Abstract
Trypanosoma brucei evades host acquired immunity through differential activation of its large archive of silent variant surface glycoprotein (VSG) genes, most of which are pseudogenes in subtelomeric arrays. We have analyzed 940 VSGs, representing one half to two thirds of the arrays. Sequence types A and B of the VSG N-terminal domains were confirmed, while type C was found to be a constituent of type A. Two new C-terminal domain types were found. Nearly all combinations of domain types occurred, with some bias to particular combinations. One-third of encoded N-terminal domains, but only 13% of C-terminal domains, are intact, indicating a particular need for silent VSGs to gain a functional C-terminal domain to be expressed. About 60% of VSGs are unique, the rest occurring in subfamilies of two to four close homologs (>50%–52% peptide identity). We found a subset of VSG-related genes, differing from VSGs in genomic environment and expression patterns, and predict they have distinct function. Almost all (92%) full-length array VSGs have the partially conserved flanks associated with the duplication mechanism that activates silent genes, and these sequences have also contributed to archive evolution, mediating most of the conversions of segments, containing ≥1 VSG, within and between arrays. During infection, intact array genes became activated by duplication after two weeks, and mosaic VSGs assembled from pseudogenes became expressed by week three and predominated by week four. The small subfamily structure of the archive appears to be fundamental in providing the interacting donors for mosaic formation.
African trypanosomes are single-cell eukaryotes that cause human sleeping sickness in Africa and a variety of livestock diseases in developing countries. Trypanosoma brucei proliferates extracellularly in the blood and tissue fluids of mammals, protected by a densely packed coat composed of variant surface glycoprotein (VSG) (Barry and McCulloch 2001; Donelson 2003; Pays 2005). The parasites survive antibody responses, and go on to establish chronic infections, by undergoing antigenic variation, in which individuals switch to expression of alternative, immunologically distinct VSGs and then proliferate (Lythgoe et al. 2007). VSGs consist of a hypervariable N-terminal domain of 350–400 residues and a more conserved C-terminal domain of 40–80 residues that is glycosyl phosphatidyl inositol (GPI)-anchored to the plasma membrane (Carrington et al. 1991). Despite the sequence divergence, N-terminal domains can adopt a very similar alpha-helical coiled-coil higher order structure (Blum et al. 1993) containing exposed surface loops thought to bear the variable epitopes (Miller et al. 1984; Hsia et al. 1996). The VSG family is large and diverse, comprising several sequence types in both domains (Carrington et al. 1991). Determination of the sequence of the genome of T. brucei TREU 927 has now greatly expanded the VSG sequence data set, enabling more global analysis of sequence variability, which might eventually allow assessment of the balance between structural constraint and epitope diversification. This can also help assessing whether some sequences have evolved to neofunction, as often occurs for duplicated genes (Lynch and Katju 2004; Aury et al. 2006).
Antigenic variation in T. brucei relies on an archive of silent VSG genes (Barry et al. 2005; Berriman et al. 2005; Taylor and Rudenko 2006), analogous to systems in other bacterial and protozoan pathogens (Barry et al. 2003). VSG genes occur at the subtelomeres of all chromosome classes in the T. brucei genome, which comprises 11 conventional diploid “megabase” chromosomes, an estimated 100 minichromosomes, and several intermediate chromosomes (Melville et al. 1998). Archival VSGs are effectively the only genes on minichromosomes, immediately adjacent to the telomere (Williams et al. 1982), and another, large, set is tandemly arrayed in megabase subtelomeres (Berriman et al. 2005; Callejas et al. 2006). Each trypanosome expresses only one VSG, and antigen switching replaces the expressed VSG by another. During infection, VSGs are transcribed from specialized loci, bloodstream expression sites (BESs), which are terminal on megabase and intermediate chromosomes (Becker et al. 2004). In switching, archive VSGs are duplicated into a BES, replacing the VSG previously resident there. Antigenic variation produces complex mixtures of variant subpopulations, which nevertheless display order in expression (Lythgoe et al. 2007). Order is thought to be important for prolongation of infection, by ensuring new variants appear as a trickle and by limiting to a sublethal level the number of extant variants. The different locus types occupied by archive VSGs help determine order, a key factor being the inherent activation probability of each VSG (Pays 1989; Morrison et al. 2005). Thus, telomere-proximal genes, such as those on minichromosomes, are expressed early, array VSGs are expressed later, and two late variants have been observed to express mosaic VSGs, assembled from fragments of pseudogenes (Thon et al. 1990; Barbet and Kamper 1993). Very little is known about how or when mosaic genes are assembled, or their prominence in antigenic variation. It has been proposed recently that antigenic variation might be far more extensive than suspected, due to the large size of the archive and the combinatorial nature of mosaic formation, and that mosaic formation is the predominant switch mode in natural infections (Barry et al. 2005; Marcello and Barry 2007). Although subtelomeres are preferential sites for the evolution of eukaryotic multigene families, little is known about mechanisms involved, and the VSG family could be an informative paradigm.
Many of the features of antigenic variation, from molecules to populations, are intimately linked; features at the molecular level are selected by what occurs at the host and parasite population levels, and in turn contribute to the molecular and population phenotypes (Marcello and Barry 2007). A major challenge is to reveal the functional nature of these links. With the recent near-completion of the genome sequence of T. brucei revealing the array archive, here we analyze bioinformatically and experimentally the nearly 1000 available VSG gene sequences (Berriman et al. 2005), to address the questions outlined above.
Results and Discussion
Archive composition and coverage
We analyzed 940 VSGs in the main contigs of chromosomes (chr) 1–11 (GENEdb v4), in the putative chr 8 homolog left array and in unordered contigs of chr 9, 10, and 11; all are retrievable from the VSGdb database (http://www.vsgdb.org). The archive is larger than this, as sequencing and assembly are incomplete and VSG array haplotypes on chromosome homologs remain to be characterized. The assembled diploid megabase chromosome core genome is ∼44 Mb and pulsed field gel electrophoretic analysis (Melville et al. 1998) suggests these chromosomes in the genome strain total 53.5 Mb. Allowing for ∼1.5 Mb comprising the five-to-nine bloodstream expression sites (M. Becker and E. Louis, pers. comm.) and other repetitive subtelomeric regions and telomeres, the remaining ∼8 Mb difference could be VSG arrays. As the arrays appear to be haploid, and with the observed average gene density of one VSG every 5 kb, it is predicted conservatively that there are ∼1600 array VSGs in the diploid genome. The 940 genes analyzed here therefore represent between half and two thirds of the array archive.
The VSG protein family
There are four categories of VSG (Berriman et al. 2005): “functional” (encodes all recognizable features of known functional VSGs), “atypical” (complete genes possibly encoding proteins with inconsistent VSG folding or post-translational modification), “pseudogene” (with frameshifts and/or in-frame stop codons), and “incomplete” (Berriman et al. 2005). Most of the 940 silent VSGs are “pseudogenes” (611, 65%) or “incomplete” (197, 21%), only a small percentage being “functional” (43, 4.5%) or “atypical” (89, 9.5%). Designation as “atypical” is necessarily conservative, 75% having small deviations from known GPI anchor signal sequences; increased knowledge of these signals possibly would lead to some being reclassified as “functional.” As the two VSG domains differ in structure, function, and evolution and are subject to independent recombination, we consider them separately.
