Abstract
While bacterial genome annotations have significantly improved in recent years, techniques for bacterial proteome annotation (including post-translational chemical modifications, signal peptides, proteolytic events, etc.) are still in their infancy. At the same time, the number of sequenced bacterial genomes is rising sharply, far outpacing our ability to validate the predicted genes, let alone annotate bacterial proteomes. In this study, we use tandem mass spectrometry (MS/MS) to annotate the proteome of Shewanella oneidensis MR-1, an important microbe for bioremediation. In particular, we provide the first comprehensive map of post-translational modifications in a bacterial genome, including a large number of chemical modifications, signal peptide cleavages, and cleavages of N-terminal methionine residues. We also detect multiple genes that were missed or assigned incorrect start positions by gene prediction programs, and suggest corrections to improve the gene annotation. This study demonstrates that complementing every genome sequencing project by an MS/MS project would significantly improve both genome and proteome annotations for a reasonable cost.
The number of sequenced bacterial genomes has been increasing rapidly, with 70 out of 250 sequenced bacterial genomes finished in the last year alone (Benson et al. 2006). Annotation of these genomes continues to be a challenging task, requiring both automated analysis and manual curation. This challenge is greatly magnified in recent meta-genomic projects, which seek to sample DNA from the environment (Venter et al. 2004; http://www.sorcerer2expedition.org).
In this article, we demonstrate the use of liquid chromatography–coupled mass spectrometry (LC-MS/MS) for both proteomic and genomic annotations of bacteria. While gene finding in prokaryotes has significantly improved, prediction of short genes, annotation of genes with unusual codon usage, as well as accurate prediction of start codons remains a challenge. Moreover, biologically important problems of annotating post-translational modifications (chemical modifications, signal peptides, proteolytic events), unusual stop codons, programmed frameshifts, quantifying protein expression, etc., still cannot be solved with genomic techniques. MS/MS is a key technology for proteomic analysis (Aebersold and Mann 2003; Jensen 2006) that fragments individual peptides and uses the resulting tandem mass-spectra as a “fingerprint” to identify the protein of origin. Chemical modifications at specific residues are detectable as a change in the fragmentation pattern (e.g., shifts in the masses of fragments containing the modification). These modifications are often critical to protein function, as in the case of regulating binding partners, subcellular localization, the three-dimensional structure of the protein or in modifying activity of a catalytic site. Protein modifications in prokaryotes are of great biological interest but are not yet well understood.
The idea of querying MS/MS data set against a genome to identify protein coding genes has been used earlier in different settings (Yates et al. 1995; Kuster et al. 2001; Oshiro et al. 2002; Fermin et al. 2006). Bacterial genomes, with a simple gene structure, are a particularly attractive target for such methods. The identified peptides validate the predicted genes, correct erroneous gene annotations, and reveal some completely missed genes. Church and colleagues used proteomic data for genome analysis of relatively small bacterium, Mycoplasma pneumoniae (Jaffe et al. 2004a), and later on the newly sequenced Mycoplasma mobile, in which 26 genes were predicted exclusively based on proteomic data (Jaffe et al. 2004b). Similar efforts have been made for other bacterial genomes (Kalume et al. 2005; Wang et al. 2005). Nevertheless, many significant technological challenges remain in using MS/MS for gene annotation, post-translational (proteolytic) processing, and modifications. For example, only one post-translational event was discovered in the whole proteome of M. pneumoniae (Jaffe et al. 2004a). In Jaffe et al. (2004b), the investigators expressed disappointment on not being able to find any post-translational modification, in spite of their ability to detect expression of 88% of all proteins with high residue coverage. This exemplifies the difficulty in studying post-translational events in large-scale studies.
This study further develops the methods of proteogenomic annotations from Yates et al. (1995), Kuster et al. (2001), Oshiro et al. (2002), Jaffe et al. (2004a, b), Kalume et al. (2005), Wang et al. (2005), and Fermin et al. (2006) and provides the first comprehensive analysis of post-translational modifications in complete genomes. Until very recently, such analysis was not feasible since the whole genome search for mutated and modified peptides was prohibitively time-consuming. The search becomes particularly time-consuming in the case of nontryptic peptides that enable identification of proteolytic events (see below). Moreover, our analysis revealed at least 25 modification types present in the sample, a significantly larger number than the existing database search tools can practically handle under realistic parameters. We capitalize on the recently developed database filtration tool Inspect (Tanner et al. 2005) and the blind database search tool MS-Alignment (Tsur et al. 2005) to overcome these difficulties.
Here we use the Gram-negative bacterium Shewanella oneidensis MR-1 as a test case for our LC-MS/MS–based approach to proteome annotation. It is an aero-tolerant anaerobe able to reduce heavy metal ions and remove them from solution, making it a potential agent for bioremediation (Nealson et al. 2002). The genome of this strain was sequenced in 2002 (Heidelberg et al. 2002) with 4931 predicted protein-coding genes, of which only a few have been experimentally verified. The revised number of genes now stands at 4928 according to the TIGR Comprehensive Microbial Resource (Peterson et al. 2001). We will refer to these as TIGR genes in this article. The organism has been extensively studied by the Shewanella Federation (http://shewanella.org/). The difficulties in gene prediction are illustrated by the fact that just a year after S. oneidensis was first annotated, it was reannotated by Daraselia et al. (2003). Some recent studies have attempted to use LC-MS/MS and LC-MS to further improve the annotations of S. oneidensis (Romine et al. 2004; Elias et al. 2005, 2006). Recently, Kolker et al. (2005) used microarray and MS data to analyze the expression of predicted genes in S. oneidensis.
We remark that even if bacterial gene predictions were 100% accurate, they would still provide incorrect protein sequences for ∼50% of bacterial genes! It is estimated that a single post-translational modification (N-terminal methionine cleavage, or NME) alters roughly half of proteins in Escherichia coli. NME is the process of cleaving N-terminal methionine residue by methionyl amino peptidase (MAP) or amino peptidase P (AmpP) from a number of cytosolic proteins. NME has important implications for protein half-life (Tobias et al. 1991), and knowledge of NME is crucial for many applications in food safety, infection diagnostics, and counter-terrorism (it improves the quality of MS-based microorganism detection by an order of magnitude; Fenselau and Demirev 2001). The role of NME remains poorly understood, but the process is recognized to be the major source of N-terminal amino acid diversity. The recognition rules for NME remain elusive, resulting in a number of conflicting studies (Link et al. 1997; Wasinger and Humphery-Smith 1998; Fenselau and Demirev 2001; Frottin et al. 2006). Frottin et al. (2006) recently estimated that the existing ambiguities in NME recognition rules make reliable proteome annotation difficult for ∼30% of bacterial proteins. This renders the production of recombinant proteins of therapeutic interest risky, given the high anti-genicity of the N terminus if incorrectly processed, the problem originally encountered in the production of human hemoglobin (Olson et al. 1981; Ben-Bassat et al. 1987). We therefore argue that MS studies are necessary to complement the existing gene prediction tools by accurate annotations of NME and many other post-translational modifications.
In this work we exploit a data set containing 14.5 million tandem mass spectra for S. oneidensis. These data include samples from 17 cell culture conditions and comprise the largest LC-MS/MS data set ever reported for a bacterium. Using very conservative cutoffs for peptide identifications, we confirm the protein expression of 1992 out of 4928 predicted TIGR genes. We correct or redefine gene boundaries of 38 genes, eight of which were not included in the TIGR predictions, and provide evidence for expression of 13 genes previously annotated as pseudogenes.
The peptides identified by MS/MS give important insights into post-translational processing, including proteolytic events. For example, cleavage of initial methionine was observed with specificity similar to that observed in vitro with bacterial enzymes. Our analysis provides significantly refined annotations for secreted proteins by confirming signal peptide processing for 94 proteins while rejecting imprecise or incorrect predictions by existing software for 119 proteins. We further performed blind search for modifications, resulting in the comprehensive map of modifications for a bacterial proteome for the first time. We argue that complementing a sequencing project by a LC-MS/MS projects critically improves both genome and proteome annotations. The data used in this study are available at http://peptide.ucsd.edu/ShewanellaOneidensis/ and http://ober-proteomics.pnl.gov/data, and the software tools, including Inspect, MS-Alignment and other scripts, are available at http://peptide.ucsd.edu/.
Results
Peptide identification
As described in the Methods section, we searched the spectra against the six-frame translation of the Shewanella genome. Some previous proteogenomic studies (Jaffe et al. 2004a, b) did not attempt to measure the rate of false peptide identification, thus raising doubts about the reliability of new gene annotations. To quantify the false discovery rate of peptide identifications, we searched all spectra against a reversed sequence database. We selected a match score cutoff that limits the number of peptides annotated in the reverse database to 5% of the number of peptides identified in the valid database (1417 distinct peptide identifications in the reversed database, compared with 28,377 peptide identifications in the valid database). Peptide matches of length less than eight amino acids were discarded to minimize the number of false positive predictions. In total, 1.4 million spectra were annotated while searching 14.5 million spectra against the forward database. In contrast, only 14,523 spectra had scores above the threshold in the reversed database, giving a spectrum-level false discovery rate below 1% on the forward database search.
Peptide coverage
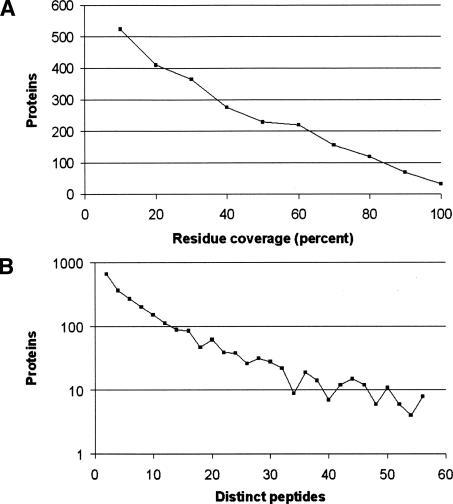
Some peptides are encoded at multiple locations within the six-frame translation. Therefore, the identified peptide list was filtered to 27,946 peptides that map to unique locations in the translated genome, removing this possible source of ambiguity. We compare our findings with genes predicted de novo by GeneMark (Besemer and Borodovsky 2005) and with the curated TIGR annotations (Peterson et al. 2001). We note that the fully automated gene predictions from GeneMark include only 4692 genes. Table 1 analyzes the locations of the identified peptides relative to the positions of the TIGR genes. Of the 331 peptides not covered by a TIGR gene, 126 were covered by GeneMark predictions. This demonstrates the utility of LC-MS/MS data in resolving discrepancies between various gene prediction tools. For each TIGR gene, we looked at all identified peptides within the gene and determined the coverage (fraction of protein residues covered by the identified peptides). Figure 1 shows an example of a protein whose entire amino acid sequence was covered by identified peptides.
Table 1.
Comparison of peptides identified by MS/MS analysis with TIGR and GeneMark genes
Some peptides are covered by a gene but in a different reading frame compared with the annotated genes. These may represent a programmed frameshift or an insertion/deletion type sequencing error in the genomic sequence.
Figure 1.
The ribosomal protein L31 (SO_4120) is entirely covered by identified peptides (all peptides longer than six amino acids are shown). The protein sequence is shown at the top in red, and the identified peptides are shown below in blue. Tryptic peptides are shown in bold.
Figure 2, A and B gives the distribution of number of identified peptides and the residue coverage for all TIGR genes. We consider protein expression to be confirmed if at least two peptides were identified from that protein (in the reversed database search, we observe only 51 pairs of peptides occurring within 200 bp of each other in the same reading frame). We verify expression of 1992 proteins in this way, while observing 402 proteins with a single identified peptide. The naïve way to estimate the number of single-hit proteins that are correct is to use the peptide false discovery rate (5% of 402 is ∼20). However, this is overly optimistic. The false discovery rate for multiple-hit proteins is lower than average, therefore the false discovery rate for single-hit proteins will be higher than average. Given an estimated 1397 false peptides (5% of 27,946), if we assume they are randomly distributed throughout the database, 98 of them should hit unconfirmed proteins. Therefore, as a conservative approximation, we estimate that 304 of the single-hit proteins were correctly identified. Thus we conclude that at least 47% of the predicted proteome (2296 TIGR genes) is expressed at a detectable level in this series of experiments.
