Abstract
Evolutionary constraints on gene regulatory elements are poorly understood: Little is known about how the strength of transcription factor binding correlates with DNA sequence conservation, and whether transcription factor binding sites can evolve rapidly while retaining their function. Here we use the model of the NFKB/Rel-dependent gene regulation in divergent Drosophila species to examine the hypothesis that the functional properties of authentic transcription factor binding sites are under stronger evolutionary constraints than the genomic background. Using molecular modeling we compare tertiary structures of the Drosophila Rel family proteins Dorsal, Dif, and Relish and demonstrate that their DNA-binding and protein dimerization domains undergo distinct rates of evolution. The accumulated amino acid changes, however, are unlikely to affect DNA sequence recognition and affinity. We employ our recently developed microarray-based experimental platform and principal coordinates statistical analysis to quantitatively and systematically profile DNA binding affinities of three Drosophila Rel proteins to 10,368 variants of the NFKB recognition sequences. We then correlate the evolutionary divergence of gene regulatory regions with differences in DNA binding affinities. Genome-wide analyses reveal a significant increase in the number of conserved Rel binding sites in promoters of developmental and immune genes. Significantly, the affinity of Rel proteins to these sites was higher than to less conserved sites and was maintained by the conservation of the DNA binding site sequence (static conservation) or in some cases despite significantly diverged sequences (dynamic conservation). We discuss how two types of conservation may contribute to the stabilization and optimization of a functional gene regulatory code in evolution.
Despite the availability of whole-genome sequences of related species, the evolution of transcriptional regulation is still poorly understood (Ludwig 2002; Xie et al. 2005). Transcription is controlled by the binding of “regulatory proteins” (i.e., transcription factors, TFs) to “DNA regulatory elements” (i.e., promoters and transcription enhancers). Functionally important regulatory sequences are usually conserved among related species. Indeed, in a few examples of well-characterized transcriptional enhancers, such as the “even-skipped” stripe 2 enhancer (Stanojevic et al. 1991), most but not all functionally important binding sites are conserved in 13 Drosophila species (Ludwig et al. 1998, 2000). On the other hand, the preservation of an optimal level of gene expression may allow and even support changes in regulatory sequences, where there are compensatory changes in transcription factors and the regulatory sequences (Landry et al. 2005). Such compensatory changes can include amino acid substitutions in the DNA-binding domains of transcription factors that alter the pattern of DNA sequence recognition and reciprocal changes in DNA regulatory sequences (Juarez et al. 2000). Another factor that may lead to changes in DNA regulatory sequences is “fuzziness” of a TF binding site itself. Certain transcription factors can bind to a number of closely related DNA sequences with similar affinities; this may allow for neutral evolution of binding sites without significant effects on TF binding (Gerland and Hwa 2002).
A major impediment to understanding the contribution of binding site sequence “fuzziness” in the evolution of regulatory DNA sequences is the lack of systematic, accurate, and quantitative measurements of binding affinities to binding site sequence variants. Recently we and others used a high-throughput microarray-based assay to address this problem (Bulyk et al. 2001; Linnell et al. 2004; Mukherjee et al. 2004). We have also developed the principal coordinate (PC) statistical analysis for analyzing protein–DNA interaction data (Udalova et al. 2002). The PC analysis accurately predicts the effect of nucleotide variations within the binding motif on protein binding affinity and automatically incorporates the effects of interactions between base pair positions in the binding site. The resulting comprehensive tables of binding affinities improve on traditional position-weight-matrix models that may fail to depict true binding specificities because they assume that each nucleotide in a binding site exerts an independent effect (Benos et al. 2002; Bulyk et al. 2002).
The Drosophila Dorsal, Dif, and Relish proteins are three members of the Rel Homology Domain (RHD) containing class of transcription factors represented in humans by the NFKB family (Silverman and Maniatis 2001). During Drosophila development, Dorsal has a key role in initiating the dorsal-ventral patterning pathway, and its target genes and their enhancers have been well studied in this context (Stathopoulos et al. 2002; Papatsenko and Levine 2005; Biemar et al. 2006). Most notably, from an evolutionary standpoint, Papatsenko and Levine (2005) have analyzed Dorsal binding sites in 18 target gene enhancers, across four species of Drosophila, and demonstrated that 80% of optimal (high affinity) binding motifs are within evolutionally conserved sequence blocks.
Dorsal, Dif, and Relish also play a critical role in the innate immune response, a function they share with NFKB in mammals. The innate immune response is an ancient evolutionary defense mechanism against microbial pathogens that is conserved from Drosophila to mammals (Hoffmann et al. 1999; Silverman and Maniatis 2001). When challenged by microbes, insects discriminate between various classes of microorganisms by activating specific intracellular signaling pathways that lead to production of antimicrobial peptides and other effector molecules (Hoffmann 2003). The Toll signaling pathway is activated in response to Gram-positive bacteria and fungal infection, and leads to the nuclear translocation of Dorsal and Dif. These transcription factors bind to DNA sequences upstream from genes encoding a large number of antifungal peptides such as drosomycin and metchnikowin. An alternate signaling pathway is activated in response to Gram-negative bacteria and results in the proteolytic cleavage of the precursor for Relish. Processed Relish translocates into the nucleus and activates expression of anti-bacterial peptides, such as diptericin and attacins. Little is known about the conservation of Rel binding sites in the orthologous enhancers of the innate immunity genes.
Here we investigate the molecular evolution of RHD-containing proteins and their associated binding sites using genome sequence assemblies of seven Drosophila species (D. melanogaster, D. simulans, D. yakuba, D. ananassae, D. pseudoobscura, D. virilis, D. mojavensis). We model the structures of Dorsal, Dif, and Relish based on available crystallographic structures of the mammalian c-Rel protein, and examine the possibility that changes in amino acid sequences in the DNA-binding domains during evolution alter the pattern of DNA sequence recognition and binding affinity. We generate quantitative binding affinity data for these proteins using a microarray-based binding assay and the PC analysis (Udalova et al. 2002) and examine the changes in DNA sequences and binding affinities of putative Rel binding sites on a genome-wide basis. We demonstrate that the Rel binding sites in the vicinity of innate immunity and developmental genes are under strong functional constraints. Our work extends that of Papatsenko and Levine (2005) by investigating the evolutionary dynamics within particular conserved Rel binding sites, and by examining the differences between those sites that are associated with target and nontarget genes. We address two key questions: (1) Does binding site affinity correlate with DNA sequence conservation and (2) is binding site conservation “dynamic”—that is, can it show high levels of nucleotide substitution while maintaining functionality?