N-terminal domain
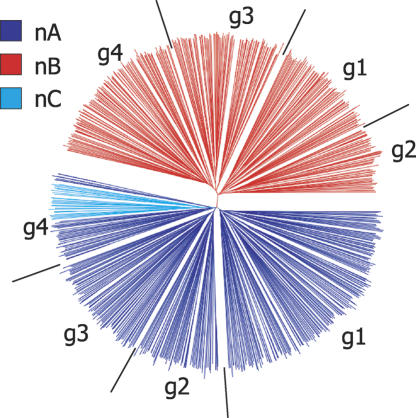
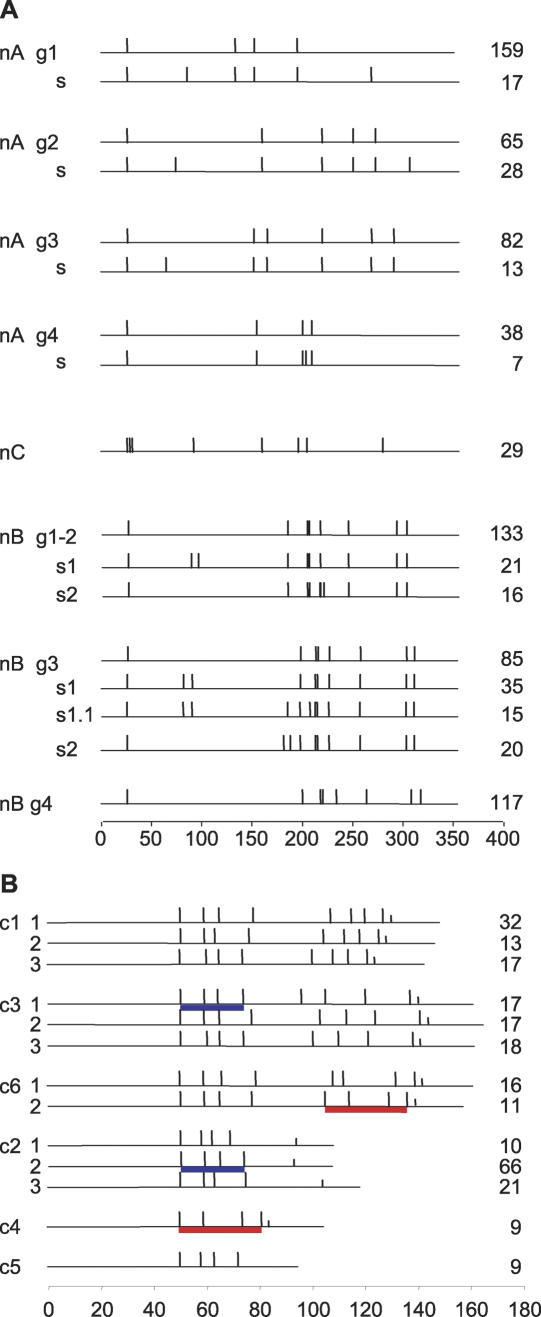
In all the 771 full-length N-terminal domains, no novel types were found. Domain types A and B (Carrington et al. 1991), hereafter referred to as nA and nB, are about equal in number, while type nC forms a small cluster within nA (Fig. 1). For each N-terminal type, about one in three domains is theoretically “functional” (Supplemental Fig. S1), suggesting that the basis for a high proportion (94.6%) of the 743 full-length genes being degenerate is C termini being defective, through presence of stop codon(s), frameshift(s), or disruption of cysteine pattern. This means that one third of N-terminal domains possibly can be utilized directly, by combining with a functional C-terminal domain from, for example, the VSG already present in the expression site. More detailed analysis of 361 nA and 29 nC domains revealed several distinct clusters, distinguished by significant differences in the number and spacing of conserved cysteines (Fig. 2A). Group 1 is the most common (44%) and has been characterized biochemically (Carrington et al. 1991). As nC resembles group 4, we propose that it be reclassified into nA. In 335 nB sequences analyzed, the cysteine pattern is much more conserved, displaying only slight variation in inter-cysteine distance (Fig. 2A). It is important now to determine more structures, one reason being to enable identification of possible epitope-bearing substructures.
Figure 1.
Diversity of VSG N-terminal domain. Neighbor-joining tree based on ClustalX-generated multiple sequence alignment of predicted peptide sequences of 725 sequences. The three domain types are colored individually (by HyperTree) as shown in the key. g represents each group depicted in Fig. 2A.
Figure 2.
Cysteine pattern of VSG domain types. Cys residue positions are shown as vertical bars, and the number of sequences in each category is shown to the right. Scale bars (number of residues) are shown for each panel. (A) N-terminal domain. g, group; s, subgroup. (B) Examples of C-terminal domain. Cys subdomains shared between different domain types are indicated by underlines of the same color. Two to three examples of domain type subgroups are given for the main domain types. Half-length vertical bars represent putative GPI signal sequences.
C-terminal domain
Amongst the 651 sequences analyzed, more structural diversity and complexity (presence of one or two subdomains, presence of indels) were detected for the C-terminal domain than for the N-terminal domain. The C-terminal domain is described by the presence of one or two four-cysteine subdomains, the spacing of cysteines, and the type of GPI anchor signal sequence. Two new domain types were found: c5 (one or two four-cysteine subdomains) and c6 (two such subdomains). c1 is the most abundant (235), followed by c2 (186), then c3 (152). c4, c5, and c6 are much less common (10, 30, and 38, respectively). Only 13% (85/651) of encoded C-terminal domains are predicted to be functional, ranging from 23% (43/186) of c2 to 7% of c1 (Supplemental Fig. S1B). A notable difference from N-terminal domains is a higher incidence of likely indels, making cysteine spacing much more variable and hampering identification of subgroups. Phylogenetic analysis (data not shown) suggests that there is extensive mixing of domain types in the region of the first four cysteines, but much less so toward the carboxy end of the domain, where the pattern of the last four cysteines and the sequence of the GPI anchor signal are more type-specific (Fig. 2B). The solved structure of a c2 domain reveals a novel, compact motif spanning the four cysteines, flanked by disorder (at least in solution) (Chattopadhyay et al. 2005). Our observation of extensive mixing of the first four-cysteine region (see also Fig. 2B) is compatible with its observed greater sequence conservation and provides the context for the documented case of recombination between sequences encoding the third and fourth cysteines of a c1 and a c2 domain (Hutchinson et al. 2003). It thus appears that the 5′ region encoding the C-terminal domain provides a relative hotspot for recombination, yielding expressed interdomain hybrids (Pays et al. 1985). Such modular use of domains appears to have led to a high level of degeneracy of C-terminal domains.
Full-length VSGs
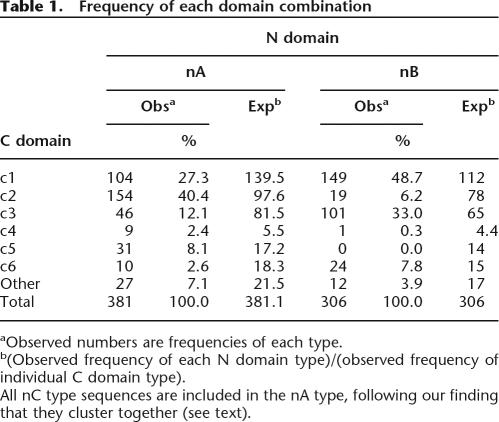
Full-length genes and pseudogenes were analyzed phylogenetically and for domain combinations. Whereas global phylogeny of predicted full-length VSGs matches that produced by aligning N-terminal domains alone (Supplemental Fig. S2), analysis of domain combinations revealed bias in combination of domain types, toward nAc2, nBc1, nBc3, and nBc6 (Table 1). However, the propensity for formation of hybrid genes during duplicative activation, allied with the requirement for most genes to gain a functional C-terminal encoding sequence, means that these biases may not be reflected in what occurs in expressed VSGs. Analysis of cDNAs shows no apparent restriction on combination of N- and C-terminal domains, and the same N-terminal domain can be expressed with different C-terminal types (Hutchinson et al. 2003). This promiscuity in domain combinations underlines the importance of the shared four-cysteine motif in the C-terminal domain in promoting N-terminal domain recombination at the DNA level.
Table 1.
Frequency of each domain combination
aObserved numbers are frequencies of each type.
b(Observed frequency of each N domain type)/(observed frequency of individual C domain type).
All nC type sequences are included in the nA type, following our finding that they cluster together (see text).
