Abstract
A recent study of a large number of genes in a panel of breast and colorectal cancers identified somatic mutations in 1149 genes. To identify potential biological processes affected by these genes, we examined their putative roles based on sequence similarity, membership in known functional groups and pathways, and predicted interactions with other proteins. These analyses identified functional groups and pathways that were enriched for mutated genes in both tumor types. Additionally, the results pointed to differences in molecular mechanisms that underlie breast and colorectal cancers, including various intracellular signaling and metabolic pathways. These studies provide a multidimensional framework to guide further research and help identify cellular processes critical for malignant progression and therapeutic intervention.
Cancer arises through the gradual accumulation of alterations in oncogenes and tumor suppressor genes. In an effort to identify such genes on a genomic scale, we have recently performed a systematic sequencing study of the majority of human genes in breast and colorectal cancers (Sjöblom et al. 2006). Analysis of 13,023 genes in 11 samples of each tumor type identified 1307 somatic (i.e., tumor-specific) mutations in 1149 genes. Using a statistical model that incorporated the mutation type, frequency, and sequence context, we identified a set of nearly 200 candidate cancer genes (CAN-genes) that were likely to play a driving role in tumorigenesis. In addition to the CAN-genes, there were additional mutated genes that may have been selected for during tumorigenesis, but which were mutated at a frequency that would not allow them to be distinguished from unselected passenger changes. The genes mutated in breast cancers were quite different from those mutated in colorectal cancers. Moreover, there were substantial differences in the mutated gene complement among any two samples of the same tumor type. Overall, this effort has identified a plethora of novel genes that are likely to play a role in human cancer. However, the study also suggested a higher level of complexity, in terms of both the number and type of genes involved, than previously thought to underlie the tumorigenic process.
Given this complexity, a systems biological approach could be useful to identify patterns among the mutated genes and to help interpret the genetic landscape of the two tumor types. An optimal approach of this sort would not only examine the individual roles of the mutated gene products, but would also explore their relationships, interactions, and network properties. Understanding this interplay could provide insight into mechanisms of tumorigenesis and prioritize specific pathways and processes for future genetic and biochemical research.
In this study, we take advantage of existing genomic and proteomic databases to highlight different aspects of the genes that are mutated in breast and colorectal cancers. Our analysis uses four different system-level perspectives: (1) sequence similarity, (2) functional annotation (including cellular function, biochemical processes, and subcellular localization), (3) protein–protein interactions, and (4) molecular pathways. At each of these levels, we identify specific gene groups that were enriched for genetic alterations, revealing potentially aberrant cellular processes in the tumors.
Results
Protein sequence similarity
We first evaluated the proteins encoded by the 1149 mutated genes through sequence-similarity analyses. This approach provides an unbiased means to group proteins based on their encoded information content. Two complementary methods were used: pairwise basic local alignment search tool (BLAST) analysis and comparison of protein domains using information from existing databases. Sequence comparisons via BLAST facilitated examination of entire coding regions, while analyses of protein domains identified motifs and sequence relationships that would not be evident through whole gene comparisons.
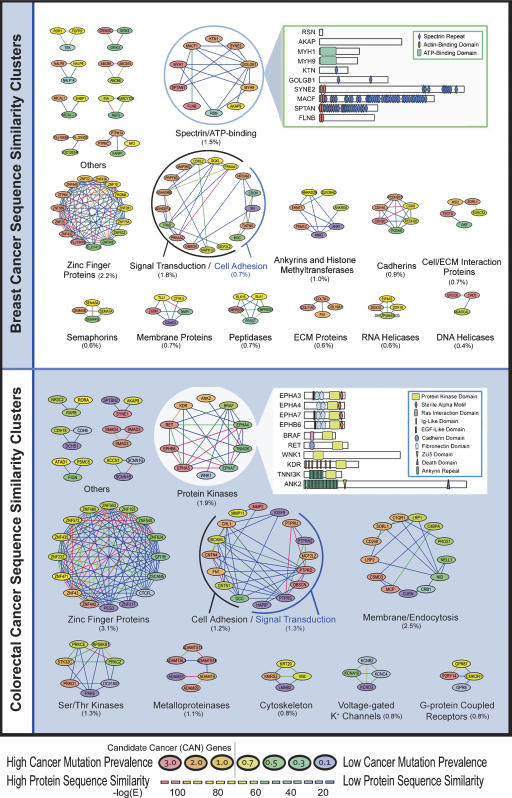
To compare entire coding regions we used BLASTP (Altschul et al. 1990) to compare sequences of all mutant proteins and constructed protein networks based on high-sequence similarity (Fig. 1). These networks identified clusters of proteins, each labeled according to the predominant functional role of the proteins contained. From a global perspective, breast and colorectal cancers shared many common clusters, including zinc finger proteins, cadherins, and genes involved in cell adhesion and signal transduction. Clusters that were mutated in one tumor type but not the other included semaphorins, RNA helicases, and DNA helicases in breast cancers and metalloproteinases, voltage-gated K+ channels, and orphan G-protein coupled receptors in colorectal cancers.
Figure 1.
Sequence similarity among mutated genes in breast and colorectal cancers. Each cluster represents genes that are mutated in breast (top) or colorectal cancers (bottom). Each node represents a gene that is colored according to the Cancer Mutation Prevalence Score (CaMP score), and each line represents a sequence-similarity relationship that is colored according to degree of sequence similarity. CAN-genes identified by Sjöblom et al. (2006) have a CaMP score >1 and are colored in orange and red. Clusters are named according to the predominant genes contained within each cluster, and those containing only two genes are not shown. The percentage of the total mutated genes contained within each cluster is showed in parentheses. The inset highlights local similarity within protein domains of genes in a specific cluster.