Results
Conservation of DNA-contacting residues in Rel Homology Domains of Dorsal, Dif, and Relish
Dorsal, Dif, and Relish proteins share a homologous region, the Rel Homology Domain (RHD), of ∼300 amino acids (aa), that is responsible for DNA-binding and dimerization (this region is covered by the Pfam domains RHD and TIG [Finn et al. 2006], but, in common with others, we use RHD to refer to this entire region). We investigated the likely functional consequences of amino acid changes in the RHD during evolution of drosophilids, by first predicting the gene sequences of Dorsal, Dif, and Relish orthologs in each species, as described in the Methods, and then mapping protein sequence divergence within ortholog and paralog sets to known 3D structures.
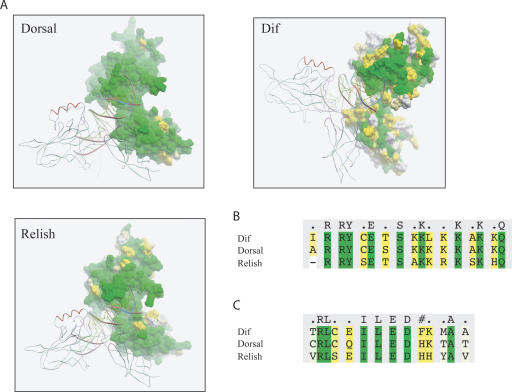
No structures of complete insect RHD-containing proteins, i.e., including both the Pfam RHD and TIG domains, are currently available (the structure of Gambif1 from mosquito does not cover the TIG; Cramer et al. 1999). Consequently we modeled the 3D structure of Dorsal, Dif, and Relish homodimers bound to DNA using the structure of mammalian c-Rel as a template (PDB accession no. 1gji; Huang et al. 2001) (see Methods). Figure 1A depicts one subunit of Drosophila Rel dimer as molecular surface, while the second subunit and DNA are shown as ribbons. The molecular surface is colored according to amino acid residue conservation. The analysis of amino acid sequence conservation showed that Dorsal is the most conserved RHD-containing protein, with none of the 11 variable sites in its RHD affecting DNA-binding or dimerization residues, suggesting that the DNA-binding specificities of Dorsal are likely to be identical across drosophilid species. RHDs of both Relish and Dif are less conserved: 47 and 124 aligned sites were variable in the RHD of Relish and Dif orthologs, respectively. However, nonconserved residues of Relish were not contacting DNA or involved in dimerization, and thus are unlikely to significantly affect its binding specificities.
Figure 1.
Conservation of amino acid residues involved in DNA recognition and protein dimerization. (A) Molecular modeling of Drosophila Rel homodimers bound to DNA. One subunit of Rel homodimer is represented as a molecular surface, while the second subunit and DNA are shown as ribbons. (B) Alignment of DNA-binding residues of Dorsal, Dif, and Relish from D. melanogaster. (C) Alignment of dimerization residues of Dorsal, Dif, and Relish from D. melanogaster. Molecular surface and amino acid residues are colored according to their conservation: green color indicates fully conserved residues; yellow marks conservative substitutions, and white, nonconserved residues.
These results suggest that DNA-binding specificity data obtained for Dorsal and Relish proteins are likely to be directly applicable to the other species studied. While the majority of DNA-binding residues are also conserved within orthologs of Dif, the sequence variation that does occur suggests that more caution needs to be applied when making inferences between species for this gene.
Rel homodimers have different DNA-binding preferences
Alignment of the RHD domains of the D. melanogaster Dorsal, Dif, and Relish paralogs (Fig. 1B,C) shows significant differences in their dimerization interfaces, most notably at M236/T236/Y252 (Dif/Dorsal/Relish) position (Fig. 1C). These differences may result in different preferences for the formation of particular homo- and heterodimers. Moreover, the loop that connects DNA-binding and dimerization domains of Dorsal has two extra amino acids compared to Relish, and the loop of Dif has four extra residues. This may result in preference for longer binding sites for Dorsal and Dif.
To examine the functional effect of the observed differences on protein–DNA recognition, we profiled DNA-binding specificities of Dorsal, Dif, and Relish homodimers to thousands of DNA variants. Oligonucleotide duplexes corresponding to 182 variants of the minimal spanning set uniformly covering the extended NFKB/Rel GGRDNNHHBS consensus, derived from the published examples of binding for mammalian NFKB and insect Rel proteins, were spotted in quadruplicate onto Codelink slides. Binding of each dimer to DNA sequences was monitored in three independent experiments. When experimental binding affinities were compared between the three proteins, we found that Dorsal had overlapping binding specificities to both Dif (correlation coefficient 0.70) and Relish (0.73), despite the limited degree of similarity between Dif and Relish binding specificities (0.29). The observed differences in DNA-binding preferences of Dif and Relish (summarized as binding sequence logos in Supplemental Fig. S1) are likely to contribute into preferential activation of specific antimicrobial peptide genes by one TF or another, consistent with the results of previously published SELEX-based analysis (Senger et al. 2004).
To extrapolate the binding affinity predictions to all 5184 variants of the GGRDNNHHBS consensus we employed the PC statistical analysis, which considers variant DNA sequences as points in a high-dimensional Euclidian space, with coordinates that reflect on the sequence composition. The binding affinity of a TF to different DNA sequences is then modeled as a function of these coordinates. The model incorporates the effects of interactions between base pair positions in the binding site and it is sensitive to subtle differences in binding specificities of homologous TFs (Udalova et al. 2002). The 15 largest PCs were used to explain the variance of the GGRDNNHHBS space, of which 10 had significant coefficients (P-value < 0.05) for Dorsal, seven for Dif, and 11 for Relish (Supplemental Table S1). The 5184 sequence variants were ranked from 0 to 1 based on their predicted binding affinity to corresponding Rel proteins. The PC predictions were sufficiently accurate over the wide range of binding affinities and explained ∼75% of total binding variance (see Methods). To incorporate the recent SELEX data for Dorsal, Dif, and Relish (Senger et al. 2004), we generated an additional set of scores for GGRDNNHHBN by averaging the N = [C,G] scores for instances where N = [A,T], giving a set of 10,368 Extended Scored Binding Sites (ESBSs) (Supplemental Table S2).
