primary MEF from different genotype mice using two-color chromosome orientation FISH (CO-FISH) (Bailey et al. 2004; Gonzalo et al. 2006). Of note, CO-FISH allows detection of recombination events between sister telomeres but not between subtelomeric repeats or telomeres at different chromosomes. The strand-specific nature of the CO-FISH typically yields two telomeric signals of each color (red: lagging; green: leading) per chromosome in the absence of recombination events (Fig. 6). A sister chromatid exchange within telomeric DNA (T-SCE) leads to the mixture of red and green fluorescence (Fig. 6). We counted as positive T-SCE events only those that were detected both with the leading and the lagging telomere probes, as well as involving an unequal exchange of telomeric signal (Fig. 6). Interestingly, PMS2 deficiency led to significantly increased T-SCE frequencies compared with wild-type cells (P < 0.001; Fig. 6A), suggesting that PMS2 may be repressing these events at telomeres, in agreement with the known role of MMR in repressing mitotic recombination. T-SCE events were also augmented in different-generation Terc−/− mice compared with the wild-type controls (Fig. 6A; P < 0.001 for all comparisons between generations; see “statistics” in Supplemental Material), in agreement with previous reports showing increased T-SCE in the absence of telomerase (Benetti et al. 2007; Blanco et al. 2007). Simultaneous absence of both PMS2 and Terc, however, did not significantly increase T-SCE frequencies in double mutant Terc−/−/PMS2−/− mice compared with the single controls (P > 0.05 for all comparisons; Fig. 6A), suggesting that increased telomere recombination between sister telomeres is unlikely to be responsible for the rescue of survival in these mice. These results are also in agreement with the fact that Terc−/−/PMS2−/− mice did not show elongated telomeres or a rescue in end-to-end fusions compared with the single mutant controls, which would have been expected from increased telomere recombination frequencies (Fig. 4). We cannot rule out, however, that PMS2 deficiency may be having an effect on the frequencies of recombination events between subtelomeres or telomeres on different chromosomes, although this effect is not sufficient to rescue telomere shortening and the elevated frequencies of end-to-end fusions and critically short telomeres in PMS2/Terc-deficient cells.
Figure 6.
Telomere recombination in Terc−/−/PMS2−/− MEF is equivalent to that of single mutant Terc−/− MEF. (A) Quantification of T-SCE frequencies in the indicated genotypes. The number of T-SCE events out of the total number of chromosomes analyzed is indicated on top of each bar. Statistical calculations using the χ2 test are shown. (B) Representative CO-FISH images of metaphases hybridized with probes against the leading (green fluorescence) and lagging (red fluorescence) telomere. (Yellow arrows) T-SCE events. A T-SCE was considered positive when it was observed with both the leading and lagging strand probes and involved an unequal exchange of telomere signal.
Microsatellite instability in Terc−/−/PMS2−/− mice
PMS2 deficiency results in increased microsatellite instability as the result of the defective MMR pathway (Prolla et al. 1998). Since microsatellite instability has been proposed to be one of the causes of increased cancer in these mice, we addressed here whether simultaneous PMS2 and telomerase deficiencies affected the frequency of microsatellite instability in Terc−/−/PMS2−/− mice compared with the single mutant controls, which could explain the lower cancer incidence of double mutant Terc−/−/PMS2−/− mice compared with the single PMS2−/− controls. To this end, we measured microsatellite instability in tail DNA from the different mouse genotypes (Supplemental Fig. 8). In particular, we determined microsatellite instability using the CA repeat loci D6Mit59 previously shown to be unstable in PMS2-deficient cells (Baker et al. 1995), as well as the D1Mit62 stable locus as a control (see Materials and Methods). As shown in Supplemental Figure 8, no alterations were found at the stable locus in any of the genotypes. In contrast, PMS2−/− deficient cells showed 71% novel alleles at the D6Mit59 locus compared with only 36% in the wild-type controls (Supplemental Fig. 8). Increasing generations of Terc-deficient mice also resulted in a progressive increase of novel alleles at the D6Mit59 locus, going from 44% in G1, 62% in G2, and 100% in G3 Terc−/− mice, which opens the intriguing possibility that telomere shortening may lead to microsatellite instability (Supplemental Fig. 8). Of note, simultaneous Terc and PMS2 deficiencies in double mutant Terc−/−/PMS2−/− mice resulted in similarly elevated frequencies of novel D6Mit59 alleles (Supplemental Fig. 8), suggesting a similar microsatellite instability in these double mutant mice compared with the single mutant PMS2−/− controls.