Genes that have high sequence identity often participate in similar intracellular roles, either through related biochemical functions, protein dimerization, genetic interactions, or more complex relationships. Within the clusters shown in Figure 1 there were several instances of patterns suggesting common functions during tumorigenesis. For example, mutations in ephrin receptors EPHA3, EPHA4, EPHA7, or EPHB6 affected 10 of the 35 colorectal tumors examined, but no tumor contained mutations in more than a single ephrin receptor, suggesting mutual exclusivity among mutations in these genes. Global analyses of sequence-similarity clusters in both breast and colorectal cancers identified nine and four clusters that showed mutual exclusivity, respectively. While the genes within some pathways act in series, and mutation of one member of the pathway is sufficient to disrupt function, clusters of sequence similarity may also include members that act in parallel pathways. For example, mutations in the TGF-beta pathway mediators SMAD2, SMAD3, or SMAD4 occurred in seven of 35 tumors. While mutations in SMAD4 did not occur in tumors with other SMAD mutations, both SMAD2 and SMAD3 were co-mutated in colorectal tumors Mx30 and Hx5 (Supplemental Fig. 1). Interestingly, SMAD2 or SMAD3 can separately heterodimerize with SMAD4 transcription factors upon pathway activation and mediate transcriptional responses (Jayaraman and Massague 2000). These results suggest that inactivation of either SMAD4 alone or SMAD2 and SMAD3 together have similar effects on the TGF-beta receptor pathway.
A complementary method for analysis of sequence similarity takes advantage of information from existing databases. Instead of determining relatedness solely using BLASTP, other methods such as Hidden Markov Models and consensus sequences have facilitated in-depth comparisons of protein sequences. The Integrated Resource of Protein Families, Domains, and Sites (InterPro) database incorporates information from 16 protein databases, including Pfam, ProDom, PRINTS, PROSITE, and SMART (Apweiler et al. 2001). Using the annotation provided by InterPro 13.0, we examined the protein sequences of all mutated genes for the presence of specific domains. A total of 13,147 possible domains were examined in 1149 mutated proteins, resulting in a total of 1029 proteins that were found to have 3549 domain assignments.
We examined these data in two ways to determine whether gene groups containing specific domains were more likely to be mutated than predicted by chance alone. First, we determined whether the number of mutations in gene groups containing specific domains reflected a mutation prevalence that was significantly higher than the passenger mutation prevalence. We performed these calculations for breast and colorectal cancers separately, using the conservative assumption that the observed mutation frequencies of 2.5 and 3.3 mutations per million base pairs, respectively, constituted the passenger rates. Note that this criterion is highly conservative, as the observed mutations actually represent the sum of passenger mutations and those mutations selected for during tumorigenesis (i.e., pathogenic mutations). The resulting Group CaMP score is similar to that used to derive the Cancer Mutation Prevalence (CaMP) score for individual genes. The Group CaMP score incorporated the total number of mutations from all genes within each group, the combined lengths of the genes in each group, and the total number of tumors examined. The P-value of observing at least the number of mutations in a binomial distribution was calculated and corrected with the Benjamini–Hochberg algorithm (Benjamini and Hochberg 1995).
Second, we examined whether the distributions of individual CaMP scores of mutated genes containing domains of interest were different from mutated genes not containing such domains. To compare such distributions, we adapted the Gene Set Enrichment Analysis (GSEA) algorithm, using CaMP scores of individual genes instead of summaries of gene-expression values (Subramanian et al. 2005). The CaMP GSEA approach incorporates the set of individual CaMP scores from all genes within each group, accounting for the number, type and context of mutations observed, gene length, as well as the total number of tumors examined. This approach is complementary to the group CaMP score described above; while the former approach is more sensitive to the overall mutation prevalence in a group of genes, the latter would be expected to identify more subtle differences among the mutated genes within such groups.
After identification of candidate groups that were significantly enriched for mutations using these approaches, we filtered the results to identify those groups that were also enriched for an increased number of mutant genes. Specifically, we determined whether the ratio of the number of mutant genes containing each specific domain to all genes containing that domain was statistically higher than the ratio of the total number of mutant genes (1149) to the number of all the genes (13,023) analyzed. This filtering step ensured that multiple genes within each gene group must be affected in order for the entire group to be considered of interest. A gene group that had contained only one highly mutated gene (e.g., mutations only in TP53) would thereby be excluded.
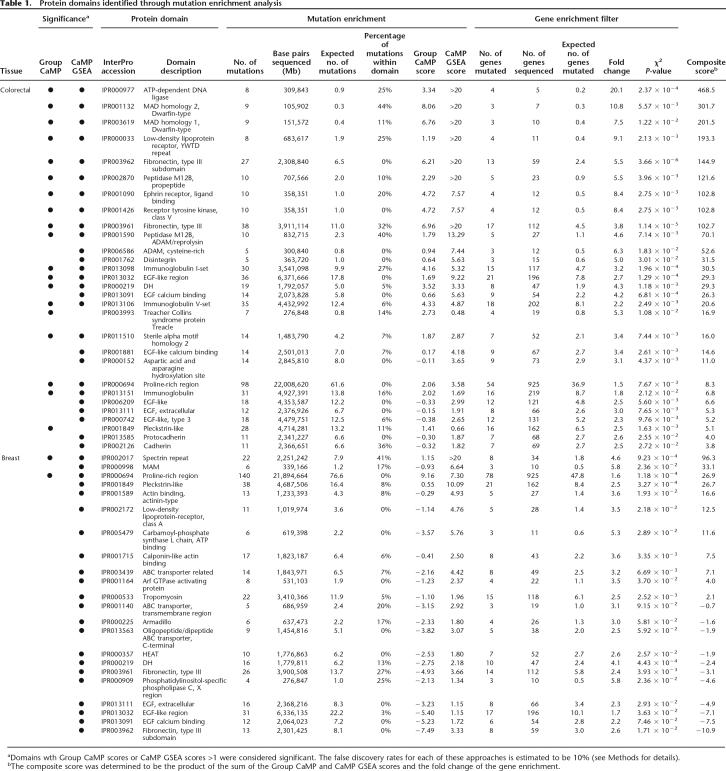
Using these two analysis approaches (Group CaMP and CaMP GSEA), a total of 31 and 22 InterPro domains were significantly associated with colorectal and breast cancers, respectively (Table 1; Supplemental Table 1). In colorectal cancers, the majority were determined to be significant by both methods and involved several related protein domains. For example, 14 of the identified domains are in proteins that have extracellular regions or are involved in cell–cell interactions (e.g., four immuloglobulin-related domains, two fibronectin domains, six EGF-related domains, and two cadherin-related domains). An additional five domains (e.g., pleckstrin-like domain, DH domain, Ephrin receptor ligand-binding domain, Sterile alpha motif homology 2, and receptor tyrosine kinase domain) are known to be involved in protein kinase or G protein signal transduction pathways. Domains identified that were associated with metalloproteases include reprolysin, peptidase M12B propeptide, cysteine-rich ADAM, and disintegrin. Finally, domains present in TGF-beta pathway transcription mediators SMAD (MAD homology 1 and MAD homology 2 domains) were also identified as significantly associated with colorectal cancer. Interestingly, proteins containing MAD homology, ephrin receptor, and Treacher Collins Syndrome protein domains were found to be exclusively mutated in colorectal cancers, while members of the other domains were mutated in both tumor types. Other domains shared by both cancer types include three of the extracellular EGF-related domains, as well as two domains involved in signaling, the DH domain and the pleckstrin-like domain. In breast cancers, two motifs were detected by both the Group CaMP and GSEA methods: one was the spectrin repeat domain that is present in various cytoskeletal proteins, while the second was the relatively nonspecific proline-rich region domain that was also associated with colorectal cancers. Three domains related to ABC transporters and two domains involved in actin binding were preferentially identified in breast tumors.