A high frequency of conserved Rel binding sites in the promoters of immune and developmental genes
To map putative Rel binding sites on a genome-wide basis, we screened the aligned genomes of seven Drosophila species for the presence of binding motifs within the GGRDNNHHBN consensus. A total of 320,701 sites (excluding those that overlapped with protein-coding exons) were found within 2 kb of the start sites of predicted genes (as defined by Ensembl). A large percentage of these sites (61%, 195,293 sites) did not have a counterpart in other genomes, with the remainder aligning in at least one more species; 3335 sites or 1% of these sites aligned in all seven species. A list of these sites is presented in Supplemental Table S3, which shows that known Dorsal-regulated developmental genes (e.g., snail, twist, zen, etc.) represent <2% of all the genes with Dorsal binding sites conserved in all seven species in their promoters: (12 genes described in Papatsenko and Levine 2005 + 21 novel genes identified in Stathopoulos et al. 2002 + 16 novel genes identified in Biemar et al. 2006)/2639 total number of genes. Other Rel binding sites conserved in all seven species were identified in the promoters of genes involved in innate immunity (e.g., attacin D, cecropin C; De Gregorio et al. 2001, 2002), other cellular events (e.g., actin, zeelin, Ets21C, etc.), or whose functions have not been analyzed (e.g., CG7313, CG10555, CG33308, etc.).
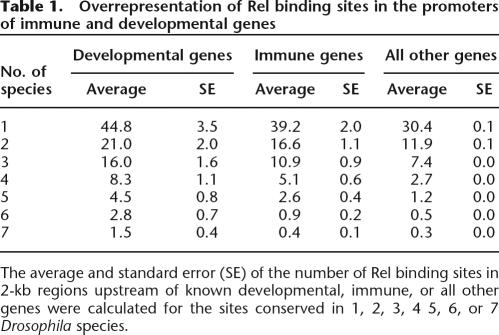
However, when we analyzed the location of Rel binding sites, we found that promoters of developmental and immune genes have significantly more sites than the genome average number of conserved Dorsal binding sites (Table 1). This trend was observed for the complete range of species (two to seven species). Our results support previously published studies in which clusters of Dorsal binding sites were used to identify putative target genes involved in the development of Drosophila embryo (Stathopoulos et al. 2002).
Table 1.
Overrepresentation of Rel binding sites in the promoters of immune and developmental genes
The average and standard error (SE) of the number of Rel binding sites in 2-kb regions upstream of known developmental, immune, or all other genes were calculated for the sites conserved in 1, 2, 3, 4 5, 6, or 7 Drosophila species.
Strong functional constraints on Rel binding sites are detected at immune and developmental loci
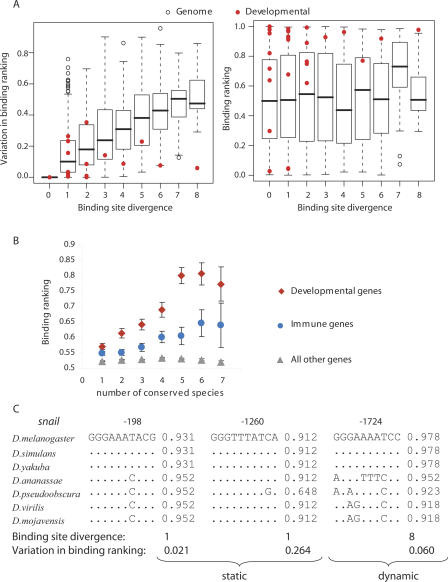
To examine the evolutionary constraints on putative Rel binding sites, we used three measures: (1) the sequence divergence of binding sites; (2) the Dorsal binding ranking (S) of the D. melanogaster site (SD_mel); (3) the range of Dorsal binding ranking among the seven species (Δs = Smax − Smin). Sequence divergence was taken as the number of historical nucleotide substitutions that occur within each evolving binding site, given the alignment and known species phylogeny (i.e., the maximum parsimony score; see Methods for details). The Dorsal binding ranking (SD_mel) was determined from the interpolated binding site data (see Methods), according to Supplemental Table S2. The range of binding ranking (Δs) was defined as the maximum difference in Dorsal binding affinities between the species variants of the site (e.g., 0.952–0.931 = 0.021 for site at −183 nt or 0.978–0.918 = 0.060 for site at −1724 nt of the snail promoter; see Fig. 2C). The 3335 Dorsal binding sites conserved in all seven species were analyzed. The left panel of Figure 2A shows boxplots of Dorsal variation in binding ranking (Δs) for the range of observed binding site divergences; the right panel shows boxplots of D. melanogaster Dorsal binding ranking (SD_mel) for given sequence divergence values. The distribution of binding ranking scores for the genomic background gene set is insensitive to the evolutionary divergence of binding sites (Fig. 2A, right panel), whereas the variation in binding site ranking shows an upward trend with increasing evolutionary divergence of binding sites (Fig. 2A, left panel)
Figure 2.
High binding affinity to more conserved Rel binding sites in developmental and immune loci. (A) The relationship between a binding site sequence divergence and range in binding ranking (Δs) (left panel) or site binding ranking of the D. melanogaster site (SD_mel) (right panel). Red dots show the scores for known Dorsal-regulated developmental genes (Papatsenko and Levine 2005). Boxplots show distributions for all other genes. Boxplot parameters are defaults of the “R” package. (B) The relationship between the sequence conservation of Rel binding sites and their average binding affinity to Dorsal. For A and B each dot represents a site aligned in seven Drosophilla species. The sites were binned into three separate subsets according to their location: (1) within 2 kb of the start of a Dorsal-regulated developmental gene (red diamonds); (2) within 2 kb of the start of a gene involved in immune response (blue circles); and (3) within 2 kb of the start of all other genes not included in either of sets 1 or 2 (gray triangles) (see Methods). (C) Static (sequence) and dynamic (binding affinity) conservation of putative Rel binding sites in the upstream regions of the snail gene involved in developmental processes.