PMS2 deficiency results in an attenuated p21 induction in response to short telomeres
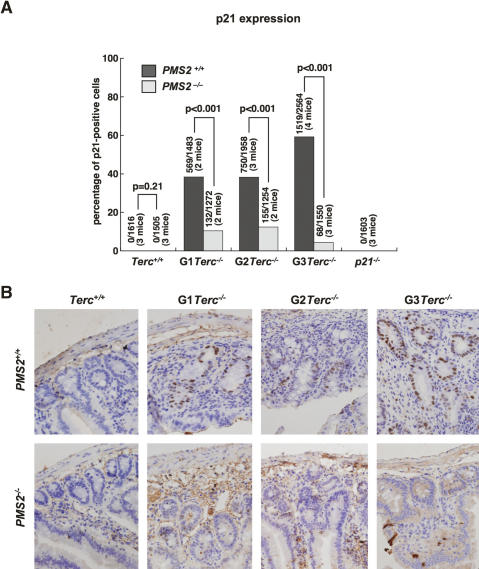
A role for MMR proteins in DNA damage signaling, which in turn could affect tumorigenesis, has also been proposed; in particular, MMR proteins have been directly involved in signaling DNA damage through the p53 pathway (Duckett et al. 1999; Peters et al. 2003; Luo et al. 2004). In addition, PMS2 has been identified as a direct target for p53 (Chen and Sadowski 2005). To address whether the rescue of Terc−/− mice survival mediated by PMS2 deficiency could be due to a role for PMS2 in response to telomere dysfunction, and given the similarity of this phenotype with that observed in the absence of p21 (Choudhury et al. 2007), we first determined p21 levels by immunohistochemistry directly on small intestine sections from different genotype mice (see Materials and Methods). Increasing generations of single Terc−/− mice showed elevated amounts of p21 in the GI tract (Fig. 7A,B), in agreement with decreased proliferation rates in these mice (Fig. 3A,B). Interestingly, PMS2 deficiency dramatically decreased p21 accumulation in the small intestine of different-generation Terc−/−/PMS2−/− mice (Fig. 7A,B), in agreement with the concomitant rescue in proliferation defects (Fig. 3A,B). These results demonstrate an attenuated p21 induction in vivo in response to telomere shortening in the absence of PMS2.
Figure 7.
A role for PMS2 in p21 induction in reponse to telomeric dysfunction. (A) Quantification of p21 levels by immunohistochemistry in small intestine sections from mice of the indicated genotypes. Statistical comparisons using the χ2 test are also shown. More than 1200 nuclei from a total of two to four mice per genotype were used for the analysis. (B) Representative images showing the dramatic decrease of p21-positive cells in Terc−/−/PMS2−/− small intestine sections compared with the single Terc−/− controls.
Next, we confirmed these results by measuring p21 protein amounts by Western blot in different-genotype MEF, both in basal conditions, as well as after ionizing radiation (Supplemental Fig. 9A,B). Again, we detected increased p21 protein levels in G2 Terc−/− mice compared with wild-type controls, while single PMS2-deficient cells showed normal p21 levels (Supplemental Fig. 9A,B). Interestingly, this accumulation of p21 protein associated with short telomeres was largely abrogated by the simultaneous absence of PMS2 in G2 Terc−/−/PMS2−/− MEF (Supplemental Fig. 9A,B). As a positive control for p21 induction in response to DNA damage, we show increased p21 protein levels in response to 10 Gy of γ-irradiation (Supplemental Fig. 9A). Quantification of p53 levels by immunofluorescence in the GI tract also showed elevated p53 protein levels in G2 Terc−/− mice compared with the Terc+/+ controls, which were largely rescued in the absence of PMS2 (P < 0.001; Supplemental Fig. 9C), in agreement with the fact that p53 is one of the key regulators of p21.
All together, these results indicate an attenuated p21 induction in response to short telomeres in the absence of PMS2, similarly to that recently shown for p21 deficiency in double mutant Terc−/−/p21−/− mice (Choudhury et al. 2007). Finally, this attenuated p21 induction in the absence of PMS2 is not likely to be due to a significantly decreased DNA damage in the absence of PMS2 as indicated both by the similarly elevated frequencies of chromosomal aberrations (end-to-end fusions and signal free ends in Supplemental Fig. 7A) and of γH2AX-positive cells (Fig. 5) in single and double mutant Terc−/−/PMS2−/− mice, although we cannot rule out that the decreased number of both breaks and fragments and bivalent recombination figures associated with PMS2 deficiency could be having an effect on p21 induction. All together, these results are in agreement with a role for PMS2 in mediating the cellular response to short and dysfunctional telomeres by attenuating p21 induction.