Table 1.
Protein domains identified through mutation enrichment analysis
aDomains wth Group CaMP scores or CaMP GSEA scores >1 were considered significant. The false discovery rates for each of these approaches is estimated to be 10% (see Methods for details).
bThe composite score was determined to be the product of the sum of the Group CaMP and CaMP GSEA scores and the fold change of the gene enrichment.
Functional annotation and gene ontology
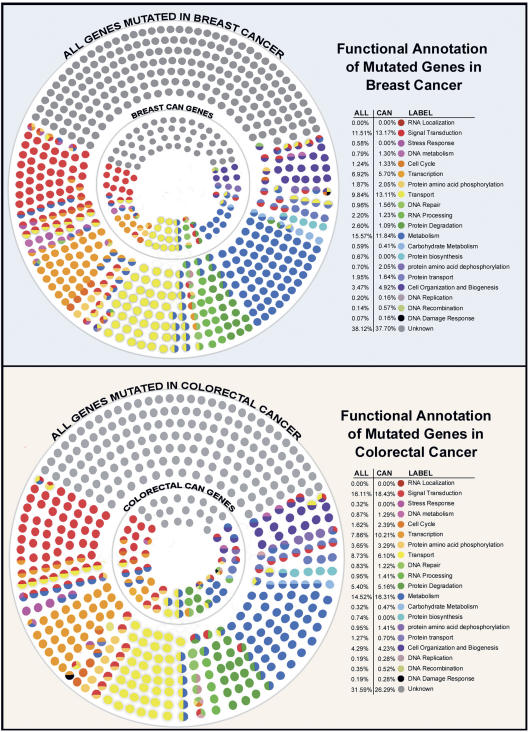
In addition to analyses based on sequence content, the mutated genes were categorized according to their annotated biological roles. The Gene Ontology (GO) Consortium has devised a controlled vocabulary for describing molecular functions and biological processes of genes based on information obtained from the literature and from sequence and biological databases (Ashburner et al. 2000). These are represented in hierarchical levels of directed acyclic relationships that progress from general descriptions to progressively more specific descriptions. In general, a gene can have multiple descriptions, and functional descriptions for any gene using GO can be complex. To simplify such descriptions, methods have been designed to summarize the GO relationships into fewer general categories (Camon et al. 2004; Martin et al. 2004). For these analyses, we first used a general approach to examine the broad functional categories of all mutated genes in breast and colorectal cancers (Fig. 2), and then identified the specific GO groups that were preferentially associated with each of these tumor types (Table 2).
Figure 2.
General functional categorization of genes mutated in breast and colorectal cancers. Each small circle represents a mutated gene in breast or colorectal cancer and is colored according to the general functional categories shown in the legend (for details, see Methods). The entire set of circles represents all the genes mutated in each cancer type, while the interior subset is comprised of the genes with the highest CaMP Scores (the CAN-genes). The percentage of genes that belong to each functional category is shown in the legend.
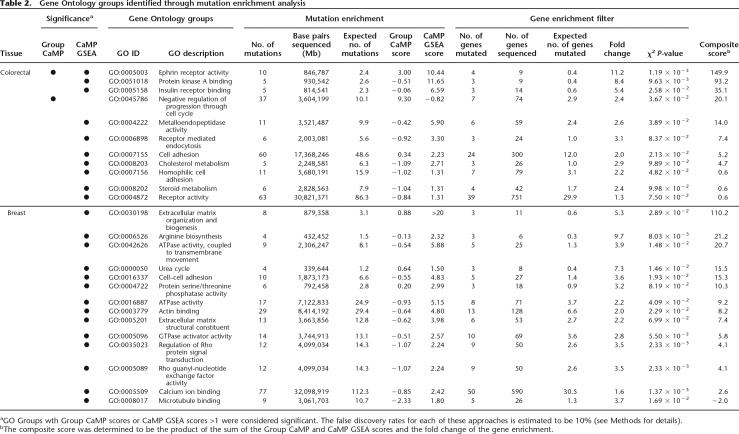
Table 2.
Gene Ontology groups identified through mutation enrichment analysis
aGO Groups wth Group CaMP scores or CaMP GSEA scores >1 were considered significant. The false discovery rates for each of these approaches is estimated to be 10% (see Methods for details).
bThe composite score was determined to be the product of the sum of the Group CaMP and CaMP GSEA scores and the fold change of the gene enrichment.
Classification of the mutated genes into general functional categories was visualized using OSPREY (Fig. 2) (Breitkreutz et al. 2003). In accord with our initial analysis of a small subset of mutated genes (Sjöblom et al. 2006), the comprehensive analyses of all mutated genes resulted in similar compositions of functional categories for both breast and colorectal cancers. The fractions of different functional categories were largely comparable between the two cancer types, with the two largest comprising signal transduction and metabolism. Importantly, the individual genes that were included in these categories were different in the different tumor types, and individual tumors had a varying composition of genes belonging to these functional categories (Supplemental Fig. 2). Noticeably, over a third of the mutant genes were not assigned to any functional category using the current annotation, a fraction that would be expected to decrease as additional biological data are obtained. A similar analysis of the subset of genes most highly mutated in breast and colorectal cancers identified subtle differences in composition of functional categories (Fig. 2). These analyses suggest that genes that are selected for mutation in human cancer may come from a variety of different functional categories, but that such broad categories may not be helpful in accurately capturing specific functional aspects of genes that are preferentially mutated in tumors.