We noticed, however, that Rel binding sites in the promoters of known Dorsal-regulated developmental genes (defined as in Papatsenko and Levine 2005 and marked in red in Fig. 2A) tend to be of a higher affinity than putative Rel binding sites in the promoters of all other genes. Moreover, their high affinity to Dorsal appeared to be maintained over evolution, with some sites displaying stable protein–DNA binding despite evolving nucleotide sequence (note low values of variation in binding ranking [Δs] for red data points at higher values of binding site divergence). Analysis of variance comparing linear multiple regression models showed a significant (P < 0.01) interaction between the binding site divergence and the set of genes (i.e., developmental or not), suggesting that the dependence of variation in binding ranking on sequence divergence is different for the two sets.
In addition, we found that the affinity of Rel proteins to the binding sites at both developmental and immune loci was increasing with site sequence conservation (Fig. 2B). Thus, Rel binding sites in the promoters of developmental and immune genes stand out from the bulk of putative binding site motifs by having the highest degree of sequence and binding affinity conservation, suggesting that these sites have evolved under functional constraints.
Static and dynamic components contribute to functional conservation
For ∼90% of sites the value of site sequence divergence did not exceed three independent substitutions since the common ancestor of the drosophilid species. Most of the binding affinity conservation was, thus, due to the conserved underlying sequence of the Rel binding site (static conservation). However, although even a single mutation can significantly alter binding affinity, there were four instances in which multiple sequence mutations did not substantially affect predicted binding to Rel proteins. We refer to this latter phenomenon as dynamic conservation and noted that it was mainly observed in the vicinity of genes involved in developmental processes. For example, the dynamically conserved Rel binding site at −1724 nt upstream of the snail gene is presented in Figure 2C along with two statically conserved Rel binding sites at −198 nt and −1260 nt. We also identified three other dynamically conserved sites: GGAGTTC CCC at −785 nt upstream of the twist gene (four substitutions vs. variation in binding ranking of 0.087), GGAGAAACCC at −1250 nt upstream of the zen gene (six substitutions vs. variation in binding ranking of 0.076), and GGAAAAACCA at −676 nt upstream of the zen2 gene (six substitutions vs. variation in binding ranking of 0.122). The dynamically conserved sites in the promoters of snail and zen genes correspond to DNase I footprint loci for Dorsal identified in the FlyReg database, which provides a nonredundant set of high-quality binding loci information for 87 transcription factors and 101 target genes for D. melanogaster (Bergman et al. 2005). This indicates that the dynamically conserved sites in the promoters of snail and zen genes are likely to be real functional sites, rather than regions showing coincidental patterns of nucleotide conservation.
In summary, a quantitative profiling of Rel protein–DNA interactions led to the detection of atypical examples of binding sites in which sequence of the site changes significantly, while the overall functional fitness is maintained.
Systematic quantitative analysis of DNA binding affinities identifies new putative functional Rel binding sites of low and moderate affinity
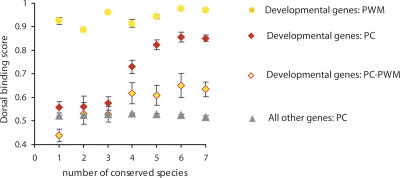
In order to assess the consistency of our results with prior analyses, we examined the enhancer regions of developmental genes described by Papatsenko and Levine (2005) using our scoring scheme and identified 370 putative Rel binding sites (PC sites), which included 136 sites that overlapped with the D. melanogaster sites identified by Papatsenko and Levine (2005) using a standard position-weight-matrix (PWM sites). The PWM sites consistently have a ranking score >0.9 in our classification (Supplemental Table S4), irrespective of the number of species in which the binding site is conserved. In contrast, our PC sites show an upward trend in ranking scores with increasing sequence conservation, reaching a plateau with five conserved species (Fig. 3). This effect is still observed if we exclude the 136 sites that overlap with PWM sites (PC–PWM), indicating that the upward trend is not solely due to conservation of high-scoring PWM sites.
Figure 3.
PC, but not PWM, method of binding site scoring detects the relationship between the binding site sequence conservation and its binding affinity to Dorsal. The Rel binding sites in enhancer regions of developmental genes described by Papatsenko and Levine (2005) were defined by either the principal coordinate (PC) analysis (red diamonds) or position-weight-matrix (PWM) analysis as in Ref (Papatsenko and Levine 2005) (orange circles). The relationship between the sequence conservation of Rel binding sites and their average binding affinity to Dorsal is shown. The sites defined by the PC analysis and not overlapping with the PWM are shown in red diamonds filled with orange (PC–PWM); the sites defined by the PC analysis in front of other genes are shown in gray triangles.
These results suggest that the scoring of PWM sites by Papatsenko and Levine (2005) was set at a relatively high threshold relative to our analysis, effectively excluding all putative Rel binding sites of low and moderate affinity to Dorsal. At the same time, some of our high-scoring Dorsal binding sequences were not scored by Papatsenko and Levine (2005), but nonetheless may be functional (as their sequences, on average, are more conserved than the genomic background; see PC–PWM data points in Fig. 3). In turn, there are 34 PWM sites that we cannot score, owing to the limited number of sequences defined by our ESBS; thus, the full extent of sequence variability at high-scoring sites has yet to be fully captured. Taken together, these results indicate broad agreement between the PWM and our PC methods of scoring binding sites, but, importantly, they show that specific genomic features can only be detected by not excluding low- and moderate-affinity binding sites.
Relish- and Dif-regulated immune genes may be under distinct functional constraints
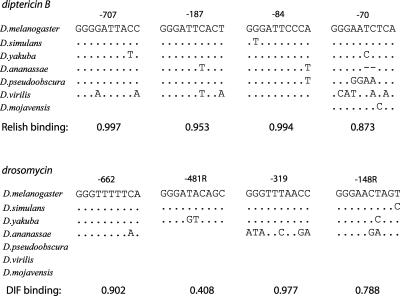
Overall, high-affinity Rel binding sites conserved in all seven species were considerably rarer in the vicinity of immune genes compared to developmental genes (Table 1; Fig. 2B). For this reason it is worth noting that two such sites were mapped to the upstream region of the CG9080 gene, encoding a 121-aa polypeptide of unknown function. Of interest, CG9080 gene expression was strongly induced in response to bacterial and fungal infections (De Gregorio et al. 2001), and the predicted protein product includes a signal peptide, suggesting it may belong to a new class of antimicrobial peptides. We also noticed that Rel binding sites in the vicinity of the immune genes known to be preferentially regulated by Relish (such as antimicrobial peptide genes diptericin and cecropin) appeared to be more conserved than those in the vicinity of genes preferentially targeted by Dif (such as drosomycin and metchikowin). Moreover, most of the sites in the diptericin promoters were within the top 10% of binding affinity ranks to Relish, whereas half of the sites in the drosomycin promoter (−481 nt and −148 nt) fell within the middle range of Dif binding affinities (Fig. 4). A higher mutation rate of amino acids in the Dif protein described in this study is consistent with the highest nucleotide sequence turnover in Rel binding sites located in Dif-regulated promoters and may, therefore, reveal an ongoing coevolution of cognate transcription factors and regulatory sequences.