Discussion
Here, we studied for the first time in a mammalian in vivo model the interaction between telomere function and MMR by generating mice doubly mutant for telomerase and PMS2. We found that telomerase deficiency rescues survival of single PMS2-deficient mice coincidental with decreased tumorigenesis in these mice, extending the observation that short telomeres act as potent tumor suppressors to carcinogenesis driven by defects in MMR. These findings suggest that telomerase inhibitors would be effective in the treatment of tumors bearing PMS2 mutations and microsatellite instability. Secondly, we show that, in an analogous manner to that previously described for telomerase-deficient yeast strains, PMS2 deficiency rescues survival as well as proliferative defects in telomerase-deficient mice with dysfunctional telomeres. We further demonstrate that this rescue occurs in the absence of telomere length changes and without significantly decreasing the DNA damage load associated with severe telomere dysfunction, as indicated by similar frequencies of end-to-end fusions and signal-free ends, as well as of γH2AX-positive cells in vivo. Similarly, in agreement with short telomeres and severe telomere dysfunction in these mice, we do not find elevated sister telomere recombination frequencies in mice doubly deficient for telomerase and PMS2 that could explain the rescue in survival. This is unlike the previously proposed role for MMR genes in controlling telomere recombination and telomere length at yeast telomeres, which in turn could rescue proliferation defects in telomerase-deficient yeast strains. Instead, the data presented here suggest a novel role for PMS2 in the cellular response to short and dysfunctional telomeres. In particular, PMS2 deficiency results in an attenuated p21 induction in response to dysfunctional telomeres both in vivo and in vitro. This attenuated p21 induction coincides with a rescue of proliferation but not of apoptosis in Terc−/−/PMS2−/− mice compared with the single Terc−/− controls, in the absence of increased tumorigenesis. This rescue of proliferation defects could explain the increased survival of Terc−/−/PMS2−/− mice compared with the single Terc−/− controls. This situation is analogous to that recently described for mice doubly deficient for Terc and p21. In particular, Terc−/−/p21−/− mice show a rescue of organismal survival independently of telomere length and telomere dysfunction, which is not accompanied by increased tumorigenesis (Choudhury et al. 2007). Furthermore, similarly to Terc−/−/PMS2−/− mice, Terc−/−/p21−/− mice show a rescue of proliferative defects but not of apoptosis (Choudhury et al. 2007). These results suggest that PMS2 and p21 may be in the same genetic pathway that has evolved to signal cell cycle arrest associated with telomere dysfunction. Finally, these results also suggest that cell cycle arrest may be dominant over apoptosis in eliciting age-related pathologies in the telomerase-deficient mouse model. It is important to point out that, in marked contrast to p21 and PMS2 deficiencies, p53 deficiency does not rescue survival of telomerase-deficient mice in spite of the fact p53 deficiency rescues both proliferative defects and apoptosis associated with telomere dysfunction (Chin et al. 1999). A possible explanation for this discrepancy is the fact that p53 deficiency leads to rapid tumor development, which is not impaired by telomerase deficiency (Chin et al. 1999). Instead, telomere dysfunction accelerates p53-induced carcinogenesis by promoting chromosomal instability (Artandi et al. 2000). In this regard, it is interesting to note that, in contrast to PMS2 and p21 deficiencies, p53 deficiency rescues apoptosis associated with telomere dysfunction, suggesting that this differential effect on apoptosis may be of relevance to explain the different carcinogenesis outputs.
Materials and methods
Generation and genotyping of mice
To generate PMS2−/−/Terc−/− mice, PMS2−/− females were crossed with Terc−/− males (Baker et al. 1995; Blasco et al. 1997). Genotyping was performed as described (Baker et al. 1995; Blasco et al. 1997).
Mice and primary mouse cells
Mouse colonies of successive generations (G1 to G3) of single Terc−/− and of double Terc−/−/PMS2−/− mice were generated in a C57BL6 background and maintained at the CNIO under specific-pathogen-free conditions in accordance with the recommendations of the Federation of European Laboratory Animal Science Associations.
Primary MEFs were prepared from day 13.5 embryos, as previously described (Blasco et al. 1997). Serial 3T3 cultivation was done as described (Todaro and Green 1963). Briefly, 106 cells were plated on 10-cm diameter dishes; 3 d later the total number of cells in the dish was counted, and 106 cells were replated again. The increase in population doubling level (ΔPDL) was calculated according to the formula PDL = log(nf/n0)/log2, where n0 is the initial number of cells and nf is the final number of cells. For calculation of PD at which the culture escaped senescence, we considered a valid change in the slope of the curve when, after the inflection point, the following six data points (passage numbers) increased the number of PDLs by at least two.
Telomere length analysis by quantitative FISH
Exponentially growing primary MEFs were incubated with 0.1 μg/mL colcemide (Gibco) for 4 h at 37°C and then fixed in methanol:acetic acid (3:1). Quantitative FISH was performed as described (Herrera et al. 1999; Samper et al. 2000). To correct for lamp intensity and alignment, images from fluorospheres (fluorescent beads; Molecular Probes) were analyzed using the TFL-Telo software (gift from Dr. Peter Lansdorp). Telomere fluorescence values were extrapolated from the telomere fluorescence of lymphoma cell lines LY-R (R cells) and LY-S (S cells) with known telomere lengths of 80 and 10 kb, respectively. There was a linear correlation (r2 = 0.999) between the fluorescence intensity of the R and S telomeres. We recorded the images using a COHU CCK camera on a fluorescence microscope (DMRb, Leica). A mercury vapor lamp (CS 100 W-2; Philips) was used as a source. We captured the images using the Leica Q-FISH software in a linear acquisition mode to prevent oversaturation of fluorescence intensity. We used the TFL-Telo software (Zijlmans et al. 1997) to quantify the fluorescence intensity of telomeres from at least 10 metaphases for each data point.