In order to identify more specific molecular functions for the mutant genes, we examined the full set of 18,740 GO groups. Using approaches similar to that used in the analysis of protein domains, we identified GO groups that were enriched for the number of mutations or distribution of CaMP scores using CaMP GSEA and Group CaMP approaches. In colorectal cancer, we identified 11 GO groups to be significant by either method (Table 2; Supplemental Table 2). Groups such as ephrin receptor activity as well as metalloendopeptidase activity corroborated results identified above through the analysis of protein domains. Two of the largest functional groups, cell adhesion and receptor activity, had 24 and 39 mutated genes and 60 and 63 mutations, respectively. More specific subgroups from these groups included insulin receptor binding and homophilic cell adhesion.
In breast cancers, 15 functional groups were identified, none overlapping precisely with those of colon. The most closely related ones involved functional groups that were involved in cell adhesion. The largest group identified was calcium ion binding, which included 50 mutated genes and 77 mutations. Five groups were associated with the extracellular matrix, including extracellular matrix organization and biogenesis, extracellular matrix structural constituent, microtubule binding, actin binding, and cell–cell adhesion. Interestingly, two metabolic groups were affected in breast tumors: the overlapping groups of the urea cycle and arginine biosynthesis. Finally, there were three groups related to G protein signaling: GTPase activator activity and two Rho protein modulating groups. These analyses clearly show that while overall functional patterns may be similar between breast and colorectal cancers, the specific group constituents of these general categories are quite different.
Protein interactions
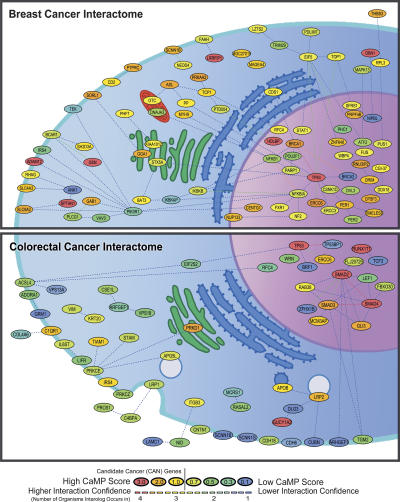
As part of their biologic roles, many proteins physically interact with other proteins; such interactions can form protein complexes or may represent functional components of molecular pathways. An additional perspective on gene function can therefore be realized through the analysis of physical interactions. Efforts to identify interacting proteins at a genome-wide scale, termed interactomes, have been performed for yeast (Uetz et al. 2000; Ito et al. 2001), fly (Giot et al. 2003), and worm (Li et al. 2004). Although the human interactome is incomplete, human protein–protein interactions can be inferred from data on model organisms using interologs (human protein orthologs that are known to interact in other organisms). Such interologs have differing levels of reliability, in part based on the strength of the interaction, the number of organisms in which the interaction is observed, and the sequence similarity of the interacting proteins to their human counterparts. Using this approach, we examined the predicted interactions of mutated proteins with other proteins. Overall, this analysis showed that the mutant proteins interacted with more partners than typical human proteins. Proteins encoded by CCDS genes had an average of 11.8 predicted interactions each, while the set of the 189 most highly mutated CAN-genes had an average of 26.0 and 16.4 interactions in colon and breast cancers, respectively (P = 0.02 and 0.12, Student’s t-test). Proteins with a large number of interactions have been suggested to serve as essential hubs of molecular pathways such as those that are disrupted in cancers (Jonsson and Bates 2006).
To identify networks of interacting proteins that were preferentially altered in cancers, we analyzed the predicted interactions of mutated proteins in each tumor type (Fig. 3>; Supplemental Figs. 3, 4). In breast cancers, over half of the mutated proteins (59 of 83) were predicted to participate in a large interaction cluster driven by links to TP53, BRCA1, PIK3R1, and NFKB. In contrast, the largest interaction cluster in colorectal cancers involved SMAD proteins and contained only 12 proteins, and the only cluster containing more than five proteins included TP53. These analyses emphasize how mutation studies coupled with systems analysis can provide information useful for understanding the pathways through which the mutant proteins function. For example, the mutation interactome highlighted three interacting SMAD proteins in colorectal cancers and a cluster of circadian rhythm proteins (PER1, PER2, and TIMELESS) in breast cancers. The proteins encoded by the latter three genes are thought to control cell cycle progression, and genetic inactivation of one of the genes (Per2) has been shown to lead to tumor predisposition in mice (Fu et al. 2002; Chen et al. 2005; Lee 2006).
Figure 3.
Interaction among proteins mutated in breast and colorectal cancers. Each node represents a mutated protein that is colored according to Cancer Mutation Prevalence (CaMP) Score, and each line represents an interaction confidence. CAN-genes identified by Sjöblom et al. (2006) have a CaMP score >1 and are colored in orange and red. The genes are placed within cellular compartments as annotated in Gene Ontology.
Molecular pathways
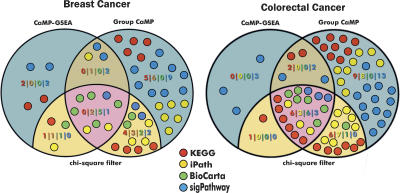
Pathways can be defined as the stepwise interaction of multiple proteins designed to achieve a defined cellular process. A variety of signaling, metabolic, and other pathways have been cataloged by the Kyoto Encylopedia of Genes and Genomes (KEGG) (Ogata et al. 1999), the iPATH (http://escience.invitrogen.com/ipath/), BioCarta (http://www.biocarta.com/), and sigPathways (Tian et al. 2005) databases. To determine whether certain pathways may be preferentially targeted by genetic alterations in human cancer, we compared the genes found to be mutated in Sjöblom et al. (2006) to the constituents of the 825 pathways contained in these four sources. We then determined whether the number of mutations and the distribution of CaMP scores within each pathway is statistically significant using Group CaMP and CaMP GSEA methods. The intersections of the two methods for the four different databases are shown in Figure 4.