Figure 4.
Higher nucleotide sequence turnover in binding sites located in Dif-regulated promoters. Binding affinity to Relish and Dif was assigned to conserved sites in the promoters of diptericin (top panel) and drosomycin (bottom panel) genes.
Discussion
Previous studies demonstrated that many Rel binding sites in the promoters of Dorsal-regulated developmental genes are within evolutionarily conserved sequence blocks (Papatsenko and Levine 2005). Here we investigate parameters of Rel binding site conservation across the entire Drosophila genome and show that sites at immune and developmental loci are under strong functional constraints. Specifically, binding affinities of Rel proteins to these sites are maintained in some case by the conservation of the DNA binding site sequence, and in others to sites with significantly diverged sequences (Fig. 2).
Scored Site Conservation (SSC) measured by the number of species that share a homologous scorable binding site at a particular aligned location shows a strong correlation with binding site strength for promoters of developmental and immune genes, a phenomenon that, to the best of our knowledge, has not been previously reported (Fig. 2B). The most obvious explanation for this phenomenon is that high-affinity binding sites are more likely to be functional and are, therefore, more likely to be conserved between different species. This explanation is not entirely satisfactory when considered alongside models of readouts of Dorsal gradient thresholds during the fly development, where nonoptimal Rel binding sites might be expected to be just as indispensable and necessary for correct function (Stathopoulos and Levine 2002), and, as such, we would expect them to be conserved. However, we note that threshold gradients are interpreted by the enhancer as a whole, with a modular architecture that may allow modification of individual binding sites (Ludwig et al. 2005). As an alternative explanation, we considered whether the effect we observe is an artifact caused by our scoring scheme containing sites that bind TFs so weakly as to be undetected by selection, but so numerous that they dominate scoring. According to this hypothesis the increase of average score with increased SSC would be due principally to low-scoring (biologically irrelevant) sites being increasingly excluded. However, when we introduce a cutoff allowing only scores >0.8 to be included, we still observe the same effect (Supplemental Fig. S2A). Moreover, when we examined the experimentally identified functional Rel binding sites listed in the FlyReg database, which were mainly located in front of developmental genes, we observe a similar increase in binding affinity to Rel proteins for more conserved sites (Supplemental Fig. S2B).
The static conservation of Rel binding sites, i.e., conservation of nucleotide sequence, is consistent with the idea that functionally important elements have a slower rate of base substitutions (Jukes and Kimura 1984). We analyzed genomes of seven diverged Drosophila species to maximize the discovery of Rel binding motifs in regions of the genome with varying evolutionary rates (Nobrega and Pennacchio 2004). Less than 40% of the putative Rel binding motifs mapped within 2 kb of predicted gene start sites align in at least two species, with only 1% of the sites aligning in all seven species. We found that the conserved Rel binding sites are more likely to be situated in the vicinity of developmental and immune genes. Of interest, when we analyzed Rel binding sites in the FlyReg database, the percentage of sites aligned in seven species increased 15-fold (six out of 42 putative Rel binding sites scored by us were conserved either statically or dynamically). This was consistent with the vast majority of the FlyReg sites located in the vicinity of developmental genes and our genome-wide observation. In addition, it further highlighted the relationship between the binding site functional properties and its conservation in evolution.
The presence of multiple, high-affinity well-conserved sites may aid in identification of putative targets of Dorsal (Stathopoulos et al. 2002). For instance, the wnt8 gene has five putative Rel binding sites conserved in seven species, with a binding ranking above a 0.8 cutoff (Supplemental Fig. S3). The wnt8 protein binds to a family of frizzled seven-transmembrane receptors and acts through a cascade of genes on the nucleus. WNT8 (WNTD) has recently been described as a feedback inhibitor of Dorsal in development and immunity, but the molecular mechanisms involved in the activation of this gene by Dorsal are not understood (Ganguly et al. 2005; Gordon et al. 2005). Taken together with the cluster of Rel binding sites, this suggests wnt8 could be a direct target of Dorsal. Another interesting candidate gene is schnurri, with four Rel binding sites conserved in seven species. The expression of schnurri is restricted to the dorsal ectoderm and the ventral mesoderm. Schnurri is believed to act as a repressor of the ind gene and possibly other pan-neurotic genes, which are not active in the ventral mesoderm and dorsal ectoderm (Stathopoulos and Levine 2005).
Dynamic conservation, where the DNA sequence of the binding motif mutates without significant effect on its binding affinity, is likely to relate to the “fuzziness” of the binding sites due to the permissiveness of transcription factor–DNA interactions. Although the plasticity of DNA binding sites is often emphasized, and may have entropic or selective advantages in evolution (Gerland and Hwa 2002), we detected only a few examples of dynamic conservation of Rel binding sites (Fig. 2C). This is perhaps unexpected, given that the standard explanation for the “fuzziness” of transcription factor binding specificity is to enable a degree of robustness to mutation in transcription factor binding sites. The result is unlikely to be due to the evolutionary proximity of the drosophilids as, in general, there is little sequence conservation in intergenic regions between the more distantly related species studied here, implying that the more commonly observed “static” conservation of binding sites is due to selection rather than an absence of mutation. However, it is also plausible that the prevalence of static conservation observed in these binding sites is a specific aspect of the Dorsal gradient response of target gene enhancers, and would not apply to the targets of transcription factors not showing concentration gradient sensitivity.