Figure 2.
(A) Distribution of the number of identified peptides observed for TIGR genes. Protein counts are plotted on a logarithmic scale. (B) Distribution of the residue coverage of TIGR genes by identified peptides. A total of 102 genes had coverage of ≥90%. Genes were grouped into percentage bins of size 10 percentage points based on their coverage.
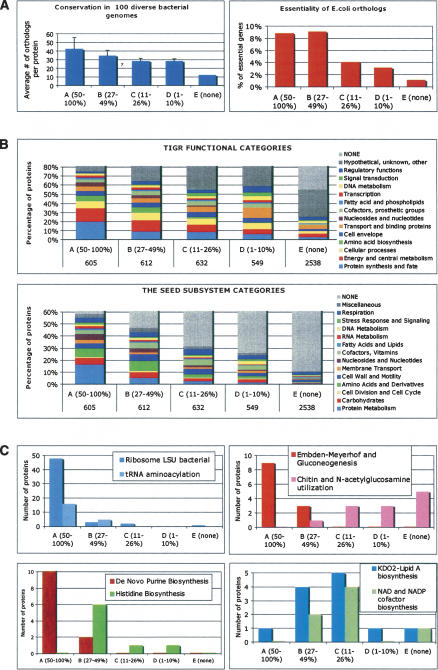
A distribution of the observed coverage of individual proteins in the expressed proteome by MS-detected peptides could be perceived as somewhat random with most biases coming from technical factors such as efficiency of extraction and existence of poorly detectable peptides (Tang et al. 2006). On the other hand, one may expect this distribution to be substantially affected by the relative protein abundance reflecting cell physiology under given experimental conditions (Lu et al. 2006). The latter hypothesis, if confirmed, would open new opportunities to exploring cellular pathways and networks. To validate this hypothesis, we used several indirect tests, as the proteome-scale data on protein abundances are not readily available. We examined whether there is any correlation between coverage and (1) conservation of orthologs in a range of sequenced microbial genomes, (2) essentiality of such orthologs in a model system of E. coli (Baba et al. 2006), and (3) functional categories inferred by gene annotations and pathway reconstruction in public genomic resources TIGR (http://cmr.tigr.org/) and SEED (http://theseed.uchicago.edu/FIG/index.cgi). Whereas none of these three features should correlate with technological constraints, one may expect them to reflect relative protein abundances. The latter expectation is quite straightforward for functional categories or pathways and may be less obvious for conservation and essentiality. Conservation and essentiality are known to correlate with each other (for discussion, see Gerdes et al. 2003; Koonin 2003), implicating proteins that constitute a Core of Life. It is plausible that a substantial fraction of these proteins would be expressed at appreciable (and even high) level in a variety of growth conditions. Particularly in the context of a global survey of proteome data acquired at different experimental conditions, conserved and essential proteins may prevail due to the universal nature of their expression.
For the purpose of this analysis S. oneidensis proteome was split to five arbitrary coverage groups: the first four groups (A–D) of a similar size (605, 612, 632, and 549 proteins, respectively) with a range of protein coverage 50%–100%, 27%–49%, 11%–26%, and 1%–10% (for details, see Supplemental Table S1A). The last group E contained proteins not covered by any peptide and represented more than half of all predicted proteins. The results of this analysis are summarized in Figure 3. One may notice that proteins in group A (coverage >50%) are on average more broadly conserved (as computed for a set of 100 diverse bacterial genomes) than proteins with lower coverage. However, the difference in conservation is particularly significant when compared with group E (no coverage). As no genome-scale essentiality data are available for S. oneidensis, we used the data published for E. coli, a close model organism within the same order of Proteobacteria. A projection of these data to the respective orthologs in S. oneidensis coverage groups revealed largely similar trends (see Fig. 3A). While we use E. coli for comparisons, we note that there may be some differences in the gene usage in a S. oneidensis pathway, such as those observed in carbon metabolism (Serres and Riley 2006).
Figure 3.
Correlations between coverage of individual proteins by MS-peptides and their biological features deduced from comparative genomics. (A) Conservation and essentiality. Conservation index for every protein was computed as a number of putative orthologs within a set of ∼100 diverse bacterial genomes (list provided in Supplemental Table S1B). A bar diagram on the left panel shows the corresponding values averaged for each coverage group. The right panel shows the fraction of E. coli orthologs (456, 419, 363, 322, and 738 in groups A–E, respectively) that were deemed essential in the published study (Baba et al. 2006) plotted for each coverage group. (B) Functional categories. The upper panel shows a distribution of proteins within each coverage group by main functional categories according to TIGR annotations (to avoid redundancy only one category was chosen for each protein). In the lower panel, a similar distribution reflects inclusion of proteins in a collection of categorized subsystems (pathways) in The SEED database (restricted to one subsystem per protein). (C) Examples of individual subsystems (pathways). A distribution of proteins between coverage groups is illustrated for eight subsystems selected from the six major functional categories (protein metabolism; carbohydrates; nucleosides and nucleotides; amino acids and derivatives; fatty acids and lipids; cofactors and vitamins).
To roughly assess a functional content within the same coverage groups we used two types of functional categories: (1) assigned in a TIGR database (main categories), and (2) deduced from a categorized collection of annotated subsystems (pathways) provided by SEED genomic resource (Overbeek et al. 2005). While differing in details of classification and coverage (TIGR nonhypothetical categories cover a larger fraction of proteome, whereas SEED collection reflects a more detailed reconstruction of the major pathways), both graphs in Figure 3B allowed us to make similar observations. The difference in the overall functional content of coverage groups (enrichment by proteins with defined functional roles) is more pronounced in case of SEED subsystem collection, consistent with its bias toward well-studied aspects of the core cellular pathways. The group E is especially enriched with proteins of unknown functions, in congruence with the results obtained by Wang et al. (2005), which may be partially due to a relatively higher content of possible pseudogenes. However, this group likely contains a substantial fraction of genuine protein encoding genes that are expressed at very low level and/or in specific environmental conditions implementing a number of unique (and yet unexplored) aspects of this organism’s life style. Not surprisingly, respective genes would be less conserved and less essential (as illustrated in Fig. 3A).
Some of the individual functional categories (e.g., those related to metabolism of proteins [pale blue], amino acids [green], and nucleotides [brown]) display consistent downward trends when compared between all coverage groups in both graphs in Figure 3B. However, the actual dependencies are stronger and more informative when analyzed for individual subsystems (pathways). Several examples are illustrated in Figure 3C. One may notice a very sharp distribution of protein components of a (1) ribosome, (2) tRNA aminoacylation machinery, (3) purine biosynthesis, and (4) central carbon metabolism, skewed toward the highest coverage group A. An example of histidine biosynthesis illustrates a shift toward moderate coverage maximized in group B without a single component in group A (whereas the biosynthetic pathways of some other amino acids, e.g., aromatic and branch-chained, are predominantly within group A). Even bigger shift to lower coverage (maximized in group C) is illustrated by the biosynthesis of NAG cofactor and Lipid A, pathways that are expected to be relatively less active. A subsystem involved with the utilization of chitin and N-acetyglucosamine provides a remarkable example of correlation between the observed coverage and expected phenotype. Most of the genes recently implicated in the respective regulon in S. oneidensis (Yang et al. 2006) display very low or no coverage, consistent with the notion that this nutrient, highly abundant in the natural marine habitat, was not a part of growth media used in this study. Notably, a predicted transcriptional repressor of this regulon (SO_3516) was expressed at an appreciable level (16% coverage).
Overall, this preliminary analysis provides us with substantial evidence confirming the hypothesis that protein coverage by MS-detected peptides may provide a reasonable approximation of relative abundance of proteins at the whole proteome scale. Further studies aimed to validate this hypothesis and to apply this principle to assess differential expression of genes and pathways as a function of growth conditions are currently in progress. One should keep in mind that pathways may be regulated at various levels, and different genes may have different levels of correlation with the pathway expression. It must be noted that while sequence coverage correlates with protein abundance in general, the sequence coverage of any specific protein may also be significantly affected by its physicochemical properties. For example, we looked carefully at various transporters and found that usually the soluble components and outer membrane proteins are observed, while the integral membrane proteins are underrepresented. Developing models to take the effect of these physicochemical properties into consideration is expected to be a part of future studies in this direction.
Improving genome annotation
Detection and mapping of multiple peptides to a single gene is evidence that it encodes a protein that is expressed. Similarly, multiple matches to a genomic region outside the boundary of genes can be used to detect new genes missed during genome annotation or to suggest that gene boundaries should be expanded.
To detect such cases, we examined the identified peptides falling outside the TIGR genes. Such peptides are combined into putative coding segments if they are located within 200 nucleotides from each other, with compatible reading frame. These coding segments point to new genes and extensions of the TIGR genes. By analyzing the location of these segments, we identified eight new genes and extended the 5′ boundaries for 30 genes (see Supplemental Table S2A,B). Of the eight new genes, all but two had been previously suggested in a second version of the annotation of the MR-1 genome (Daraselia et al. 2003). The remaining two, which we have designated SO_4799 and SO_A0180, were similar to proteins found in other bacteria. Furthermore, our data provide the necessary information required to validate that these genes, six of which would otherwise have been annotated as hypothetical, encode proteins.
Based on comparative protein sequence alignment, extensions of the 5′ ends of all 30 genes were consistent with predicted N termini of proteins identified in other bacteria, including other species of Shewanella. Figure 4 illustrates an example of how peptides were used to identify an earlier gene start position for SO_1175. Five frequently observed peptides (Fig. 4A) align upstream to and within the same reading frame as the original SO_1175 gene (Fig. 4B). The proposed new gene start is further validated by the detection of four peptides that span the original suggested start codon. Had this GTG codon occurred at the translational start position, it would have been translated as a methionine rather than a valine (Sussman et al. 1996). Furthermore, the proposed new gene start is the preferred ATG codon and is found downstream of a Shine-Dalgarno like site (Shine and Dalgarno 1974). As added proof, the alignment of SO_1175 with similar proteins deduced from other Shewanella genome sequences (Fig. 4C) is consistent with the proposed new 5′ extension of the gene.
Figure 4.
(A) Alignment of identified peptides (colored blue) and the hypothetical TIGR protein SO_1175 (colored red). Numbers on the right show the number of times the peptide was identified in MS/MS spectra (spectral count). The start codon, at position 1,218,574 on the forward strand of the chromosome, is normally read as valine. These peptide identifications demonstrate that translation begins upstream of the annotated start site. (B) Alignment of identified peptides (blue) with the nucleotide sequence of N-terminal region of SO_1175, relative to proposed new start codon (single arrow) and to the original proposed start codon (double arrow). A Shine Dalgarno-like site (underlined) is found upstream only to the proposed new start site. (C) Multiple sequence alignment of SO_1175 of S. oneidensis (red) with orthologs in other Shewanella strains.
Figure 5 illustrates how peptides upstream of SO_2300, which encodes translation initiation factor IF-3 (infC) led to the prediction of a nontraditional start codon. Three peptides map upstream to and within the same reading frame as SO_2300 (Fig. 5A). As in the previous example, one of these peptides spans the original predicted GTG start codon, validating that it is translated as a valine rather than a methionine. However, none of the more common ATG, GTG, or TTG start codons were found upstream to the region spanned by these three peptides. A survey of available literature on translation initiation factor IF-3 revealed that the infC gene from a variety of other bacteria is initiated at a rare ATT start codon, thereby serving as a basis for autoregulation (Sacerdot et al. 1982; Pon et al. 1989; Hu et al. 1993; Liveris et al. 1993). While no ATT codon was found, we speculate that SO_2300 starts at an ATA codon (Fig. 5B). ATA is known to function as a rare translation initiator (Sussman et al. 1996; Schoenhals et al. 1998) and, in this case, is adjacent to a strong ribosome binding signal. Mutagenesis of the E. coli infC ATT start to ATA (but not ATG) was shown not to affect its functionality (Sacerdot et al. 1996), suggesting that the same may be true for the MR-1 infC. One final line of evidence is shown in Figure 5C, where results of TBLASTN analysis reveal conservation in sequences spanning the entire proposed N-terminal extension of 11 Shewanella species as well as a large portion of this region in more distantly related bacteria.