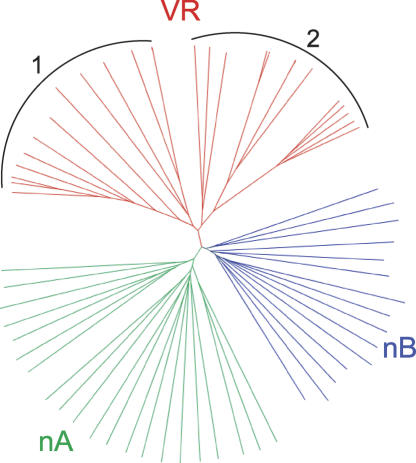
VSG-related proteins
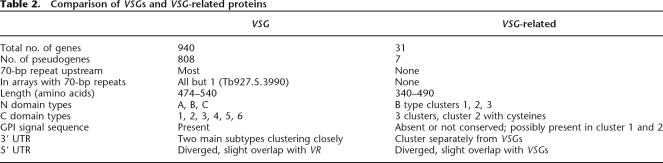
Thirty one genes, termed VSG-related (VR) (Berriman et al. 2005), cluster separately from VSGs (Fig. 3) and have several features suggesting they are not authentic VSGs. All VR N-terminal domains form a discrete subgroup of nB, but the C-terminal domain is distinct from all of the VSG C-terminal domains. The VR family is as divergent as the much larger VSG archive but, unlike the VSGs, appears not to have assortment between putative N- and C-terminal domain types, suggesting a different mode of evolution. Only seven of 31 (22%) VRs are pseudogenes, in contrast to the ≥86% in the VSG archive. VRs are organized as single genes or in small arrays at strand switch regions in chromosome cores, rather than in subtelomeric VSG arrays. Over the examined 300 bp, VR flanks cluster separately from those of VSGs, and VRs do not have the upstream 70-bp repeats common to most VSGs. Features of VSG and VR genes are compared in Table 2.
Figure 3.
Tree of amino acid alignment of putative VSG-related proteins. The tree compares predicted products of 35 full-length, functional VSGs with those of 29 VR genes. The VR set falls into two clusters, denoted by 1 and 2.
Table 2.
Comparison of VSGs and VSG-related proteins
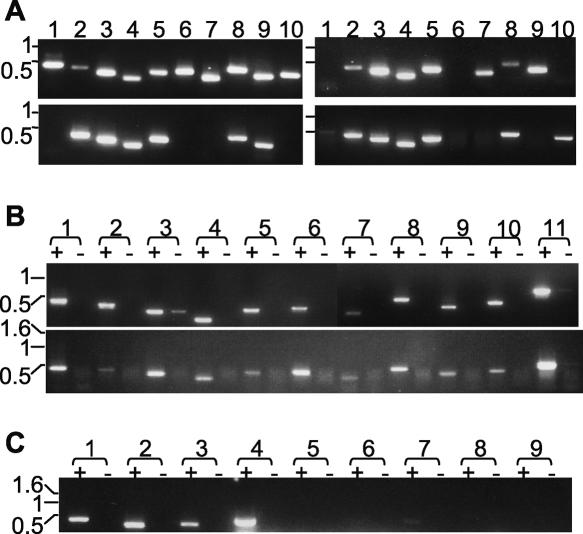
If VRs are functionally distinct from VSGs, they might be predicted to be conserved between strains, unlike VSGs (Van Meirvenne et al. 1977; Frasch et al. 1982). PCR analysis revealed that about half of the 10 tested VR sequences are present in genomic DNA from three other T. brucei strains (Fig. 4A). To test whether VRs have different expression control from that of VSGs, which yield mRNA in bloodstream stage trypanosomes in a clone-specific pattern, but no mRNA in the procyclic stage parasite from the tsetse midgut, we applied the RT-PCR. Using the same primers as for genomic DNA analysis, we found at least eight of the 10 VRs, including four of the five ubiquitous genes (see Fig. 4A), expressed as RNA in both stages of the genome strain (Fig. 4B). We next tested whether VR expression is not clonally restricted in bloodstream trypanosomes, through analysis of VR expression in a T. brucei Lister 427 strain in which the HYG gene is in tight linkage with the 221 VSG (McCulloch et al. 1997). RT-PCR analysis of this strain, grown under hygromycin selection, showed transcripts of all three tested VR genes (VR2, VR4, and VR15) and of 221 VSG, but not of five other VSGs to which 221 commonly switches (Fig. 4C). We conclude that these VRs are coexpressed with the 221 VSGs. Overall, the expression patterns of VRs are inconsistent with VSG function and are consistent with novel function.
Figure 4.
Analysis of VR genes. (A) Presence of VR sequences in several T. brucei strains. PCR analysis of genomic DNA was undertaken with primers specific for: lanes 1–10, VR 1, 2, 4, 5, 8, 9, 11, 13, 15, and 18. T. brucei genomic DNA sources: top left, TREU 927; top right, Lister 427; bottom left, STIB 247; bottom right, EATRO 795. (B) Lack of stage-specificity of VR transcripts. RT-PCR analysis of VR transcripts represented in oligo[dT]-primed cDNA from bloodstream stage (top) and procyclic stage (bottom) TREU 927. Lane pairs 1–10: primers for VR 1, 2, 4, 5, 8, 9, 11, 13, 15, and 18; lane pairs 11: tubulin primers. In each lane pair, “+” denotes presence, and “−” denotes absence, of reverse transcriptase at the cDNA synthesis step. (C) Analysis of whether transcripts of alleles of VR and VSG genes are mutually exclusive. RT-PCR analysis of Lister 427 bloodstream trypanosome clone expressing linked HYG and 221 VSG genes, grown in hygromycin. oligo[dT]-primed cDNA was subjected to PCR using (lane pairs 1–9) primers for VR 2, VR 5, VR 15, 221 VSG, 118 VSG, VO2 VSG, 121 VSG, G4 VSG, S8 VSG. All images are of ethidium bromide-stained gels, and DNA size markers (kb) are indicated to the left of each.
Despite this, there are no specific sequence features conserved amongst the predicted VR proteins. Several functional trypanosomal surface proteins homologous to VSGs have been proposed to form the distinctive coiled-coil “VSG fold” (Carrington and Boothroyd 1996). One is the heterodimeric transferrin receptor, both subunits of which resemble VSGs, although it is unknown whether they evolved from, or share an ancestor with, VSGs (Salmon et al. 1997). A clearer case for evolution from the VSG has been argued for the serum resistance protein (Campillo and Carrington 2003). Although the VR putative products may have evolved from VSGs in T. brucei, they could have evolved from an earlier precursor, as they have an nB N-terminal domain, which is the only type demonstrated for VSGs of Trypanosoma congolense (Bussler et al. 1998).
VSG gene array architecture and evolution
All of the VSGs analyzed here are located in subtelomeric arrays, with a sole exception. Tb927.5.3990 is interstitial, has none of the upstream 70-bp imperfect repeats characteristic of VSGs, and has a nonconserved C-terminal domain. Within the arrays, as elsewhere in the genome, the basic VSG unit is a 3–4-kb cassette, flanked upstream by the 70-bp repeats and downstream by the partially conserved region from within the 3′ end of the coding sequence to elements in the 3′ untranslated region (UTR) (Michels et al. 1983). The 70-bp repeats form the upstream boundary of the duplication events of VSG switching. Some 92% of full-length array VSGs (687/743) are flanked upstream by at least one such repeat, 74% having one copy, 14% having two, and 5% having between three and 15 copies. The degeneracy amongst these repeats is much greater than previously recognized, especially in the case of section 3 of the repeat (Aline et al. 1985), which sometimes is absent or incomplete.