For Q-FISH analysis on small intestine sections, deparaffinated sections were hybridized with a PNA-telomeric probe, and telomere fluorescence was determined as described (González-Suárez et al. 2000; Muñoz et al. 2005). More than 60 nuclei from each mouse and condition were captured at 100× magnification by using a Leica CTR MIC microscope and a COHU High Performance CCD camera. Telomere fluorescence was integrated using spot IOD analysis in the TFL-TELO program (kindly provided by Dr. Landsdorp, Vancouver, Canada) (Zijlmans et al. 1997).
As median length, we refer to the telomere length value, which divides the entire population of telomeres in two equal parts, i.e., the value at which 50% of the telomeres are shorter than the median telomere length value and 50% are longer.
TRF-based telomere length analysis of MEF
Primary MEF of the indicated genotypes were included in agarose plugs following instructions provided by the manufacturer (Bio-Rad), and TRF analysis was performed as previously described (Blasco et al. 1997).
Cytogenetic analysis using telomere Q-FISH on metaphases
For analysis of chromosomal aberrations, at least 50 metaphases per genotype were analyzed by superimposing the telomere image on the DAPI image using the TFL-telo software.
Telomere recombination measurements using chromosome orientation FISH (CO-FISH)
Exponentially growing primary MEFs were subcultured in the presence of 5′-bromo-2′-deoxyuridine (BrdU; Sigma) at a final concentration of 1 × 10−5 M, and then allowed to replicate their DNA once for 24 h at 37°C. Colcemide was added at a concentration of 1 μg/mL during the last 4 h. Cells were then recovered and metaphases prepared as described (Samper et al. 2000). CO-FISH was performed as described (Bailey et al. 2004; Gonzalo et al. 2006) using first a (TTAGGG)7 probe labeled with Cy3 and then a second (CCCTAA)7 probe labeled with Rhodamine Green (Applied Biosystems). Metaphase spreads were captured on a Leitz Leica DMRB fluorescence microscope.
Histopathology and immunohistochemistry
Small intestine sections were fixed in 10% buffered formalin, embedded in paraffin wax, and sectioned at 2–3 μm. Immunohistochemistry was performed on d-paraffinated intestine sections processed with 10 mM sodium citrate (pH 6.5) cooked under pressure for 2 min. Slides were washed in water, washed in Buffer TBS Tween 20 0.5%, blocked with peroxidase, washed with TBS Tween 20 0.5% again, and blocked with fetal bovine serum followed by another wash. Then the slides were incubated with the primary antibodies: rabbit monoclonal to Ki-67 antibody (prediluted, SP6, Master Diagnostica), active caspase 3 at 1:200 (R&D Systems), or p21 at 1:15 (C-19-G, Santa Cruz). Slides were then incubated with secondary antibodies conjugated with peroxidase from DAKO, goat anti-rabbit (1:50) in the case of Ki-67 and active caspase 3 and rabbit anti-goat in the case of p21. For signal development, DAB (DAKO) was used as a substrate. Sections were lightly counterstained with hematoxylin and analyzed by light microscopy.
Phosphorylated H2AX foci (γ-H2AX) were detected using a mouse monoclonal anti-phospho-histone H2AX antibody (1:500; Upstate Biotechnology). After incubation with Cy3-goat anti-mouse antibody (1:400; Jackson ImmunoResearch Laboratories, Inc.) for 30 min at room temperature, slides were mounted in Vectashield with 4′,6-diamino-2 phenylindole (DAPI). Images were obtained using a fluorescence microscope (Leica DMRB).
Intestinal lesions were classified as mild, moderate, and severe lesions according to the pathological findings. Mild lesions were characterized by mild muscular atrophy of the small and/or large intestine. Moderate lesions showed a partial atrophy of the small and/or large intestine. Severe lesions were those characterized by (1) severe muscular atrophy, (2) severe inflammation and/or ulceration, enteritis/peritonitis, or (3) by intestinal atrophy (independent of its grade) in combination with other pathologies indicative of severe intestinal dysfunction (inflammation and/or ulceration, enteritis peritonitis).
Western blots
Whole-cell extracts were prepared from nonirradiated and irradiated primary MEF as described (Blanco et al. 2007). Protein concentration was determined using the Bio-Rad DC Protein Assay. Twenty-five micrograms of each extract was separated in 4%–20% gradient SDS-polyacrylamide gels by electrophoresis. After transfer, the membranes were incubated with an anti-p21 C-19 polyclonal antibody (1:250; Santa Cruz Biotechnology) and anti-β-actin monoclonal (1:10.000; Sigma). Antibody binding was detected after incubation with a secondary antibody coupled to horseradish peroxidase using enhanced chemiluminescence.
γ-Irradiation of primary MEF
A total of 3 × 105 MEFs were seeded in 60-mm plates and incubated overnight at 37°C. Then, MEFs were treated with 10 Gy by using a 137Cs source (MARK 1–30 irradiator; Shepherd & Associates) at a rate of 2.11 Gy/min. After removing the medium, cells were washed and fresh medium was added. To analyze the effect of γ-irradiation on protein expression, cells were incubated for 3–18 h at 37°C and then recovered and frozen for further analysis by Western blot.