Figure 5.
(A) Alignment of identified peptides (blue) with the intergenic region between TIGR proteins SO_2299 and SO_2300. The starred positions indicate the last codon of SO_2299 (left, chromosome position 2,412,384) and the first codon of SO_2300 (right, chromosome position 2,412,499). The arrow points to the newly postulated translational start site for SO_2300 (infC gene). (B) Nucleotide sequence of the chromosome region between SO_2299 and SO_2300. The first red segment is the C-terminal end of SO_2299, and the second red segment is the N-terminal region of SO_2300. The region covered by the three identified peptides is underlined, and the arrow indicates our suggested start position for SO_2300. (C) A TBLASTN comparison of the proposed new S. oneidensis MR-1 infC N terminus to genome sequences for S. baltica OS155, Shewanella sp. MR-4, S. putrefaciens strains CN32, S. loihica PV-4, Sodalis glossinidius (gi 84778498), and Photobacterium profundum (gi 46913734). The original start position is indicated by the arrow.
To our surprise, we also detected peptides that mapped to 13 genes originally annotated as pseudogenes (Supplemental Table S2C). SO_0991 encodes peptide chain release factor 2, PrfB. The orthologous gene from other bacteria has been shown to undergo programmed frameshifting (Baranov et al. 2002). The occurrence of the expected recoding signals in the MR-1 prfB gene, as described in the RECODE database (Baranov et al. 2003), suggests that the seven peptides that we detected after the frameshift position are a consequence of programmed frameshifting of SO_0991. Alignments of the deduced products for two genes (SO_4538 and SO_4809) suggest that their original annotation as pseudogenes was inaccurate, since numerous orthologs of similar size were found. Alignments of proteins deduced from the remaining 10 genes were in agreement with the original suggestion that they were encoded by pseudogenes. However, examination of the trace archives available at NCBI for the MR-1 genome revealed mistakes in base calling that, when repaired, yield genes with an open reading frame of a significant length. Representative examples of the latter two cases are illustrated in Figure 6. In Supplemental Table S2, A–C, if the identification was based on only one peptide, we also provide the peptide sequence, corresponding F-Score from one representative spectrum and the precursor ion mass and charge. The corresponding DTA spectrum and a labeled image are provided in Supplemental Data S2D (files names are same as the peptide; precursor masses of spectra are in the spreadsheet PrecursorMass_onePeptideID.xls included in S2D).
Figure 6.
(A) The positioning of two identified peptides (boxed) relative to the three frame translation of the SO_0590 pseudogene and to homologous protein from Shewanella W3–18–1. The nucleotide sequence is shown on top (in red), and the three translated frames are shown below (in black), with stop codons translations shown as asterisk (*). The locus containing the extra A identified by re-evaluation of sequence trace TI|202865473 available in the NCBI trace archives is indicated by the arrow. (B) Alignment of identified peptides (blue) with the gene SO_4538, annotated as a degenerate M16 protease. Alignment is shown to homologous sequences in S. baltica OS155 and Shewanella sp. ANA-3.
Proteolytic sites
Proteolytic cleavage through cellular proteases is extremely important for many biological functions. While such cleavage is often specific and tightly regulated, protease activity in cells is relatively unexplored, primarily due to the lack of effective high-throughput technology to detect proteolytic events. Large-scale efforts are currently underway to address this technological bottleneck (e.g., at the NIH Center for Proteolytic Pathways at Burnham Institute for Medical Research). However, these approaches mainly rely on labeling protocols and thus face a number of chemical and computational challenges. We show that at least some proteolytic events can be reliably identified by label-free analysis, as illustrated below by our analysis of high-throughput MS/MS data.
Large MS/MS data sets offer an unprecedented opportunity to study in vivo cleavage specificity by looking at over-represented nontryptic peptides that may be manifestations of proteolytic events. Since all protein samples were digested with trypsin, we expect the majority of the peptide endpoints to correspond to tryptic cleavage sites (after arginine or lysine, but not before proline). Given trypsin’s high specificity (Olsen et al. 2004), it is natural to consider that nontryptic endpoints may reveal proteolytic events (Mann and Pandey 2001). We also consider the natural N and C termini of any protein as fully consistent with conventional tryptic cleavage. A peptide endpoint that is not tryptic according to either of the above definitions is considered “nontryptic.” Nontryptic endpoints suggest the possibility of a proteolytic event, either in vivo or in vitro. Our peptide identifications include 21,297 tryptic peptides (75%), 6670 peptides with one nontryptic endpoint (24%), and 409 peptides with two nontryptic endpoints (1%). However, caution is needed while analyzing peptides with nontryptic endpoints, since they can also reflect post-digestion trimming of tryptic peptides. Figure 7 shows the N-terminal portion of a well-covered TIGR protein whose first 26 amino acids are, surprisingly, not covered by any peptides. The hypothesis that these initial 26 amino acids represent a signal peptide is supported by the fact that the first two peptides mapped to the protein (starting at residue 26A) have a nontryptic N terminus. However, it is not the only nontryptic endpoint observed for this peptide; for example, the peptide SIGTDTLLQIK is also nontryptic. Below, we present an approach to distinguishing between proteolytic events and post-digestion trimming.
Figure 7.
Peptides from the N-terminal portion of conserved hypothetical protein SO_3842. The peptide breakage before the starred residue is produced when the signal peptide is cleaved and degraded. The other nontryptic peptides are properly contained in observed tryptic peptides and are most likely generated by post-digestion breakup. The N-terminal “ladder” observed for the tryptic peptide QMSIGTDTLLQIK is a likely result of aminopeptidase-driven trimming or in-source fragmentation.
We note that 96% of nontryptic peptides fall within confirmed TIGR proteins, similar to tryptic peptides (97%). Since confirmed TIGR proteins make up only 7% of the whole genome database size (translated in six frames), incorrect identifications (which are randomly distributed) are unlikely to fall within confirmed proteins. Thus, we argue that our false discovery rate for nontryptic peptides is not significantly larger than that for tryptic peptides. In this section, we consider only those peptides contained within confirmed proteins.
Detecting proteolytic events via MS/MS analysis
Nontryptic peptides may arise from post-digestion breakup, due to hydrolysis (driven by endogenous or exogenous peptidases or by harsh chemical conditions in course of the sample preparation) or in-source decay (Olsen et al. 2004). Of the 7079 nontryptic peptides, 5474 (77%) are properly contained in a longer observed tryptic peptide, and 1605 (23%) are not. It is likely that the majority of nontryptic peptides contained within other observed peptides result from post-digestion breakup, particularly when the longer peptide is more abundant (as estimated by spectrum count), although some of them may result from the partial proteolytic processing in vivo. To examine this in further detail, we measure the distance from these nontryptic termini to the tryptic endpoint of the containing peptide (Fig. 8).
Figure 8.
Most nontryptic peptides are contained in an identified tryptic peptide. The plot gives the distribution of residue distances from a nontryptic endpoint to the endpoint in the containing tryptic peptide. The plot describes 2178 peptides with a nontryptic C terminus and 3508 peptides with a nontryptic N terminus. Surprisingly, elimination of two residues (as opposed to a single residue) from the C terminus is particularly common. Peaks at even positions (2, 4, and 6) from the C terminus may reflect peptidyl dipeptidase activity that digest two amino acids at a time; dipeptidases acting at the N terminus are not known in Shewanella.
Surprisingly, the distance from a nontryptic C terminus to the containing peptide’s tryptic endpoint is often precisely two amino acids (31% of peptides). Moreover, the plot has peaks at even distances (two, four, and six amino acids), suggesting an increased propensity to deleting two amino acids at a time. This observation may be explained by the action of a peptidyl dipeptidase (such as dcp in E. coli) (Yaron 1976), which cleaves off dipeptides from the C termini of oligopeptides. S. oneidensis indeed has two dcp orthologs: SO_3142, dcp-1 (46% identity with E. coli dcp) and SO_3564, dcp-2 (47% identity with E. coli dcp). In addition to carboxypeptidase (and dipeptidase), the observed trimming patterns point to a likely presence of aminopeptidase activity (Fig. 7).
We now focus on the 1605 nontryptic peptides that are not contained within tryptic peptides. We further reduce this set to 1372 peptides that are not contained within any other peptide and that are located within confirmed TIGR genes (such peptides are called “noncovered peptides”). While the 688 proteins containing noncovered peptides may be potential proteolytic targets, one can argue that these peptides may also represent (1) erroneous peptide identifications or (2) instances where the containing tryptic peptide does not generate any observed MS/MS spectra, perhaps due to extreme length (Tang et al. 2006). In the following section, we demonstrate that while it is a valid concern, many noncovered peptides indeed correspond to proteolytic events. To prove that this is the case, we point to the extremely nonuniform distribution of starting positions of these peptides along the protein (Fig. 9). If these peptides were artifacts, we would expect to see a relatively uniform distribution of starting positions. Instead we see two pronounced peaks at positions 2 and 20, both reflecting two well-known biological phenomena: N-terminal methionine cleavage (position 2) and signal peptide (average length of signal peptides in Gram-negative bacteria is ∼25 amino acids) (Nielsen et al. 1997; Paetzel et al. 2002). It should be noted that although our signal peptide peak is at 20, the distribution is skewed toward right, with average signal peptide length around 26, in strong agreement with the previously reported average of 25.
Figure 9.
Distribution of the N termini of all noncovered peptides, and of those which also have no upstream coverage. Two peaks are observed at two and ∼20 amino acids. These correspond to N-terminal methionine cleavage and to cleavage of signal peptides.
To focus on these two phenomena, N-terminal methionine and signal peptide cleavages, we limit our attention to proteins in which the leftmost identified peptide is a noncovered peptide, i.e., proteins with no peptide coverage upstream of the first noncovered peptide. A total of 366 N-terminal peptide endpoints are obtained accordingly.
N-terminal methionine cleavage
The N-terminal methionine residue is cleaved by MAP or AmpP from a number of cytosolic proteins. Methionine, which is important during translation, may not be required (or actually be detrimental) for the function of the protein. An earlier study (Hirel et al. 1989) measured the efficiency of cleavage between initial methionine and various second residues in vitro. This study genetically engineered and expressed 20 mutants of a gene in E. coli differing only at the second position. The expressed proteins were purified and subjected to MAP activity. Using Edman degradation to analyze the N-terminal sequences of the resultants, they showed that methionine cleavage is more efficient if the second residue has a smaller side chain.
In our analysis of noncovered peptides, we observed many peptides starting at the second residue of a protein. These peptides confirm cleavage of N-terminal methionine in 218 proteins (Supplemental Table S3A contains the list). To check whether the effect of second residue on cleavage (Hirel et al. 1989) is also seen in our data, we computed a cleavage efficiency factor for each of the 20 amino acids. For a given amino acid, we identify all peptides that contain the second residues of proteins having the particular amino acid at that position. If X is the number of such peptides that begin at residue 1 of a protein (indicating no cleavage) and Y is the number of peptides beginning at residue 2 (indicating a cleavage), the cleavage efficiency for that amino acid is defined as Y/(X + Y). The relative levels of these efficiencies of different amino acids are similar to the results observed in vitro for E. coli (Fig. 10). It appears that activity in vivo may be more specific than that seen in vitro. These observations are also in close agreement with recent results independently obtained at the Burnham Institute in the E. coli model using a novel labeling technology (G. Salvesen, pers. comm.).
Figure 10.
Fraction of peptides undergoing cleavage of N-terminal methionine, for a given second-position residue. Amino acids are arranged in increasing order by size of side chain. The in vitro data come from measurements of E. coli MAP enzyme efficiency (Hirel et al. 1989). The rates in vivo were estimated by counting the number of peptides. If X is the number of peptides that begin at residue 1 of a protein (indicating no cleavage) and Y is the number of peptides beginning at residue 2 (indicating a cleavage), the cleavage efficiency for that amino acid is defined as Y/(X + Y). Some amino acids are rarely used as the second residue of any of the TIGR genes (or GeneMark predictions). For instance, 308 protein sequences have serine at the second residue, while only nine have tryptophan. Because of this, our identifications contain relatively few N-terminal peptides for some amino acids. For the starred residues, 10 or fewer N-terminal peptides were observed with that residue at the second position.