The Rel proteins evolve with different mutation rates. Relish and Dif have a four- and 10-fold higher number of diverging amino acids than Dorsal, respectively (Fig. 1). We also detect a somewhat faster sequence turnover of Rel binding sites at immune loci, especially at the promoters of preferentially Dif-regulated genes. We speculate that this may be linked to a high number of evolving amino acid residues in the DNA-binding domain of Dif, which may induce reciprocal changes in Rel binding sites to adapt to changing trans-acting environments. Further investigation is required to test the level of expression of reporter constructs driven by orthologous Dif-regulated promoters in divergent Drosophila species (Gompel et al. 2005; Prud’homme et al. 2006). Ludwig et al. (2005) suggested that the coevolution of cis- and trans-acting elements may involve changes in expression patterns or levels rather than changes in the protein sequences of trans-acting factors. In the case of TFs, amino acid mutations that alter the affinity of TF–DNA recognition could effectively mimic the changes in the level of TF expression, as both factors (affinity and TF concentration) contribute to the efficient TF recruitment to DNA. Thus, DNA-binding domains of TFs may provide an additional genetic substrate for evolution of a gene regulatory code.
The studies reported here further our understanding of the genome organization and functional conservation of transcription factor binding sites. They also highlight the importance of quantitative approaches to analyzing the genome regulatory code, as they provide a more sensitive tool for the annotation of putative binding sites and discerning structure-function relationships, i.e., the correlation between the binding affinity and DNA sequence conservation.
Methods
Identifying orthologs of Dorsal, Dif, and Relish and phylogenetic analysis
We used the Drosophila melanogaster protein sequences of Dorsal, Dif, and Relish to search the genomes of other drosophilids, downloaded from the UCSC genome Web site (http://genome.ucsc.edu) using TBLASTN. We then isolated top matching regions and used GeneWise (Birney et al. 2004) to obtain protein predictions for each species. Other RHD-containing sequences were identified via protein BLAST searches of the NCBI database (Altschul et al. 1997). We confirmed orthology relationships via phylogenetic analysis of the aligned Rel Homology Domains (defined as the region covered by the PFAM RHD [Finn et al. 2006] and SMART IPT [Letunic et al. 2006]) using PHYML (Guindon et al. 2005)
Protein modeling
Multiple sequence alignments as well as homology models of Dif, Relish, and Dorsal were built in Internal Coordinate Mechanics (ICM) using homology modeling based on the available structure of mammalian c-Rel (PDB 1gji) as a template. Briefly, the sequence-structure alignment was done using the ICM alignSS algorithm that optimizes the sequence-structure match using residue accessibilities, secondary structures, and functional sites of the template and sequence. Loop predictions and model refinement were done using local global energy optimization strategy (http://www.molsoft.com).
Protein–DNA binding assay and PC model
RHD domains of Rel proteins of D. melanogaster origin (Dorsal: aa 16–384; Dif: aa 17–378; Relish: aa 117–434) were cloned into pET21d vector (Novagen) and their sequences were verified by DNA sequencing. The proteins were expressed in BL21(DE3) bacterial cells. The proteins were purified by DNA-binding affinity chromatography using biotin-labeled oligoduplexes comprising either NFKB site GGGGGATTCC or GGGAATTTCC essentially as described in Udalova et al. (1998). Oligonucleotide duplexes corresponding to 182 variants of the minimal spanning subset of motifs that uniformly covers the GGRDNNHHBS consensus binding space were spotted in quadruplicate onto Codelink slides as previously described (Linnell et al. 2004). Three independent binding experiments were performed for each Rel protein. The protein–DNA binding was detected with anti-HIS antibodies (H-15, sc-308, Santa Cruz Biotechnology, Inc.) followed by the secondary Cy5-conjugated anti-rabbit IgG antibodies (Jackson Immunoresearch Laboratories). Slides were scanned using an Axon 4000B scanner (Axon Instruments, Inc.) and analyzed with GenePix 4.1. Protein binding signal was normalized against DNA concentration in the corresponding spot, using Sybr Green (Molecular Probes), according to the manufacturer's instructions. The average of four binding values per slide was ascribed to each sequence variant. Binding signals were normalized within each slide, compared to the fluorescent readings for the GGGGTTC CCC motif, which was given a value of 1000. Affinity weighted sequence logos were generated by producing a sequence list containing as many copies of each representative sequence as the normalized experimental binding value for that sequence scaled by 1/100. These sequence lists were then used as input for the WebLogo program (version 2.8.2) (Crooks et al. 2004) (available at http://weblogo.berkeley.edu/logo.cgi).
Extrapolation of the binding affinity predictions to all DNA motifs was achieved by fitting Principal Coordinate models to experimental data, essentially as described in Udalova et al. (2002). The PC model was fitted to the logarithms of three replicated measurements for Rel binding and included extra terms to account for between-microarrays effects. The 15 largest PCs were used to explain the variance of the GGRDNNHHBS space. In the regression out of 15 PCs, 10 had significant coefficients (P-value < 0.05) for Dorsal, seven for Dif, and 11 for Relish (Supplemental Table 1). The correlation coefficients between the affinities predicted by the PC model and experimentally observed affinities were 0.53 for Dif, 0.53 for Dorsal, and 0.55 for Relish and explained 74%, 74%, and 75% of total binding variance, respectively. Binding affinity predictions were extrapolated to all 5184 variants of the consensus and the sites were ranked. We further generated a set of scores for GGRDNNHHBN by averaging the N = [C,G] scores for instances where N = [A,T], giving a set of 10,368 Extended Scored Binding Sites (ESBSs) (Supplemental Table S2).
Genome analysis
We identified all instances of ESBSs in the D. melanogaster genome. We then mapped these onto alignments of drosophilid genomes (D. melanogaster, D. simulans, D. yakuba, D. ananassae, D. virilis, D. mojavensis, D. pseudoobscura) taken from the UCSC Web server (http://genome.ucsc.edu/multiz9way “maf” files), and assigned each aligned sequence the D. melanogaster score for that particular 10mer. Species in which the 10mer was not present in the ESBS set were counted as nonconserved. The range of binding site strengths was calculated by subtracting the lowest binding ranking from the highest for each scoring binding site. Sequence variability at a given binding site was calculated using the baseml program from the PAML software package—the “maximum parsimony score” from the mlb output file (http://abacus.gene.ucl.ac.uk/software/paml.html).
RHD binding sites that were within 2 kb of the start of an Ensembl gene (http://www.ensembl.org) and that did not overlap with protein coding exons were analyzed. Ensembl genes were partitioned into three sets: (1) those used by Papatsenko and colleagues as known targets of Dorsal in dorsoventral patterning (Papatsenko and Levine 2005); (2) those identified by De Gregorio and colleagues as being involved in the immune response (De Gregorio et al. 2001); (3) all other genes not included in either of sets 1 or 2.