Quantification of p53 expression by immunofluorescence
High-throughput quantitative image analysis of p53 expression levels in intestine sections was performed on fluorescence microscope images using the Metamorph platform (version 6.3r6; Molecular Devices). The DAPI image was used to define the nuclear area and the Cy3 image for quantification of p53 fluorescence. The DAPI images were signal-intensity thresholded, segmented, and converted to a 1-bit binary image. The binary DAPI mask was applied to the matching Cy3 to obtain a combined image with p53 fluorescence information for each nucleus. Cy3 fluorescence intensity (p53 fluorescence) was measured as “average gray values” units (arbitrary units of fluorescence). Finally, p53 fluorescence values for each histological region were exported to Microsoft Excel, and the frequency histograms were generated. p53-deficient mice were used as a negative control for p53 expression.
Microsatellite instability (MSI)
Microsatellite instability in tail DNA was assayed by PCR amplification (Baker et al. 1995) with primers for D6Mit59 (forward primer: 5′-TGTGCCATGACAGAGGGAAA-3′ and reverse primer: 5′-GAAGAAGCTGCCATCCTTTGTAATAA-3′) and D1Mit62, the control primer set (forward primer: 5′-CCTGA GTTCAGTTATCAGCGC-3′ and reverse primer: 5′-GAGAC CAGAAGAGCGTGTCC-3′). Then the products were loaded onto a 3% high-resolution agarose gel to determine allelic variants (MetaPhor Agarose, Cambrex).
Statistical analysis
A log rank test was used to calculate statistical differences in survival and median survival of the different mouse cohorts.
A t-student test was used to calculate the statistical significance of the observed differences in mean lifespan.
The Wilcoxon–Mann–Whitney rank sum test was used for statistical comparisons of the mean telomere length in MEFs and small intestine sections.
A χ2 test was used to calculate statistical differences in pathologies, Q-FISH analysis of MEF (signal free ends, telomeres <5 kb), chromosomal aberrations, γH2AX, sister chromatid exchange, apoptosis, proliferation, cell loss, p21, and p53 expression.
Acknowledgments
We thank E. Santos and R. Serrano for mouse care and genotyping. I.S.-C. is a predoctoral fellow founded by the Spanish Ministry of Education and Culture (MEC). P.M. is a Ramon y Cajal senior scientist. M.A. Blasco’s laboratory is funded by the MEC (SAF2001-1869, GEN2001-4856-C13-08), by the CAM (08.1/0054/01), European Union (TELOSENS FIGH-CT-2002-00217, INTACT LSHC-CT-2003-506803, ZINCAGE FOOD-CT-2003-506850, RISC-RAD FI6R-CT-2003-508842, MOL CANCER MED LSHC-CT-2004-502943), and the Josef Steiner Award 2003.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.430107
References
- Artandi S.E., Chang S., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Chang S., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Gottlieb G.J., Chin L., DePinho R.A., Chin L., DePinho R.A., DePinho R.A. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- Bailey S.M., Brenneman M.A., Goodwin E.H., Brenneman M.A., Goodwin E.H., Goodwin E.H. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 2004;32:3743–3751. doi: 10.1093/nar/gkh691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.M., Bronner C.E., Zhang L., Plug A.W., Robatzek M., Warren G., Elliott E.A., Yu J., Ashley T., Arnheim N., Bronner C.E., Zhang L., Plug A.W., Robatzek M., Warren G., Elliott E.A., Yu J., Ashley T., Arnheim N., Zhang L., Plug A.W., Robatzek M., Warren G., Elliott E.A., Yu J., Ashley T., Arnheim N., Plug A.W., Robatzek M., Warren G., Elliott E.A., Yu J., Ashley T., Arnheim N., Robatzek M., Warren G., Elliott E.A., Yu J., Ashley T., Arnheim N., Warren G., Elliott E.A., Yu J., Ashley T., Arnheim N., Elliott E.A., Yu J., Ashley T., Arnheim N., Yu J., Ashley T., Arnheim N., Ashley T., Arnheim N., Arnheim N., et al. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- Bechter O.E., Zou Y., Walker W., Wright W.E., Shay J.W., Zou Y., Walker W., Wright W.E., Shay J.W., Walker W., Wright W.E., Shay J.W., Wright W.E., Shay J.W., Shay J.W. Telomeric recombination in mismatch repair deficient human colon cancer cells and telomerase inhibition. Cancer Res. 2004;64:3444–3451. doi: 10.1158/0008-5472.CAN-04-0323. [DOI] [PubMed] [Google Scholar]