We observed several cases of an apparent cleavage before the second methionine in proteins starting with double methionine. A comparative genomics analysis of other Shewanella strains revealed that a large portion of them (e.g., SO_4343 and SO_2364) have orthologous proteins with a single methionine, rather than a double methionine. Therefore, we speculate that many proteins starting with double methionine in TIGR may represent a misannotation of the translation start site.
We note that aside from the noncovered peptides, we observed 32 nontryptic peptides, starting at residue 2 where a peptide containing the N-terminal methionine was also observed. These peptides may be explained by partial processing by MAP activity or, more likely, by the N-terminal trimming in course of sample preparation.
Signal peptides
A signal peptide is a short N-terminal region of a protein that targets a protein for secretion or for transportation to a desired cellular location. Signal peptides are cleaved and quickly degraded to produce the mature protein sequence. The average length of a signal targeting proteins to the Sec pathway in Gram-negative bacteria is estimated to be 25 amino acids, with most signal peptides in the range from 20–30 amino acids (Nielsen et al. 1997; Paetzel et al. 2002). The distribution of starting positions of noncovered peptides has a pronounced peak at 20 amino acids, with a longer tail on right side (bigger lengths) as shown in Figure 9. Of the noncovered peptides that are not explained by N-terminal methionine cleavage, 55% start at positions 21–30. Figure 9 suggests that most peptides in 21- to 30-amino-acid window reflect signal peptides, since other 10-amino-acid-long windows have very few peptides.
While knowledge of signal peptides is important for understanding protein function, they are difficult to confirm experimentally, and computational predictions are used to fill the gap. SignalP (Bendtsen et al. 2004) is popular signal peptide prediction software which uses neural network (NN) and hidden Markov model (HMM) models of known signal peptides. Another such program, PrediSi (Hiller et al. 2004), uses a position weight matrix (PWM) to predict signal peptides. It is important to note that the bulk of protein secretion in Shewanella, as well as in other Gram-negative bacteria, occurs to the periplasmic space; therefore, the corresponding processed proteins can be experimentally observed in the whole-cell extract.
There have been some concerns regarding the quality of signal peptide predictions (Antelmann 2001) since these methods consider a generalized signal motif for all proteins and may not identify interesting cases that are limited to a few proteins. Also, these tools make predictions based on a rather small signal peptide database, since experimental data about signal peptides are limited. For example, SignalP v3 made predictions for Shewanella based on a data set of only 334 experimentally confirmed signal peptides in all Gram-negative bacteria. The number of experimentally confirmed signal peptides in Gram-positive bacteria is half as large (Bendtsen et al. 2004). It is clear that LC-MS/MS evidence can greatly increase the number of experimentally confirmed signal peptides and confidence of signal peptide predictions.
We analyzed our peptide annotations in order to confirm or refute signal predictions, and possibly to discover new signal cleavage sites. We examined peptides from the confirmed proteins with nontryptic N termini. From this set, we selected peptides with no upstream coverage (see Methods). The list of 117 predictions is provided in Supplemental Table S3B. A clear sequence motif (Crooks et al. 2004) emerges when we examine the sequence immediately upstream of these 117 putative signal peptides predicted by MS/MS analysis (Fig. 11). This motif closely matches motifs used by SignalP and PrediSi, thus providing additional support for using noncovered peptides for signal peptide identification.
Figure 11.
(Top) Sequence logo for the amino acid sequence motif of all signal peptides identified by MS/MS analysis. Position −1 correspond to the last residue of the signal peptide. (Middle) Sequence logo for Gram-negative bacteria employed by PrediSi (Hiller et al. 2004). (Bottom) Sequence logo for Gram-negative bacteria employed by SignalP (Nielsen et al. 1997).
SignalP and PrediSi predict 370 and 403 proteins with signal peptides. However, there is a substantial discrepancy between these tools—only 211 signals are predicted by both tools. LC-MS/MS evidence provides a possibility to resolve the discrepancies between SignalP and PrediSi as well as to identify signal peptides missed by both tools. Figure 12A compares our predicted signal peptides with the predictions made by SignalP and PrediSi on the 1992 confirmed proteins (see Methods). Our results confirm a total of 94 signal peptide predictions in vivo. In 31 cases, both SignalP and PrediSi predict signal cleavage on a confirmed protein but disagree as to the cleavage site. This ambiguity highlights the difficulties in computation signal prediction. In four of these cases, we are able to confirm the correct prediction with MS/MS evidence.
Figure 12.
(A) Venn diagram of all signal peptide predictions on confirmed proteins. A total of 94 signal peptide cleavage sites are validated by mass spectrometry (23 of them missed by both SignalP and PrediSi). (B) Number of signal predictions by SignalP (89) and PrediSi (38) rejected due to the observation of peptides upstream of the signal cleavage site. Eight of these sites were predicted by both tools.
On 119 of the confirmed proteins, the MS/MS results include peptides upstream of the cleavage site predicted by SignalP/PrediSi and thus represent evidence against SignalP/PrediSi predictions (Supplemental Table S3C). We call these the refuted sites. We refute 89 sites predicted by SignalP, and 38 sites predicted by PrediSi (with eight refuted sites predicted by both tools; Fig. 12B). It is conceivable that those peptides N-terminal to the signal site come from mislocalized proteins, where the signal sequence is not cleaved. If cleavage is the norm, then peptides immediately C-terminal to the refuted sites should be seen. However, they are observed for only four of the refuted sites, and each is contained in a much more abundant (by spectrum count) fully tryptic peptide that spans the refuted site. Thus, the peptide evidence suggests that these refuted signal peptide predictions are indeed incorrect.
If the predicted start site of a gene in the TIGR annotation falls before (toward 5′) the actual start site (i.e., the predicted TIGR protein is longer than the actual protein at the N terminus), a peptide covering the N terminus of this misannotated protein may also appear as a nontryptic peptide with no coverage in the upstream region. Thus it might be falsely predicted as a signal peptide, and it is likely that some of the cases where our predictions do not match SignalP or PrediSi belong to this category. Similarly, cases where the N-terminal most peptide is nontryptic and within 15 residues of the start position (too short for a signal peptide) might represent misannotated translational start sites. To investigate this further, we looked at the codon usage at the site of the first observed residue (position 0) or the one just before it (position −1) to account for N-terminal methionine cleavage. If some of these cases are indeed late translational start sites, we would expect higher frequency of start codons at these positions. We analyzed 36 proteins in all where the N-terminal most peptide was nontryptic mapping to the protein at a distance of two to 30 amino acids from the annotated start position and did not conform to SignalP or PrediSi signal predictions.
For example, while ATG is expected to appear 36/61 = 0.59 times at position −1, we observe it five times, an order of magnitude increase in frequency. Further comparative genomics analysis of these five proteins demonstrates that at least four of them are likely to be misannotated. All other codons appear zero, one, or two times in the sample, and conspicuously, the relatively rare start codons GTG, TTG, and ATT are also over-represented (each appears two times in position −1). While the size of the sample is too small to claim that this threefold over-representation reveals the new start codons, the comparative genomics analysis implies that it is likely to be the case. Figure 13 shows a fragment of SO_2760 protein (that was predicted to start with an ATG codon) and illustrates how comparative genomics analysis and MS/MS data help to correct gene annotations. The N-terminal most observed peptide for this gene starts at the ninth position, which is nontryptic, and indicates a late translational start site with N-terminal methionine cleavage. The eighth codon is GTG, and orthologs in many other Shewanella strains are also found to start with this position. Presence of a Shine-Dalgarno-like site (Shine and Dalgarno 1974), not shown here, upstream to the GTG codon further strengthens our claim for the start of translation at this codon.
Figure 13.
Sequence of phosphoribosylformylglycinamidine cyclo-ligase (SO_2760) in Shewanella oneidensis MR-1 according to TIGR annotation (red), observed nontryptic peptide (blue) and alignment to the orthologs in other Shewanella strains (green).
Chemical modifications
The current understanding of post-translational chemical modifications (PTMs) in bacteria is very limited even for well-studied organisms like E. coli, let alone for Shewanella. Any information that could be obtained about PTMs from large-scale MS/MS studies will prove to be very important toward gaining an understanding of the molecular biology of bacterial genomes. Note that while proteolytic events are also included in the term post-translational modifications, we will use the acronym PTM to specifically refer to in vivo chemical modifications of specific residues.
We analyzed the mass spectra using MS-Alignment (Tsur et al. 2005), which allows for the discovery and statistical validation of unanticipated modifications (without distinguishing between PTMs and in vitro chemical adducts). We considered all spectral annotations with one modification permitted for all possible mass-shifts of up to 250 Da. Since MS/MS annotations of modified peptides typically have high error rates, we applied a careful scoring procedure to highlight the valid modifications (see Methods). A total of 10,758 modification sites were seen with a false discovery rate of 5% (Supplemental Table S4). Some modifications types were observed on many different sites. Table 2 presents 24 common modification types, each observed on five or more distinct sites, with their likely chemical explanations. Since the false positive rate is low, it is extremely unlikely that any of these modification types represent a computational artifact. Moreover, all but two are known modification types, further reinforcing the conclusion that they are not artifact. We remark that the number of such modification types is rather large, significantly larger than the usual limit imposed by the popular restrictive PTM search tools like Mascot, SEQUEST, and X!Tandem. Many of these modifications appear to represent chemical events with low site specificity, which can result from chemical damage in vitro (Hunyadi-Gulyas and Medzihradszky 2004). Wherever possible, we cross-reference these modifications with known modifications from the UNIMOD (Creasy and Cottrell 2004) or RESID (Garavelli 2004) databases.
Table 2.
List of common mass shifts (observed on at least five sites) with possible explanations
Carbamidomethylation (abbreviated as CAM) is added to cysteine side chains by treatment with iodoacetamide, but can be attached to other sites. Most of these modifications occur in vitro. Masses are computed as the average modification mass over FT spectra (except entries shown with an asterisk, which had no corresponding FT spectra).
After filtering out the modifications that can be explained by these common modifications, we retain 4037 modification sites, corresponding to 390 distinct modification mass-shifts in 1673 proteins. While these numbers appear surprisingly large, similar diversity of PTMs has also been found in another recent study (Nielsen et al. 2006). There are no methods presently available to identify in vivo modifications from the list of all modifications, and one has to rely on comparisons with previous literature and databases. Below, we highlight several modifications of particular biological interest.
We anticipate that many biologically important PTMs in Shewanella and E. coli will be located on aligned positions in orthologous proteins. We highlight several modifications (Table 3) that are similar to those previously reported on orthologous positions in E. coli. For instance, Kowalak and Walsh (1996) reported the occurrence of β-methylthio-aspartic acid in E. coli, which is a modification of mass 46 at D88 of ribosomal protein S12p (Swiss-Prot ID: RS12_ECOLI). This is a well-conserved protein, and its ortholog, SO_0226, in S. oneidensis has almost identical amino acid sequence. This modification may be important for stabilizing the ribosome structure. We observed the same modification at D89 of the S. oneidensis protein, the homologous position to D88 of the E. coli protein, as shown below (the modified aspartate residues are shown in bold):
Table 3.
Selected PTMs supported either by studies in other bacterial genomes or by comparative genomics analysis
Some modifications are similar to previously described biological modifications in prokaryotes (typically in E. coli). Substitutions are supported by the presence of the target residue in other Shewanella strains at the orthologous residue. Hydroxylation of R on SO_0238, although not previous reported, is strongly supported by our data.
81-LIRGGRVKDLPGVRYHTVRG-100 Shewanella protein SO_0226
80-LIRGGRVKDLPGVRYHTVRG-99 E. coli ribosomal protein S12
Sometimes, the modifications we find in Shewanella differ somewhat from what has been previously reported for E. coli. For example, we observed both methylation and dimethylation at residue K83 of RplL. Single methylation of this protein was previously observed (Chang 1981) and localized to the orthologous residue in E. coli (Arnold and Reilly 1999). We observed an apparent hydroxylation (net mass shift 16 Da) at residue 54 in translation elongation factor Tu (TufB). Methylation of a nearby lysine has been reported for in E. coli (L’Italien and Laursen 1979). It is possible that the hydroxylation plays a similar regulatory or structural role.