Our initial global analysis of annotated VSGs showed almost complete dispersion of closest homologs across the arrays (Berriman et al. 2005), in keeping with earlier analysis of one array gene in different trypanosome strains (Bernards et al. 1986), which had led to a model involving deletion and duplication of VSG cassettes. We now confirm and extend this model, through analysis of recent duplications in the genome. For pairs of N-terminal domain-encoding sequences >90% nucleotide-identical, we located the physical extent of each putative gene duplication event, through pairwise alignment of their flanking regions. Scrutiny of several long (up to 30 kb) gene conversion tracts revealed localized disruption of homology, often coincident with 70-bp repeats, which in many cases displayed repeat contractions and expansions. This, and subsequent possible conversion events, complicated the identification of precise ends of conversion events. Over the 38 conversion events analyzed, nucleotide identity between putative conversion tracts ranged between 74% and 99%. The shortest putative conversion tract was 1.6 kb long, and the longest contained nine VSGs. In 89% of cases, at least one of the ends corresponded to either 70-bp repeats or the C-terminal end region; 52% of cases involved either of these sequences at both ends. To dissect the evolution on a finer scale and assess the incidence of relatively recent duplication events, we queried geneDB for the number of duplicates (defined as ≥75% nucleotide identity) of each of 31 VSGs in the chr 8 left subtelomere array (chr 8L). Five of the 31 genes are within a family of five or more members (Fig. 5). Again, duplications in general are delimited by the conserved cassette flanks and often have involved single genes, although some events will be concealed by subsequent incidents. It appears now that the cassette flanks driving VSG duplication are important not only for antigen switching, but also in array expansion and diversification.
Figure 5.
Array evolution through duplication. For 31 VSG in the chr 8L array, which are depicted by boxes (followed by an orientation indicator) in the map at the top, the number of other copies of each in the genome (≥75% nucleotide identity) is shown by color intensity in the lower graphic, according to the tone key. The ends of duplicated sequences are aligned between the two graphics, demonstrating that these ends often coincide with the ends of VSGs.
A revised model for array evolution can now be suggested: Shorter rearrangements, involving deletions and conversions of VSG cassettes, promote gene duplication and family divergence, while longer-range contractions and expansions are effected via the non-LTR retrotransposon INGI, which is found at most strand switches in VSG arrays (Berriman et al. 2005). The passive and active routes by which such transposons reshape genomes (Kazazian 2004) could contribute extensively to the large differences in archive size between trypanosome strains and, within a strain, between arrays and between chromosome homologs. Gene conversion clearly is a major mechanism in array evolution, and we detected no polarity in the duplication map (Fig. 5), which argues against a role for break-induced replication (Fischer et al. 2006). Subtelomeres of other organisms, including Kluyveromyces lactis (Fairhead and Dujon 2006) and Homo sapiens (Linardopoulou et al. 2005), are patchworks with few tandem duplications and so resemble the VSG array subtelomere regions. A reason for this common architectural theme might be evident from studies of Saccharomyces cerevisiae, where experimental selection for duplication yielded many cases of translocation, an event shown to be most likely to lead to fixation (Koszul et al. 2004, 2006), presumably because tandem duplication creates excessive, local genome instability. The extensive traffic between trypanosome subtelomeres, including gene duplication, is followed by rapid sequence diversification, which presumably limits which VSGs can readily interact in homologous recombination within their coding sequence. As we show below, that is central to ordered VSG expression after the initial phase of infection.
VSG gene subfamilies
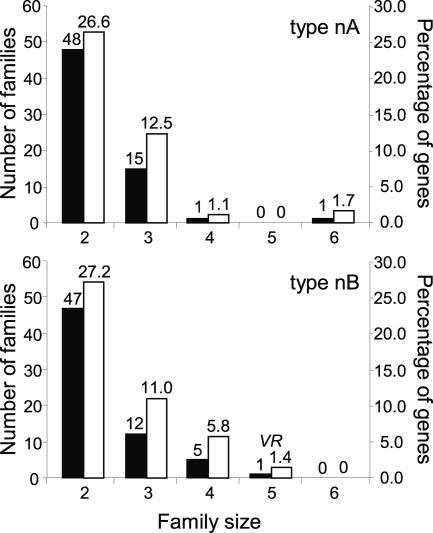
As we have proposed that mosaic VSG expression is a major mechanism in antigenic variation (Marcello and Barry 2007) and mosaic formation appears to depend on highly related donor partners, we investigated whether the archive contains VSG sequence subfamilies. We aligned N-terminal domains to avoid the complication of higher similarity amongst C-terminal sequences, and because the variable domain is more directly related to mosaic gene formation, as we show below. Systematic ClustalW pairwise analysis of 361 type nA domain sequences revealed an outlier group of pairs with >51% peptide identity. The outlier group (41.8% of genes) comprises small subfamilies, ranging in size from two to six members (Fig. 6, upper panel), with a minimum pairwise nucleotide identity of 67% (average for the least-score 10 pairs is 73.5%). Almost exclusively, the subfamilies are pairs or triplets, in the ratio ∼3:1 (Fig. 6, upper panel). The largest subfamily, of six members, is abnormal, as four unusually form a directional gene cluster demarking the core of chr 4 from the right-side VSG array. The subfamily of four illustrates the dispersion of subfamily members: They are in arrays on different subtelomeres. For type nB VSGs, comparison of 345 N-terminal domains, using an observed minimum cutoff value of 53% peptide identity, yielded similar data (Fig. 6, lower panel). Again, the largest subfamily is anomalous, consisting of an array of VR genes. Across the whole archive of an estimated 1500 VSGs, and taking account of the proportion of sequences that are full-length or encode N-terminal fragments, it might be extrapolated that ∼550 VSGs are in high-identity subfamilies, comprising ∼180 pairs, ∼50 triplets, and ∼10 quadruplets.
Figure 6.
VSG families. Pairwise comparison within N-terminal domain types. Peptide sequences encoded by 361 type nA VSGs were all scored against each other in ClustalW multiple sequence alignment, and 345 type nB VSGs were compared in the same way (bottom chart). The number of families (left scale of Y-axis) of given size (X-axis) are depicted in black, and the percentage of genes (right scale of Y-axis) within each domain type is depicted in white. VR indicates the family of VSG-related genes.
Expression of array VSGs and formation of mosaic genes
To address the significance, to antigenic variation, of mosaic gene expression and the subfamily structure, we sequenced 37 cDNAs, which corresponded to 21 different VSGs, isolated from 11 mice infected with the T. brucei genome strain 927 (Table 3). A pattern is evident in the sequential use of different donor types, as expected from previous studies (Pays 1989; Robinson et al. 1999), although it should be noted that our interpretations are partly speculative, due to the incomplete extent of genome sequencing. On day 3, we recovered only the VSG, GUTat 10.1, expressed by the infecting trypanosomes. For day 9 and in one case for day 14, no corresponding donor gene sequences are present in geneDB. Precedence suggests they arose by duplication of minichromosomal VSGs into the expression site (Liu et al. 1985; Robinson et al. 1999); the minichromosomal genes of TREU 927 have not been sequenced. Array donor genes became involved from day 14. Of the 21 distinct sequences, 14 could be attributed to array VSG donors, with 10 matching fully sequenced silent genes and four matching short, partial reads. Five of these 14 arose from “functional” array donors, although they had gained some or all C-terminal sequence from elsewhere. Two cDNAs (days 14, 24) arose from “atypical” genes, each with donation of a short stretch of sequence at the 3′ end. Mosaic genes were present as seven cDNAs from days 22 and 28 and became predominant, contributing 40% of recovered cDNAs by days 22–24 and 62.5% by day 28 (Table 3). Previously, mosaics were detected on days 23–35 in rabbits, but their prevalence was not investigated (Capbern et al. 1977; Marcello and Barry 2007). Notably, we found distinct sets of variants in different infections on day 28, which is consistent with the proposition that mosaic formation leads to distinct variant strings in different animals, due to antigenic variation being stochastic (rather than to host-related differences) (Barry et al. 2005; Marcello and Barry 2007).
Table 3.