Figure 4.
Comparison of mutation enrichment in cellular pathways using complementary statistical approaches. Venn diagrams show the number of pathways identified from four different databases in breast (left) and colorectal cancers (right) using CaMP GSEA and Group CaMP approaches. Each circle represents one pathway and is colored according to the database it belongs to. Pathways that were enriched for mutations and which were filtered for an increase in the number of genes using the χ2 test are shown in tan or pink. Blue and dark tan areas represent pathways that were excluded using the χ2 filter (for additional details, see Methods).
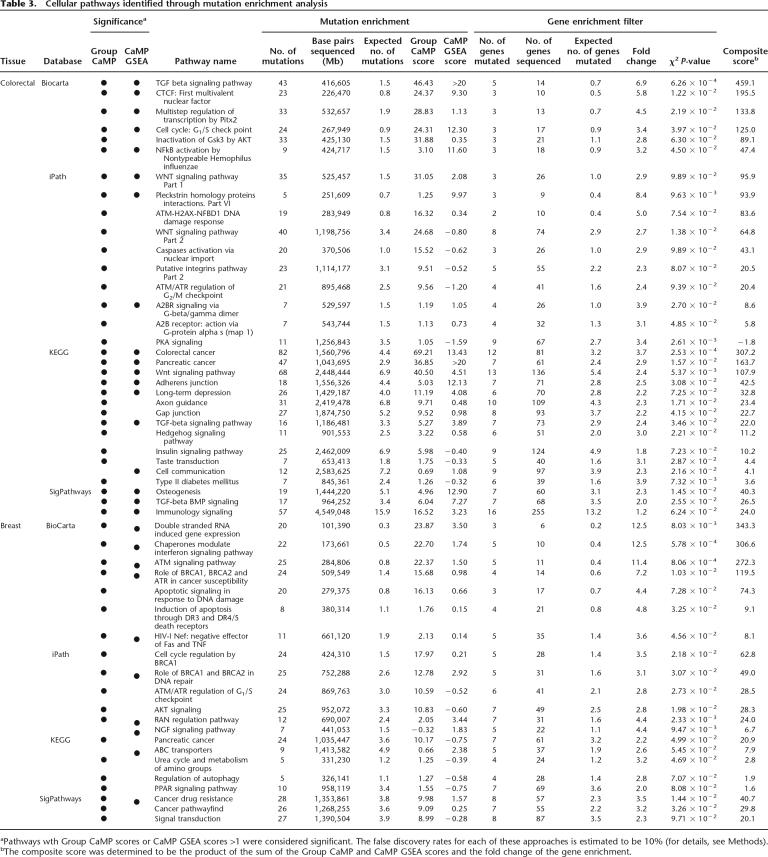
In colorectal cancer, 21 pathways were identified to be enriched for mutations from the different pathway databases (Table 3; Supplemental Table 3). Two pathways previously implicated in colorectal tumorigenesis were identified in multiple databases; TGF-beta signaling was identified in three databases and WNT signaling in two. Other signaling pathways contributing to tumorigenesis were also identified, including Insulin signaling, JAK/STAT signaling, MAP kinase signaling, and hedgehog signaling pathways. Two pathways identified were related to the cell cycle and the G1/S and G2/M checkpoints. Finally, genes in pathways thought to be important in controlling cell–cell interactions (axon guidance, adherens junctions, and gap junctions) were preferentially mutated in colorectal cancers.
Table 3.
Cellular pathways identified through mutation enrichment analysis
aPathways wth Group CaMP scores or CaMP GSEA scores >1 were considered significant. The false discovery rates for each of these approaches is estimated to be 10% (for details, see Methods).
bThe composite score was determined to be the product of the sum of the Group CaMP and CaMP GSEA scores and the fold change of the gene enrichment.
In breast cancer, several known signaling and checkpoint pathways were also identified (Table 3; Supplemental Table 3). These included those involved in AKT signaling, in BRCA1 and BRCA2 repair and cell cycle regulation processes, and in ATM/ATR checkpoint control. Although TP53 was frequently mutated in each of these pathways, many other genes were also implicated, suggesting that multiple mechanisms may exist for dysregulation of these pathways in breast cancer. Additionally, seven members of the RAN regulation pathway were found to be mutated in breast cancers, while none were mutated in colorectal cancers. The RAN pathway members included proteins involved in nuclear transport such as NUP133, NUP214, NUP98, and KPNA5. NUP98 and NUP214 have been shown to be targets of translocation in several human malignancies (Kau et al. 2004; Nakamura 2005), but no intragenic mutations of these genes have been previously observed in any cancer. In addition, a number of pathways were detected that were related to groups identified using other approaches. These included ABC transporters, which were identified through domain analyses, and the urea cycle pathway, which was identified through the functional group approach.
Integrative analysis
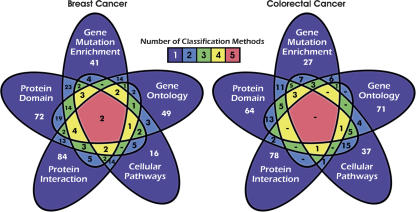
We next integrated the results obtained from the different system-level analyses described above. Although it is not possible to directly compare these disparate groups (e.g., one cannot compare pathways to protein domain groups), one can examine the overlap among the genes belonging to the groups identified through the different analysis modalities. Comparisons of these data, including the set of CAN-genes are shown in Figure 5 and in Supplemental Table 4, A and B. Overall, a large fraction (64% and 77%) of the genes in breast and colorectal cancers were uniquely identified by one of the approaches used. These results demonstrate the value of a multidimensional analysis, as each method can identify features that are missed by others. Conversely, the intersecting regions of the different dimensions can point to specific genes that may be of interest because they are implicated by several criteria. For example, in breast cancer, TP53 and SPTAN1 were identified to be significant in all five dimensional analyses, and 12 other genes were significant by four methods; in colorectal cancer, five genes lie at the intersection of four of the five analyses, including TP53, EPHA3, EPHB6, LRP2, and IRS4. Examining the specific groups that identify these and other genes to be enriched may provide insight into new combinations of ways in which these genes may be involved in tumorigenesis.