Overlapping binding sites were treated as distinct, e.g., a 12mer could potentially contain three overlapping Dorsal binding sites. Taking only the highest scoring binding site in overlapping sets does not significantly alter the results. In the analysis of enhancer regions of developmental genes, 83 D. melanogaster PWM sites defined by Papatsenko and Levine (2005) were overlapped by 128 PC sites.
Acknowledgments
We thank Dr D. Papatsenko (UC Berkeley) for providing us with the sequences of Drosophila enhancers involved in dorsoventral patterning, Dr. W. Valdar and Professor R. Mott (WTCHG) for advice on statistics, and Professor C. Ponting (MRC-FGU Oxford) for helpful comments. We also thank the Berkeley Drosophila Genome Project, Agencourt Bioscience Corporation, Genome Sequencing Center WUSTL School of Medicine, Human Genome Sequencing Center at Baylor College of Medicine, and the Flybase Consortium for providing access to the assemblies of Drosophila genome sequences. I.A.U. is supported by the MRC New Investigator Award and the ARC. R.R.C. and J.R. are supported by the Wellcome Trust.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.6490707
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benos P.V., Lapedes A.S., Stormo G.D., Lapedes A.S., Stormo G.D., Stormo G.D. Is there a code for protein-DNA recognition? Probab(ilistical)ly. Bioessays. 2002;24:466–475. doi: 10.1002/bies.10073. [DOI] [PubMed] [Google Scholar]
- Bergman C.M., Carlson J.W., Celniker S.E., Carlson J.W., Celniker S.E., Celniker S.E. Drosophila DNase I footprint database: A systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics. 2005;21:1747–1749. doi: 10.1093/bioinformatics/bti173. [DOI] [PubMed] [Google Scholar]
- Biemar F., Nix D.A., Piel J., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Nix D.A., Piel J., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Piel J., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Sementchenko V., Bell I., Manak J.R., Levine M.S., Bell I., Manak J.R., Levine M.S., Manak J.R., Levine M.S., Levine M.S. Comprehensive identification of Drosophila dorsal-ventral patterning genes using a whole-genome tiling array. Proc. Natl. Acad. Sci. 2006;103:12763–12768. doi: 10.1073/pnas.0604484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R., Clamp M., Durbin R., Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk M.L., Huang X., Choo Y., Church G.M., Huang X., Choo Y., Church G.M., Choo Y., Church G.M., Church G.M. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl. Acad. Sci. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk M.L., Johnson P.L., Church G.M., Johnson P.L., Church G.M., Church G.M. Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res. 2002;30:1255–1261. doi: 10.1093/nar/30.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., Varrot A., Barillas-Mury C., Kafatos F.C., Muller C.W., Varrot A., Barillas-Mury C., Kafatos F.C., Muller C.W., Barillas-Mury C., Kafatos F.C., Muller C.W., Kafatos F.C., Muller C.W., Muller C.W. Structure of the specificity domain of the Dorsal homologue Gambif1 bound to DNA. Structure. 1999;7:841–852. doi: 10.1016/s0969-2126(99)80107-5. [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E., Hon G., Chandonia J.M., Brenner S.E., Chandonia J.M., Brenner S.E., Brenner S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., Spellman P.T., Rubin G.M., Lemaitre B., Spellman P.T., Rubin G.M., Lemaitre B., Rubin G.M., Lemaitre B., Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., Spellman P.T., Tzou P., Rubin G.M., Lemaitre B., Spellman P.T., Tzou P., Rubin G.M., Lemaitre B., Tzou P., Rubin G.M., Lemaitre B., Rubin G.M., Lemaitre B., Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Moxon S., Marshall M., Khanna A., Durbin R., Marshall M., Khanna A., Durbin R., Khanna A., Durbin R., Durbin R., et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A., Jiang J., Ip Y.T., Jiang J., Ip Y.T., Ip Y.T. Drosophila WntD is a target and an inhibitor of the Dorsal/Twist/Snail network in the gastrulating embryo. Development. 2005;132:3419–3429. doi: 10.1242/dev.01903. [DOI] [PubMed] [Google Scholar]
- Gerland U., Hwa T., Hwa T. On the selection and evolution of regulatory DNA motifs. J. Mol. Evol. 2002;55:386–400. doi: 10.1007/s00239-002-2335-z. [DOI] [PubMed] [Google Scholar]
- Gompel N., Prud’homme B., Wittkopp P.J., Kassner V.A., Carroll S.B., Prud’homme B., Wittkopp P.J., Kassner V.A., Carroll S.B., Wittkopp P.J., Kassner V.A., Carroll S.B., Kassner V.A., Carroll S.B., Carroll S.B. Chance caught on the wing: Cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Gordon M.D., Dionne M.S., Schneider D.S., Nusse R., Dionne M.S., Schneider D.S., Nusse R., Schneider D.S., Nusse R., Nusse R. WntD is a feedback inhibitor of Dorsal/NF-κB in Drosophila development and immunity. Nature. 2005;437:746–749. doi: 10.1038/nature04073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Lethiec F., Duroux P., Gascuel O., Lethiec F., Duroux P., Gascuel O., Duroux P., Gascuel O., Gascuel O. PHYML Online—A web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann J.A. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- Hoffmann J.A., Kafatos F.C., Janeway C.A., Ezekowitz R.A., Kafatos F.C., Janeway C.A., Ezekowitz R.A., Janeway C.A., Ezekowitz R.A., Ezekowitz R.A. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- Huang D.B., Chen Y.Q., Ruetsche M., Phelps C.B., Ghosh G., Chen Y.Q., Ruetsche M., Phelps C.B., Ghosh G., Ruetsche M., Phelps C.B., Ghosh G., Phelps C.B., Ghosh G., Ghosh G. X-ray crystal structure of proto-oncogene product c-Rel bound to the CD28 response element of IL-2. Structure. 2001;9:669–678. doi: 10.1016/s0969-2126(01)00635-9. [DOI] [PubMed] [Google Scholar]