Interestingly, formylation was observed only on one N-terminal methionine residue, although translation in bacteria begins with a formylmethionine residue. This indicates that peptide deformylases (such as Def-1, Def-2, and Def-3) operate with high efficiency.
Some modifications correspond to amino acid substitutions, either due to polymorphisms or due to errors in the genomic sequence. We validate such cases by considering the sequences of other Shewanella strains. For example, a modified peptide K.QQIG+14ENPIIVYMK.G was identified from glutaredoxin domain protein (SO_2880). This 14-Da offset is readily explained by a glycine-to-alanine amino acid substitution of glycine at position 12 of the protein. Indeed, analysis of the raw sequence trace TI|202899805 from S. oneidensis available in the NCBI trace archives revealed a mistake in the genome sequence at the corresponding locus resulting in a GGT (Gly) rather than the correct GCT (Ala). Similarly, we find that Ser177 (AGU) in SO_1637 should be corrected to Gly (GGU), and Cys33 (TGT) in SO_2880 should be changed to Ser (TCT).
Labeled images of example spectra for modifications reported in Table 2 and Table 3 are presented in Supplemental Data S5. The names of these images represent the modification; for example, D46-SO_0226.png represents the +46 modification on D in SO_0226. The corresponding spectra are also provided in DTA format (named like D46-SO_0226.dta) and contain the mass and charge of precursor and the list of fragment ions (precursor mass are shown in spreadsheet precursorMass_modification Examples.xls included in Supplemental Data S5.)
While we cannot validate all the predicted modifications at this time due to lack of available experimental data about PTMs for Shewanella, future research on this organism may confirm many of these putative modifications and begin to uncover their biological function.
Discussion
With recent improvements in sequencing technology, the number of sequenced bacterial genomes has been rapidly increasing (Benson et al. 2006). There have been significant improvements in algorithms for analyzing these sequences in the last two decades, especially in the area of gene prediction. However, there is a limit to what we can learn about the biology of an organism just from its DNA sequence. For example, it is very difficult to predict the post-translational modifications from protein sequence. What is needed, besides the primary sequence, are experimental data about the expressed proteins, and MS/MS has emerged as the preferred high-throughput technology in this field.
MS has been extensively used for studying individual proteins; however, its application to whole genomes became feasible only recently with the arrival of efficient database search tools. Some groups have been successful in using MS/MS for improving gene predictions in newly sequenced organisms (Jaffe et al. 2004a, b; Kalume et al. 2005; Wang et al. 2005), but there are no previous proteomic studies to obtain information about post-translational processing (proteolysis, chemical modification) at whole proteome scale. Studies like those by Jaffe et al. (2004a, b) that attempted to find PTMs at genome level had little or no success.
In this study, for the first time, we have provided a whole-bacterial proteome map of post-translational modifications for S. oneidensis MR-1. With effective control on the false discovery rate, we exploit nontryptic peptides to detect proteolytic events. In particular, we confirm 94 signal peptide predictions from SignalP or PrediSi and provide evidence for refuting 119 of their predictions. We also detect N-terminal methionine cleavage in 218 proteins. Using the recently developed MS-Alignment algorithm (Tsur et al. 2005), we find a large number of PTMs in the MS/MS data set, some of which represent in vivo modifications. We also improved the genome annotation with 30 N-terminal corrections, eight new genes, and validation of 13 expressed genes that were misannotated as pseudogenes.
MS takes a snapshot of the expressed proteins in a cell under specific conditions. Samples were collected across multiple experimental conditions in order to sample the complete proteome. We reliably identified >40% of all predicted genes (4928) as expressed proteins. This coverage is lower than 81% and 88% coverage reported in some Mycoplasma strains (Jaffe et al. 2004a, b). One reason for this difference in coverage, besides our more stringent control over peptide and protein selection, is the fact that Mycoplasma is a simpler organism with no transcriptional control (Jaffe et al. 2004a). On the other hand, Shewanella, with a genome seven times larger than Mycoplasma, does have an expression control mechanism, and some proteins may be expressed only under very specific conditions that might not have been captured by our experiments. DNA microarrays are a complementary technology that might be helpful in verifying expressed genes, and may be used in conjunction with MS (Kolker et al. 2005). However, our primary focus in this study is post-translational modifications that cannot be observed at the RNA level.
In a large-scale computational study of this kind, it is critical to keep the false positive rates under control when making predictions about gene corrections or post-translational modifications. We have used rigorous statistical measures to quantify and control the error rates below <5% and obtain reliable predictions. At the same time, experimental validation of these results by complementary approaches will be of great importance, and we anticipate that the results presented in this article will encourage such experiments by other research groups in the field.
Application of MS for whole genome studies is a relatively unexplored territory, especially in the area of post-translational modifications. A number of interesting challenges remain to be addressed. For example, we have demonstrated an approach for confidently detecting signal peptides and N-terminal methionine cleavages in the proteome with the assumption that the N-terminal fragment is degraded after proteolysis. However, a solution to the general proteolysis detection problem where both fractions of the protein may remain functional is not yet available, and we are only beginning to make attempts toward solving it. Similarly, even though we use the current state-of-the-art methods for detection of chemical modifications, it remains a challenge to distinguish between in vivo and in vitro modifications. We are considering strategies of improving the scoring and post-processing of MS-Alignment results to address this challenge. It has been shown in this and other proteogenomic studies how one can extend N-terminal boundaries of genes by observing upstream peptides. However, there is no good solution yet to make a case for shortening the N terminus of a gene. Modifying the experimental set-up is a possible approach to these and many other unsolved problems in this field, but it is likely that some problems can be solved by developing novel algorithms for interpreting the same data sets. For example, in this study we were able to detect many signal peptides and N-terminal methionine cleavages from MS/MS data without any labeling or other special treatment of the samples. It remains to be seen if we can further extend these computational methods to address other challenges as mentioned above, and apply them successfully to other organisms. Colloquially speaking, we are only beginning to scratch the surface of what can be learnt from whole genome MS/MS data sets.
Methods
MS/MS data
The majority of the 14.5 million spectra in our data set came from ion-trap MS/MS experiments, as described in further details in the Supplemental Data S6. Some (∼2 million) come from FT-ICR instruments. In this study, we treat FT data the same as ion-trap, with the exception of PTM analysis. The whole data set involves a large number of experiments under different conditions including aerobic (mid log), aerobic (steady state), suboxic, and anaerobic conditions with different additives. This data set is expected to represent most of the proteins expressed in Shewanella under these different conditions.
Whole genome search
The S. oneidensis genome consists of a circular chromosome (4,969,803 bp) and a plasmid (161,613 bp). We searched the MS/MS data set against a six-frame translation of the entire genome. The size of the translated genome (10,262,824 amino acids) was almost seven times the total size of the TIGR genes (1,432,446 amino acids). Sequences of common contaminants, including porcine trypsin and human keratins, were also added to the query database. A database of reversed sequences was created and searched as a negative control.
Translation was made according to standard codon usage. Two exceptions to this are TGA and TAG stop codons that sometimes code for the rare amino acids Selenocysteine and Pyrrolysine, respectively (Stadtman 1996; Hao et al. 2002). We translated all TGA and TAG codons into these amino acids, while kept the third stop codon TAA as the true stop codon. It is assumed that most TAG and TGA codons are authentic stop codons, and so will not be covered by any identified peptide. The advantage of this approach is that it can allow discovery of loci where read-through of TAG and TGA codons occurs (like in methylamine methyltransferase genes of Methanosarcina barkeri; Hao et al. 2002), without predicting those sites in advance. However, we did not observe such read-through codons in Shewanella.
The restrictive database search was done using Inspect (Tanner et al. 2005), with two fixed modifications, M+16 and N-terminal Q-17, that are common chemical artifacts (see below for a description of unrestricted PTM search). For each spectrum, the peptide with the top hit (lowest P-value) was selected. It took 2 d to search all spectra against this database on a 70-node grid.
Non-tryptic peptides
We considered all non-tryptic peptide annotations from confirmed proteins. Often such a peptide is properly contained in a longer tryptic peptide observed in the sample. These “covered peptides” are assumed to arise primarily from post-digestion break of tryptic peptides and not from cleavage in vivo. We focused our attention on noncovered peptides with no upstream peptide identifications. A total of 457 peptides (corresponding to 366 distinct N-terminal endpoints) satisfy these criteria. These peptides exhibit a strong sequence motif that closely matches the known motif characteristic for signal peptides in Gram-negative bacteria. We note also that a similar filter applied to C-terminal endpoints selects only 97 endpoints. These C-terminal endpoints do not correspond to a strong sequence motif, and have no clear position bias.
Signal peptides
We ran both SignalP and PrediSi against the TIGR genes to predict signal peptides, retaining all predictions made by these tools with scores ≥0.5. We analyzed all peptides from confirmed proteins that have a non-tryptic N terminus, and are not contained in an observed tryptic peptide. We discarded those that begin at position 2 (these correspond to N-terminal methionine cleavage, which is handled separately). We also restricted our attention to peptides with no upstream coverage at all. This provides a list of 164 predictions (94 of which match predictions made by SignalP and/or PrediSi). However, since no predicted signal peptides were confirmed outside the protein residue range 17–55, we further filtered the list to only the 117 predictions within this residue range. Some nontryptic peptides outside this range may also be generated by proteolytic cleavage. For example, a cut before residue 9 of SO_4743 may be a valid (but unusually short) signal peptide, as its upstream sequence matches the consensus AXA motif.
Signal peptides are rapidly degraded after cleavage. Therefore, if a peptide was identified upstream of a predicted signal cleavage site, then we consider the SignalP/PrediSi prediction to be refuted.
PTMs
Recently, we developed the MS-Alignment unrestrictive database search algorithm for finding unanticipated modifications (Tsur et al. 2005). We applied MS-Alignment to our data set, allowing for an arbitrary single modification with mass shift up to 250 Da per peptide. We constructed a database consisting of all confirmed Shewanella proteins found by the initial search, together with a shuffled sequence corresponding to each protein in this database. In a search of this database with MS-Alignment, spurious modifications are expected to be distributed randomly throughout the database, including the shuffled sequences. After selecting a score cutoff, we obtained an empirical false discovery rate by simply counting the number of peptides from spurious proteins. A score cutoff providing a spectrum-level false discovery rate of 5% was used. Modification sites were selected from these results using the Inspect analysis scripts (Tanner et al. 2006). The search required ∼1 mo on a 64-processor cluster. Modifications with small mass shift (e.g., mass shift 1) may result from low parent mass accuracy or the presence of isotopic peaks; therefore, only modifications with mass shift 3 Da or more were considered in subsequent analysis.
Although the spectrum-level false discovery rate is 5%, the error rate at the level of modification sites may be higher. This phenomenon is similar to the increase in error rate that occurs as one moves from spectrum-level to peptide-level or protein-level analysis. Therefore, we scored and rank the modification sites, and computed the false discovery rate at the level of modification sites. For each modification site, we generated a consensus spectrum by averaging together the peaks from all spectra carrying the modification. This consensus spectrum eliminates much of the noise from individual spectra, and provides a more accurate measurement of modification masses. We then scored modification sites by computing several features and then computing a weighted sum of these features. Various features were initially considered, and a collection of features was selected by iteratively selecting the feature providing the best marginal improvement in accuracy. The features used in computing the score are as follows:
Match score of the consensus spectrum.
Delta-score between the modified annotation and the next best annotation.
Number of spectra carrying the annotation.
Length of the modified peptide.
Number of overlapping peptides containing the modification of interest. Authentic modifications are often observed as part of two or more distinct peptides due to missed tryptic cleavage or post-digestion breakup. Such overlap is quit rare for spurious modifications.
Fraction of spectra carrying the modification which are tryptic.