VSGs expressed in chronic infections
VSGs corresponding to known archive genes are given the geneDB identifier of the donor; novel VSGs do not have a geneDB code.
aInc, only incomplete sequence obtained.
The proposal that the number of 70-bp repeats upstream of the gene, and their degree of identity with those in the BES, help determine order in antigenic variation (Aline et al. 1985) is not supported by our data, as there is only minimal variation in 70-bp repeat number amongst silent cassettes. For the downstream end of duplications into the BES, our experimental analysis reveals that inheritance of at least some of the C-terminal encoding sequence from the previously expressed VSG is common. In the 14 cDNAs that arose from array VSGs, eight had gained a 3′ end (Table 3), in agreement with the observation that most array genes encode defective C-terminal domains.
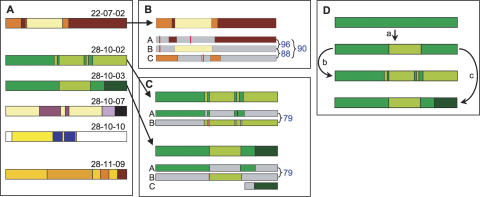
The archive’s subfamily structure appears to be central to the formation of expressed mosaic genes. A main driver for mosaic gene formation is high identity between the coding sequences of donor genes (Fig. 7). Of the seven putative mosaics detected, four (22-07-02, 22-07-04, 28-10-02, and 28-10-03) apparently were derived from fully sequenced donors. Discounting mosaic VSG 22-07-04, which is mosaic only in the C-terminal domain, the other three were assembled from donors with high full-length nucleotide identity (79%–96%) (Fig. 7). The three originally reported mosaic gene sets also showed involvement of high donor identity, in the ranges 85%–97% (Roth et al. 1991), 69% (Thon et al. 1990), and 87%–91% (Kamper and Barbet 1992). High donor identity does not necessarily mean that long stretches of homology are required for the recombination reactions. Short homology may be adequate, but longer stretches increase the opportunity for recombination. Short-stretch homologous recombination could be important perhaps later in infection, by recruiting more diverged genes for mosaic formation.
Figure 7.
Structure and origin of putative mosaic cDNAs. (A) Mosaic structure. The seven detected mosaics for which significant donor information is available. Colors indicate patches matching different donors. The white regions in mosaic 28-10-10 indicate where no donor has been identified. (B) Origin and structure of N-terminal domain mosaic 22-07-02 from mouse 7 on day 22. The three putative silent donor genes (A, B, C) are shown below the cDNA, as thinner rectangles. Colors represent patches in common between donor and cDNA (positions of junctions in cDNA are averages where donors are identical in that region). The percentage nucleotide identity between silent donors is shown to the right. Vertical red bars depict frameshifts or stop codons in donor genes. The likely donors are: A, Tb11.v4.0074; B, Tb10.v4.0161; C, Tb11.14.0001. (C) Origin and structure of related mosaics coexisting in mouse 10 on day 28. Diagram is constructed as in B. For both mosaics, likely donors A and B are Tb11.09.0005 and Tb11.13.0003. For 28-10-03, likely donor C is read AZ217061. (D) Deduced pathway for development of related mosaics in mouse 10 on day 28.
We obtained the cDNA clones by RT-PCR, which could yield an artifact, whereby mosaicism arises from template switching in PCRs. We therefore applied PCR to genomic DNA, to test the prediction that the putative mosaic genes would be absent from the genome of trypanosomes at the start of infection, or in an independent infection. These predictions were upheld for the two examples tested, VSG 28-10-02 and 28-10-03 (Supplemental Fig. S3), validating the observed sequences as mosaics.
Because these two mosaics, which were isolated simultaneously from one infection, originated from the same donor genes and share a gene conversion boundary, we have deduced their likely assembly route (Fig. 7C,D). Gene A initially was being expressed and then formed a simple mosaic with B (step a), after which mosaic 28-10-03 acquired a different C-terminal end (step c), while 28-10-02 had further interaction with gene B and the silent copy of gene A, eventually yielding seven transitions between the A and B sequences (step b). This pathway suggests two important possibilities: Mosaics are built through discrete, sequential steps, and the same donors participate repeatedly, reinforcing the importance of high donor identity in mosaicism. In theory, mosaics could arise by a stepwise process occurring in the active BES or, perhaps more likely, in a silent expression site, a route to eventual expression that would not require every step to yield a functional VSG.
Mosaicism has two, and maybe three, important functions. First, it allows the use of many archive pseudogenes. Second, it generates novel variants. Third, it might provide the possibility of formation of distinct variant strings, enabling superinfection of partially immune hosts (Barry et al. 2005). Like many biological systems, VSG mosaicism is inherently wasteful, as the steps leading to a mosaic sequence in a BES probably often yield trypanosomes doomed by attempting to express mosaic pseudogenes, or by expressing intact mosaics sharing some antigenicity with previous variants, a problem that compounds as infection proceeds. Unless there is a means for copying expressed genes back into the archive, the successful end product of repeated rounds of mosaic conversion and antibody selection can be eliminated on death of a variant. That so few archive genes are intact suggests such retrograde movement is, at best, rare. There is scope, nevertheless, for inheritance of some mosaics in silent expression sites.
In Anaplasma marginale, expressed antigen genes become more complex with time of infection (Futse et al. 2005), but our limited analysis shows no evidence for such an increase from day 21 to day 28, although it should be noted that we studied distinct trypanosome infections. There was, however, decrease in donor sequence identity between days 21 and 28, from 90%–96% to 83%–90%, similarly to the BoTat mosaics, as discussed above.
The evolution to new VSGs is reminiscent of the neofunctionalization that occurs as duplicated genes diverge in eukaryote genomes (Lynch and Katju 2004; Aury et al. 2006). There is a broad parallel between trypanosomes and yeasts, where pathogens have the most unstable genomes, presumably reflecting the strength of host-mediated selective pressure (Fischer et al. 2006). The linked architecture and differential expression of the VSG arrays provide a striking illustration of how critical subtelomeres can be to the diversity an organism requires for survival and proliferation. Taking together the observations of a marked subfamily structure and the predominance of mosaicism as infection proceeds, we propose that the evolutionary mechanics of trypanosome subtelomeres has been selected to achieve equilibrium between gene duplication and rapid diversification, satisfying needs for a subfamily structure conducive to expressed mosaic formation and for the production of novel silent VSGs.
Methods
Bioinformatic analysis of the VSG archive
The VSG annotation procedure has been described (Berriman et al. 2005). Multiple sequence alignments were conducted, via default settings, using ClustalX or ClustalW at EBI (http://www.ebi.ac.uk/clustalw/) or at the Pasteur Institute (http://bioweb.pasteur.fr/seqanal/interfaces/clustalw.html), the alignment was then edited manually with Bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and then a second alignment with the edited sequences was performed. A neighbor-joining tree produced by ClustalX was then visualized and colored using the HyperTree software (Bingham and Sudarsanam 2000). GPI signal sequences were identified initially by manual comparison with known VSG signals, then through big-PI (http://mendel.imp.ac.at/gpi/gpi_server.html) and DGPI (http://129.194.185.165/dgpi/index_en.html) prediction programs.
Trypanosomes
Most analyses were performed on the genome sequencing strain, T. brucei TREU 927/4 GUTat 10.1, either as bloodstream stage in mice or, for analysis of VR gene expression, as the procyclic stage in SDM79 medium (Brun and Schonenberger 1979). Some VR gene expression analysis was performed on T. brucei Lister 427 bloodstream stage line 3174.2, which has the hygromycin resistance gene HYG inserted upstream of the expressed 221 VSG. This line was grown in vitro in HMI-9 medium as described (McCulloch et al. 1997). Genomic DNA of T. brucei EATRO 795 and STIB 247 was also analyzed for presence of VR genes. For chronic infections with the genome strain, 11 mice were each injected with 80 μL of blood containing 1.3 × 107 trypanosomes/mL. In order to maximize recovery of parasite gDNA and RNA, each mouse was sampled only terminally. On days 3, 9, 14, 21–24, and 28, blood was harvested from mice with the highest parasitaemia (usually, 107–108 trypanosomes/mL), and RNA, genomic DNA, and stabilates were prepared.