- Benetti R., Garcia-Cao M., Blasco M.A., Garcia-Cao M., Blasco M.A., Blasco M.A. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat. Genet. 2007;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- Blanco R., Muñoz P., Klatt P., Flores J.M., Blasco M.A., Muñoz P., Klatt P., Flores J.M., Blasco M.A., Klatt P., Flores J.M., Blasco M.A., Flores J.M., Blasco M.A., Blasco M.A. Telomerase abrogation dramatically accelerates TRF2-induced epithelial carcinogenesis. Genes & Dev. 2007;21:206–220. doi: 10.1101/gad.406207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M.A. Telomeres and human disease: cancer, ageing and beyond. Nat. Rev. Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- Blasco M.A., Hahn W.C., Hahn W.C. Evolving views of telomerase and cancer. Trends Cell Biol. 2003;13:289–294. doi: 10.1016/s0962-8924(03)00085-0. [DOI] [PubMed] [Google Scholar]
- Blasco M.A., Lee H.-W., Hande P., Samper E., Lansdorp P., DePinho R., Greider C.W., Lee H.-W., Hande P., Samper E., Lansdorp P., DePinho R., Greider C.W., Hande P., Samper E., Lansdorp P., DePinho R., Greider C.W., Samper E., Lansdorp P., DePinho R., Greider C.W., Lansdorp P., DePinho R., Greider C.W., DePinho R., Greider C.W., Greider C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Bronner C.E., Baker S.M., Morrison P.T., Warren G., Smith L.G., Lescoe M.K., Kane M., Earabino C., Lipford J., Lindblom A., Baker S.M., Morrison P.T., Warren G., Smith L.G., Lescoe M.K., Kane M., Earabino C., Lipford J., Lindblom A., Morrison P.T., Warren G., Smith L.G., Lescoe M.K., Kane M., Earabino C., Lipford J., Lindblom A., Warren G., Smith L.G., Lescoe M.K., Kane M., Earabino C., Lipford J., Lindblom A., Smith L.G., Lescoe M.K., Kane M., Earabino C., Lipford J., Lindblom A., Lescoe M.K., Kane M., Earabino C., Lipford J., Lindblom A., Kane M., Earabino C., Lipford J., Lindblom A., Earabino C., Lipford J., Lindblom A., Lipford J., Lindblom A., Lindblom A., et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- Brown K.D., Rathi A., Kamath R., Beardsley D.I., Zhan Q., Mannino J.L., Baskaran R., Rathi A., Kamath R., Beardsley D.I., Zhan Q., Mannino J.L., Baskaran R., Kamath R., Beardsley D.I., Zhan Q., Mannino J.L., Baskaran R., Beardsley D.I., Zhan Q., Mannino J.L., Baskaran R., Zhan Q., Mannino J.L., Baskaran R., Mannino J.L., Baskaran R., Baskaran R. The mismatch repair system is required for S-phase checkpoint activation. Nat. Genet. 2003;33:80–84. doi: 10.1038/ng1052. [DOI] [PubMed] [Google Scholar]
- Chan S.W.-L., Blackburn E.H., Blackburn E.H. New ways not to make ends meet: Telomerase, DNA damage proteins and heterochromatin. Oncogene. 2002;21:553–563. doi: 10.1038/sj.onc.1205082. [DOI] [PubMed] [Google Scholar]
- Chen J., Sadowski I., Sadowski I. Identification of the mismatch repair genes PMS2 and MLH1 as p53 target genes by using serial analysis of binding elements. Proc. Natl. Acad. Sci. 2005;102:4813–4818. doi: 10.1073/pnas.0407069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L., Artandi S.E., Shen Q., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A., Artandi S.E., Shen Q., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A., Shen Q., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A., Gottlieb G.J., Greider C.W., DePinho R.A., Greider C.W., DePinho R.A., DePinho R.A. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- Choudhury A.R., Ju Z., Djojosubroto M.W., Schienke A., Lechel A., Schaetzlein S., Jiang H., Stepczynska A., Wang C., Buer J., Ju Z., Djojosubroto M.W., Schienke A., Lechel A., Schaetzlein S., Jiang H., Stepczynska A., Wang C., Buer J., Djojosubroto M.W., Schienke A., Lechel A., Schaetzlein S., Jiang H., Stepczynska A., Wang C., Buer J., Schienke A., Lechel A., Schaetzlein S., Jiang H., Stepczynska A., Wang C., Buer J., Lechel A., Schaetzlein S., Jiang H., Stepczynska A., Wang C., Buer J., Schaetzlein S., Jiang H., Stepczynska A., Wang C., Buer J., Jiang H., Stepczynska A., Wang C., Buer J., Stepczynska A., Wang C., Buer J., Wang C., Buer J., Buer J., et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat. Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- di d’Aadda Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Saretzki G., Carter N.P., Jackson S.P., Carter N.P., Jackson S.P., Jackson S.P. DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes & Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- de Wind N., Dekker M., Berns A., Radman M., te Riele H., Dekker M., Berns A., Radman M., te Riele H., Berns A., Radman M., te Riele H., Radman M., te Riele H., te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- Duckett D.R., Bronstein S.M., Taya Y., Modrich P., Bronstein S.M., Taya Y., Modrich P., Taya Y., Modrich P., Modrich P. hMutSα- and hMutLα-dependent phosphorylation of p53 in response to DNA methylator damage. Proc. Natl. Acad. Sci. 1999;96:12384–12388. doi: 10.1073/pnas.96.22.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham M.A., Neumann A.A., Fasching C.L., Reddel R.R., Neumann A.A., Fasching C.L., Reddel R.R., Fasching C.L., Reddel R.R., Reddel R.R. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- Espejel S., Blasco M.A., Blasco M.A. Identification of telomere-dependent “senescence-like” arrest in mouse embryonic fibroblasts. Exp. Cell Res. 2002;276:242–248. doi: 10.1006/excr.2002.5533. [DOI] [PubMed] [Google Scholar]