- Juarez K., Flores H., Davila S., Olvera L., Gonzalez V., Morett E., Flores H., Davila S., Olvera L., Gonzalez V., Morett E., Davila S., Olvera L., Gonzalez V., Morett E., Olvera L., Gonzalez V., Morett E., Gonzalez V., Morett E., Morett E. Reciprocal domain evolution within a transactivator in a restricted sequence space. Proc. Natl. Acad. Sci. 2000;97:3314–3318. doi: 10.1073/pnas.060444897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T.H., Kimura M., Kimura M. Evolutionary constraints and the neutral theory. J. Mol. Evol. 1984;21:90–92. doi: 10.1007/BF02100633. [DOI] [PubMed] [Google Scholar]
- Landry C.R., Wittkopp P.J., Taubes C.H., Ranz J.M., Clark A.G., Hartl D.L., Wittkopp P.J., Taubes C.H., Ranz J.M., Clark A.G., Hartl D.L., Taubes C.H., Ranz J.M., Clark A.G., Hartl D.L., Ranz J.M., Clark A.G., Hartl D.L., Clark A.G., Hartl D.L., Hartl D.L. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–1822. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Copley R.R., Pils B., Pinkert S., Schultz J., Bork P., Copley R.R., Pils B., Pinkert S., Schultz J., Bork P., Pils B., Pinkert S., Schultz J., Bork P., Pinkert S., Schultz J., Bork P., Schultz J., Bork P., Bork P. SMART 5: Domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnell J., Mott R., Field S., Kwiatkowski D.P., Ragoussis J., Udalova I.A., Mott R., Field S., Kwiatkowski D.P., Ragoussis J., Udalova I.A., Field S., Kwiatkowski D.P., Ragoussis J., Udalova I.A., Kwiatkowski D.P., Ragoussis J., Udalova I.A., Ragoussis J., Udalova I.A., Udalova I.A. Quantitative high-throughput analysis of transcription factor binding specificities. Nucleic Acids Res. 2004;32:e44. doi: 10.1093/nar/gnh042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M.Z. Functional evolution of noncoding DNA. Curr. Opin. Genet. Dev. 2002;12:634–639. doi: 10.1016/s0959-437x(02)00355-6. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Patel N.H., Kreitman M., Patel N.H., Kreitman M., Kreitman M. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: Rules governing conservation and change. Development. 1998;125:949–958. doi: 10.1242/dev.125.5.949. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Bergman C., Patel N.H., Kreitman M., Bergman C., Patel N.H., Kreitman M., Patel N.H., Kreitman M., Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Palsson A., Alekseeva E., Bergman C.M., Nathan J., Kreitman M., Palsson A., Alekseeva E., Bergman C.M., Nathan J., Kreitman M., Alekseeva E., Bergman C.M., Nathan J., Kreitman M., Bergman C.M., Nathan J., Kreitman M., Nathan J., Kreitman M., Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Berger M.F., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Berger M.F., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Snyder M., Young R.A., Bulyk M.L., Young R.A., Bulyk M.L., Bulyk M.L. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega M.A., Pennacchio L.A., Pennacchio L.A. Comparative genomic analysis as a tool for biological discovery. J. Physiol. 2004;554:31–39. doi: 10.1113/jphysiol.2003.050948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatsenko D., Levine M., Levine M. Quantitative analysis of binding motifs mediating diverse spatial readouts of the Dorsal gradient in the Drosophila embryo. Proc. Natl. Acad. Sci. 2005;102:4966–4971. doi: 10.1073/pnas.0409414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B., Gompel N., Rokas A., Kassner V.A., Williams T.M., Yeh S.D., True J.R., Carroll S.B., Gompel N., Rokas A., Kassner V.A., Williams T.M., Yeh S.D., True J.R., Carroll S.B., Rokas A., Kassner V.A., Williams T.M., Yeh S.D., True J.R., Carroll S.B., Kassner V.A., Williams T.M., Yeh S.D., True J.R., Carroll S.B., Williams T.M., Yeh S.D., True J.R., Carroll S.B., Yeh S.D., True J.R., Carroll S.B., True J.R., Carroll S.B., Carroll S.B. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- Senger K., Armstrong G.W., Rowell W.J., Kwan J.M., Markstein M., Levine M., Armstrong G.W., Rowell W.J., Kwan J.M., Markstein M., Levine M., Rowell W.J., Kwan J.M., Markstein M., Levine M., Kwan J.M., Markstein M., Levine M., Markstein M., Levine M., Levine M. Immunity regulatory DNAs share common organizational features in Drosophila. Mol. Cell. 2004;13:19–32. doi: 10.1016/s1097-2765(03)00500-8. [DOI] [PubMed] [Google Scholar]
- Silverman N., Maniatis T., Maniatis T. NF-κB signaling pathways in mammalian and insect innate immunity. Genes & Dev. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- Stanojevic D., Small S., Levine M., Small S., Levine M., Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A., Levine M., Levine M. Dorsal gradient networks in the Drosophila embryo. Dev. Biol. 2002;246:57–67. doi: 10.1006/dbio.2002.0652. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A., Levine M., Levine M. Localized repressors delineate the neurogenic ectoderm in the early Drosophila embryo. Dev. Biol. 2005;280:482–493. doi: 10.1016/j.ydbio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A., Van Drenth M., Erives A., Markstein M., Levine M., Van Drenth M., Erives A., Markstein M., Levine M., Erives A., Markstein M., Levine M., Markstein M., Levine M., Levine M. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell. 2002;111:687–701. doi: 10.1016/s0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- Udalova I.A., Knight J.C., Vidal V., Nedospasov S.A., Kwiatkowski D., Knight J.C., Vidal V., Nedospasov S.A., Kwiatkowski D., Vidal V., Nedospasov S.A., Kwiatkowski D., Nedospasov S.A., Kwiatkowski D., Kwiatkowski D. Complex NF-κB interactions at the distal tumor necrosis factor promoter region in human monocytes. J. Biol. Chem. 1998;273:21178–21186. doi: 10.1074/jbc.273.33.21178. [DOI] [PubMed] [Google Scholar]
- Udalova I.A., Mott R., Field D., Kwiatkowski D., Mott R., Field D., Kwiatkowski D., Field D., Kwiatkowski D., Kwiatkowski D. Quantitative prediction of NF-κB DNA-protein interactions. Proc. Natl. Acad. Sci. 2002;99:8167–8172. doi: 10.1073/pnas.102674699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lindblad-Toh K., Lander E.S., Kellis M., Lander E.S., Kellis M., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]