Amplification and cloning of VSGs
Reverse transcription used oligo[dT] as primer, and subsequent PCR utilized Herculase proofreading DNA polymerase (Stratagene), with a forward primer annealing to the spliced leader sequence (at the 5′ end of all trypanosome processed mRNA molecules) and a reverse primer recognizing a conserved 16-mer sequence specific to the 3′ end of all VSG mRNAs. Reaction conditions were: 5 min at 95°C, followed by 30 cycles of 95°C for 1 min, 38°C for 2 min, 72°C for 2 min, and a final extension of 5 min at 72°C. Amplified products of the expected size (∼1.6 kb) were TOPO-cloned (Invitrogen) after a 20 min Taq (Thermus aquaticus) DNA polymerase (ABGene) extension at 72°C, to add A residues to the ends, in order to provide cohesive ends for the T overhangs present in the vector. Each TOPO cDNA clone recovered from the chronic mouse infections was given a three-part name (e.g., 03-01-01), indicating the day of harvest (3 to 28), the mouse (1 to11), then the specific clone number.
The full-length VSG coding sequence was assembled by overlapping sequencing reads, produced from M13 forward and reverse primers corresponding to sequence in the TOPO vector, and internal primers designed specifically to cover the central region of the gene. At least two rounds of sequencing were performed to confirm independently the sequence of the insert across its whole length, unless the first result gave 100% sequence identity to a VSG already present in the database. GenBank accession numbers are given in Table 3.
VR gene expression
For analysis of VR genes, PCRs were performed using Taq polymerase and reactions were composed of 1–3 μL cDNA or genomic DNA, 2 μL of each primer (1 μM final concentration), 0.2 μL Taq, 2 μL custom buffer mix (yielding final concentrations of 45 mM Tris-HCl at pH 8.8, 11 mM [NH4]2SO4, 4.5 mM MgCl2, 6.7 mM 2-mercaptoethanol, 4.4 μM EDTA at pH 8.0, 113 μg/mL BSA, 1 mM each of the four dNTPs), and made up to 20 μL with water. Reaction conditions were 30 cycles of 95°C for 50 sec, 55°C–63°C for 50 sec, and 65°C for 1 min per kb of expected product.
Acknowledgments
We thank colleagues at the Sanger Institute (Matt Berriman, Christiane Hertz-Fowler, Hubert Renauld) and The Institute for Genome Research (Najib El-Sayed, Gaelle Blandin) for extensive discussions during annotation of the VSG arrays. We thank the Wellcome Trust for funding; J.D.B. is a Wellcome Trust Principal Research Fellow.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6421207
References
- Aline R., Macdonald G., Brown E., Allison J., Myler P., Rothwell V., Stuart K., Macdonald G., Brown E., Allison J., Myler P., Rothwell V., Stuart K., Brown E., Allison J., Myler P., Rothwell V., Stuart K., Allison J., Myler P., Rothwell V., Stuart K., Myler P., Rothwell V., Stuart K., Rothwell V., Stuart K., Stuart K. (TAA)n within sequences flanking several intrachromosomal variant surface glycoprotein genes in Trypanosoma brucei. Nucleic Acids Res. 1985;13:3161–3177. doi: 10.1093/nar/13.9.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury J.M., Jaillon O., Duret L., Noel B., Jubin C., Porcel B.M., Segurens B., Daubin V., Anthouard V., Aiach N., Jaillon O., Duret L., Noel B., Jubin C., Porcel B.M., Segurens B., Daubin V., Anthouard V., Aiach N., Duret L., Noel B., Jubin C., Porcel B.M., Segurens B., Daubin V., Anthouard V., Aiach N., Noel B., Jubin C., Porcel B.M., Segurens B., Daubin V., Anthouard V., Aiach N., Jubin C., Porcel B.M., Segurens B., Daubin V., Anthouard V., Aiach N., Porcel B.M., Segurens B., Daubin V., Anthouard V., Aiach N., Segurens B., Daubin V., Anthouard V., Aiach N., Daubin V., Anthouard V., Aiach N., Anthouard V., Aiach N., Aiach N., et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Barbet A.F., Kamper S.M., Kamper S.M. The importance of mosaic genes to trypanosome survival. Parasitol. Today. 1993;9:63–66. doi: 10.1016/0169-4758(93)90039-i. [DOI] [PubMed] [Google Scholar]
- Barry J.D., McCulloch R., McCulloch R. Antigenic variation in trypanosomes: Enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 2001;49:1–70. doi: 10.1016/s0065-308x(01)49037-3. [DOI] [PubMed] [Google Scholar]
- Barry J.D., Ginger M.L., Burton P., McCulloch R., Ginger M.L., Burton P., McCulloch R., Burton P., McCulloch R., McCulloch R. Why are parasite contingency genes often associated with telomeres? Int. J. Parasitol. 2003;33:29–45. doi: 10.1016/s0020-7519(02)00247-3. [DOI] [PubMed] [Google Scholar]
- Barry J.D., Marcello L., Morrison L.J., Read A.F., Lythgoe K., Jones N., Carrington M., Blandin G., Bohme U., Caler E., Marcello L., Morrison L.J., Read A.F., Lythgoe K., Jones N., Carrington M., Blandin G., Bohme U., Caler E., Morrison L.J., Read A.F., Lythgoe K., Jones N., Carrington M., Blandin G., Bohme U., Caler E., Read A.F., Lythgoe K., Jones N., Carrington M., Blandin G., Bohme U., Caler E., Lythgoe K., Jones N., Carrington M., Blandin G., Bohme U., Caler E., Jones N., Carrington M., Blandin G., Bohme U., Caler E., Carrington M., Blandin G., Bohme U., Caler E., Blandin G., Bohme U., Caler E., Bohme U., Caler E., Caler E., et al. What the genome sequence is revealing about trypanosome antigenic variation. Biochem. Soc. Trans. 2005;33:986–989. doi: 10.1042/BST20050986. [DOI] [PubMed] [Google Scholar]
- Becker M., Aitcheson N., Byles E., Wickstead B., Louis E., Rudenko G., Aitcheson N., Byles E., Wickstead B., Louis E., Rudenko G., Byles E., Wickstead B., Louis E., Rudenko G., Wickstead B., Louis E., Rudenko G., Louis E., Rudenko G., Rudenko G. Isolation of the repertoire of VSG expression site containing telomeres of Trypanosoma brucei 427 using transformation-associated recombination in yeast. Genome Res. 2004;14:2319–2329. doi: 10.1101/gr.2955304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A., Van der Ploeg L.H.T., Gibson W.C., Leegwater P., Eijgenraam F., de Lange T., Weijers P., Calafat J., Borst P., Van der Ploeg L.H.T., Gibson W.C., Leegwater P., Eijgenraam F., de Lange T., Weijers P., Calafat J., Borst P., Gibson W.C., Leegwater P., Eijgenraam F., de Lange T., Weijers P., Calafat J., Borst P., Leegwater P., Eijgenraam F., de Lange T., Weijers P., Calafat J., Borst P., Eijgenraam F., de Lange T., Weijers P., Calafat J., Borst P., de Lange T., Weijers P., Calafat J., Borst P., Weijers P., Calafat J., Borst P., Calafat J., Borst P., Borst P. Rapid change of the repertoire of variant surface glycoprotein genes in trypanosomes by gene duplication and deletion. J. Mol. Biol. 1986;190:1–10. doi: 10.1016/0022-2836(86)90070-7. [DOI] [PubMed] [Google Scholar]
- Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Lennard N.J., Caler E., Hamlin N.E., Haas B., Caler E., Hamlin N.E., Haas B., Hamlin N.E., Haas B., Haas B., et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Bingham J., Sudarsanam S., Sudarsanam S. Visualizing large hierarchical clusters in hyperbolic space. Bioinformatics. 2000;16:660–661. doi: 10.1093/bioinformatics/16.7.660. [DOI] [PubMed] [Google Scholar]
- Blum M.L., Down J.A., Gurnett A.M., Carrington M., Turner M.J., Wiley D.C., Down J.A., Gurnett A.M., Carrington M., Turner M.J., Wiley D.C., Gurnett A.M., Carrington M., Turner M.J., Wiley D.C., Carrington M., Turner M.J., Wiley D.C., Turner M.J., Wiley D.C., Wiley D.C. A structural motif in the variant surface glycoproteins of Trypanosoma brucei. Nature. 1993;362:603–609. doi: 10.1038/362603a0. [DOI] [PubMed] [Google Scholar]
- Brun R., Schonenberger M., Schonenberger M. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- Bussler H., Linder M., Linder D., Reinwald E., Linder M., Linder D., Reinwald E., Linder D., Reinwald E., Reinwald E. Determination of the disulfide bonds within a B domain variant surface glycoprotein from Trypanosoma congolense. J. Biol. Chem. 1998;273:32582–32586. doi: 10.1074/jbc.273.49.32582. [DOI] [PubMed] [Google Scholar]
- Callejas S., Leech V., Reitter C., Melville S., Leech V., Reitter C., Melville S., Reitter C., Melville S., Melville S. Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length. Genome Res. 2006;16:1109–1118. doi: 10.1101/gr.5147406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillo N., Carrington M., Carrington M. The origin of the serum resistance associated (SRA) gene and a model of the structure of the SRA polypeptide from Trypanosoma brucei rhodesiense. Mol. Biochem. Parasitol. 2003;127:79–84. doi: 10.1016/s0166-6851(02)00306-7. [DOI] [PubMed] [Google Scholar]
- Capbern A., Giroud C., Baltz T., Mattern P., Giroud C., Baltz T., Mattern P., Baltz T., Mattern P., Mattern P. Trypanosoma equiperdum: Etude des variations antigeniques au cours de la trypanosomose experimentale du lapin. Exp. Parasitol. 1977;42:6–13. doi: 10.1016/0014-4894(77)90055-8. [DOI] [PubMed] [Google Scholar]
- Carrington M., Boothroyd J., Boothroyd J. Implications of conserved structural motifs in disparate trypanosome surface proteins. Mol. Biochem. Parasitol. 1996;81:119–126. doi: 10.1016/0166-6851(96)02706-5. [DOI] [PubMed] [Google Scholar]
- Carrington M., Miller N., Blum M., Roditi I., Wiley D., Turner M., Miller N., Blum M., Roditi I., Wiley D., Turner M., Blum M., Roditi I., Wiley D., Turner M., Roditi I., Wiley D., Turner M., Wiley D., Turner M., Turner M. Variant specific glycoprotein of Trypanosoma brucei consists of two domains each having an independently conserved pattern of cysteine residues. J. Mol. Biol. 1991;221:823–835. doi: 10.1016/0022-2836(91)80178-w. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay A., Jones N.G., Nietlispach D., Nielsen P.R., Voorheis H.P., Mott H.R., Carrington M., Jones N.G., Nietlispach D., Nielsen P.R., Voorheis H.P., Mott H.R., Carrington M., Nietlispach D., Nielsen P.R., Voorheis H.P., Mott H.R., Carrington M., Nielsen P.R., Voorheis H.P., Mott H.R., Carrington M., Voorheis H.P., Mott H.R., Carrington M., Mott H.R., Carrington M., Carrington M. Structure of the C-terminal domain from Trypanosoma brucei variant surface glycoprotein MITat1.2. J. Biol. Chem. 2005;280:7228–7235. doi: 10.1074/jbc.M410787200. [DOI] [PubMed] [Google Scholar]
- Donelson J.E. Antigenic variation and the African trypanosome genome. Acta Trop. 2003;85:391–404. doi: 10.1016/s0001-706x(02)00237-1. [DOI] [PubMed] [Google Scholar]
- Fairhead C., Dujon B., Dujon B. Structure of Kluyveromyces lactis subtelomeres: Duplications and gene content. FEM. Yeast Res. 2006;6:428–441. doi: 10.1111/j.1567-1364.2006.00033.x. [DOI] [PubMed] [Google Scholar]
- Fischer G., Rocha E.P., Brunet F., Vergassola M., Dujon B., Rocha E.P., Brunet F., Vergassola M., Dujon B., Brunet F., Vergassola M., Dujon B., Vergassola M., Dujon B., Dujon B. Highly variable rates of genome rearrangements between hemiascomycetous yeast lineages. PLoS Genet. 2006;2:e32. doi: 10.1371/journal.pgen.0020032.eor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch A.C., Borst P., Van den Burg J., Borst P., Van den Burg J., Van den Burg J. Rapid evolution of genes coding for variant surface glycoproteins in trypanosomes. Gene. 1982;17:197–211. doi: 10.1016/0378-1119(82)90073-7. [DOI] [PubMed] [Google Scholar]
- Futse J.E., Brayton K.A., Knowles D.P., Palmer G.H., Brayton K.A., Knowles D.P., Palmer G.H., Knowles D.P., Palmer G.H., Palmer G.H. Structural basis for segmental gene conversion in generation of Anaplasma marginale outer membrane protein variants. Mol. Microbiol. 2005;57:212–221. doi: 10.1111/j.1365-2958.2005.04670.x. [DOI] [PubMed] [Google Scholar]
- Hsia R., Beals T., Boothroyd J.C., Beals T., Boothroyd J.C., Boothroyd J.C. Use of chimeric recombinant polypeptides to analyse conformational, surface epitopes on trypanosome variant surface glycoproteins. Mol. Microbiol. 1996;19:53–63. doi: 10.1046/j.1365-2958.1996.351878.x. [DOI] [PubMed] [Google Scholar]
- Hutchinson O.C., Smith W., Jones N.G., Chattopadhyay A., Welburn S.C., Carrington M., Smith W., Jones N.G., Chattopadhyay A., Welburn S.C., Carrington M., Jones N.G., Chattopadhyay A., Welburn S.C., Carrington M., Chattopadhyay A., Welburn S.C., Carrington M., Welburn S.C., Carrington M., Carrington M. VSG structure: Similar N-terminal domains can form functional VSGs with different types of C-terminal domain. Mol. Biochem. Parasitol. 2003;130:127–131. doi: 10.1016/s0166-6851(03)00144-0. [DOI] [PubMed] [Google Scholar]
- Kamper S.M., Barbet A.F., Barbet A.F. Surface epitope variation via mosaic gene formation is potential key to long-term survival of Trypanosoma brucei. Mol. Biochem. Parasitol. 1992;53:33–44. doi: 10.1016/0166-6851(92)90004-4. [DOI] [PubMed] [Google Scholar]
- Kazazian H.H. Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Koszul R., Caburet S., Dujon B., Fischer G., Caburet S., Dujon B., Fischer G., Dujon B., Fischer G., Fischer G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004;23:234–243. doi: 10.1038/sj.emboj.7600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Dujon B., Fischer G., Dujon B., Fischer G., Fischer G. Stability of large segmental duplications in the yeast genome. Genetics. 2006;172:2211–2222. doi: 10.1534/genetics.105.048058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou E.V., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J., Fan Y., Friedman C., Young J.M., Trask B.J., Friedman C., Young J.M., Trask B.J., Young J.M., Trask B.J., Trask B.J. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A.Y.C., Michels P.A.M., Bernards A., Borst P., Michels P.A.M., Bernards A., Borst P., Bernards A., Borst P., Borst P. Trypanosome variant surface glycoprotein genes expressed early in infection. J. Mol. Biol. 1985;182:383–396. doi: 10.1016/0022-2836(85)90198-6. [DOI] [PubMed] [Google Scholar]