Figure 5.
Comparison of genes annotated though different mutation enrichment classification methods. Five-way Venn diagrams (Grünbaum 1975) show the number of genes annotated through the indicated methods for breast and colorectal cancers. The “Gene Mutation Enrichment” set are the CAN-genes defined by Sjöblom et al. (2006). Each region indicates the number of genes that are detected by the different analytical methods, and is colored according to the number of methods that identify those genes. The genes detected by each classification method are listed in Supplemental Table 4, A and B.
Discussion
Interpreting the large and complex datasets that arise from genome-wide mutational analyses of cancer is challenging. Given the improvements in bioinformatics and sequencing technologies, we expect that many such projects will come to fruition over the next several years. In the first study of this type, Sjöblom et al. (2006) primarily focused on individual genes and attempted to identify those whose mutation frequencies reflected rates that were higher than the passenger mutation frequencies. This initial analysis was unable to discern important genes that were mutated at relatively low frequencies and did not analyze relationships among the mutated genes. The multidimensional analysis performed herein was designed for just these purposes. While the initial study identified individual genes that were mutated, this current analysis identifies pathways, functional groups, and interacting networks that are mutated in colorectal and breast cancers. Although our analyses are at present incomplete and depend on the fidelity and extent of current annotation databases, several conclusions have already begun to emerge from these studies.
The first is that the distribution of mutations observed in the Sjöblom et al. (2006) study is clearly nonrandom. This was initially suggested by the overall number of mutations observed and the prevalence of mutations in specific genes. In this multidimensional analysis, we have identified 51 protein domains, 25 functional groups, and 53 pathways that are enriched for somatic mutations. The Group CaMP and CaMP GSEA approaches used to delineate these gene groups were performed in a rigorous manner, requiring that both the fraction of genes within a particular group and the number or distribution of mutations be higher than what would be expected in the absence of selection. As the Group CaMP studies are sensitive to the passenger mutation prevalence, we performed our analyses using the most conservative estimate available for this prevalence by assuming that all mutations detected in the Sjöblom et al. (2006) study were simply passengers. This approach is clearly an overestimate of the true passenger mutation frequency because it includes mutations of genes that play a causal role in tumorigenesis as well as true passenger mutations. Nevertheless, this process ensured that the groups identified in this study were statistically significant even if the passenger mutation rate was higher than estimated by previous experimental studies.
A second conclusion is that there is substantial value in examining these datasets from different dimensions. Enrichment in protein domains reveals groups of highly related proteins, each of which may be mutated at low levels. Although there is a clear relationship between sequence and function, analysis of enriched functional annotation can allow for abstraction of important biological processes shared by disparate proteins that may not be similar on a sequence level. Examination of protein–protein interactions can provide a more global view of networks that are enriched for mutations. Finally, pathways reveal organizing structures that may not be determined from the other three dimensions. Together, these four complementary views can provide a global view of mutated gene groups and processes.
What are the gene groups and processes that are enriched in these cancer types and what do they tell us about the mechanisms underlying tumorigenesis? For both tumor types, our results pointed to the importance of alterations in intercellular interactions. These included proteins with extracellular domains involved in adhesion (e.g., fibronectin and cadherins), functional groups involved in cell adhesion and extracellular matrix generation and biogenesis, and pathways implicated in cell–cell communication. A multitude of mutated genes are contained in these groups and are delineated in Supplemental Tables 1–3. These observations are generally consistent with the hypothesis that in order for tumor cells to proliferate and invade, they must alter their adhesion dependence to other cells and to the basement membrane and escape control by contact inhibition (Gupta et al. 2007).
The enriched groups and pathways also suggested that certain aspects of intracellular signaling, cell cycle control, and metabolism may be important for tumorigenesis. Two known signaling pathways, involving AKT and ATM/ATR, were enriched in both colorectal and breast tumors, reflecting the important role these play in these tumor types (Vogelstein and Kinzler 2004). However, many of the remaining groups and pathways were specific to one of the tumor types, suggesting that there may be distinct cellular processes underlying breast or colorectal cancer. This is in part exemplified by the dramatic differences of the number and type of interacting mutant proteins present in either breast or colorectal cancers (Fig. 3). For colorectal cancer, such groups included MAD domain-containing proteins, ephrin receptors, metalloproteases, and the TGF-beta and WNT pathways. The latter two of these are consistent with known tumor-related signaling pathways in colorectal cancers (Vogelstein and Kinzler 2004). While the function of metalloproteases and ephrin receptors remains to be further elucidated, one intriguing possibility is that these proteins may be related to the late stage of the tumors examined. All of the colorectal tumors examined in the Sjöblom et al. (2006) study were derived from metastatic lesions, and expression changes (but not mutations) of metalloproteases and ephrin receptors have previously been associated with late stage tumors (Deryugina and Quigley 2006). Breast tumors, on the other hand, demonstrated an enrichment of mutated genes involved in BRCA1 and BRCA2 DNA repair processes, nuclear and cell-surface transporters, urea and arginine biosynthesis, ATPase and GTPase activity, and PPAR signaling pathways. None of these gene groups, except for BRCA genes, had been definitively linked to breast tumorigenesis and raise interesting hypotheses about the role of these genes in tumor development and progression. For example, the mutations in the urea pathway affected four proteins (ACY1, ASL, CPS1, and OTC) that impact directly or indirectly on the production ornithine, a key precursor of polyamine synthesis (Casero and Marton 2007). As polyamines have substantial effects on cellular proliferation and apoptosis, mutations of this pathway may represent a novel mechanism of polyamine dysregulation in human cancers.