- Fishel R., Lescoe M.K., Rao M.R., Copeland N.G., Jenkins N.A., Garber J., Kane M., Kolodner R., Lescoe M.K., Rao M.R., Copeland N.G., Jenkins N.A., Garber J., Kane M., Kolodner R., Rao M.R., Copeland N.G., Jenkins N.A., Garber J., Kane M., Kolodner R., Copeland N.G., Jenkins N.A., Garber J., Kane M., Kolodner R., Jenkins N.A., Garber J., Kane M., Kolodner R., Garber J., Kane M., Kolodner R., Kane M., Kolodner R., Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;77:167. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- González-Suárez E., Samper E., Flores J.M., Blasco M.A., Samper E., Flores J.M., Blasco M.A., Flores J.M., Blasco M.A., Blasco M.A. Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nat. Genet. 2000;26:114–117. doi: 10.1038/79089. [DOI] [PubMed] [Google Scholar]
- Gonzalo S., Jaco I., Fraga M.F., Chen T., Li E., Esteller M., Blasco M.A., Jaco I., Fraga M.F., Chen T., Li E., Esteller M., Blasco M.A., Fraga M.F., Chen T., Li E., Esteller M., Blasco M.A., Chen T., Li E., Esteller M., Blasco M.A., Li E., Esteller M., Blasco M.A., Esteller M., Blasco M.A., Blasco M.A. DNA methytransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- Goytisolo F.A., Blasco M.A., Blasco M.A. Many ways to telomere dysfunction: In vivo studies using mouse models. Oncogene. 2002;21:584–591. doi: 10.1038/sj.onc.1205085. [DOI] [PubMed] [Google Scholar]
- Greenberg R.A., Chin L., Femino A., Lee K.H., Gottlieb G.J., Singer R.H., Greider C.W., DePinho R.A., Chin L., Femino A., Lee K.H., Gottlieb G.J., Singer R.H., Greider C.W., DePinho R.A., Femino A., Lee K.H., Gottlieb G.J., Singer R.H., Greider C.W., DePinho R.A., Lee K.H., Gottlieb G.J., Singer R.H., Greider C.W., DePinho R.A., Gottlieb G.J., Singer R.H., Greider C.W., DePinho R.A., Singer R.H., Greider C.W., DePinho R.A., Greider C.W., DePinho R.A., DePinho R.A. Short dysfunctional telomeres impair tumorigenesis in the INK4a(delta2/3) cancer-prone mouse. Cell. 1999;97:515–525. doi: 10.1016/s0092-8674(00)80761-8. [DOI] [PubMed] [Google Scholar]
- Herrera E., Samper E., Martin-Caballero J., Flores J.M., Lee H.W., Blasco M.A., Samper E., Martin-Caballero J., Flores J.M., Lee H.W., Blasco M.A., Martin-Caballero J., Flores J.M., Lee H.W., Blasco M.A., Flores J.M., Lee H.W., Blasco M.A., Lee H.W., Blasco M.A., Blasco M.A. Disease states associated to telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 1999;18:2950–2960. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Leach F.S., Nicolaides N.C., Papadopoulos N., Liu B., Jen J., Parsons R., Peltomaki P., Sistonen P., Aaltonen L.A., Nystrom-Lahti M., Nicolaides N.C., Papadopoulos N., Liu B., Jen J., Parsons R., Peltomaki P., Sistonen P., Aaltonen L.A., Nystrom-Lahti M., Papadopoulos N., Liu B., Jen J., Parsons R., Peltomaki P., Sistonen P., Aaltonen L.A., Nystrom-Lahti M., Liu B., Jen J., Parsons R., Peltomaki P., Sistonen P., Aaltonen L.A., Nystrom-Lahti M., Jen J., Parsons R., Peltomaki P., Sistonen P., Aaltonen L.A., Nystrom-Lahti M., Parsons R., Peltomaki P., Sistonen P., Aaltonen L.A., Nystrom-Lahti M., Peltomaki P., Sistonen P., Aaltonen L.A., Nystrom-Lahti M., Sistonen P., Aaltonen L.A., Nystrom-Lahti M., Aaltonen L.A., Nystrom-Lahti M., Nystrom-Lahti M., et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- Lee H.-W., Blasco M.A., Gottlieb G.J., Greider C.W., DePinho R.A., Blasco M.A., Gottlieb G.J., Greider C.W., DePinho R.A., Gottlieb G.J., Greider C.W., DePinho R.A., Greider C.W., DePinho R.A., DePinho R.A. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Luo Y., Lin F.T., Lin W.C., Lin F.T., Lin W.C., Lin W.C. ATM-mediated stabilization of hMutL DNA mismatch repair proteins augments p53 activation during DNA damage. Mol. Cell. Biol. 2004;24:6430–6444. doi: 10.1128/MCB.24.14.6430-6444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesti M., Kanaar R., Kanaar R. DNA repair: Spot(light)s on chromatin. Curr. Biol. 2002;11:229–232. doi: 10.1016/s0960-9822(01)00112-9. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mismatch repair, genetic stability, and cancer. Science. 1994;266:1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- Modrich P., Lahue R., Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Muñoz P., Blanco R., Flores J.M., Blasco M.A., Blanco R., Flores J.M., Blasco M.A., Flores J.M., Blasco M.A., Blasco M.A. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 2005;10:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- Muntoni A., Reddel R.R., Reddel R.R. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 2005;14:191–196. doi: 10.1093/hmg/ddi266. [DOI] [PubMed] [Google Scholar]