- Lynch M., Katju V., Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004;20:544–549. doi: 10.1016/j.tig.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Lythgoe K.A., Morrison L.J., Read A.F., Barry J.D., Morrison L.J., Read A.F., Barry J.D., Read A.F., Barry J.D., Barry J.D. Parasite-intrinsic factors can explain ordered progression of trypanosome antigenic variation. Proc. Natl. Acad. Sci. 2007;104:8095–8100. doi: 10.1073/pnas.0606206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello L., Barry J.D., Barry J.D. From silent genes to noisy populations-dialogue between the genotype and phenotypes of antigenic variation. J. Eukaryot. Microbiol. 2007;54:14–17. doi: 10.1111/j.1550-7408.2006.00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch R., Rudenko G., Borst P., Rudenko G., Borst P., Borst P. Gene conversions mediating antigenic variation in Trypanosoma brucei can occur in variant surface glycoprotein expression sites lacking 70 base-pair repeat sequences. Mol. Cell. Biol. 1997;17:833–843. doi: 10.1128/mcb.17.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville S.E., Leech V., Gerrard C.S., Tait A., Blackwell J.M., Leech V., Gerrard C.S., Tait A., Blackwell J.M., Gerrard C.S., Tait A., Blackwell J.M., Tait A., Blackwell J.M., Blackwell J.M. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol. Biochem. Parasitol. 1998;94:155–173. doi: 10.1016/s0166-6851(98)00054-1. [DOI] [PubMed] [Google Scholar]
- Michels P.A.M., Liu A.Y.C., Bernards A., Sloof P., Vanderbijl M.M.W., Schinkel A.H., Menke H.H., Borst P., Veeneman G.H., Tromp M.C., Liu A.Y.C., Bernards A., Sloof P., Vanderbijl M.M.W., Schinkel A.H., Menke H.H., Borst P., Veeneman G.H., Tromp M.C., Bernards A., Sloof P., Vanderbijl M.M.W., Schinkel A.H., Menke H.H., Borst P., Veeneman G.H., Tromp M.C., Sloof P., Vanderbijl M.M.W., Schinkel A.H., Menke H.H., Borst P., Veeneman G.H., Tromp M.C., Vanderbijl M.M.W., Schinkel A.H., Menke H.H., Borst P., Veeneman G.H., Tromp M.C., Schinkel A.H., Menke H.H., Borst P., Veeneman G.H., Tromp M.C., Menke H.H., Borst P., Veeneman G.H., Tromp M.C., Borst P., Veeneman G.H., Tromp M.C., Veeneman G.H., Tromp M.C., Tromp M.C., et al. Activation of the genes for variant surface glycoprotein-117 and glycoprotein-118 in Trypanosoma brucei. J. Mol. Biol. 1983;166:537–556. doi: 10.1016/s0022-2836(83)80283-6. [DOI] [PubMed] [Google Scholar]
- Miller E.N., Allan L.M., Turner M.J., Allan L.M., Turner M.J., Turner M.J. Topological analysis of antigenic determinants on a variant surface glycoprotein of Trypanosoma brucei. Mol. Biochem. Parasitol. 1984;13:67–81. doi: 10.1016/0166-6851(84)90102-6. [DOI] [PubMed] [Google Scholar]
- Morrison L.J., Majiwa P., Read A.F., Barry J.D., Majiwa P., Read A.F., Barry J.D., Read A.F., Barry J.D., Barry J.D. Probabilistic order in antigenic variation of Trypanosoma brucei. Int. J. Parasitol. 2005;35:961–972. doi: 10.1016/j.ijpara.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Pays E. Pseudogenes, chimaeric genes and the timing of antigen variation in African trypanosomes. Trends Genet. 1989;5:389–391. doi: 10.1016/0168-9525(89)90181-9. [DOI] [PubMed] [Google Scholar]
- Pays E. Regulation of antigen gene expression in Trypanosoma brucei. Trends Parasitol. 2005;21:517–520. doi: 10.1016/j.pt.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Pays E., Houard S., Pays A., Van Assel S., Dupont F., Aerts D., Huet-Duvillier G., Gomes V., Richet C., Degand P., Houard S., Pays A., Van Assel S., Dupont F., Aerts D., Huet-Duvillier G., Gomes V., Richet C., Degand P., Pays A., Van Assel S., Dupont F., Aerts D., Huet-Duvillier G., Gomes V., Richet C., Degand P., Van Assel S., Dupont F., Aerts D., Huet-Duvillier G., Gomes V., Richet C., Degand P., Dupont F., Aerts D., Huet-Duvillier G., Gomes V., Richet C., Degand P., Aerts D., Huet-Duvillier G., Gomes V., Richet C., Degand P., Huet-Duvillier G., Gomes V., Richet C., Degand P., Gomes V., Richet C., Degand P., Richet C., Degand P., Degand P., et al. Trypanosoma brucei: The extent of conversion in antigen genes may be related to the DNA coding specificity. Cell. 1985;42:821–829. doi: 10.1016/0092-8674(85)90278-8. [DOI] [PubMed] [Google Scholar]
- Robinson N.P., Burman N., Melville S.E., Barry J.D., Burman N., Melville S.E., Barry J.D., Melville S.E., Barry J.D., Barry J.D. Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol. Cell. Biol. 1999;19:5839–5846. doi: 10.1128/mcb.19.9.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C., Jacquemot C., Giroud C., Bringaud F., Eisen H., Baltz T., Jacquemot C., Giroud C., Bringaud F., Eisen H., Baltz T., Giroud C., Bringaud F., Eisen H., Baltz T., Bringaud F., Eisen H., Baltz T., Eisen H., Baltz T., Baltz T. Antigenic variation in Trypanosoma equiperdum. Res. Microbiol. 1991;142:725–730. doi: 10.1016/0923-2508(91)90087-q. [DOI] [PubMed] [Google Scholar]
- Salmon D., Hanocq-Quertier J., Paturiaux-Hanocq F., Pays A., Tebabi P., Nolan D.P., Michel A., Pays E., Hanocq-Quertier J., Paturiaux-Hanocq F., Pays A., Tebabi P., Nolan D.P., Michel A., Pays E., Paturiaux-Hanocq F., Pays A., Tebabi P., Nolan D.P., Michel A., Pays E., Pays A., Tebabi P., Nolan D.P., Michel A., Pays E., Tebabi P., Nolan D.P., Michel A., Pays E., Nolan D.P., Michel A., Pays E., Michel A., Pays E., Pays E. Characterization of the ligand-binding site of the transferrin receptor in Trypanosoma brucei demonstrates a structural relationship with the N-terminal domain of the variant surface glycoprotein. EMBO J. 1997;16:7272–7278. doi: 10.1093/emboj/16.24.7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.E., Rudenko G., Rudenko G. Switching trypanosome coats: What's in the wardrobe? Trends Genet. 2006;22:614–620. doi: 10.1016/j.tig.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Thon G., Baltz T., Giroud C., Eisen H., Baltz T., Giroud C., Eisen H., Giroud C., Eisen H., Eisen H. Trypanosome variable surface glycoproteins: Composite genes and order of expression. Genes & Dev. 1990;4:1374–1383. doi: 10.1101/gad.4.8.1374. [DOI] [PubMed] [Google Scholar]
- Van Meirvenne N., Magnus E., Vervoort T., Magnus E., Vervoort T., Vervoort T. Comparisons of variable antigenic types produced by trypanosome strains of the subgenus Trypanozoon. Ann. Soc. Belg. Med. Trop. 1977;57:409–423. [PubMed] [Google Scholar]
- Williams R.O., Young J.R., Majiwa P.A.O., Young J.R., Majiwa P.A.O., Majiwa P.A.O. Genomic environment of T. brucei VSG genes: Presence of a minichromosome. Nature. 1982;299:417–421. doi: 10.1038/299417a0. [DOI] [PubMed] [Google Scholar]