After sorting modification sites by these scores, we obtained 10,758 modification sites with a false discovery rate of 5%. Supplemental Table 4 summarizes these modifications. For each row of this spreadsheet, a representative spectrum (DTA format) and the corresponding image with b and y ions labeled are available at http://peptide.ucsd.edu/ShewanellaOneidensis/ (the files are named based on their row number in Supplemental Table 4, and precursor masses are in the file precursorMass_allPTMSpectra. xls). These modified annotations include delta-correct annotations (see Tsur et al. 2005) where the modification is localized to an incorrect (typically adjacent) residue or has the wrong mass (due to limited parent mass accuracy). We use an automated analysis procedure to suggest alternatives for each modification. It considers edits, such as shifting the modification to the adjacent residue, which have minor effects on the theoretical fragmentation pattern. If these edits generate a candidate peptide that contains only common modifications (such as oxidized methionine) and whose score comparable to the initial annotation, then we consider the modification to be satisfactorily explained. For example, the putative annotation “S.F+115SVEAPKT.K” from rplD was replaced with “E.S+28FSVEAPKT.K,” since the latter annotation invokes a common chemical modification (formylation of the N terminus) and explains the spectrum just as well. Modification types were added to the collection of “common” modifications after successful manual validation of five or more sites carrying the same modification mass. After these automated procedures were run, modification sites were curated, and a putative annotation assigned to each site.
Acknowledgments
We thank Mark Borodovsky and Gary Olsen for useful comments and help with gene annotations. S.T. is supported by NSF IGERT training grant DGE0504645. This research was supported in part by the UCSD FWGrid Project, NSF Research Infrastructure grant number EIA-0303622. Part of this investigation was supported using the computing facility made possible by the Research Facilities Improvement Program grant no. C06 RR017588 awarded to the Whitaker Biomedical Engineering Institute, and the Biomedical Technology Resource Centers Program grant no. P41 RR08605 awarded to the National Biomedical Computation Resource, UCSD, from the National Center for Research Resources, National Institutes of Health. This project was supported by U.S. National Institutes of Health grant NIGMS 1-R01-RR16522. Shewanella genome sequences were kindly provided by the Joint Genome Institute. Part of this research at Pacific Northwest National Laboratory was supported by the Genomics:GtL Program, Office of Biological and Environmental Research, U.S. Department of Energy. Pacific Northwest National Laboratory is operated for the DOE by Battelle Memorial Institute under Contract DE-AC06-76RLO 1830.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6427907
References
- Aebersold R., Mann M., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Antelmann H., Tjalsma H., Voight B., Ohlmeier S., Bron S., van Dijl J.M., Hecker M., Tjalsma H., Voight B., Ohlmeier S., Bron S., van Dijl J.M., Hecker M., Voight B., Ohlmeier S., Bron S., van Dijl J.M., Hecker M., Ohlmeier S., Bron S., van Dijl J.M., Hecker M., Bron S., van Dijl J.M., Hecker M., van Dijl J.M., Hecker M., Hecker M. A proteomic view on genome-based signal peptide predictions. Genome Res. 2001;11:1484–1502. doi: 10.1101/gr.182801. [DOI] [PubMed] [Google Scholar]
- Arnold R.J., Reilly J.P., Reilly J.P. Observation of Escherichia coli ribosomal proteins and their posttranslational modifications by mass spectrometry. Anal. Biochem. 1999;269:105–112. doi: 10.1006/abio.1998.3077. [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B., Mori H., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B., Mori H., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B., Mori H., Takai Y., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B., Mori H., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B., Mori H., Baba M., Datsenko K., Tomita M., Wanner B., Mori H., Datsenko K., Tomita M., Wanner B., Mori H., Tomita M., Wanner B., Mori H., Wanner B., Mori H., Mori H., et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006;2:E1–E11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P., Gesteland R., Atkins J., Gesteland R., Atkins J., Atkins J. Recoding: Translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- Baranov P., Gurvich O., Hammer A., Gesteland R., Atkins J., Gurvich O., Hammer A., Gesteland R., Atkins J., Hammer A., Gesteland R., Atkins J., Gesteland R., Atkins J., Atkins J. RECODE 2003. Nucleic Acids Res. 2003;31:87–89. doi: 10.1093/nar/gkg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Bassat A., Bauer K., Chang S., Myambo K., Boosman A., Chang S., Bauer K., Chang S., Myambo K., Boosman A., Chang S., Chang S., Myambo K., Boosman A., Chang S., Myambo K., Boosman A., Chang S., Boosman A., Chang S., Chang S. Processing of the initiation methionine from proteins: Properties of the Escherichia coli methionine aminopeptidase and its gene structure. J. Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen J.D., Nielsen H., von Heijne G., Brunak S., Nielsen H., von Heijne G., Brunak S., von Heijne G., Brunak S., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L., Lipman D.J., Ostell J., Wheeler D.L., Ostell J., Wheeler D.L., Wheeler D.L. GenBank. Nucleic Acids Res. 2006;34:16–20. [Google Scholar]
- Besemer J., Borodovsky M., Borodovsky M. GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:451–454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boja E.S., Fales H.M., Fales H.M. Overalkylation of a protein digest with iodoacetamide. Anal. Chem. 2001;73:3576–3582. doi: 10.1021/ac0103423. [DOI] [PubMed] [Google Scholar]
- Chang F.N. Methylation of ribosomal proteins during ribosome assembly in Escherichia coli. Mol. Gen. Genet. 1981;183:418–421. doi: 10.1007/BF00268759. [DOI] [PubMed] [Google Scholar]
- Creasy D.M., Cottrell J.S., Cottrell J.S. Unimod: Protein modifications for mass spectrometry. Proteomics. 2004;4:1534–1536. doi: 10.1002/pmic.200300744. [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E., Hon G., Chandonia J.-M., Brenner S.E., Chandonia J.-M., Brenner S.E., Brenner S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Zhang Y., Wang J., Li X., Lu Z., Cai Y., Qian X., Zhang Y., Wang J., Li X., Lu Z., Cai Y., Qian X., Wang J., Li X., Lu Z., Cai Y., Qian X., Li X., Lu Z., Cai Y., Qian X., Lu Z., Cai Y., Qian X., Cai Y., Qian X., Qian X. Identification of degradation products formed during performic oxidation of peptides and proteins by high-performance liquid chromatography with matrix-assisted laser desorption/ionization and tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2005;19:1130–1138. doi: 10.1002/rcm.1901. [DOI] [PubMed] [Google Scholar]
- Daraselia N., Dernovoy D., Tian Y., Borodovsky M., Tatusov R., Tatusova T., Dernovoy D., Tian Y., Borodovsky M., Tatusov R., Tatusova T., Tian Y., Borodovsky M., Tatusov R., Tatusova T., Borodovsky M., Tatusov R., Tatusova T., Tatusov R., Tatusova T., Tatusova T. Reannotation of Shewanella oneidensis genome. OMICS. 2003;7:171–176. doi: 10.1089/153623103322246566. [DOI] [PubMed] [Google Scholar]
- Elias D.A., Monroe M.E., Marshall M.J., Romine M.F., Belieav A.S., Fredrickson J.K., Anderson G.A., Smith R.D., Lipton M.S., Monroe M.E., Marshall M.J., Romine M.F., Belieav A.S., Fredrickson J.K., Anderson G.A., Smith R.D., Lipton M.S., Marshall M.J., Romine M.F., Belieav A.S., Fredrickson J.K., Anderson G.A., Smith R.D., Lipton M.S., Romine M.F., Belieav A.S., Fredrickson J.K., Anderson G.A., Smith R.D., Lipton M.S., Belieav A.S., Fredrickson J.K., Anderson G.A., Smith R.D., Lipton M.S., Fredrickson J.K., Anderson G.A., Smith R.D., Lipton M.S., Anderson G.A., Smith R.D., Lipton M.S., Smith R.D., Lipton M.S., Lipton M.S. Global detection and characterization of hypothetical proteins in Shewanella oneidensis MR-1 using LC-MS based proteomics. Proteomics. 2005;5:3120–3130. doi: 10.1002/pmic.200401140. [DOI] [PubMed] [Google Scholar]
- Elias D.A., Monroe M.E., Smith R.D., Fredrickson J.K., Lipton M.S., Monroe M.E., Smith R.D., Fredrickson J.K., Lipton M.S., Smith R.D., Fredrickson J.K., Lipton M.S., Fredrickson J.K., Lipton M.S., Lipton M.S. Confirmation of the expression of a large set of conserved hypothetical proteins in Shewanella oneidensis MR-1. J. Microbiol. Methods. 2006;66:223–233. doi: 10.1016/j.mimet.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Fenselau C., Demirev P., Demirev P. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 2001;20:157–171. doi: 10.1002/mas.10004. [DOI] [PubMed] [Google Scholar]
- Fermin D., Allen B., Blackwell T., Menon R., Adamski M., Xu Y., Ulintz P., Omenn G., States D., Allen B., Blackwell T., Menon R., Adamski M., Xu Y., Ulintz P., Omenn G., States D., Blackwell T., Menon R., Adamski M., Xu Y., Ulintz P., Omenn G., States D., Menon R., Adamski M., Xu Y., Ulintz P., Omenn G., States D., Adamski M., Xu Y., Ulintz P., Omenn G., States D., Xu Y., Ulintz P., Omenn G., States D., Ulintz P., Omenn G., States D., Omenn G., States D., States D. Novel gene and gene model detection using a whole genome open reading frame analysis in proteomics. Genome Biol. 2006;7:R35. doi: 10.1186/gb-2006-7-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frottin F., Martinez A., Peynot P., Mitra S., Holz R., Giglione C., Meinnel T., Martinez A., Peynot P., Mitra S., Holz R., Giglione C., Meinnel T., Peynot P., Mitra S., Holz R., Giglione C., Meinnel T., Mitra S., Holz R., Giglione C., Meinnel T., Holz R., Giglione C., Meinnel T., Giglione C., Meinnel T., Meinnel T. The proteomics of N-terminal methionine cleavage. Mol. Cell. Proteomics. 2006;5:2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- Garavelli J.S. The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics. 2004;4:1527–1533. doi: 10.1002/pmic.200300777. [DOI] [PubMed] [Google Scholar]
- Geoghegan K.F., Hoth L.R., Tan D.H., Borzilleri K.A., Withka J.M., Boyd J.G., Hoth L.R., Tan D.H., Borzilleri K.A., Withka J.M., Boyd J.G., Tan D.H., Borzilleri K.A., Withka J.M., Boyd J.G., Borzilleri K.A., Withka J.M., Boyd J.G., Withka J.M., Boyd J.G., Boyd J.G. Cyclization of N-terminal S-carbamoylmethylcysteine causing loss of 17 Da from peptides and extra peaks in peptide maps. J. Proteome Res. 2002;1:181–187. doi: 10.1021/pr025503d. [DOI] [PubMed] [Google Scholar]