Finally, the results lead to a deeper understanding of the mutational data and its implications for neoplasia. In the Sjöblom et al. (2006) study, it was noted that there were a great number of genes mutated in each tumor and that the genetic alterations in tumors of the same type were quite heterogeneous. Thus, no two breast cancers shared more than a few mutated genes. This heterogeneity may indeed account for the large biologic differences among breast cancers noted by clinicians, and suggests that a large number of novel therapeutic approaches will be needed to combat these cancers. In contrast, our studies suggest that, though the precise genes mutated in different cancers are heterogeneous, there are a more limited number of genetic groups and pathways through which these genes act. Thus, the complexity at the gene level is likely to be substantially reduced at the pathway level. Future work will be needed to fully elucidate how the gene groups identified herein operate to initiate or accelerate cancerous growth. However, one can at least envision development of therapeutic strategies that inhibit downstream components of a relatively small number of pathways and that would be applicable to a large number of patients. For this vision to be realized, additional sequencing studies as well as more systems biologic analyses, coupled with functional studies of the mutated genes, will all be essential.
Methods
Genomic sequence similarity calculation
The nucleotide and amino acid sequences of all 14,795 CCDS entries were downloaded from NCBI (ftp://ftp.ncbi.nlm.nih.gov/pub/CCDS/current) according to the March 2, 2005 release based on the 35.1 genome annotation build. Using formatdb, we created a CCDS blast database and analyzed each of the CCDS entries using blastp with a minimum E-value cut-off of 0.05 and a score of 100. In total, 1639 and 2118 sequences for breast and colon, respectively, were identified and visualized with Cytoscape 2.3.1.
Protein domain comparisons
The InterPro database release 13.0 was downloaded in August 2006 (http://www.ebi.ac.uk/interpro/project_outlines.html) and cross-referenced with the CCDS entries. In total, 12,781 CCDS IDs matched one of 4151 IPR domains. We identified all domains contained within the 1149 mutant genes identified, considering all transcripts of these genes.
Significance of gene sets
Group CaMP
For each group of genes we determined the total number of mutations observed for the tissue of interest, as well as the number of base pairs that were successfully sequenced. We then computed the P-value as the probability of a group having at least as many mutations as were observed, given the numbers of base pairs sequenced and the background passenger frequencies, using the binomial distribution in R (http://www.r-project.org). The background passenger frequencies were conservatively estimated as the total numbers of mutations observed in each tumor type in Sjöblom et al. (2006) divided by the total number of base pairs sequenced in the study (i.e., assuming that all of the mutations observed were passengers). The P-values were then corrected for multiple comparisons according to the method described by Benjamini and Hochberg (1995) with an FDR of 10%. The Group CaMP score is the negative log of the corrected P-value.
CaMP GSEA
We also identified gene sets that were statistically significant in their distribution of CaMP scores (CaMP GSEA) when compared with the whole CCDS set. For each gene, we calculated the CaMP score as described previously (Sjöblom et al. 2006). First, for each of the seven predefined mutational contexts, we computed the probability of observing the number of mutations found using the binomial distribution and the passenger mutation rate. We took the product of these values and divided by the relative rank of this statistic. Using the CaMP score as a metric, we implemented the GSEA algorithm using the R statistical environment. In a list of all the mutated genes sorted by CaMP score, we compared the ranking of the set of genes that contained each domain with those that did not, using the Wilcoxon test, as implemented by the Limma package in Bioconductor (Smyth 2005). Then, the statistical significance of deviation from the null hypothesis of random distribution was calculated and corrected for multiplicity by the Benjamini-Hochberg algorithm (Benjamini and Hochberg 1995) with an FDR of 10%.
For the groups identified by either the Group CaMP or CaMP GSEA approaches, we further focus on those that were also enriched for an increased number of mutant genes. For each group, we computed the total number of genes observed to be mutated and sequenced, taking into consideration multiple CCDS entries for some genes. For each group, the expected number of mutated genes was calculated to be the product of the number of sequenced genes in the group and the proportion of genes mutated in the entire study. Although this is a post-test filter and not a test in itself, we report a P-value calculated using the Pearson χ2 test in the R statistical environment.
Functional annotation and Gene Ontology
All mutated genes for both colorectal and breast cancers were categorized to general functional categories and visualized using Osprey 1.2.0.
Biological process and molecular function categories were obtained from the GO Consortium website (http://www.geneontology.org). These contained 11,295 biological processes and 7445 molecular functions, as of August 2006. The cross-reference to CCDS entries resulted in 22,705 and 26,430 assignments, respectively. For each GO category, similar calculations were performed for the total number of mutations observed and the total number of base pairs sequenced, as described above for the protein domains. The Group CaMP and GSEA CaMP scores were calculated as described above.
Predicted protein–protein interactions
Predicted protein–protein interactions in humans were downloaded from the Online Predicted Human Interaction Database (OPHID). As of August, 2006, the database contained 49,008 interactions involving 10,682 proteins. CCDS name translation was performed with both RefSeq and SWISS-PROT identifiers as well as via manual curation. Protein–protein interactions between genes mutated in either cancer type were abstracted. In total, 196 and 134 interactions (involving 80 and 59 genes) were identified in breast and colorectal cancers, respectively.
Cellular component data was obtained from Gene Ontology. As of August, 2006, 1802 cellular component terms were available. For network visualization, we first generated the initial network with Cytoscape 2.3.1. Each individual gene was then placed into an appropriate cellular component based on the Gene Ontology data.
Molecular pathways analysis
Pathway assignment data were downloaded from the KEGG (Ogata et al. 1999), Invitrogen iPath (http://escience.invitrogen.com/ipath), BioCarta (http://www.biocarta.com), and sigPathway databases (http://www.chip.org/~ppark/Supplements/PNAS05.html). For KEGG, we used release 39.0 downloaded in August, 2006, which included 41,689 pathways generated from 303 reference pathways. We used NCBI gene IDs to cross-referencing these pathways to CCDS genes, resulting in 7787 assignments. For iPath, we used the online interactive tool to obtain 171 signal transduction and 54 metabolic pathways in August 2006. We identified 4027 assignments of CCDS genes to at least one of the iPath groups. For sigPathway and BioCarta, we used version 1.1.4 from April 2006. We specifically used the 50 pathways identified as humanpaths and 308 pathways identified with BioCarta. In total, 378 and 350 assignments were made to sigPathway and BioCarta. The Group CaMP and GSEA CaMP scores were calculated as described above in the Methods section for the protein domain comparisons to identify cellular pathways that were enriched for mutations in breast or colorectal cancers.