- Nicolaides N.C., Papadopoulos N., Liu B., Wei Y.F., Carter K.C., Ruben S.M., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Papadopoulos N., Liu B., Wei Y.F., Carter K.C., Ruben S.M., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Liu B., Wei Y.F., Carter K.C., Ruben S.M., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Wei Y.F., Carter K.C., Ruben S.M., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Carter K.C., Ruben S.M., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Ruben S.M., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Haseltine W.A., Fleischmann R.D., Fraser C.M., Fleischmann R.D., Fraser C.M., Fraser C.M., et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N., Nicolaides N.C., Wei Y.F., Ruben S.M., Carter K.C., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Adams M.D., Nicolaides N.C., Wei Y.F., Ruben S.M., Carter K.C., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Adams M.D., Wei Y.F., Ruben S.M., Carter K.C., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Adams M.D., Ruben S.M., Carter K.C., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Adams M.D., Carter K.C., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Adams M.D., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Adams M.D., Haseltine W.A., Fleischmann R.D., Fraser C.M., Adams M.D., Fleischmann R.D., Fraser C.M., Adams M.D., Fraser C.M., Adams M.D., Adams M.D., et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- Peters A.C., Young L.C., Maeda T., Tron V.A., Andrew S.E., Young L.C., Maeda T., Tron V.A., Andrew S.E., Maeda T., Tron V.A., Andrew S.E., Tron V.A., Andrew S.E., Andrew S.E. Mammalian DNA mismatch repair protects cells from UVB-induced DNA damage by facilitating apoptosis and p53 activation. DNA Repair (Amst.) 2003;2:427–435. doi: 10.1016/s1568-7864(03)00003-x. [DOI] [PubMed] [Google Scholar]
- Prolla T.A., Baker S.M., Harris A.C., Tsao J.L., Yao X., Bronner C.E., Zheng B., Gordon M., Reneker J., Arnheim N., Baker S.M., Harris A.C., Tsao J.L., Yao X., Bronner C.E., Zheng B., Gordon M., Reneker J., Arnheim N., Harris A.C., Tsao J.L., Yao X., Bronner C.E., Zheng B., Gordon M., Reneker J., Arnheim N., Tsao J.L., Yao X., Bronner C.E., Zheng B., Gordon M., Reneker J., Arnheim N., Yao X., Bronner C.E., Zheng B., Gordon M., Reneker J., Arnheim N., Bronner C.E., Zheng B., Gordon M., Reneker J., Arnheim N., Zheng B., Gordon M., Reneker J., Arnheim N., Gordon M., Reneker J., Arnheim N., Reneker J., Arnheim N., Arnheim N., et al. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat. Genet. 1998;18:276–279. doi: 10.1038/ng0398-276. [DOI] [PubMed] [Google Scholar]
- Rizki A., Lundblad V., Lundblad V. Defects in mismatch repair promote telomerase-independent proliferation. Nature. 2001;411:647–648. doi: 10.1038/35079641. [DOI] [PubMed] [Google Scholar]
- Samper E., Goytisolo F.A., Slijepcevic P., van Buul P.P., Blasco M.A., Goytisolo F.A., Slijepcevic P., van Buul P.P., Blasco M.A., Slijepcevic P., van Buul P.P., Blasco M.A., van Buul P.P., Blasco M.A., Blasco M.A. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J.W., Wright W.E., Wright W.E. Telomerase therapeutics for cancer: Challenges and new directions. Nat. Rev. Drug Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- Shibata D., Peinado M.A., Ionov Y., Malkhosyan S., Perucho M., Peinado M.A., Ionov Y., Malkhosyan S., Perucho M., Ionov Y., Malkhosyan S., Perucho M., Malkhosyan S., Perucho M., Perucho M. Genomic instability in repeated sequences is an early somatic event in colorectal tumorigenesis that persists after transformation. Nat. Genet. 1994;6:273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]
- Todaro G.J., Green H., Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska A., de Lange T., Smogorzewska A., de Lange T., de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Zijlmans J.M., Martens U.M., Poon S.S., Raap A.K., Tanke H.J., Ward R.K., Lansdorp P.M., Martens U.M., Poon S.S., Raap A.K., Tanke H.J., Ward R.K., Lansdorp P.M., Poon S.S., Raap A.K., Tanke H.J., Ward R.K., Lansdorp P.M., Raap A.K., Tanke H.J., Ward R.K., Lansdorp P.M., Tanke H.J., Ward R.K., Lansdorp P.M., Ward R.K., Lansdorp P.M., Lansdorp P.M. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]