- Gerdes S., Scholle M., Campbell J., Balazsi G., Ravasz E., Daugherty M., Somera A., Kyrpides N., Anderson I., Gelfand M., Scholle M., Campbell J., Balazsi G., Ravasz E., Daugherty M., Somera A., Kyrpides N., Anderson I., Gelfand M., Campbell J., Balazsi G., Ravasz E., Daugherty M., Somera A., Kyrpides N., Anderson I., Gelfand M., Balazsi G., Ravasz E., Daugherty M., Somera A., Kyrpides N., Anderson I., Gelfand M., Ravasz E., Daugherty M., Somera A., Kyrpides N., Anderson I., Gelfand M., Daugherty M., Somera A., Kyrpides N., Anderson I., Gelfand M., Somera A., Kyrpides N., Anderson I., Gelfand M., Kyrpides N., Anderson I., Gelfand M., Anderson I., Gelfand M., Gelfand M., et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B., Gong W., Ferguson T.K., James C.M., Krzycki J.A., Chan M.K., Gong W., Ferguson T.K., James C.M., Krzycki J.A., Chan M.K., Ferguson T.K., James C.M., Krzycki J.A., Chan M.K., James C.M., Krzycki J.A., Chan M.K., Krzycki J.A., Chan M.K., Chan M.K. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- Heidelberg J.F., Paulsen I.T., Nelson K.E., Gaidos E.J., Nelson W.C., Read T.D., Eisen J.A., Seshadri R., Ward N., Methe B., Paulsen I.T., Nelson K.E., Gaidos E.J., Nelson W.C., Read T.D., Eisen J.A., Seshadri R., Ward N., Methe B., Nelson K.E., Gaidos E.J., Nelson W.C., Read T.D., Eisen J.A., Seshadri R., Ward N., Methe B., Gaidos E.J., Nelson W.C., Read T.D., Eisen J.A., Seshadri R., Ward N., Methe B., Nelson W.C., Read T.D., Eisen J.A., Seshadri R., Ward N., Methe B., Read T.D., Eisen J.A., Seshadri R., Ward N., Methe B., Eisen J.A., Seshadri R., Ward N., Methe B., Seshadri R., Ward N., Methe B., Ward N., Methe B., Methe B., et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 2002;20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- Heurgue-Hamard V., Champ S., Engstrom A., Ehrenberg M., Buckingham R.H., Champ S., Engstrom A., Ehrenberg M., Buckingham R.H., Engstrom A., Ehrenberg M., Buckingham R.H., Ehrenberg M., Buckingham R.H., Buckingham R.H. The hemK gene in Escherichia coli encodes the N5-glutamine methyltransferase that modifies peptide release factors. EMBO J. 2002;21:769–778. doi: 10.1093/emboj/21.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller K., Grote A., Scheer M., Munch R., Jahn D., Grote A., Scheer M., Munch R., Jahn D., Scheer M., Munch R., Jahn D., Munch R., Jahn D., Jahn D. PrediSi: Prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004;32:375–379. doi: 10.1093/nar/gkh378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel P.H., Schmitter M.J., Dessen P., Fayat G., Blanquet S., Schmitter M.J., Dessen P., Fayat G., Blanquet S., Dessen P., Fayat G., Blanquet S., Fayat G., Blanquet S., Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Wang R., Shih J., Lo S., Wang R., Shih J., Lo S., Shih J., Lo S., Lo S. Identification of a putative infC-rpmI-rplT operon flanked by long inverted repeats in Mycoplasma fermentans (incognitus strain) Gene. 1993;127:79–85. doi: 10.1016/0378-1119(93)90619-e. [DOI] [PubMed] [Google Scholar]
- Hunyadi-Gulyas E., Medzihradszky K., Medzihradszky K. Factors that contribute to the complexity of protein digests. DDT: Targets—Mass Spectrom. Proteomics Suppl. 2004;3:S3–S10. [Google Scholar]
- Jaffe J., Berg H., Church G., Berg H., Church G., Church G. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics. 2004a;4:59–77. doi: 10.1002/pmic.200300511. [DOI] [PubMed] [Google Scholar]
- Jaffe J., Stange-Thomann N., Smith C., DeCaprio D., Fisher S., Butler J., Calvo S., Elkins T., FitzGerald M., Hafez N., Stange-Thomann N., Smith C., DeCaprio D., Fisher S., Butler J., Calvo S., Elkins T., FitzGerald M., Hafez N., Smith C., DeCaprio D., Fisher S., Butler J., Calvo S., Elkins T., FitzGerald M., Hafez N., DeCaprio D., Fisher S., Butler J., Calvo S., Elkins T., FitzGerald M., Hafez N., Fisher S., Butler J., Calvo S., Elkins T., FitzGerald M., Hafez N., Butler J., Calvo S., Elkins T., FitzGerald M., Hafez N., Calvo S., Elkins T., FitzGerald M., Hafez N., Elkins T., FitzGerald M., Hafez N., FitzGerald M., Hafez N., Hafez N., et al. The complete genome and proteome of mycoplasma mobile. Genome Res. 2004b;14:1447–1461. doi: 10.1101/gr.2674004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen O.N. Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell Biol. 2006;7:391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- Joshi A.B., Kirsch L.E., Kirsch L.E. The relative rates of glutamine and asparagine deamidation in glucagon fragment 22-29 under acidic conditions. J. Pharm. Sci. 2002;91:2331–2345. doi: 10.1002/jps.10213. [DOI] [PubMed] [Google Scholar]
- Kalume D., Peri S., Reddy R., Zhong J., Okulate M., Kumar N., Pandey A., Peri S., Reddy R., Zhong J., Okulate M., Kumar N., Pandey A., Reddy R., Zhong J., Okulate M., Kumar N., Pandey A., Zhong J., Okulate M., Kumar N., Pandey A., Okulate M., Kumar N., Pandey A., Kumar N., Pandey A., Pandey A. Genome annotation of Anopheles gambiae using mass spectrometry-derived data. BMC Genomics. 2005;2005:128. doi: 10.1186/1471-2164-6-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanto S., Okino H., Aizawa S., Yamaguchi S., Okino H., Aizawa S., Yamaguchi S., Aizawa S., Yamaguchi S., Yamaguchi S. Amino acids responsible for flagellar shape are distributed in terminal regions of flagellin. J. Mol. Biol. 1991;219:471–480. doi: 10.1016/0022-2836(91)90187-b. [DOI] [PubMed] [Google Scholar]
- Kehry M.R., Engstrom P., Dahlquist F.W., Hazelbauer G.L., Engstrom P., Dahlquist F.W., Hazelbauer G.L., Dahlquist F.W., Hazelbauer G.L., Hazelbauer G.L. Multiple covalent modifications of Trg, a sensory transducer of Escherichia coli. J. Biol. Chem. 1983;258:5050–5055. [PubMed] [Google Scholar]
- Khandke K.M., Fairwell T., Chait B.T., Manjula B.N., Fairwell T., Chait B.T., Manjula B.N., Chait B.T., Manjula B.N., Manjula B.N. Influence of ions on cyclization of the amino terminal glutamine residues of tryptic peptides of streptococcal PepM49 protein. Resolution of cyclized peptides by HPLC and characterization by mass spectrometry. Int. J. Pept. Protein Res. 1989;34:118–123. doi: 10.1111/j.1399-3011.1989.tb01499.x. [DOI] [PubMed] [Google Scholar]
- Kleene S.J., Toews M.L., Adler J., Toews M.L., Adler J., Adler J. Isolation of glutamic acid methyl ester from an Escherichia coli membrane protein involved in chemotaxis. J. Biol. Chem. 1977;252:3214–3218. [PubMed] [Google Scholar]
- Kolker E., Picone A.F., Galperin M.Y., Romine M.F., Higdon R., Makarova K.S., Kolker N., Anderson G.A., Qiu X., Auberry K.J., Picone A.F., Galperin M.Y., Romine M.F., Higdon R., Makarova K.S., Kolker N., Anderson G.A., Qiu X., Auberry K.J., Galperin M.Y., Romine M.F., Higdon R., Makarova K.S., Kolker N., Anderson G.A., Qiu X., Auberry K.J., Romine M.F., Higdon R., Makarova K.S., Kolker N., Anderson G.A., Qiu X., Auberry K.J., Higdon R., Makarova K.S., Kolker N., Anderson G.A., Qiu X., Auberry K.J., Makarova K.S., Kolker N., Anderson G.A., Qiu X., Auberry K.J., Kolker N., Anderson G.A., Qiu X., Auberry K.J., Anderson G.A., Qiu X., Auberry K.J., Qiu X., Auberry K.J., Auberry K.J., et al. Global profiling of Shewanella oneidensis MR-1: Expression of hypothetical genes and improved functional annotations. Proc. Natl. Acad. Sci. 2005;102:2099–2104. doi: 10.1073/pnas.0409111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- Kowalak J.A., Walsh K.A., Walsh K.A. β-Methylthio-aspartic acid: Identification of a novel posttranslational modification in ribosomal protein S12 from Escherichia coli. Protein Sci. 1996;5:1625–1632. doi: 10.1002/pro.5560050816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuster B., Mortensen P., Andersen J.S., Mann M., Mortensen P., Andersen J.S., Mann M., Andersen J.S., Mann M., Mann M. Mass spectrometry allows direct identification of proteins in large genomes. Proteomics. 2001;1:641–650. doi: 10.1002/1615-9861(200104)1:5<641::AID-PROT641>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Lagerwerf F.M., de van Weert M., Heerma W., Haverkamp J., de van Weert M., Heerma W., Haverkamp J., Heerma W., Haverkamp J., Haverkamp J. Identification of oxidized methionine in peptides. Rapid Commun. Mass Spectrom. 1996;10:1905–1910. doi: 10.1002/(SICI)1097-0231(199612)10:15<1905::AID-RCM755>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Lapko V.N., Smith D.L., Smith J.B., Smith D.L., Smith J.B., Smith J.B. Identification of an artifact in the mass spectrometry of proteins derivatized with iodoacetamide. J. Mass Spectrom. 2000;35:572–575. doi: 10.1002/(SICI)1096-9888(200004)35:4<572::AID-JMS971>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Link A., Robison K., Church G., Robison K., Church G., Church G. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis. 1997;18:1259–1313. doi: 10.1002/elps.1150180807. [DOI] [PubMed] [Google Scholar]
- Lippincott J., Apostol I., Apostol I. Carbamylation of cysteine: A potential artifact in peptide mapping of hemoglobins in the presence of urea. Anal. Biochem. 1999;267:57–64. doi: 10.1006/abio.1998.2970. [DOI] [PubMed] [Google Scholar]
- L’Italien J.J., Laursen R.A., Laursen R.A. Location of the site of methylation in elongation factor Tu. FEBS Lett. 1979;107:359–362. doi: 10.1016/0014-5793(79)80407-x. [DOI] [PubMed] [Google Scholar]
- Liveris D., Schwartz J., Geertman R., Schwartz I., Schwartz J., Geertman R., Schwartz I., Geertman R., Schwartz I., Schwartz I. Molecular cloning and sequencing of encoding translation initiation factor enterobacterial species. FEMS Microbiol. Lett. 1993;112:211–216. doi: 10.1111/j.1574-6968.1993.tb06450.x. [DOI] [PubMed] [Google Scholar]
- Lu P., Vogel C., Wang R., Yao X., Marcotte E., Vogel C., Wang R., Yao X., Marcotte E., Wang R., Yao X., Marcotte E., Yao X., Marcotte E., Marcotte E. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 2006;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- Mann M., Pandey A., Pandey A. Use of mass spectrometry-derived data to annotate nucleotide and protein sequence databases. Trends Biochem. Sci. 2001;26:54–61. doi: 10.1016/s0968-0004(00)01726-6. [DOI] [PubMed] [Google Scholar]
- Nealson K.H., Belz A., McKee B., Belz A., McKee B., McKee B. Breathing metals as a way of life: Geobiology in action. Antonie Van Leeuwenhoek. 2002;81:215–222. doi: 10.1023/a:1020518818647. [DOI] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., von Heijne G., Engelbrecht J., Brunak S., von Heijne G., Brunak S., von Heijne G., von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nielsen M.L., Savitski M.M., Zubarev R.A., Savitski M.M., Zubarev R.A., Zubarev R.A. Extent of modifications in human proteome samples and their effect on dynamic range of analysis in shotgun proteomics. Mol. Cell. Proteomics. 2006;5:2384–2391. doi: 10.1074/mcp.M600248-MCP200. [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Ong S.-E., Mann M., Ong S.-E., Mann M., Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- Olson K., Fenno J., Lin N., Harkins R., Snider C., Kohr W., Ross M., Fodge D., Prender G., Stebbing N., Fenno J., Lin N., Harkins R., Snider C., Kohr W., Ross M., Fodge D., Prender G., Stebbing N., Lin N., Harkins R., Snider C., Kohr W., Ross M., Fodge D., Prender G., Stebbing N., Harkins R., Snider C., Kohr W., Ross M., Fodge D., Prender G., Stebbing N., Snider C., Kohr W., Ross M., Fodge D., Prender G., Stebbing N., Kohr W., Ross M., Fodge D., Prender G., Stebbing N., Ross M., Fodge D., Prender G., Stebbing N., Fodge D., Prender G., Stebbing N., Prender G., Stebbing N., Stebbing N., et al. Purified human growth hormone from E. coli is biologically active. Nature. 1981;293:408–411. doi: 10.1038/293408a0. [DOI] [PubMed] [Google Scholar]