Acknowledgments
This study was supported by The Virginia and D.K. Ludwig Fund for Cancer Research, NIH grants CA 121113, CA 43460, CA 57345, CA105090-03 and CA62924, NSF grant DMS034211, The Pew Charitable Trusts, The Clayton Fund, The Blaustein Foundation, and the NCI Division of Cancer Prevention contract HHSN261200433002C.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6431107
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Apweiler R., Attwood T.K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D., Attwood T.K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D., Bucher P., Cerutti L., Corpet F., Croning M.D., Cerutti L., Corpet F., Croning M.D., Corpet F., Croning M.D., Croning M.D., et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Dolinski K., Dwight S.S., Eppig J.T., Dwight S.S., Eppig J.T., Eppig J.T., et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. [Ser A] 1995;57:289–300. [Google Scholar]
- Breitkreutz B.J., Stark C., Tyers M., Stark C., Tyers M., Tyers M. Osprey: A network visualization system. Genome Biol. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camon E., Barrell D., Lee V., Dimmer E., Apweiler R., Barrell D., Lee V., Dimmer E., Apweiler R., Lee V., Dimmer E., Apweiler R., Dimmer E., Apweiler R., Apweiler R. The Gene Ontology Annotation (GOA) Database–an integrated resource of GO annotations to the UniProt Knowledgebase. In Silico Biol. 2004;4:5–6. [PubMed] [Google Scholar]
- Casero R.A., Marton L.J., Marton L.J. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- Chen S.T., Choo K.B., Hou M.F., Yeh K.T., Kuo S.J., Chang J.G., Choo K.B., Hou M.F., Yeh K.T., Kuo S.J., Chang J.G., Hou M.F., Yeh K.T., Kuo S.J., Chang J.G., Yeh K.T., Kuo S.J., Chang J.G., Kuo S.J., Chang J.G., Chang J.G. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- Deryugina E.I., Quigley J.P., Quigley J.P. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- Fu L., Pelicano H., Liu J., Huang P., Lee C., Pelicano H., Liu J., Huang P., Lee C., Liu J., Huang P., Lee C., Huang P., Lee C., Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Ooi C.E., Godwin B., Vitols E., Godwin B., Vitols E., Vitols E., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Grünbaum B. Venn diagrams and Independent Families of Sets. Mathematics Mag. 1975;48:12–23. [Google Scholar]
- Gupta G.P., Nguyen D.X., Chiang A.C., Bos P.D., Kim J.Y., Nadal C., Gomis R.R., Manova-Todorova K., Massague J., Nguyen D.X., Chiang A.C., Bos P.D., Kim J.Y., Nadal C., Gomis R.R., Manova-Todorova K., Massague J., Chiang A.C., Bos P.D., Kim J.Y., Nadal C., Gomis R.R., Manova-Todorova K., Massague J., Bos P.D., Kim J.Y., Nadal C., Gomis R.R., Manova-Todorova K., Massague J., Kim J.Y., Nadal C., Gomis R.R., Manova-Todorova K., Massague J., Nadal C., Gomis R.R., Manova-Todorova K., Massague J., Gomis R.R., Manova-Todorova K., Massague J., Manova-Todorova K., Massague J., Massague J. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Yoshida M., Hattori M., Sakaki Y., Hattori M., Sakaki Y., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman L., Massague J., Massague J. Distinct oligomeric states of SMAD proteins in the transforming growth factor-beta pathway. J. Biol. Chem. 2000;275:40710–40717. doi: 10.1074/jbc.M005799200. [DOI] [PubMed] [Google Scholar]
- Jonsson P.F., Bates P.A., Bates P.A. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006;22:2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau T.R., Way J.C., Silver P.A., Way J.C., Silver P.A., Silver P.A. Nuclear transport and cancer: From mechanism to intervention. Nat. Rev. Cancer. 2004;4:106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- Lee C.C. Tumor suppression by the mammalian Period genes. Cancer Causes Control. 2006;17:525–530. doi: 10.1007/s10552-005-9003-8. [DOI] [PubMed] [Google Scholar]
- Li S., Armstrong C.M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Armstrong C.M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Vidalain P.O., Han J.D., Chesneau A., Hao T., Han J.D., Chesneau A., Hao T., Chesneau A., Hao T., Hao T., et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D., Brun C., Remy E., Mouren P., Thieffry D., Jacq B., Brun C., Remy E., Mouren P., Thieffry D., Jacq B., Remy E., Mouren P., Thieffry D., Jacq B., Mouren P., Thieffry D., Jacq B., Thieffry D., Jacq B., Jacq B. GOToolBox: Functional analysis of gene datasets based on Gene Ontology. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T. NUP98 fusion in human leukemia: Dysregulation of the nuclear pore and homeodomain proteins. Int. J. Hematol. 2005;82:21–27. doi: 10.1532/IJH97.04160. [DOI] [PubMed] [Google Scholar]
- Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M., Sato K., Fujibuchi W., Bono H., Kanehisa M., Fujibuchi W., Bono H., Kanehisa M., Bono H., Kanehisa M., Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Mandelker D., Leary R.J., Ptak J., Silliman N., Leary R.J., Ptak J., Silliman N., Ptak J., Silliman N., Silliman N., et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. limma: Linear models for microarray data. In: Gentleman R., et al., editors. Bioinformatics and computational biology solutions using R and bioconductor. Springer; London, UK: 2005. pp. 397–420. [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Pomeroy S.L., Golub T.R., Lander E.S., Golub T.R., Lander E.S., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Greenberg S.A., Kong S.W., Altschuler J., Kohane I.S., Park P.J., Greenberg S.A., Kong S.W., Altschuler J., Kohane I.S., Park P.J., Kong S.W., Altschuler J., Kohane I.S., Park P.J., Altschuler J., Kohane I.S., Park P.J., Kohane I.S., Park P.J., Park P.J. Discovering statistically significant pathways in expression profiling studies. Proc. Natl. Acad. Sci. 2005;102:13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Lockshon D., Narayan V., Srinivasan M., Pochart P., Narayan V., Srinivasan M., Pochart P., Srinivasan M., Pochart P., Pochart P., et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Kinzler K.W., Kinzler K.W. Cancer genes and the pathways they control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]