- Oshiro G., Wodicka L., Washburn M., Yates J., Lockhart D., Winzeler E., Wodicka L., Washburn M., Yates J., Lockhart D., Winzeler E., Washburn M., Yates J., Lockhart D., Winzeler E., Yates J., Lockhart D., Winzeler E., Lockhart D., Winzeler E., Winzeler E. Parallel identification of new genes in Saccharomyces cerevisiae. Genome Res. 2002;12:1210–1220. doi: 10.1101/gr.226802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.-Y., Cohoon M., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.-Y., Cohoon M., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., Butler R.M., Choudhuri J.V., Chuang H.-Y., Cohoon M., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., Choudhuri J.V., Chuang H.-Y., Cohoon M., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., Chuang H.-Y., Cohoon M., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., Cohoon M., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., Diaz N., Disz T., Edwards R., Disz T., Edwards R., Edwards R., et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetzel M., Karla A., Strynadka N.C.J., Dalbey R.E., Karla A., Strynadka N.C.J., Dalbey R.E., Strynadka N.C.J., Dalbey R.E., Dalbey R.E. Signal peptidases. Chem. Rev. 2002;102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- Peterson J.D., Umayam L.A., Dickinson T., Hickey E.K., White O., Umayam L.A., Dickinson T., Hickey E.K., White O., Dickinson T., Hickey E.K., White O., Hickey E.K., White O., White O. The comprehensive microbial resource. Nucleic Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon C., Brombach M., Thamm S., Gualerzi C., Brombach M., Thamm S., Gualerzi C., Thamm S., Gualerzi C., Gualerzi C. Cloning and characterization of a gene cluster from Bacillus stearothermophilus comprising infC, rpmI and rplT. Mol. Genet. Genomics. 1989;218:355–357. doi: 10.1007/BF00331290. [DOI] [PubMed] [Google Scholar]
- Rice M.S., Dahlquist F.W., Dahlquist F.W. Sites of deamidation and methylation in Tsr, a bacterial chemotaxis sensory transducer. J. Biol. Chem. 1991;266:9746–9753. [PubMed] [Google Scholar]
- Romine M.F., Elias D.A., Monroe M.E., Auberry K., Fang R., Fredrickson J.K., Anderson G.A., Smith R.D., Lipton M.S., Elias D.A., Monroe M.E., Auberry K., Fang R., Fredrickson J.K., Anderson G.A., Smith R.D., Lipton M.S., Monroe M.E., Auberry K., Fang R., Fredrickson J.K., Anderson G.A., Smith R.D., Lipton M.S., Auberry K., Fang R., Fredrickson J.K., Anderson G.A., Smith R.D., Lipton M.S., Fang R., Fredrickson J.K., Anderson G.A., Smith R.D., Lipton M.S., Fredrickson J.K., Anderson G.A., Smith R.D., Lipton M.S., Anderson G.A., Smith R.D., Lipton M.S., Smith R.D., Lipton M.S., Lipton M.S. Validation of Shewanella oneidensis MR-1 small proteins by AMT tag-based proteome analysis. OMICS. 2004;8:239–254. doi: 10.1089/omi.2004.8.239. [DOI] [PubMed] [Google Scholar]
- Sacerdot C., Fayat G., Dessen P., Springer M., Plumbridge J., Grunberg-Manago M., Blanquet S., Fayat G., Dessen P., Springer M., Plumbridge J., Grunberg-Manago M., Blanquet S., Dessen P., Springer M., Plumbridge J., Grunberg-Manago M., Blanquet S., Springer M., Plumbridge J., Grunberg-Manago M., Blanquet S., Plumbridge J., Grunberg-Manago M., Blanquet S., Grunberg-Manago M., Blanquet S., Blanquet S. Sequence of a 1.26-kb DNA fragment containing the structural gene for E. coli initiation factor IF3: Presence of an AUU initiator codon. EMBO J. 1982;1:311–315. doi: 10.1002/j.1460-2075.1982.tb01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdot C., Chiaruttini C., Engst K., Graffe M., Milet M., Mathy N., Dondon J., Springer M., Chiaruttini C., Engst K., Graffe M., Milet M., Mathy N., Dondon J., Springer M., Engst K., Graffe M., Milet M., Mathy N., Dondon J., Springer M., Graffe M., Milet M., Mathy N., Dondon J., Springer M., Milet M., Mathy N., Dondon J., Springer M., Mathy N., Dondon J., Springer M., Dondon J., Springer M., Springer M. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol. Microbiol. 1996;21:331–346. doi: 10.1046/j.1365-2958.1996.6361359.x. [DOI] [PubMed] [Google Scholar]
- Sarioglu H., Lottspeich F., Walk T., Jung G., Eckerskorn C., Lottspeich F., Walk T., Jung G., Eckerskorn C., Walk T., Jung G., Eckerskorn C., Jung G., Eckerskorn C., Eckerskorn C. Deamidation as a widespread phenomenon in two-dimensional polyacrylamide gel electrophoresis of human blood plasma proteins. Electrophoresis. 2000;21:2209–2218. doi: 10.1002/1522-2683(20000601)21:11<2209::AID-ELPS2209>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Schoenhals G.J., Kihara M., Macnab R.M., Kihara M., Macnab R.M., Macnab R.M. Translation of the flagellar gene fliO of Salmonella typhimurium from putative tandem starts. J. Bacteriol. 1998;180:2936–2942. doi: 10.1128/jb.180.11.2936-2942.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serres M., Riley M., Riley M. Genomic analysis of carbon source metabolism of Shewanella oneidensis MR-1: Predictions versus experiments. J. Bacteriol. 2006;188:4601–4609. doi: 10.1128/JB.01787-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L., Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen H.H., Thomsen J., Bayne S., Hojrup P., Roepstorff P., Thomsen J., Bayne S., Hojrup P., Roepstorff P., Bayne S., Hojrup P., Roepstorff P., Hojrup P., Roepstorff P., Roepstorff P. Strategies for determination of disulphide bridges in proteins using plasma desorption mass spectrometry. Biomed. Environ. Mass Spectrom. 1990;19:713–720. doi: 10.1002/bms.1200191110. [DOI] [PubMed] [Google Scholar]
- Stadtman T.C. Selenocysteine. Annu. Rev. Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- Sussman J.K., Simons E.L., Simons R.W., Simons E.L., Simons R.W., Simons R.W. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 1996;21:347–360. doi: 10.1046/j.1365-2958.1996.6371354.x. [DOI] [PubMed] [Google Scholar]
- Swiderek K.M., Davis M.T., Lee T.D., Davis M.T., Lee T.D., Lee T.D. The identification of peptide modifications derived from gel-separated proteins using electrospray triple quadrupole and ion trap analyses. Electrophoresis. 1998;19:989–997. doi: 10.1002/elps.1150190614. [DOI] [PubMed] [Google Scholar]
- Tang H., Arnold R., Alves P., Xun Z., Clemmer D., Novotny M., Reilly J., Rejivojac P., Arnold R., Alves P., Xun Z., Clemmer D., Novotny M., Reilly J., Rejivojac P., Alves P., Xun Z., Clemmer D., Novotny M., Reilly J., Rejivojac P., Xun Z., Clemmer D., Novotny M., Reilly J., Rejivojac P., Clemmer D., Novotny M., Reilly J., Rejivojac P., Novotny M., Reilly J., Rejivojac P., Reilly J., Rejivojac P., Rejivojac P. A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics. 2006;22:e481–e488. doi: 10.1093/bioinformatics/btl237. [DOI] [PubMed] [Google Scholar]
- Tanner S., Shu H., Frank A., Wang L., Zandi E., Mumby M., Pevzner P., Bafna V., Shu H., Frank A., Wang L., Zandi E., Mumby M., Pevzner P., Bafna V., Frank A., Wang L., Zandi E., Mumby M., Pevzner P., Bafna V., Wang L., Zandi E., Mumby M., Pevzner P., Bafna V., Zandi E., Mumby M., Pevzner P., Bafna V., Mumby M., Pevzner P., Bafna V., Pevzner P., Bafna V., Bafna V. Inspect: Fast and accurate identification of post-translationally modified peptides from tandem mass spectra. Anal. Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- Tanner S., Pevzner P., Bafna V., Pevzner P., Bafna V., Bafna V. Unrestrictive identification of post-translational modifications through peptide mass spectrometry. Nat. Protoc. 2006;1:67–72. doi: 10.1038/nprot.2006.10. [DOI] [PubMed] [Google Scholar]
- Tobias J., Shrader T., Rocap G., Varshavsky A., Shrader T., Rocap G., Varshavsky A., Rocap G., Varshavsky A., Varshavsky A. The N-end rule in bacteria. Science. 1991;254:1374. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- Tronick S.R., Martinez R.J., Martinez R.J. Methylation of the flagellin of Salmonella typhimurium. J. Bacteriol. 1971;105:211–219. doi: 10.1128/jb.105.1.211-219.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsur D., Tanner S., Zandi E., Bafna V., Pevzner P., Tanner S., Zandi E., Bafna V., Pevzner P., Zandi E., Bafna V., Pevzner P., Bafna V., Pevzner P., Pevzner P. Identification of post-translational modifications via blind search of mass-spectra. Nat. Biotechnol. 2005;23:1562–1567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- Venter J.C., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Wu D., Paulsen I., Nelson K.E., Nelson W., Paulsen I., Nelson K.E., Nelson W., Nelson K.E., Nelson W., Nelson W., et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Volkin D.B., Mach H., Middaugh C.R., Mach H., Middaugh C.R., Middaugh C.R. Degradative covalent reactions important to protein stability. Mol. Biotechnol. 1997;8:105–122. doi: 10.1007/BF02752255. [DOI] [PubMed] [Google Scholar]
- Wang R., Prince J., Marcotte E., Prince J., Marcotte E., Marcotte E. Mass spectrometry of the M. smegmatis proteome: Protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005;15:1118–1126. doi: 10.1101/gr.3994105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasinger V., Humphery-Smith I., Humphery-Smith I. Small genes/gene-products in Escherichia coli K-12. FEMS Microbiol. Lett. 1998;169:375–382. doi: 10.1111/j.1574-6968.1998.tb13343.x. [DOI] [PubMed] [Google Scholar]
- Wilkins M.R., Gasteiger E., Gooley A.A., Herbert B.R., Molloy M.P., Binz P.A., Ou K., Sanchez J.C., Bairoch A., Williams K.L., Gasteiger E., Gooley A.A., Herbert B.R., Molloy M.P., Binz P.A., Ou K., Sanchez J.C., Bairoch A., Williams K.L., Gooley A.A., Herbert B.R., Molloy M.P., Binz P.A., Ou K., Sanchez J.C., Bairoch A., Williams K.L., Herbert B.R., Molloy M.P., Binz P.A., Ou K., Sanchez J.C., Bairoch A., Williams K.L., Molloy M.P., Binz P.A., Ou K., Sanchez J.C., Bairoch A., Williams K.L., Binz P.A., Ou K., Sanchez J.C., Bairoch A., Williams K.L., Ou K., Sanchez J.C., Bairoch A., Williams K.L., Sanchez J.C., Bairoch A., Williams K.L., Bairoch A., Williams K.L., Williams K.L., et al. High-throughput mass spectrometric discovery of protein post-translational modifications. J. Mol. Biol. 1999;289:645–657. doi: 10.1006/jmbi.1999.2794. [DOI] [PubMed] [Google Scholar]
- Yang C., Rodionov D., Li X., Laikova O., Gelfand M., Zagnitko O., Romine M., Obraztsova A., Nealson K., Osterman A., Rodionov D., Li X., Laikova O., Gelfand M., Zagnitko O., Romine M., Obraztsova A., Nealson K., Osterman A., Li X., Laikova O., Gelfand M., Zagnitko O., Romine M., Obraztsova A., Nealson K., Osterman A., Laikova O., Gelfand M., Zagnitko O., Romine M., Obraztsova A., Nealson K., Osterman A., Gelfand M., Zagnitko O., Romine M., Obraztsova A., Nealson K., Osterman A., Zagnitko O., Romine M., Obraztsova A., Nealson K., Osterman A., Romine M., Obraztsova A., Nealson K., Osterman A., Obraztsova A., Nealson K., Osterman A., Nealson K., Osterman A., Osterman A., et al. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J. Biol. Chem. 2006;281:29872–29875. doi: 10.1074/jbc.M605052200. [DOI] [PubMed] [Google Scholar]
- Yaron A. Dipeptidyl carboxypeptidase from Escherichia coli. Methods Enzymol. 1976;45:599–610. doi: 10.1016/s0076-6879(76)45053-x. [DOI] [PubMed] [Google Scholar]
- Yates J., Eng J., McCormack A., Eng J., McCormack A., McCormack A. Mining genomes: Correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal. Chem. 1995;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]